Abstract
Mitochondrial DNA (mtDNA) deletions are a primary cause of mitochondrial disease and are believed to contribute to the aging process and to various neurodegenerative diseases. Despite strong observational and experimental evidence, the molecular basis of the deletion process remains obscure. In this study, we test the hypothesis that the primary cause of mtDNA vulnerability to breakage resides in the formation of non-B DNA conformations, namely hairpin, cruciform and cloverleaf-like elements. Using the largest database of human mtDNA deletions built thus far (753 different cases), we show that site-specific breakage hotspots exist in the mtDNA. Furthermore, we discover that the most frequent deletion breakpoints occur within or near predicted structures, a result that is supported by data from transgenic mice with mitochondrial disease. There is also a significant association between the folding energy of an mtDNA region and the number of breakpoints that it harbours. In particular, two clusters of hairpins (near the D-loop 3′-terminus and the L-strand origin of replication) are hotspots for mtDNA breakage. Consistent with our hypothesis, the highest number of 5′- and 3′-breakpoints per base is found in the highly structured tRNA genes. Overall, the data presented in this study suggest that non-B DNA conformations are a key element of the mtDNA deletion process.
INTRODUCTION
It is undeniable that the complementary strands (named the L- and H-strands) of human mitochondrial DNA (mtDNA) are primarily organized into the canonical right-handed double-helical structure of B-form DNA. Unfortunately, our knowledge of the higher order topology of mtDNA and its interaction with the mitochondrial environment remains quite limited. It is plausible that many deformations to the canonical B-form of DNA occur in the mitochondrial genome and have important biological consequences, as has been unequivocally shown in many other genetic systems (1–3). In recent years, several studies have demonstrated that non-B DNA structures (often called non-canonical, unusual, alternative or secondary DNA structures) (4–6) occur, at least transiently, in the mitochondrial genome. For instance, a stem-loop structure is required to activate the initiation of DNA replication in the L-strand origin of replication (OL) (7,8). DNA bending in the L-strand promoter (LSP) induced by the mitochondrial transcription factor A (TFAM) is necessary for transcription initiation (9,10), whereas DNA unwinding and base eversion at the tRNA-Leu(UUA/G) gene by the mitochondrial transcription termination factor 1 (MTERF1) is critical for transcription termination (11). The structures with no clear biological function vary according to alterations in the primary DNA sequence in a random way and are only removed by purifying selection if their formation interferes with any relevant genomic function. In contrast, those structures with functional relevance are probably maintained under strong selective pressures as previously suggested (8,12).
The formation of most non-B DNA structures is favoured by the local unwinding of the DNA double helix, which is associated with negative supercoiling (4,13,14). As is most DNA in vivo, mtDNA is believed to be predominantly negatively supercoiled (i.e. the torsional tension diminishes the DNA helicity and facilitates strand separation) and is subject to dynamic processes that constantly alter the canonical conformation of the double helix. Unlike nuclear DNA, mtDNA is continuously replicated and transcribed during the entire cell cycle, and according to the available models, a large portion of the mtDNA is single-stranded for a significant period of time during such processes (15–17). This phenomenon provides an opportunity for structures with intra-strand base pairing to form and to persist for a relatively long period. In addition, several proteins are continuously tracking through the mtDNA (e.g. the movement of an RNA polymerase during transcription), resulting in a redistribution of the local supercoiling characteristics, which can be counterbalanced by the action of topoisomerases or the formation of non-B DNA structures (18,19). The conformational flexibility of the mtDNA is also affected by the packaging factor TFAM, which induces negative supercoiling upon binding to mtDNA (10,20). Thus, the binding of proteins may restrict the transmission of the superhelical tension throughout the DNA, although this possibility remains to be determined for the mitochondrial genome. Similarly, stable and partially hybridized RNA molecules (R-loops) were found to be associated with the mammalian mtDNA and are thought to maintain the genome in a more open conformation (21).
The versatile nature of the mitochondrial genome is also evident in the multiple forms of gene organization and structural diversity resulting from numerous genomic rearrangements that occur through insertions, deletions, duplications, inversions or translocations of DNA segments (22). Among the different types of rearrangements, the loss of a section of the mitochondrial genome (an mtDNA deletion) has attracted the attention of researchers. The reason for such concern is that mtDNA deletions are associated with the multifactorial aging process and with a variety of progressive disorders that cause substantial disability and can lead to premature death (17,23,24). The loss of mtDNA-encoded proteins and/or tRNA genes required for protein synthesis results in mitochondrial dysfunction and cell death due to their crucial role in energy metabolism. Individual deleted mtDNA molecules will have no discernible effect on cellular function. In order to cause mitochondrial dysfunction, a deleted form of mtDNA must accumulate above a critical threshold that varies from tissue to tissue based on energy requirements. The expansion of a deleted mtDNA molecule to the detriment of other variants might occur due to the genetic bottleneck for mtDNA transmission in the germline or through the unequal portioning of molecules in daughter cells (mitotic segregation). The level of different types of mtDNA molecules within a cell (heteroplasmy) will also change over long time scales randomly (intracellular drift) and independently of the cell cycle by a process of relaxed replication (25–28).
However, the exact mechanism(s) underlying the formation of mtDNA deletions remain elusive. What is undeniable is that most mtDNA deletions occur in the major arc of the mtDNA, between the two proposed origins of replication (OH and OL), and that they present sequence homologies at the boundaries of their breakpoints (29–31). In addition, several authors have noticed that mtDNA deletion breakpoints are often located in regions with the potential to adopt non-B DNA conformations, raising the possibility of a mechanistic role of alternative DNA structures in the generation of deletions (12,31–38). In this study, we describe a comprehensive survey of non-B DNA conformations across the complete human mitochondrial genome and provide multiple lines of evidence for their association with mtDNA deletions. Our study was prompted by the clear recognition that certain DNA sequences adopt structural configurations that are more prone to breakage (39–43) and the emerging understanding that non-B DNA structures are inherently hypermutable (1–3,44).
MATERIALS AND METHODS
MtDNA deletion breakpoints
We started by collecting information about all of the available mtDNA deletions (5′- and 3′-breakpoints) from the MITOMAP (http://www.mitomap.org) and MitoTool (45) databases and from 83 peer-reviewed papers published from 1989 to 2010 (Supplementary Figure S1). The 929 different deletions that were initially retrieved have been identified in (i) patients with an mtDNA deletion syndrome (chronic progressive external ophthalmoplegia, Kearns–Sayre syndrome or Pearson syndrome) or with a complex multi-system disorder that did not fit into any of the preceding categories, (ii) patients with autosomal disorders of mtDNA maintenance or mitochondrial nucleotide metabolism and (iii) post-mitotic tissues as part of normal aging. We decided to combine deletions from different sources because there is no evidence so far that different molecular mechanisms cause deletions in the different clinical scenarios that would justify a separate analysis. It has been suggested that a similar mechanism generates mtDNA deletions in all clinical situations (30).
Each deletion was defined by a unique combination of two breakpoints and was only included once in our database. This procedure was used to avoid the ascertainment bias that is caused by the regular use of methods that only identify a restricted group of deletions, which would lead to an over-representation of some deletions in our database if frequency values were considered. Moreover, we attempted to minimize the noise inherent to the presence of the same deletion in different databases or publications (sometimes even with a different nomenclature) that would cause an artificial duplication of data. If different deletions shared the same breakpoint at one end, then the shared breakpoint was counted once for each deletion in most analyses. The deletion breakpoints were always numbered according to the conventional L-strand positions of the revised Cambridge reference mtDNA sequence (rCRS, NC_012920). We always considered, in this study, that ‘breakpoints’ are the mtDNA positions that are retained in the deleted mtDNA sequence and that flank the deleted region. In other words, 5′-breakpoints are upstream of the 5′-break and 3′-breakpoints are downstream of the 3′-break, considering the L-strand numbering.
In several cases, we observed that mtDNA deletions are described in the literature by an interval of values as breakpoint positions. This type of nomenclature is used because of the existence of equal sequence motifs in the breakpoint areas (e.g. 8016–8019:15 516–15 519), which renders the precise identification of the break sites impossible. In such situations, we have retained the smallest number for each breakpoint in the interval (in the previous example, 8016:15 516). With this correction, a few deletions were found to be repeated in our original database and were removed, leaving a total of 788 deletions. We then generated the 788 deleted mtDNA sequences by removing the region between the 5′- and 3′-breakpoints in the rCRS, retrieved from the NCBI Entrez Nucleotide database (http://www.ncbi.nlm.nih.gov). Each deleted sequence was aligned with the full-length rCRS using python scripts (Python v.2.6, www.python.org/) from the third party application ‘Muscle’ (46) available on the PyCogent v1.5 package (47). The existence of equal sequence motifs in breakpoint areas implies that different sequence alignments are possible for the same deletion: certain bases might be equally aligned upstream or downstream of the deletion area, in the equal sequence motifs (Supplementary Figure S2). Therefore, we have corrected the limits of the deletion when necessary by keeping all possible matches at the 5′-breakpoint. This adjustment revealed that several deletions with different reported breakpoints were in fact equal. As a result, 753 unique deletions remained and were used in all of the analyses.
In addition, we collected all of the available information on the mtDNA breakpoints of transgenic mice expressing an altered Twinkle mtDNA helicase (48). The breaking sites were plotted on the Mus musculus mtDNA reference sequence (NC_005089), positions 15 150–15 469.
Prediction of non-B DNA conformations
In this work, we use four expressions to describe the predicted structural alterations to the orthodox right-handed Watson–Crick B-form of mtDNA: hairpin, cruciform, cloverleaf-like elements and other secondary structures. All of these terms refer to deviations from the conventional B-form of DNA, collectively known as non-B DNA conformations. A hairpin or stem–loop structure is a section of single-stranded DNA that folds back on itself to form a paired double helix that ends in an unpaired loop. The cruciform structure consists of a pair of hairpin structures in complementary DNA strands forming a four-way junction with a cross-shaped configuration. The cloverleaf structure is a single-strand DNA arrangement with four stems and three terminal loops, usually used to describe the secondary structure of tRNA molecules. All other types of structural arrangements that do not belong to the preceding categories are designated here as ‘other secondary structures’. The terms are used regardless of the length and folding energy of the stem and loop regions. It should be taken into consideration that other types of non-B DNA elements exist and were not addressed here (e.g. triplexes, G4-tetrad, slipped structures, left-handed Z-DNA, bent DNA and sticky DNA) (2).
The folding prediction for single-stranded DNA was performed with the hybrid-ss-min core programme of the UNAFold 3.8 software package (49). Python scripts were written to run automatic executables from UNAfold. The prediction is based on free energy minimization using nearest neighbour thermodynamic rules and dynamic programming algorithms (50). The folding of the single-stranded DNA was carried out using the default parameters, including predictions at a temperature of 37°C, sodium concentration of 1 M and magnesium concentration of 0 M. The hybrid-ss-min programme predicts the thermodynamically most stable secondary structure that a single-stranded DNA segment can form and calculates the variation in the free energy of the folding (ΔG, expressed in kcal/mol). The magnitudes of the ΔG values indicate the relative stabilities of the structures formed by each segment: the greater the variation in the ΔG value (more negative value), the more likely it is that a stable secondary structure will form. We always used the predicted lowest free energy structure (the structure with the most negative ΔG value) in the various analyses, although other suboptimal structures were sometimes predicted by the programme. The graphical representations of DNA secondary structures were obtained from the sir-graph programme, which belongs to the mfold-util software v4.6 (51). The circular maps of the human mtDNA were produced using the Circos software, version 0.52 (52).
Statistical analyses
The descriptive statistics for the different datasets, the Student’s t-test (independent samples with separate variance estimates) and the Fisher’s exact test for contingency tables were obtained with the STATISTICA v7 software (StatSoft, Inc., Tulsa, OK). All reported P-values are two-sided and a significance level of 0.05 was used.
RESULTS
Deletion breakpoints are not randomly distributed throughout the mitochondrial genome
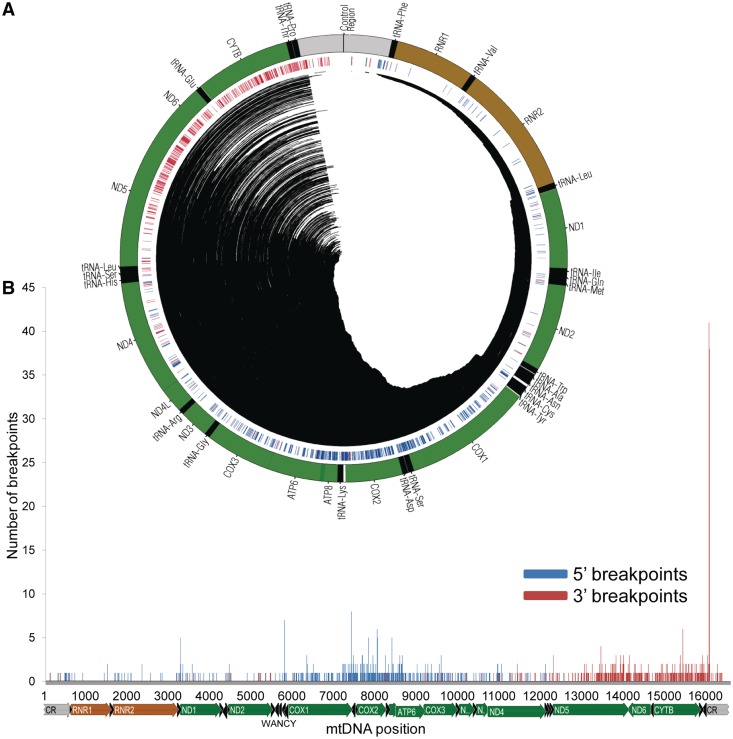
We started by collecting data from all of the available mtDNA deletions in public databases and peer-reviewed publications (Supplementary Figure S1) that have been identified in pathological and non-pathological situations. The 753 unique mtDNA deletions that fulfilled our selection criteria (Supplementary Figure S3 and see ‘Materials and Methods’ section) are defined by 620 and 497 different 5′- and 3′-breakpoints, respectively (1117 different breakpoint in total). The number of breakpoints is lower than 1506 (i.e. twice the total number of 753 deletions) because different deletions sometimes share the same breakpoint at one end. The distributions of the 5′- and 3′-breakpoints across the mtDNA are clearly different from each other (Figure 1, Supplementary Figure S4 and S5). This difference has been previously noted in smaller deletion datasets (29,31,53).
Figure 1.
Deletion breakpoints are not randomly distributed throughout the mitochondrial genome. (A) A circular representation of the human mtDNA with annotated tRNA (black), rRNA (brown) and protein-coding (green) genes (outer track). The central track depicts the location of 5′- (blue) and 3′- (red) breakpoints. The central lines indicate the deleted region in the 753 reported cases. (B) The distribution of 5′- (blue bars) and 3′- (red bars) deletion breakpoints in the human mtDNA. The locations of the mitochondrial genes are shown below the x-axis.
The mean value of the distribution of the 5′-breakpoints is position 7658 with a standard deviation of 2296 (Supplementary Figure S4). The mode is position 7402 with a total of eight breakpoints, although it is not clearly distinct from the values at other positions (e.g. positions 5787 and 8032, with seven and six breakpoints, respectively). The histogram of the distribution of 5′-breakpoints suggests a multimodal distribution with major peaks around and within COX2 and in the WANCY cluster of tRNA genes (Figure 1 and Supplementary Figure S4). The mean value of the distribution of the 3′-breakpoints is position 14 503, with a standard deviation of 2185 (Figure 1 and Supplementary Figure S5). There is a clear mode in the distribution: position 16 071 has noticeably the highest number of 3′-breakpoints, with 41 out of 753 total breakpoints (5.44%). The flanking sites of the 16 071 hotspot (positions 16 065–16 080) harbour 25.09% of all of the 3′-breakpoints (189 out of 753 breakpoints) (Supplementary Figure S6). The remaining deletions are mainly found in the ND5 and CYTB genes, although there is a sudden decrease in the number of 3′-breakpoints between them, in the region of the ND6 and tRNA-Glu genes (Figure 1 and Supplementary Figure S5). Overall, the mtDNA deletions are not randomly distributed, and their breakpoints do not follow a normal distribution around any specific mtDNA position (Figure 1).
A completely different pattern would be expected if the deletions were random. As a simple way to estimate the frequency of deletions per site, we generated a dataset of 20 000 random deletions with no restrictions. The distribution of mtDNA breakage hotspots in the real data contrasts with the distribution observed in simulated deletions. The most common breakpoints have frequency values [e.g. mtDNA positions 16 071 (5.4 × 10−2), 7402 (1.1 × 10−2), 5787 (9.3 × 10−3) and 15 435 (8.0 × 10−3)] that are considerably higher than those estimated for 20 000 random deletions, where the highest breakpoint frequency at any site is 4.0 × 10−4 (8 occurrences out of 20 000 breakpoints) (Supplementary Figure S7). The most frequent real and simulated breakpoints have a significantly different proportion in our datasets (P-value < 1.00 × 10−4, Student’s t-test): 8/753 = 0.011 and 41/753 = 0.054 for 5′- and 3′-breakpoints, respectively, 8/20 000 = 0.0004 for random deletions.
The most frequent deletion breakpoints occur within or near predicted hairpins
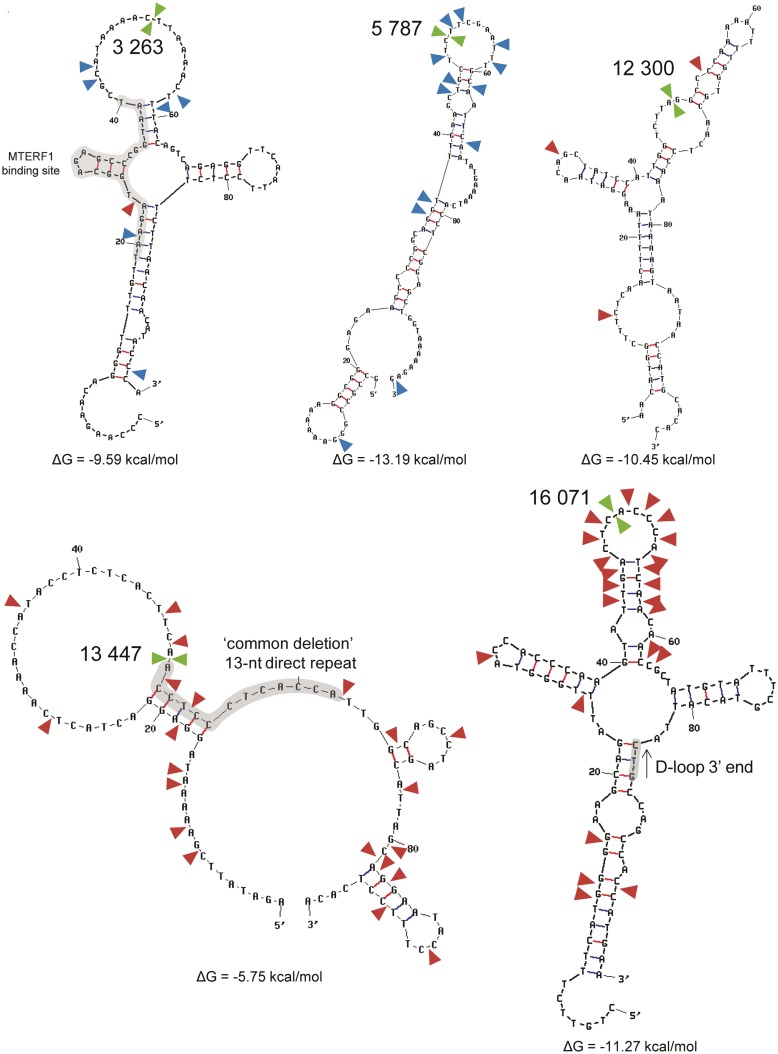
We started by investigating the locations of the 10 main mtDNA breakage hotspots in the predicted secondary structure of the 100-nt windows (L- and H-strands) enclosing each of these breakpoints (selected as window midpoints, i.e. each window extends 50 nt upstream and downstream of the break) (Figure 2 and Supplementary Figures S7–S9). The free energy of folding of the 100-nt windows varied from −13.19 to −3.05 kcal/mol in the L-strand and from −14.47 to −1.34 kcal/mol in the H-strand. Although all of the 100-nt windows present at least one stem element, four of these regions (centred on sites 3263, 5787, 12 300 and 16 071) stand out as being highly structured, with several stem elements and a folding energy <−9 kcal/mol in the L- and H-strands (Figure 2). In most cases, similar hairpins were predicted for both mtDNA strands, which is compatible with the formation of cruciform structures. Cloverleaf structures were predicted for the mtDNA regions of breakpoints 3263 (L-strand) and 16 071 (L- and H-strands). Similar DNA structures were predicted using 300-nt windows (also with breakpoints as window midpoints), suggesting that short-distance interactions are more stable than long-distance base interactions (data not shown). The exact location of the 3′-breakpoint of the ‘common deletion’ (13 447) is impossible to ascertain due to the presence of a 13-nt direct repeat at the breakpoint regions. The 100-nt window around this breakpoint folds differently in L- and H-strands, with four and three hairpins, respectively. The breaking of the DNA might occur in a stem or loop element. Other structures predicted for the L- and H-strands also differ in the number and location of hairpins. For instance, the 7402 and 8032 breakpoints are located in hairpin elements predicted only for the H-strand (Figure 2, Supplementary Figures S8 and S9).
Figure 2.
The most frequent deletion breakpoints occur within or near predicted hairpins. Five of the most frequent breakpoint sites (mtDNA positions 3263, 5787, 12 300, 13 447 and 16 071) are indicated by a green arrow in the predicted structure (L-strand) of the 100-nt flanking region (breakpoints were used as window midpoints). The blue and red arrows indicate less frequent 5′- and 3′-breakpoints, respectively. Highlighted in grey are the binding sites of the MTERF1, the 13-nt direct repeat at the 3′-breakpoint of the ‘common deletion’ and the D-loop 3′-terminus.
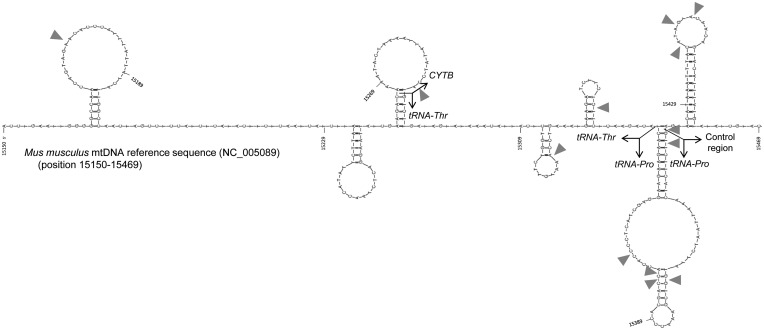
To verify whether the association between breakpoints and hairpins was not only a feature of human mitochondrial genome, we analysed the distribution of breakpoints in the mtDNA of transgenic mice with mitochondrial disease (48). The distribution of the deletion breakpoints in the Mus musculus mtDNA (position 15 150–15 469) is biased towards hairpins, with all of the breakpoints occurring in this category of non-B DNA (Figure 3). The difference between the proportion of breakpoints located inside (13 breakpoints in a total of 229 nt) and outside (0 breakpoints in a total of 91 nt) hairpins attains statistical significance (P-value = 0.023, Fisher’s exact test).
Figure 3.
Breakage sites in the mouse mitochondrial genome are associated with hairpin elements. All of the deletion breakpoints described in transgenic mice with mitochondrial disease (grey arrows) are indicated in the Mus musculus mtDNA L-strand reference sequence (NC_005089), from position 15 150 to 15 469. The secondary structures were obtained in the mfold-util software v4.6.
Two stable clusters of hairpins are hotspots for mtDNA breakage
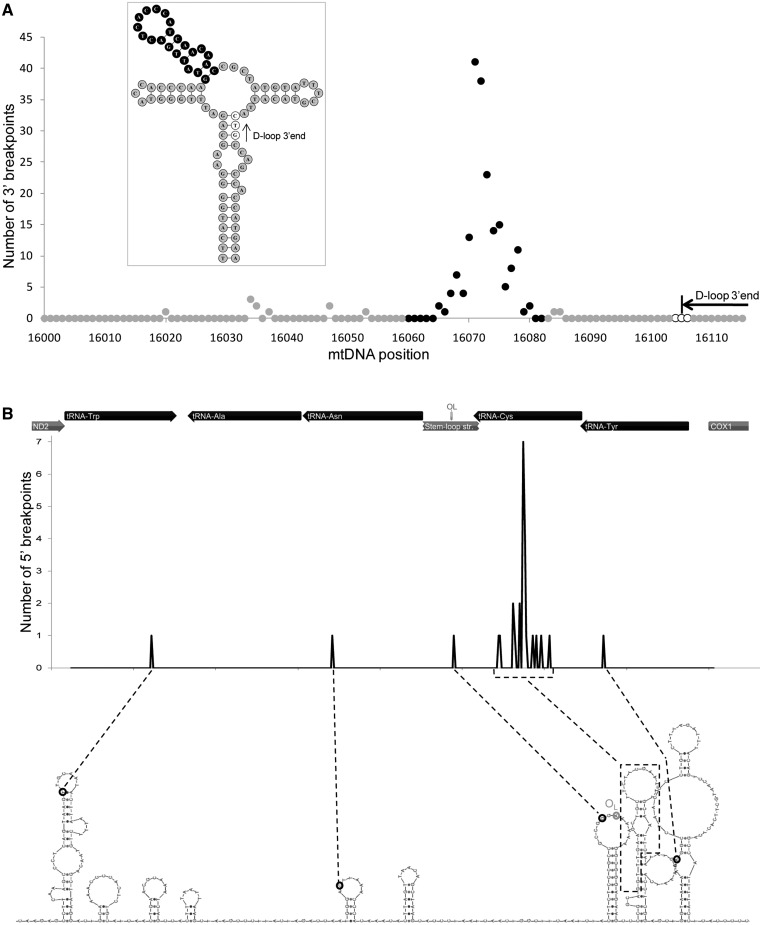
The major mtDNA deletion breakpoint (16 071) is located in a 93-nt cloverleaf-like structure (positions 16 028–16 120) that we previously identified and named structure A (12). The central hairpin of this structure (16 060–16 082) is a hotspot of DNA breakage, with 189 reported 3′-breakpoints occurring in this short region (Figure 4A). The central hairpin has a higher proportion of sites with breakpoints (16 sites out of 23) than the remaining structure (8 sites out of 70, P-value < 1.00 × 10−4, Fisher’s exact test). Of these 189 breakpoints, 144 (19% of all 3′-breakpoints) are located on the 8-nt terminal loop (P-value < 1.00 × 10−4, Fisher’s exact test). The 16,071 hotspot is located upstream of the trinucleotide stop point (16 104–16 106) for the premature arrest of the H-strand synthesis (D-loop 3′-end), according to the numbering of the mtDNA, or downstream according to the direction of the H-strand synthesis. The 16 071 hotspot is not within the three-stranded D-loop structure (Figure 4A). It could be hypothesized that the 16 071 3′-breakpoint hotspot is over-represented in our database by being associated with certain particular types of 5′-breakpoints in a narrow region of the mitochondrial genome. However, we observed that deletions with a 3′-breakpoint in the 16 071 hotspot (16 075–16 080) have 5′-breakpoints in very different mtDNA regions (Supplementary Figure S10).
Figure 4.
Two stable clusters of hairpins are hotspots for mtDNA breakage. (A) A total of 189 reported 3′-breakpoints occur in the central hairpin (black circles) of a large cloverleaf-like structure (enclosed image) predicted for a 93-nt stretch (positions 16 028 to 16 120) of the control region near the tRNA-Pro (L-strand). Inside the hairpin, 144 breakpoints (19% of all of the 3′-breakpoints) are located on the 8-nt terminal loop. This deletion hotspot is located near the trinucleotide stop point (16 104–16 106; white circles) for the premature arrest of the H-strand synthesis responsible for forming a three-stranded DNA structure known as the displacement loop (D-loop). (B) All of the 5′-deletion breakpoints (n = 27) identified in the WANCY cluster of tRNAs are located in hairpin elements (L-strand). Most of them (n = 23) are located in a single stem–loop element predicted for the tRNA-Cys gene, downstream of the stem–loop structure that is associated with the origin of L-strand replication (OL).
One of the most relevant 5′-breakpoint hotspots is located in the WANCY region (Figures 1 and 4B). This region comprises a cluster of five tRNA genes (tRNA-Trp, tRNA-Ala, tRNA-Asn, tRNA-Cys and tRNA-Tyr) that is located between the ND2 and COX1 genes and that includes the OL. We found that this mtDNA segment (5512–5903) has a very high folding potential (ΔG = −36.41 kcal/mol for the L-strand and ΔG = −40.49 kcal/mol for the H-strand), comprising several hairpin structures (Figure 4B and Supplementary Figure S11). We discovered that all of the 5′-ends of the deletions identified in this region (n = 27) are located in five of these hairpin elements (Figure 4B and Supplementary Figure S11). We observed that the difference in the proportion of breakpoints inside (16 in a total of 295 nt) and outside (0 in a total of 97 nt) hairpins is statistically significant (P-value = 0.015, Fisher’s exact test). In particular, 23 of the deletion breakpoints are located in a single stem–loop element predicted for positions 5772–5803 in the tRNA-Cys gene. This element is only five bases downstream of the previously identified stem–loop structure that is associated with the origin of L-strand replication (7,8).
Mitochondrial tRNA genes are hotspots for mtDNA breakage
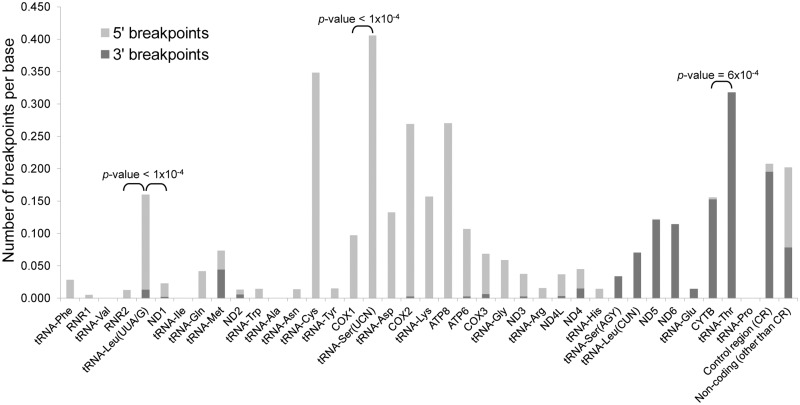
We investigated the distribution of the deletion breakpoints according to the coding/non-coding features of mtDNA. The mtDNA regions with the highest number of 5′-breakpoints are the COX2 and COX1 genes, with 182 and 149 5′-breakpoints, respectively (Supplementary Figures S12 and S13). The control region (the largest non-coding region of mtDNA located between tRNA-Pro and tRNA-Phe genes) and the ND5 gene have the highest number of 3′-breakpoints (219 breakpoints each). When considering both 5′- and 3′-breakpoints, the control region is the location where more breaks occur (15.47% of the total breakpoints), followed by the ND5 (14.74%), COX2 (12.22%) and CYTB (11.82%) genes.
Strikingly, when we take into account the lengths of the different coding or non-coding mtDNA regions, those with the highest number of 5′- and 3′-breakpoints per base (number of deletions/region length) are the tRNA genes (Figure 5). tRNA-Ser(UCN) and tRNA-Cys are the genes with more 5′-breakpoints per base (0.406 and 0.348, respectively) (Supplementary Figure S13). Although they have the highest absolute number of 5′-breakpoints, the COX1 and COX2 genes only have 0.097 and 0.266 5′-breakpoints per base, respectively. The tRNA-Ser(UCN) gene has a higher proportion of sites with breakpoints than its adjacent COX1 gene (P-value < 1.00 × 10−4, Student’s t-test). Inside the minor arc, the tRNA-Leu(UUA/G) gene, which encodes the MTERF1-binding site, stands out as having a significantly higher number of 5′-breakpoints per base (0.147) than its flanking genes (RNR2 and ND1 with 0.013 and 0.021, respectively, P-values < 1.00 × 10−4, Student’s t-test). Similarly, the gene with the highest number of 3′-breakpoints per base is tRNA-Thr (0.318) (Supplementary Figure S13). The other regions with the highest number of 3′-breakpoints per base are the control region (0.195) and the CYTB gene (0.152). Nevertheless, the proportion of sites with breakpoints is significantly higher in the tRNA-Thr gene than in the adjacent CYTB (P-value = 6.00 × 10−4, Student’s t-test, Figure 5).
Figure 5.
Mitochondrial tRNA genes are hotspots for mtDNA breakage. The graph displays the number of deletion breakpoints per base (number of breakpoints/region length) according to the coding features of the mitochondrial genome (5′- and 3′-breakpoints in light and dark grey, respectively). The significance of the difference between the number of breakpoints in some tRNA and their flanking genes is shown (two-sided P-values; Student’s t-test).
Overall, the tRNA gene sequences fold into structures quite different from the common tRNA cloverleaf structure with four stems and three loops (Supplementary Figures S14 and S15). In several cases, the predicted structures lack complete domains of the cloverleaf structure such as the acceptor arm (e.g. tRNA-Ala, tRNA-Arg or tRNA-Asn). The differences between structures are explained by the different folding proprieties of DNA and RNA molecules. Another important factor is that some post-transcriptional modifications must be made in the RNA molecules before the cloverleaves can be formed (54). We also found that the variation in folding energies among tRNA genes is not sufficient to explain the difference in the frequency of breakpoints (Supplementary Figure S16). The mtDNA regions with more breakpoints per base (5′ and 3′) are the tRNA-Ser(UCN) (0.406), tRNA-Cys (0.348) and tRNA-Thr (0.318) genes (Figure 5). These three breakpoint-prone tRNA genes fold with the formation of at least two stem elements. The tRNA-Thr gene is one of the few cases where the DNA is predicted to form the classical cloverleaf structure (Supplementary Figures S14, and S15).
Deletion breakpoints are located in mtDNA regions with high folding potentials
We performed a sliding-window analysis of the folding potentials (100-nt windows with 1 nt of overlap) throughout the entire mitochondrial genome (L- and H- strands) to capture the folding energy of all of the possible conformational transitions in which every mtDNA position is involved (Supplementary Figures S17 and S18). The mean ΔG value in the 16 569 100-nt mtDNA windows is −5.504 kcal/mol (SD = 3.387) for the L-strand and −6.366 kcal/mol (SD = 3.403) for the H-strand. There are marked variations in the folding energies across the mtDNA, with sudden increases and decreases in the ΔG values (Supplementary Figure S18). The mtDNA region with the highest folding potential is the WANCY cluster of tRNAs, matching the second-highest peak in the number of 5′ breakpoints (and the third of all breakpoints).
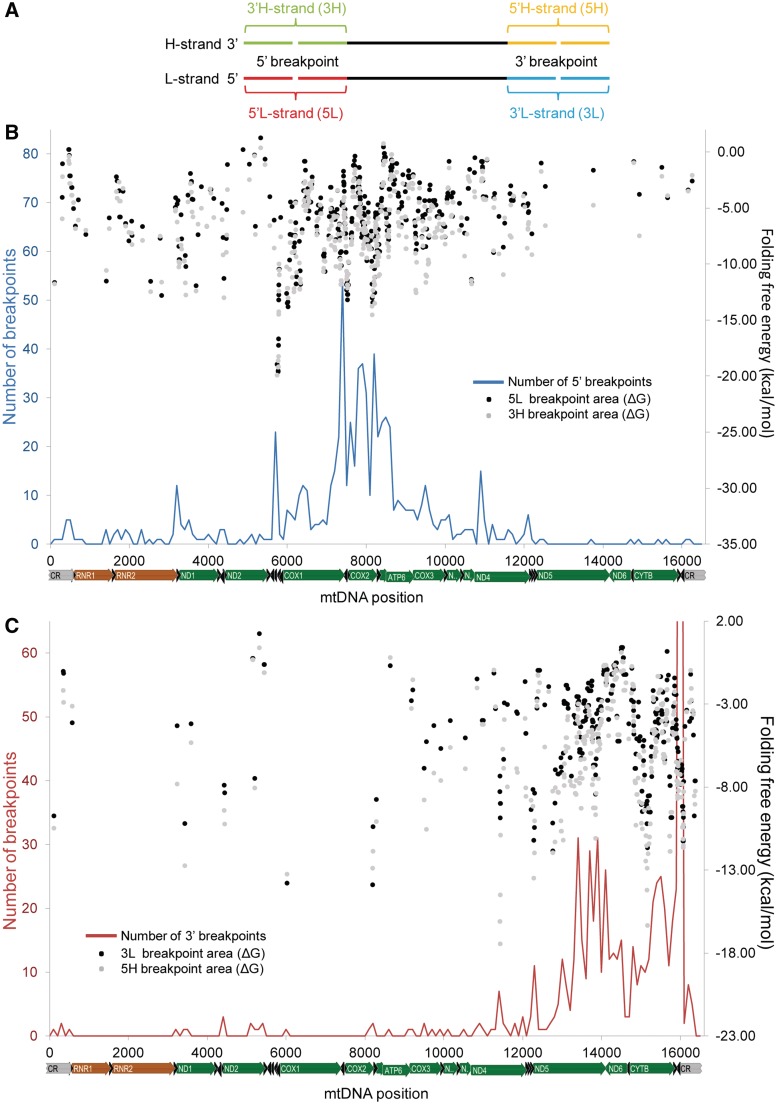
To investigate how the local DNA sequence environment might contribute to the formation of deletions, we extracted and folded all of the 100-nt segments from both the H- and L-strands that enclosed a 5′- or 3′-breakpoint as the midpoint (Figure 6A). The code used to identify each region is composed of a number (‘5’ or ‘3’) according to the type of breakpoint and a letter (‘H’ or ‘L’) for the mtDNA strand. The mean observed free energy values were −5.66 (5L), −5.93 (3L), −6.58 (3H) and −7.01 (5H) kcal/mol (Supplementary Figure S19).
Figure 6.
Genome-wide distribution of folding potentials in the breakpoint areas of mtDNA deletions. (A) The nomenclature and schematic representation of the location of the genomic region enclosing 5′- and 3′-deletion breakpoints. (B, C) Black and grey dots indicate the free energy of folding (kcal/mol) of 100-nt windows around the 5′- (B) and 3′- (C) breakpoints (black and grey dots for L-strand and H-strand segments, respectively). The blue (B) and red (C) lines indicate the distribution of 5′- and 3′-breakpoints, respectively (measured in 100-nt sliding windows with an overlap of 1 nt). The peak at the 16 071 hotspot reaches 201 deletions but is not completely shown to facilitate the visualization of smaller peaks.
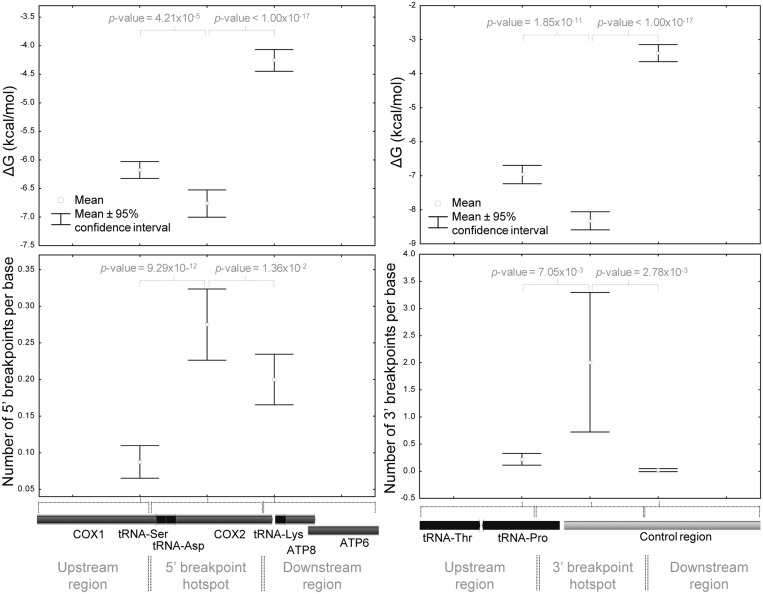
The distribution of free energies of folding along the mtDNA considering all breakpoint areas is represented in Figure 6B, C and Supplementary Figures S20 and S21. In order to test if there is an association between the number of deletion breakpoints and the folding energy of the breakpoint area, we compared the two mtDNA segments where 5′- and 3′-breakpoints are more frequent with their upstream and downstream flanking segments with the same length. In both cases, we found that there is a significantly higher number of breakpoints and folding potential (more negative ΔG values) in the target region than in both flanking segments (Figure 7 and Supplementary Figure S22). For instance, the mean ΔG value (−6.76 kcal/mol) of the sliding windows with midpoints from positions 7401 to 8200 (the hotspot of 5′-breakpoints upstream and within COX2) is significantly lower than the estimated for its upstream (mean ΔG = −6.18 kcal/mol; P-value = 4.21 × 10−5; Student’s t-test) and downstream (mean ΔG = −4.26 kcal/mol; P-value < 1.00 × 10−17; Student’s t-test) regions. A significant difference was also found between the 97-nt segment defined by the hairpin element around OL and the adjacent tRNA-Cys gene (positions 5730–5826) and their flanking regions, for both ΔG and the number of 5′-breakpoints parameters (data not shown). Similarly, the 16 001–16 100 mtDNA region (around the 16 071 hotspot) has a significantly higher number of 3′-breakpoints (n = 201) than its upstream (n = 22; P-value = 7.05 × 10−3; Student’s t-test) and downstream (n = 2; P-value = 2.78 × 10−3; Student’s t-test) regions, together with a significantly higher folding potential (Figure 7 and Supplementary Figure S22).
Figure 7.
The main hotspots of mtDNA breakage have higher folding potentials than adjacent regions. The two mtDNA segments where 5′- and 3′-breakpoints are more frequent (positions 7401–8200 and 16 001–16 100, respectively) were compared with their upstream and downstream flanking segments. We estimated for each segment the mean number of breakpoints per base (and the 95% confidence interval for the mean) and the average folding potential of the 100-nt windows with a midpoint position in that region. There is a significant higher folding potential (more negative ΔG values; top graphs) and higher number of breakpoints per base (bottom graphs) in the hotspot regions than in their flanking segments. The results of the statistical tests (Student’s t-test; two-sided P-values) to evaluate the differences in means (ΔG values and number of breakpoints) between adjacent regions are indicated.
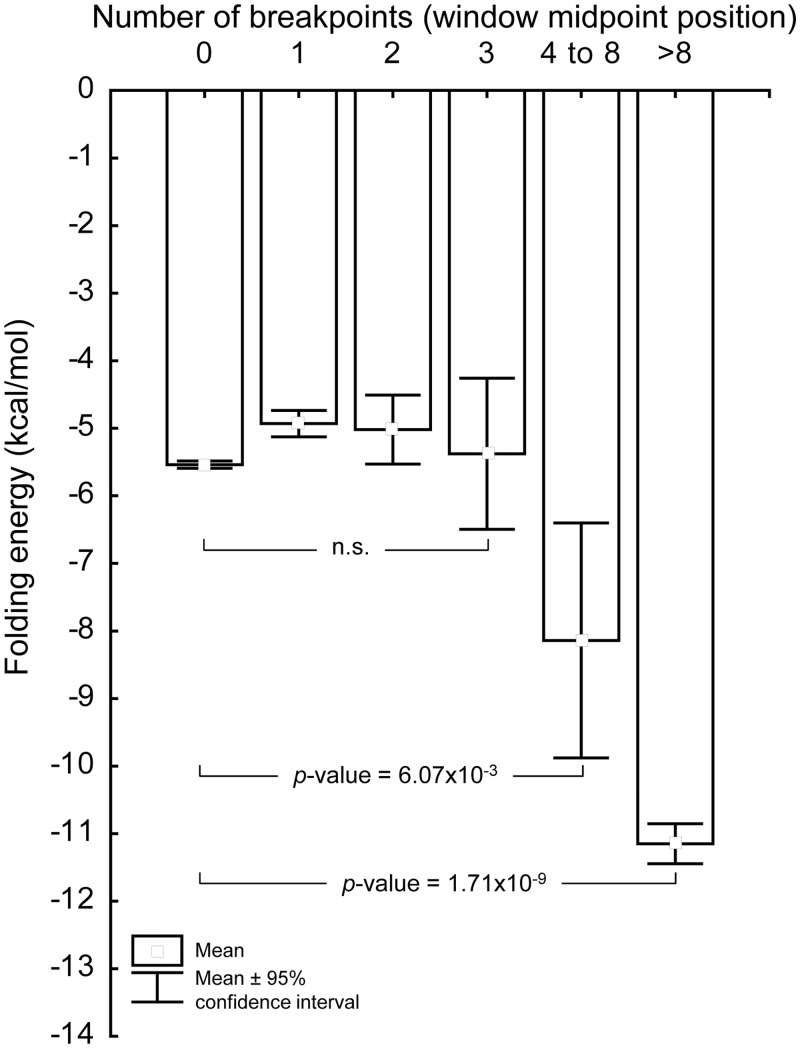
We next detailed the relationship between the folding potential of each 100-nt mtDNA sliding window and the number of breakpoints that it harbours. For this purpose, we estimated the number of breakpoints in the window midpoint position, i.e. only position 50 in each 100-nt window was considered. The distribution of 100-nt windows according to the number of breakpoints at the midpoint position shows that a total of 1114 window midpoint positions have at least one deletion breakpoint (6.7%). In general, windows with more deletion breakpoints have a significantly higher folding potential. For example, windows with more than 8 reported breakpoints have an average folding energy (mean = −11.15 kcal/mol) significantly different (P-value = 1.71 × 10−9; Student’s t-test) from that of windows with no breakpoints (mean = −5.54 kcal/mol) (Figure 8). Similarly, windows with four to eight breakpoints in the midpoint positions have an average folding energy (mean = −8.14 kcal/mol) that is significantly different (P-value = 6.07 × 10−3; Student's t-test) from that of windows with no breakpoints (mean = −5.54 kcal/mol). The average folding energy of windows with one, two or three breakpoints are not significantly different from windows with no breakpoints.
Figure 8.
Deletion breakpoints are located in mtDNA regions with high folding potentials. The bar chart depicts the relationship between the mean folding potential (ΔG values) and the number of breakpoints in the midpoint position of 100-nt sliding windows covering the entire mitochondrial genome. The results of the statistical tests (Student’s t-test; two-sided P-values) show that windows with more breakpoints have a higher folding potential than windows where breakpoints are rare.
DISCUSSION
Since the first reports of mtDNA deletions, scientists have noticed the presence of non-B DNA structures encompassing or in the vicinity of the breakpoints. Just 1 year after the first description of deletions in human mtDNA (37,55) called the attention to the fact that the human mtDNA contains long regions with the potential to form ‘bent DNA’, including around and within the 13-nt repeats that flank the 4977-bp ‘common deletion’ (8470–8482 to 13 447–13 459). The authors noticed that polypyrimidine tracts and AT-rich regions around breakpoints may render such regions susceptible to the formation of single-stranded DNA on supercoiling. Consistent with these observations, we found that the 3′-breakpoint of the ‘common deletion’ is one of the most frequent breaking sites (position 13 447) of mtDNA and occurs within or near a stable hairpin (Figure 2 and Supplementary Figure S9). No hairpin element has been detected associated with the 5′-breakpoint of the ‘common deletion’, although it has been shown by 2D gel electrophoresis that this genomic region exhibits retarded mobility due to the putative formation of bent DNA structures (32,33).
In the first description of an autosomal disorder causing multiple mtDNA deletions (38), a hotspot for deletion formation was identified near the D-loop 3′-termini (at positions 16 068–16 079). This region is now recognized as a preferential location of mtDNA breakage in both pathological and non-pathological conditions (31,56,57). In their pioneering study, Zeviani and colleagues identified two stable hairpins around this 3′-breakpoint hotspot. We have recently discovered that these hairpins are part of a highly conserved 93-nt cloverleaf-like structure (12). In fact, more than one quarter (201 out of 753) of all of the 3′-breakpoints occur at this cluster of hairpins, most of them at the 8-nt terminal loop (Figures 2 and 3A). The preference for breakage at single-stranded regions exposed by stable stem elements is also common to other mtDNA regions (Figure 2, Supplementary Figure S8 and S9). The deletion hotspot around position 16 071 is located near the trinucleotide stop point (16 104–16 106) that is associated with the premature arrest of the H-strand synthesis that forms a three-stranded DNA structure known as D-loop or displacement loop (58) (Figure 4A). Its location outside of the three-stranded D-loop structure, but still very close to its 3′-end, points to a possible relationship between the process of deletion formation and the functional/structural features of the D-loop. Indeed, the formation of the D-loop induces superhelical tension to the mtDNA in solution, at least in Xenopus laevis oocytes (59), which might cause distortions of the canonical B-DNA. The complete elucidation of the function(s) of the D-loop under normal physiological conditions will help to understand such potential relationships. Its involvement in mtDNA organization, segregation and replication have been hypothesized (60,61). The mtDNA breakage hotspot at the D-loop 3′-end is not only a feature of human mtDNA. It has been repeatedly found in the homologous region of the mtDNA of transgenic mice with mitochondrial disease (48,62,63). We found that all breaks reported between the CYTB gene and the control region of this transgenic mice (48) occur in hairpin elements (Figure 3). As suggested by (48), our data also indicate that the general mechanism underlying multiple deletions with site-specific breakpoints is not due to the primary sequence itself but to its secondary structure or functional location.
The association between hairpin structures and breakpoints is also clear in the WANCY cluster of tRNAs that surrounds the main OL (Figure 4B). Our different computational analyses demonstrated that this region stands out as having the highest folding potential of the mitochondrial genome (Figure 6B, C, Supplementary Figures S20 and S21), with all of the reported 5′-deletion breakpoints occurring in hairpin elements (Figure 4B and Supplementary Figure S11). The genomic instability at the WANCY cluster is also perceptible when comparing vertebrate mitochondrial genomes: it is a hotspot of gene order rearrangements by tandem duplication and random loss of genes (64). In fact, the breakpoints of such rearrangements observed in different phylogenetic groups are often thought to involve hairpin elements (65).
Our large dataset revealed that deletion breakpoints are over-represented in tRNA genes (Figure 5 and Supplementary Figure S13). The structural elements of tRNA molecules, which are well known for playing important biological roles, explain the high folding potential observed in the DNA regions encoding them [e.g. ΔG = −5.1 kcal/mol predicted for the tRNA-Ser(UCN) gene]. It is therefore likely that the formation of secondary structures at tRNA genes, for instance, during transcription or replication when the mtDNA is single-stranded, may contribute to the formation of the deletions, as previously suggested (34,35). The ability of tRNA genes to mediate genomic rearrangements is well documented in the mitochondrial and other genomes (65–67). A notable example detected in our study is the breakpoint-prone tRNA-Leu(UUA/G) gene, which has a significantly higher number of breakpoints than its flanking genes (Figure 5). Although it is located in the minor arc, it surrounds 1 of the top 10 mtDNA breakage hotspots, at position 3263 (Figure 2).
The non-random genomic distribution of the deletion breakpoints indicates that the root cause of mtDNA vulnerability to breakage resides either in specific characteristics of the local DNA sequence environment or in higher order features of the genomic architecture. We found that the number of deletion breakpoints in a particular mtDNA position is associated with the folding capacity of the region where it occurs. The genomic regions with more breakpoints have a significant higher folding potential than regions with a low number of breakpoints (Figures 7 and 8). In agreement with this observation, we detected several mtDNA breakage hotspots (e.g. mtDNA positions 16 071, 7402, 5787 or 15 435), with cases of different deletions sharing the same breakpoint at one end (Figures 1, 2, Supplementary Figures S4 and S5). Similarly, our data shows that site-specific breakpoint hotspots exist in the mitochondrial genome, as demonstrated by the significant difference of two orders of magnitude observed between the highest frequencies of breakage in real and simulated data (Supplementary Figure S7). There is now abundant evidence showing that non-B DNA is more prone to DNA breaks than B-DNA (39–43). For example, the formation of secondary structure intermediates between DNA ends at translocation or gross deletion breakpoints is common in human inherited diseases and cancer (68) and non-B DNA-forming sequences are enriched in breakpoints of copy number variations (69) and chromosomal rearrangements (40–42). It is possible that such extruded bases are more prone to breakage by mechanical or chemical stress. Such conformations can also be the template for the action of trans-acting factors, such as structure-specific nucleases (70). Intriguingly, it has been shown that replication pausing in the vicinity of the highly structured WANCY tRNA cluster in a mouse model expressing a mutant mitochondrial polymerase leads to the generation of linear deleted mtDNA molecules (62). Thus the secondary structures have the capacity in vivo to induce breaks in mtDNA that, if rejoined incorrectly, would lead to the mtDNA deletions.
The breaking of the DNA is only one of the many factors that shape the distribution of mtDNA deletions. There are several constraints to the formation of circular deleted mtDNA molecules and their subsequent proliferation in cells that might explain why genomic regions with very high folding potentials (and thus mutational prone) are devoid of detectable breakpoints. It is likely that many breakage sites in mtDNA are not detected just because they are not involved in the formation of circular deleted mtDNA molecules with an efficient replication capacity. Although other constraints might act on mtDNA deletions, the presence of homology at breakpoints and the removal of replication origins are believed to significantly influence their distribution (29–31).
The presence of homology at the edges of mtDNA deletions, including both perfect and imperfect short repeats, is a well-known feature of this type of genomic rearrangement (31,34,37,71). In particular, two 13-nt direct repeats were found associated with most human mtDNA deletions (31). However, an important breakthrough was recently made by Guo et al. (29) by showing that deletion breakpoints coincide with distant segments of mtDNA that are capable of forming stable imperfect duplexes with each other, rather than with the presence of perfect repeats, as previously assumed. In other words, 5′- and 3′-deletion breakpoints tend to have large regions of partial or interrupted homology with, at least, 100 nt (29), which is possibly related with the joining of the distant mtDNA regions rather than with the breakage of the DNA strand. Therefore, only those breakage sites near regions with long stretches of high or moderate sequence homology result in mtDNA deletions. This feature might explain why some putative breakpoint-rich areas are not detected in deletions. For instance, the tRNA-Pro gene (devoid of breakpoints) is located in a region that apparently does not form stable duplexes, as suggested by the white or clear ‘stripes’ around position 16 000 that run across the matrix of free energies made by Guo et al. (29). It is possible that breakpoints in the tRNA-Pro gene remain undetectable just because they do not continue the process of the formation of circular deleted mtDNA molecules through duplex formation at nearby regions with sufficient homology. The development of accurate methods to directly detect breakage sites in the mtDNA will clarify this issue.
The mtDNAs that lose an origin of replication have a limited replication capacity that influences their likelihood of propagation and detection. This feature clearly influences the distribution of mtDNA deletions, which are mostly located inside the major arc (88.8% of the cases) without removing the replication origins (Figure 1). In fact, we only identified 71 deletions (9.43% of all cases) without the OL and one deletion without the OH (0.13% of all cases), as defined by the strand-asynchronous model of mtDNA replication (15,72). It is probably because of their location that the rRNA and tRNA genes inside the minor arc and near OH, do not display a high number of breakpoints [the tRNA-Leu(UUA/G) gene being a noticeable exception]. Large deletions with a breakpoint in this area are likely to remove an origin of replication, ensuring that these molecules cannot be subsequently replicated and thus would be very rare within the cells. Moreover, the profile of folding energy across the rRNA genes is not different from that observed in the rest of the genome (Supplementary Figure S18). The highest folding potential is reached at the end of the RNR1 gene, with a ΔG of −18.19 kcal/mol (L-strand sequence) below the folding capacity of the WANCY region (ΔG of −20.56 kcal/mol, window with midpoint at position 5732). Further work is necessary to uncover all the constraints to the distribution of deletions. Nevertheless, we were able to find a clear association with non-B DNA conformations even taking into account that many breakage hotspots might remain undetectable.
Various hypotheses have been advanced in recent years to explain the formation of mtDNA deletions, each one giving a different prominence to the roles of DNA replication (57,71), recombination (34,37) and repair (30) in the deletion process. Despite the intense debate, there is no compelling evidence unequivocally demonstrating which mechanism (if only one) generates deletions in the various physiological situations. The data presented here indicate that, whatever the process underlying deletions, non-B DNA conformations (intra-strand hairpins and cloverleaf-like elements) should be considered an important piece in the complex puzzle of mitochondrial genomic rearrangements. Unfortunately, our poor knowledge of the in vivo dynamics and organization of mtDNA makes it very difficult to understand exactly how such alternative DNA structures cause deletions. It was recently suggested that mtDNA deletions are most likely to occur during repair of damaged mtDNA (30). The formation of non-B DNA might trigger this process by increase the rate of single and double-strand lesions (39–43). In contrast, several models posit that mtDNA deletions are a replication-associated phenomenon (57,71,73). Previous in vivo and in vitro experiments have shown that non-B DNA acts as a preferential pausing site for DNA polymerases (74,75), which may be an obstacle to fork progression or a target for nucleolytic attack, thus permitting DNA breakage and deletion formation (76–79). The preferential location of 5′- and 3′-breakpoints in the large arc between OH and OL suggests that a strand-asynchronous replicative process is involved in the generation of deletions. Consistent with this assumption, a linear, deleted mtDNA molecule is continuously generated in mice expressing a defective mtDNA polymerase. The polymerase displays elevated replication pausing and breakage at fragile sites just after passing OL, assuming a strand-asynchronous replication mode (62,80). A different mechanism suggests that the formation of a transient DNA triple helix in pyrimidine-rich sequences might guide the slipped mispairing of the replication complex, causing mtDNA rearrangements (36). In addition, the accumulation of multiple mtDNA deletions in individuals with defects in the replicative helicase Twinkle and DNA polymerase γ have been used to support the idea that deletion formation might be induced by replication stalling (57). This process might be related to the breakpoint hotspot in the cluster of hairpins near the D-loop 3′-end (Figure 2A), a putative replication fork barrier.
The formation of hypermutable non-B DNA in certain regions of the mitochondrial genome might indeed be the link between the high incidence of mtDNA deletions in individuals with defects in proteins that move, organize or replicate the mtDNA (which collectively control the mtDNA topology). The inefficient activity of the altered versions of such DNA-interacting proteins might induce considerable changes in the topology and supercoil-state of mtDNA, whose tensional stress might be released by the formation of non-B DNA and the breakage of DNA. In fact, even the normal movement of RNA and DNA polymerases through the mtDNA may generate regions of superhelical tension and other topological alterations, which may be associated with the genesis of mtDNA rearrangements (81). More detailed biochemical and computational studies are needed to verify all of these conjectures. Although the mechanisms remains elusive, our analyses suggest that DNA structure-induced genomic instability seems to be at the heart of the mtDNA deletion process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–22.
FUNDING
Portuguese Foundation for Science and Technology (FCT) [SFRH/BPD/44637/2008]; [PTDC/CVT/100881/2008]; IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partially supported by FCT. Funding for open access charge: Portuguese Foundation for Science and Technology (FCT) [PTDC/CVT/100881/2008].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Cooper DN, Bacolla A, Ferec C, Vasquez KM, Kehrer-Sawatzki H, Chen JM. On the sequence-directed nature of human gene mutation: the role of genomic architecture and the local DNA sequence environment in mediating gene mutations underlying human inherited disease. Hum. Mutat. 2011;32:1075–1099. doi: 10.1002/humu.21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates A, Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005. [Google Scholar]
- 5.Mirkin SM. Encyclopedia of Life Sciences (ELS) Chichester, West Sussex, UK: John Wiley & Sons, Ltd; 2001. DNA topology: fundamentals. [Google Scholar]
- 6.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuste JM, Wanrooij S, Jemt E, Granycome CE, Cluett TJ, Shi Y, Atanassova N, Holt IJ, Gustafsson CM, Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Hixson JE, Wong TW, Clayton DA. Both the conserved stem-loop and divergent 5'-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J. Biol. Chem. 1986;261:2384–2390. [PubMed] [Google Scholar]
- 9.Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, Coll M, Bernado P, Solà M. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 11.Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira F, Soares P, Carneiro J, Pereira L, Richards MB, Samuels DC, Amorim A. Evidence for variable selective pressures at a large secondary structure of the human mitochondrial DNA control region. Mol. Biol. Evolut. 2008;25:2759–2770. doi: 10.1093/molbev/msn225. [DOI] [PubMed] [Google Scholar]
- 13.Lilley DM. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc. Natl Acad. Sci. USA. 1980;77:6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panayotatos N, Wells RD. Cruciform structures in supercoiled DNA. Nature. 1981;289:466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- 15.Clayton DA. Mitochondrial DNA replication: what we know. IUBMB Life. 2003;55:213–217. doi: 10.1080/1521654031000134824. [DOI] [PubMed] [Google Scholar]
- 16.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 17.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 18.Dayn A, Malkhosyan S, Mirkin SM. Transcriptionally driven cruciform formation in vivo. Nucleic Acids Res. 1992;20:5991–5997. doi: 10.1093/nar/20.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc. Natl Acad. Sci. USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 21.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J. Biol. Chem. 2008;283:36743–36751. doi: 10.1074/jbc.M806174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 24.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 25.Cree LM, Samuels DC, Chinnery PF. The inheritance of pathogenic mitochondrial DNA mutations. Biochim. Biophys. Acta Mol. Basis Disease. 2009;1792:1097–1102. doi: 10.1016/j.bbadis.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiMauro S, Hirano M. Mitochondrial DNA deletion syndromes. In: Pagon R, Bird T, Dolan C, Stephens K, editors. GeneReviews. Seattle: University of Washington; 2003. [Google Scholar]
- 27.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat. Rev. Genet. 2008;9:657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 28.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X, Popadin KY, Markuzon N, Orlov YL, Kraytsberg Y, Krishnan KJ, Zsurka G, Turnbull DM, Kunz WS, Khrapko K. Repeats, longevity and the sources of mtDNA deletions: evidence from ‘deletional spectra’. Trends Genet. 2010;26:340–343. doi: 10.1016/j.tig.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. What causes mitochondrial DNA deletions in human cells? Nat. Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 31.Samuels DC, Schon EA, Chinnery PF. Two direct repeats cause most human mtDNA deletions. Trends Genet. 2004;20:393–398. doi: 10.1016/j.tig.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Hou JH, Wei YH. The unusual structures of the hot-regions flanking large-scale deletions in human mitochondrial DNA. Biochem. J. 1996;318 (Pt 3):1065–1070. doi: 10.1042/bj3181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou JH, Wei YH. AT-rich sequences flanking the 5′-end breakpoint of the 4977-bp deletion of human mitochondrial DNA are located between two bent-inducing DNA sequences that assume distorted structure in organello. Mutat. Res. 1998;403:75–84. doi: 10.1016/s0027-5107(98)00054-2. [DOI] [PubMed] [Google Scholar]
- 34.Mita S, Rizzuto R, Moraes CT, Shanske S, Arnaudo E, Fabrizi GM, Koga Y, DiMauro S, Schon EA. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 1990;18:561–567. doi: 10.1093/nar/18.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishigaki Y, Marti R, Hirano M. ND5 is a hot-spot for multiple atypical mitochondrial DNA deletions in mitochondrial neurogastrointestinal encephalomyopathy. Hum. Mol. Genet. 2004;13:91–101. doi: 10.1093/hmg/ddh010. [DOI] [PubMed] [Google Scholar]
- 36.Rocher C, Letellier T, Copeland WC, Lestienne P. Base composition at mtDNA boundaries suggests a DNA triple helix model for human mitochondrial DNA large-scale rearrangements. Mol. Genet. Metab. 2002;76:123–132. doi: 10.1016/s1096-7192(02)00024-0. [DOI] [PubMed] [Google Scholar]
- 37.Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeviani M, DiMauro S. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science. 1989;244:346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- 38.Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 39.Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O'Connell CD, Cooper DN, Wells RD. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darmon E, Eykelenboom JK, Lincker F, Jones LH, White M, Okely E, Blackwood JK, Leach DR. E. coli SbcCD and RecA control chromosomal rearrangement induced by an interrupted palindrome. Mol. Cell. 2010;39:59–70. doi: 10.1016/j.molcel.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inagaki H, Ohye T, Kogo H, Kato T, Bolor H, Taniguchi M, Shaikh TH, Emanuel BS, Kurahashi H. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 43.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc. Natl Acad. Sci. USA. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mani RS, Chinnaiyan AM. Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat. Rev. Genet. 2010;11:819–829. doi: 10.1038/nrg2883. [DOI] [PubMed] [Google Scholar]
- 45.Fan L, Yao YG. MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion. 2011;11:351–356. doi: 10.1016/j.mito.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC, Eaton M, Hamady M, Lindsay H, Liu Z, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl Acad. Sci. USA. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 50.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 52.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadikovic B, Wang J, El-Hattab A, Landsverk M, Douglas G, Brundage EK, Craigen WJ, Schmitt ES, Wong LJ. Sequence homology at the breakpoint and clinical phenotype of mitochondrial DNA deletion syndromes. PLoS One. 2010;5:e15687. doi: 10.1371/journal.pone.0015687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 56.Kajander OA, Rovio AT, Majamaa K, Poulton J, Spelbrink JN, Holt IJ, Karhunen PJ, Jacobs HT. Human mtDNA sublimons resemble rearranged mitochondrial genomes found in pathological states. Hum. Mol. Genet. 2000;9:2821–2835. doi: 10.1093/hmg/9.19.2821. [DOI] [PubMed] [Google Scholar]
- 57.Wanrooij S, Luoma P, van GG, van BC, Suomalainen A, Spelbrink JN. Twinkle and POLG defects enhance age-dependent accumulation of mutations in the control region of mtDNA. Nucleic Acids Res. 2004;32:3053–3064. doi: 10.1093/nar/gkh634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kasamatsu H, Robberson DL, Vinograd J. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc. Natl Acad. Sci. USA. 1971;68:2252–2257. doi: 10.1073/pnas.68.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Callen JC, Tourte M, Dennebouy N, Mounolou JC. Changes in D-loop frequency and superhelicity among the mitochondrial DNA molecules in relation to organelle biogenesis in oocytes of Xenopus laevis. Exp. Cell Res. 1983;143:115–125. doi: 10.1016/0014-4827(83)90114-3. [DOI] [PubMed] [Google Scholar]
- 60.Antes A, Tappin I, Chung S, Lim R, Lu B, Parrott AM, Hill HZ, Suzuki CK, Lee CG. Differential regulation of full-length genome and a single-stranded 7S DNA along the cell cycle in human mitochondria. Nucleic Acids Res. 2010;38:6466–6476. doi: 10.1093/nar/gkq493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN. Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion. 2007;7:311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Bailey LJ, Cluett TJ, Reyes A, Prolla TA, Poulton J, Leeuwenburgh C, Holt IJ. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 2009;37:2327–2335. doi: 10.1093/nar/gkp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.San MD, Gower DJ, Zardoya R, Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol. Biol. Evol. 2006;23:227–234. doi: 10.1093/molbev/msj025. [DOI] [PubMed] [Google Scholar]
- 65.Stanton DJ, Daehler LL, Moritz CC, Brown WM. Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics. 1994;137:233–241. doi: 10.1093/genetics/137.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cantatore P, Gadaleta MN, Roberti M, Saccone C, Wilson AC. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature. 1987;329:853–855. doi: 10.1038/329853a0. [DOI] [PubMed] [Google Scholar]
- 67.Juhling F, Putz J, Bernt M, Donath A, Middendorf M, Florentz C, Stadler PF. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012;40:2833–2845. doi: 10.1093/nar/gkr1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chuzhanova N, Abeysinghe SS, Krawczak M, Cooper DN. Translocation and gross deletion breakpoints in human inherited disease and cancer II: Potential involvement of repetitive sequence elementsin secondary structure formation between DNA ends. Hum. Mutat. 2003;22:245–251. doi: 10.1002/humu.10253. [DOI] [PubMed] [Google Scholar]
- 69.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang YJ, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bzymek M, Saveson CJ, Feschenko VV, Lovett ST. Slipped misalignment mechanisms of deletion formation: in vivo susceptibility to nucleases. J. Bacteriol. 1999;181:477–482. doi: 10.1128/jb.181.2.477-482.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shoffner JM, Lott MT, Voljavec AS, Soueidan SA, Costigan DA, Wallace DC. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc. Natl Acad. Sci. USA. 1989;86:7952–7956. doi: 10.1073/pnas.86.20.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robberson DL, Kasamatsu H, Vinograd J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc. Natl Acad. Sci. USA. 1972;69:737–741. doi: 10.1073/pnas.69.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buroker NE, Brown JR, Gilbert TA, Ohara PJ, Beckenbach AT, Thomas WK, Smith MJ. Length heteroplasmy of sturgeon mitochondrial DNA - an illegitimate elongation model. Genetics. 1990;124:157–163. doi: 10.1093/genetics/124.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weaver DT, Depamphilis ML. Specific sequences in native DNA that arrest synthesis by DNA polymerase-alpha. J. Biol. Chem. 1982;257:2075–2086. [PubMed] [Google Scholar]
- 76.Bierne H, Ehrlich SD, Michel B. Deletions at stalled replication forks occur by two different pathways. EMBO J. 1997;16:3332–3340. doi: 10.1093/emboj/16.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bzymek M, Lovett ST. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: single-strand annealing and replication slipped mispairing. Genetics. 2001;158:527–540. doi: 10.1093/genetics/158.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glickman BW, Ripley LS. Structural intermediates of deletion mutagenesis - a role for palindromic DNA. Proc. Natl Acad. Sci. USA Biol. Sci. 1984;81:512–516. doi: 10.1073/pnas.81.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Madsen CS, Ghivizzani SC, Hauswirth WW. In-vivo and in-vitro evidence for slipped mispairing in mammalian mitochondria. Proc. Natl Acad. Sci. USA. 1993;90:10409. doi: 10.1073/pnas.90.16.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly Y, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 81.Tengan CH, Moraes CT. Duplication and triplication with staggered breakpoints in human mitochondrial DNA. Biochim. Biophys. Acta. 1998;1406:73–80. doi: 10.1016/s0925-4439(97)00087-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.