Abstract
The molecular basis of cell signal-regulated alternative splicing at the 3′ splice site remains largely unknown. We isolated a protein kinase A-responsive ribonucleic acid (RNA) element from a 3′ splice site of the synaptosomal-associated protein 25 (Snap25) gene for forskolin-inhibited splicing during neuronal differentiation of rat pheochromocytoma PC12 cells. The element binds specifically to heterogeneous nuclear ribonucleo protein (hnRNP) K in a phosphatase-sensitive way, which directly competes with the U2 auxiliary factor U2AF65, an essential component of early spliceosomes. Transcripts with similarly localized hnRNP K target motifs upstream of alternative exons are enriched in genes often associated with neurological diseases. We show that such motifs upstream of the Runx1 exon 6 also bind hnRNP K, and importantly, hnRNP K is required for forskolin-induced repression of the exon. Interestingly, this exon encodes the peptide domain that determines the switch of the transcriptional repressor/activator activity of Runx1, a change known to be critical in specifying neuron lineages. Consistent with an important role of the target genes in neurons, knocking down hnRNP K severely disrupts forskolin-induced neurite growth. Thus, through hnRNP K, the neuronal differentiation stimulus forskolin targets a critical 3′ splice site component of the splicing machinery to control alternative splicing of crucial genes. This also provides a regulated direct competitor of U2AF65 for cell signal control of 3′ splice site usage.
INTRODUCTION
Alternative splicing is a common way of gene regulation contributing greatly to the proteomic diversity particularly in humans and in the nervous system (1–8). It is often controlled by external signals, with important physiological and therapeutic implications (9–11); however, in most cases, the underlying molecular basis remains unclear.
Alternative splicing is mostly controlled by multiple cis-acting pre-messenger ribonucleic acid (mRNA) elements and their trans-acting factors in mammalian systems (3,12). Their positive (enhancers/activators) and negative (silencers/repressors) effects in combination modulate the assembly of constitutive splicing factors of the spliceosome and thus the inclusion level of an alternative exon (12,13). Such a mode of regulation requires the isolation of individual regulatory elements/factors from the endogenous genes to dissect each of their roles in splicing, particularly by upstream cell signals.
The 3′ splice sites at intron ends are generally composed of three consensus motifs: the branch point, the polypyrimidine tract and the 3′ AG (14). These motifs could also be targets of a number of trans-acting regulatory proteins (15–22), besides constitutive splicing factors. Particularly, the polypyrimidine tract, bound by the constitutive U2 auxiliary splicing factor U2AF65 in the early steps of spliceosome assembly, is a target of several regulatory factors including the Sex-lethal (Sxl) in drosophila and the polypyrimidine tract-binding proteins [PTB or heterogeneous nuclear ribonucleo-protein (hnRNP) I] in mammals (16–18,22,23). HnRNP K, another hnRNP member, also prefers pyrimidine-rich motifs [UCCC (U/A)-] (24) and plays important roles in splicing control (25–29), but its role in 3′ splice site regulation has not been observed. Moreover, whether these factors are involved in 3′ splice site regulation by upstream signals remains unknown.
We have been studying the control of alternative splicing by extracellular factors and intracellular signals (30–36). A system we have been examining is the change of pre-mRNA splicing/protein factors during neuronal differentiation of rat pheochromacytoma PC12 cells by the forskolin/protein kinase A (PKA) pathway (32,34,36,37). Observations in other systems also support that this pathway controls alternative splicing (38–41).
Here we examined forskolin/PKA-regulated splicing through the upstream 3′ splice site of the exon 5a of the synaptosomal-associated protein 25 (Snap25) gene during neuronal differentiation. This identified hnRNP K as an essential factor competing with U2AF65 for the polypyrimidine tract in forskolin/PKA-regulated splicing of Snap25 and other critical neuronal genes.
MATERIALS AND METHODS
Plasmid construction
The plasmid for HA-hnRNP K recombinant protein is a generous gift of Dr Ze’ev Ronai (Mont Sinai School of Medicine, New York, NY, USA), as described previously (42). The His-tagged hnRNP K plasmid was made by inserting the open reading frame of hnRNP K between the EcoRI and XhoI restriction sites of pET28a.
For Snap25 splicing reporter minigenes, a 1.6-kb fragment starting from 565 bp upstream of exon 5a to 602 bp downstream of exon 5b was amplified from mouse genomic deoxyribonucleic acid (DNA) and inserted between the ApaI and BglII sites of the vector DUP-175 (30). Further deletions/replacements were made based on this template (Figure 1).
Figure 1.
Isolation of a KARRE from the upstream 3′ splice site of the exon 5a of Snap25 gene based on forskolin reduction of 5a/5b ratios in PC12 cells during neuronal differentiation. (A) Forskolin-induced neuronal differentiation of PC12 cells. Shown are representative images of PC12 cells without or with forskolin addition for 18 h. (B) Diagram of the alternative splicing of the exons 5a and 5b of the Snap25 gene. Arrows: sites of restriction enzymes NdeI (5a) and AvaII (5b) used to digest the PCR products. Arrowheads: PCR primers. (C) RT-PCR products from untreated (−) and treated PC12 cells. ETOH: ethanol, vehicle for forskolin and Dex (dexamethasone); H89: a PKA-specific inhibitor; NC: PCR negative control; M: molecular size marker. (D) Diagram of the deletion/replacement mutants of Snap25 splicing reporters and their exon inclusion levels in the presence of PKAm or active PKA. Asterisks: indicating P value levels in one tail, paired Student’s t-test (***P < 0.001, *P < 0.05, n ≥ 3). (E) An agarose gel showing the PKA response of the heterologous S175NK reporter containing the 17 nt Snap25a element upstream of a stronger exon (175NK, about 75% inclusion with PKAm coexpression).
Cell culture and reverse transcriptase-polymerase chain reaction (RT-PCR)
Cultures of PC12 and human embryonic kidney (HEK) 293T cells and reverse transcriptase-polymerase chain reaction (RT-PCR) were described previously (31,37). Overnight cultures of PC12 cells were treated with cpt-cAMP (8-(4-Chlorophenylthio)adenosine 3′,5′-cyclic monophosphate sodium salt, 100 µM), forskolin (10 µM), KCl (25 mM) or dexamethasone (100 µM) for 6 hours before RNA extraction. Some cultures were pretreated with 10 µM of H89 (N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamidedihydrochloride, Sigma, #B1427) for 10 min. To differentiate products of Snap25 endogenous mRNA transcripts from exons 5a and 5b, we digested 1.0 µl of 32P-labelled-PCR products with AvaII and NdeI restriction enzymes in buffer 4 (New England Biolabs) in a 10 -µl reaction at 37°C for 1 h and run in 6% denaturing polyacrylamide gel electrophoresis (PAGE) gel. For minigene splicing reporters, 24 cycles of PCR were carried out and products resolved in 3% agarose gels stained with ethidium bromide. For Runx1 splicing analysis using RT-PCR, 35 cycles of PCR was carried out with primers Runx1S: 5′-AGTTGCCACCTACCATAGAGC-3′ and Runx1A: 5′-AGAGGCTGGTCATGCCACTG-3′.
Western blot and stripping/reprobing
This has been described by Yu et al. (31). The anti-hnRNP K/J (3C2, sc-32307) and anti-hnRNP F/H (1G11, sc-32310) monoclonal antibodies were purchased from Santa Cruz Biotech. The anti-U2AF65 monoclonal antibody (MC3, U4758) is from Sigma Aldrich Co.
Biotin-RNA pull-down assay
Biotin-RNA probes were synthesized by Integrated DNA Technologies. Biotin-Snap25 RNA pull-down assay for mass spectrometric analysis was adapted from that by Hall-Pogar et al. (43). Basically, 400 µg (∼100 µl) of HeLa nuclear extracts were precleared with 20 µl of streptavidin beads (Cat# 53113, Thermo Scientific) containing yeast transfer RNA (200 ng/µl) at room temperature and then incubated with 10 nmol of either wild type or mutant biotin-Snap25 RNA probes at 30°C for 20 min. The resulted mixture was incubated with yeast transfer RNA (200 ng/µl)-packed beads on ice for 30 min. After centrifugation, the beads were washed with 1 ml of radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris-Cl pH 7.6, 150 mM NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS) and 1 mM phenylmethanesulfonyl fluoride (PMSF)] for three times, boiled in 6 × SDS sample loading buffer for 5 min and then subject to 4–20% SDS-PAGE gel analysis followed by Coomassie Blue staining. The bands were excised and sent for mass spectrometric analysis (The Southern Alberta Mass Spectrometry centre, University of Calgary). For hnRNP K and U2AF65 binding analysis, the biotin-RNA pull-down assay was performed in a smaller scale (35 µl).
Purification of His-tagged hnRNP K and phosphorylation assay
The protein purification and in vitro phosphorylation by PKA was carried out as previously described by Xie et al. (34).
Preparation of nuclear extracts
Small scale preparation of PC12 and large scale preparation of HeLa nuclear extracts were according to Yu et al. (31).
Ultraviolet (UV) cross-linking
The basic procedure was carried out at 254 nm according to Yu et al. (31), except that 5 × 105 cpm of probe was used in each cross-linking reaction. In addition, 2′-thio-UTP (1:4 ratio to UTP) was added to the in vitro transcription reaction for higher cross-linking efficiency with hnRNP K, whose three KH-type RNA binding domains contain only two aromatic amino acid residues. However, cross-linking signals at 365 nm were not observed, likely with minimal contribution if any from the modified UTP in this experiment. Pretreatment of nuclear extracts with λ protein phosphatase (PPase) was according to Yu et al. (31).
RNA interference
This was according to the previous procedure (31). The short hairpin RNA (shRNA) targeting the rat hnRNP K nucleotides 1769–1787 (GenBank accession # BC061867): 5′-cagcagcagagtgagtgac-3′, at the 3′ untranslated region, was cloned as a hairpin sequence: 5′-GATCCCGcagcagcagagtgagtgacTTCAAGAGAgtcactcactctgctgctgTTTTTTGGAAA-3′, similarly as for hnRNP L, into the lentiviral vector FG12 (31,44).
Analysis of clustered functions of the 3′ splice site-TCCCT-containing genes
The functional analyses were generated through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Six sets of 46 genes randomly picked from 18 193 known human genes from BioMart (http://www.biomart.org/) were used as controls in the analysis against the same database.
Measurement of neurite densities
Circles around the centre of each image were drawn, and the numbers of neurites that cross with the perimeters were counted. Fold changes of the numbers were compared between circles of the same diameters of different image samples.
RESULTS
Isolation of a PKA-responsive RNA element (KARRE) based on forskolin-regulated alternative splicing of Snap25 during neuronal differentiation of PC12 cells
In studying alternative splicing during neuronal differentiation, we treated PC12 cells with ethanol or forskolin. The cells differentiate into neurons with interweaving neurites on forskolin treatment (Figure 1A), as reported previously (37). We then examined the alternative splicing of various exons of genes that are known to be important for neuronal development or function, including the mutually exclusive exons 5a and 5b of Snap25 (Figure 1B). In untreated or ethanol-treated cells, the intensity ratio of the product 5a to 5b was about 2.5, as shown by semiquantitative RT-PCR using a 32P-labelled primer (Figure 1C, lanes 2 and 3). On forskolin treatment, the 5a intensity was reduced accompanying an increase in 5b (lane 4), leading to a significant decrease in the 5a/5b ratio (from about 2.5 to 1.3), in resemblance to the critical developmental switch from 5a to 5b seen in animals (45,46). A similar decrease was observed in cpt-cAMP-treated cells (lane 6) and to a lesser extent in KCl-treated cells (lane 8), as reported previously (47), but not in dexamethasone-treated cells (lane 9). Importantly, pretreatment with H89, an inhibitor of the PKA pathway (48), significantly inhibited the effect of forskolin or cpt-cAMP (Figure 1C, lanes 5 and 7). In contrast, H89 did not inhibit the forskolin effect on a CaMKK2 exon (36). Moreover, the STREX (stress axis-regulated) exon, of the Slo gene (49), was not changed by forskolin or cpt-cAMP in these cells (data not shown). Taken together, these results suggest that forskolin/cpt-cAMP specifically regulates the alternative splicing of the Snap25 gene, likely through the PKA pathway, in PC12 cells.
Figure 2.
Identification of hnRNP K and U2AF65 as specific factors binding to the KARRE. (A) Alignment of the KARRE-containing upstream 3′ splice sites of Snap25 exon 5a from different species. The TCCCT, as well as a TCCT, (with heavy bars above) is mostly conserved within the polypyrimidine tract (boxed) among vertebrates. The 17 nt mouse KARRE sequence is above the dotted heavy line. (B) Sequence of the Biotin-RNA wild type and mutant probes used to pull down 3′ splice site-binding proteins, with the hnRNP K target consensus motifs underlined. (C) A Coomassie-stained SDS-PAGE gel of proteins pulled down from HeLa nuclear extracts using the probes in B. 1: nucleolin, 2: hnRNP K, 3: PTB, 4: YB-1, as identified in mass spectrometry with significance against random events. (D) Human hnRNP K protein sequence, with the tryptic peptide hits in MALDI-TOF mass spectrometry highlighted and underlined. (E) A Western blot of U2AF65 proteins similarly pulled down as in C.
To isolate a Snap25 gene fragment sufficient to mediate the PKA effect, we made a series of mouse Snap25 minigene splicing reporters containing either both the Snap25 exons or only single exons with partial flanking introns (Figure 1D). These reporters were transfected into HEK293T cells, and their responses to coexpressed Flag-PKA or its kinase-dead mutant Flag-PKAm, as used previously (32), were examined by semiquantitative RT-PCR. From these preliminary tests, the only observed effect that is consistent with the endogenous exon changes in PC12 cells was a repression by PKA in several reporters containing exon 5a only (Figure 1D).
By serial deletion/replacement of the flanking introns of 5a, we found that the exon inclusion was increased between reporters DUP-5a4 and DUP-5a5 or decreased between DUP-5a2 and DUP-5a3 when coexpressed with the PKAm (Figure 1D). These changes indicate that there are likely splicing silencers within nucleotides 17 to 1 or enhancers within 64 to 33 in the upstream intron, suggesting the control of exon 5a inclusion by both positive and negative regulatory elements.
We observed the PKA reduction of exon 5a from reporters DUP-5a1 to DUP-5a4 where the upstream intron was shortened to only 17 nt. Replacing the 17 nt with the corresponding vector sequence strongly promoted exon 5a inclusion to almost 100% in DUP-5a5, which was only slightly reduced by PKA. In contrast, transferring the 17 nt to the upstream of the vector 175 nt exon conferred strong repression of the heterologous exon by PKA (Figure 1D, S175, ∼60% reduction relative to the level with PKAm). The PKA effect was still kept even when the 17 nt was transferred to the upstream of a stronger heterologous exon derived from the DUP-175 (S175NK, Figure 1E). Therefore, the 17 nt at the upstream 3′ splice site is a major PKA-responsive RNA element (KARRE) that is sufficient to confer PKA response on a heterologous gene.
HnRNP K, as well as U2AF65, specifically binds the KARRE
To identify potential trans-acting factors binding the KARRE, we used biotinylated mouse RNA probes containing the 17 nt (WT) or its mutant (Mut) to pull down proteins from HeLa cell nuclear extracts (Figures 2A and B). In the resulting SDS-PAGE gel, we observed a wild type RNA-specific band close to 60 kD (Figure 2C, WT, band 2), which was almost abolished by the mutations. Matrix-assisted laser desorption/ionisation-time of flight (MALDI-TOF) mass spectrometric analysis of peptide mass fingerprints identified the protein as hnRNP K [Figure 2D, Mascot protein score = 84, a number of -10*Log(P) where P is the probability that the observed match is a random event (50)]. Consistently, within the Biotin-RNA probe, there are two hnRNP K target consensus motifs ‘UCCCU’ and ‘UCCU’, both highly conserved among vertebrates (Figure 2A), which were mutated to ‘UGGGA’ or ‘AGGA’ in the mutant (Figure 2B). In contrast, the commonly known PTB and others showed no specificity for the WT and Mut probes (Figure 2C, bands 1, 3 and 4), further supporting the specific binding of hnRNP K to the pyrimidine-rich sequences.
To examine the binding of U2AF65, which interacts with polypyrimidine tracts at the 3′ splice site, to these RNA probes, we carried out Western blots using anti-U2AF65 antibody with samples similarly pulled down from the nuclear extracts. The result showed that U2AF65 was strongly detected in the wild type probe sample as expected. In contrast, it was undetectable in the mutant sample (Figure 2E), suggesting that U2AF65 binds specifically to the polypyrimidine tract as well.
Taken together, both hnRNP K and U2AF65 bind the KARRE in a polypyrimidine tract-dependent way, raising a question whether hnRNP K functions by directly competing with U2AF65 binding to the same site.
HnRNP K is a forskolin-regulated competitor of U2AF65 and an essential splicing repressor of Snap25 exon 5a
To determine whether hnRNP K binding to the 3′ splice site is regulated by forskolin, we carried out similar RNA pull-down assays using nuclear extracts from PC12 cells treated with either ethanol or forskolin. The hnRNP K binding was detectable in the ethanol-treated samples using the wild type RNA (Figure 3A, lane 1). Interestingly, an about 65% increase (P < 0.05, n = 3) of the bound hnRNP K was observed in the forskolin-treated samples with the same input amount of proteins (lane 2). In contrast, no hnRNP K was detected to bind the mutant RNA probe in either sample (lanes 3 and 4).
Figure 3.
Regulation of hnRNP K binding to the KARRE by forskolin and its direct competition with U2AF65. (A) Western blot of hnRNP K proteins pulled down from PC12 nuclear extracts using the biotin-RNA probes in Figure 2B. Et: vehicle ethanol, Fsk: forskolin (10 µM). (B) UV cross-linking immunoprecipitation of hnRNP K in forskolin-treated PC12 nuclear extracts with the 3′ splice site of exon 5a and its sensitivity to pretreatment by PPase. The wild type RNA probe sequence is the same as in Figure 2B except that it is without biotin. The upper panel is a phosphorimage and the lower a Western blot for the hnRNP K protein in the same SDS-PAGE gel. (C) (Upper panel) a phosphorimage of recombinant His-hnRNP K incubated with active or heat-inactivated PKA in the presence of [32P-γ]ATP in in vitro kinase assay. Lower panel is a Western bot image of the same gel showing equal loading of His-hnRNP K. (D) HnRNP K interacts with the endogenous Snap25 pre-mRNA transcript. Above the gel is a diagram of the PCR target pre-mRNA region of Snap25, with thin lines as introns, boxes as exons and arrowheads as locations of PCR primers. The agarose gel shows the RT-PCR products from RNA samples isolated from the nuclear extracts of forskolin-treated PC12 cells, or from immunoprecipitates using anti-hnRNP K (anti-K) or protein G beads. Each RNA sample was treated with DNase I and one of them also with RNase (A + T1) as indicated. a: a band insensitive to either DNase or RNase treatment, probably nonspecific product from the PCR primers. (E) Western blots of hnRNP K and U2AF65 proteins pulled down from HeLa nuclear extracts with increasing amount of His-hnRNP K added, using the wild type biotin-RNA probe in Figure 2B. The blot was first probed with anti-hnRNP K antibody, stripped with SDS buffer and then reprobed with anti-U2AF65. (F) UV cross-linking of hnRNP K and U2AF65 to the 3′ splice site of exon 5a. The hnRNP K consensus motifs (underlined) and its C to G mutations (italicized) are shown above the denaturing PAGE gel. b: a protein band enhanced by the mutation, likely preferring the G tracts in the mutant, at similar size as hnRNP F/H.
Although hnRNP K binding to the RNA probe was increased, we did not detect a corresponding increase in hnRNP K protein level (see later) or its nuclear localization on forskolin treatment (Supplementary Figure S1). Interestingly, PPase pretreatment of the nuclear extracts from forskolin-induced cells consistently reduced hnRNP K binding to the RNA probe in Ultraviolet (UV) cross-linking assays (Figure 3B), suggesting that phosphorylation is required for its efficient binding to the upstream 3′ splice site. Moreover, hnRNP K was also directly phosphorylated by PKA in in vitro kinase assays (Figure 3C). Furthermore, using RT-PCR, the endogenous Snap25 pre-mRNA transcript was detected to be associated with hnRNP K immunprecipitated from forskolin-treated PC12 nuclear extracts (Figure 3D). Therefore, together these data support that hnRNP K is a regulated factor whose binding to the KARRE is enhanced by forskolin treatment in PC12 cells in a polypyrimidine tract- and phosphorylation-dependent way.
To see whether increased hnRNP K binding competes with the binding of U2AF65, we carried out similar RNA pull-down assays for U2AF65 binding by adding recombinant His-tagged hnRNP K proteins to HeLa nuclear extracts. U2AF65 binding was strongly detected in samples without His-hnRNP K (Figure 3E, lane 1). With increasing amounts of His-hnRNP K added, the U2AF65 binding was decreased in an hnRNP K dosage-dependent way (Figure 3E, lanes 2–4). The relative level of bound U2AF65 to hnRNP K ratio was reduced from 100 to 3, an about 30-fold reduction. Therefore, increased hnRNP K binding to the KARRE competes off U2AF65 binding.
To verify that hnRNP K binds directly to the KARRE in the aforementioned effect on U2AF65, we carried out UV cross-linking using HeLa nuclear extracts with the RNA probe or its mutant of the hnRNP K consensus motifs (Figure 3F). The wild type probe strongly cross-linked to a band of about 65 kD (lane 2). Interestingly, the intensity of this band was drastically reduced by mutating the C’s of the hnRNP K consensus motifs to G’s (lane 3). Immunoprecipitation (IP) showed that both hnRNP K and U2AF65 proteins, similar in sizes, were contained in this band region, supporting that hnRNP K binds directly to the KARRE as does U2AF65.
Together these data indicate that hnRNP K is a forskolin-regulated competitor of the constitutive splicing factor U2AF65, therefore implying a critical role for hnRNP K in the forskolin-regulated splicing through the KARRE.
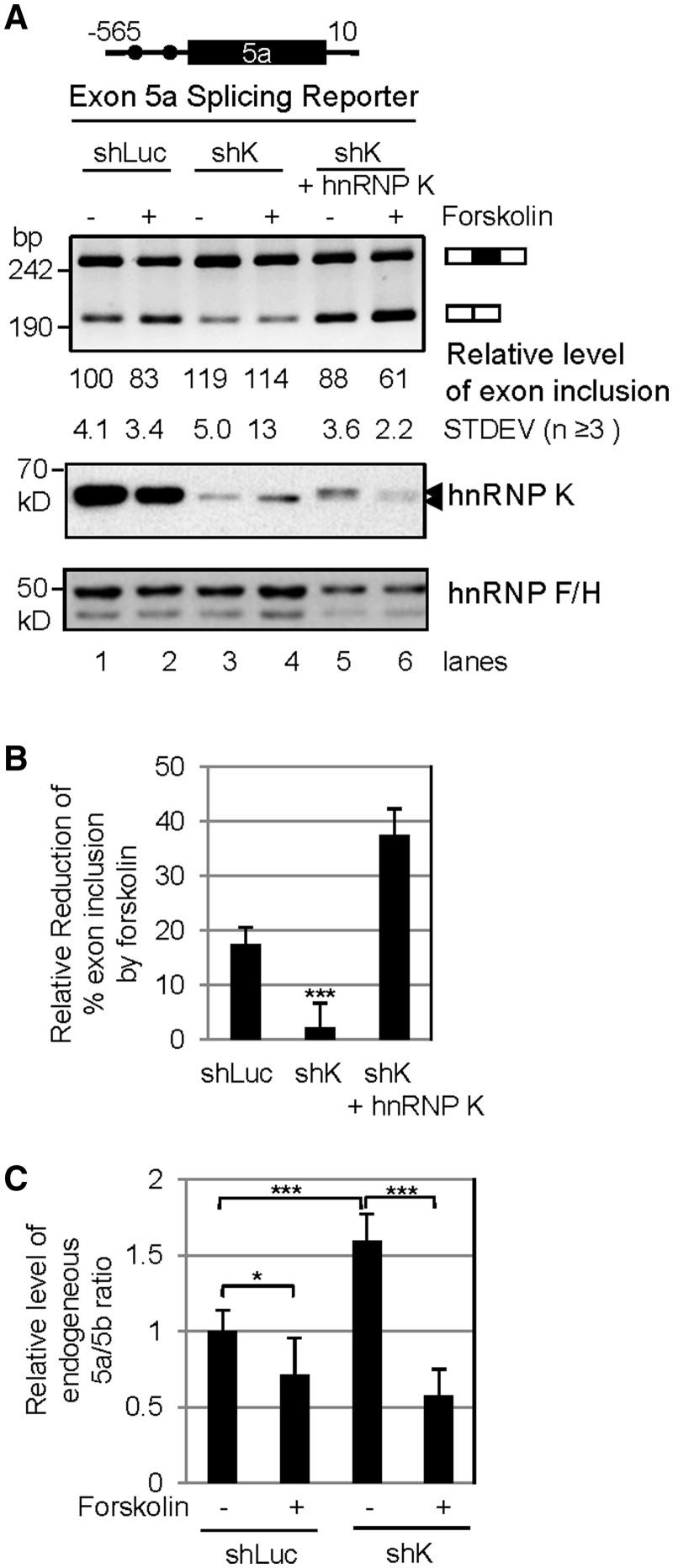
To see whether hnRNP K is required for the forskolin repression of the exon 5a, we tested an exon 5a splicing reporter that is responsive to forskolin in PC12 cells. These cells were knocked down of the hnRNP K protein by RNA interference using lentiviral vectors expressing shRNA against hnRNP K (shK) (Figure 4A). Compared with the control samples (shLuc), knocking down hnRNP K almost abolished the forskolin repression of exon 5a (Figure 4A, lanes 3 and 4, and 4B). To verify the specificity of the knockdown effect, we coexpressed an exogenous hnRNP K protein with the splicing reporter in these cells (lanes 5 and 6). We were able to detect the exogenous protein as bands with slightly slower mobility in Western blots (at an estimated transfection efficiency of 15%). Importantly, the coexpressed exogenous hnRNP K strongly repressed the exon 5a inclusion and restored its response to forskolin. Therefore, hnRNP K is an essential factor for forskolin-repression of exon 5a in the reporter.
Figure 4.
Effect of lentiviral vector-mediated expression of shRNA against hnRNP K (shK) or rescue with an exogenous HA-tagged hnRNP K on forskolin-regulated exon 5a inclusion. (A). Shown are the exon 5a insert (top), an agarose gel of the spliced products (middle) and Western blots of hnRNP K and loading control hnRNP F/H. The intron lengths of Snap25 are indicated above the insert. Dots: hnRNP K consensus motifs. shLuc: control shRNA, against luciferase. The two arrowheads point to the positions of the endogenous (lower) and exogenous (upper) hnRNP K proteins. (B) A bar graph of the relative forskolin-reduction (average ± SD, n = 3) of exon 5a inclusion of the splicing reporter in A, which is calculated as the net reduction of percent exon 5a inclusion relative to its level without forskolin (same as in the followings). (C) A bar graph of endogenous 5a/5b ratio levels in PC12 cells transduced with shK relative to that in the mock- or shLuc-transduced cells without (−) or with (+) forskolin treatment (average ± SD, n ≥ 3). *P < 0.05, ***P < 0.001.
Examination of the endogenous exon 5a in these cells indicates that the level of the exon 5a/5b ratio was increased by about 1.5-fold on hnRNP K knockdown (Figure 4C), indicating that hnRNP K is also an endogenous repressor of exon 5a. This repression is consistent with its competition with U2AF65. However, in contrast to the splicing reporter situation, the forskolin effect was further enhanced (Figure 4C), suggesting that, besides hnRNP K, there are also other elements/factors involved in the forskolin-repression of exon inclusion in PC12 cells, as indicated by our previous study (36) (see also Figures 1C and D and its text).
Taken together, these data indicate that hnRNP K is a forskolin-regulated competitor of U2AF65 and acts as a repressor of Snap25 exon 5a.
A group of transcripts containing similarly localized hnRNP K target motifs within U2AF65 binding regions are highly enriched for genes often associated with neurological diseases
To identify more transcripts containing similarly localized hnRNP K target motifs within U2AF65 binding regions, we searched for the hnRNP K consensus motif TCCCT within 50 nt of the upstream 3′ splice sites of 6189 human exons from the alternative splicing database ASAPII (51). This identified 915 (15%) 3′ splice sites containing at least one copy of the motif, with 47 (5% of the 915) containing at least two copies (Supplementary Table SI), which is sufficient to confer forskolin/PKA or hnRNP K regulation of splicing in heterologous genes (Figures 1–4). The transcripts code for proteins of various functions and subcellular localizations (Supplementary Figure S2A and B). When compared with six sets of randomly chosen human genes, the functions of the 46 genes cluster significantly for cell morphology, cell cycle, neurological diseases, drug metabolism, endocrine system development and functions and lipid metabolism (Supplementary Figure S2C). Strikingly, almost half (22) of the 46 genes are involved in neurological diseases (Figure 5A and Supplementary Tables SI and SII and Supplementary Figure S2), for example, RunX1, a critical gene in neuronal differentiation and associated with mental retardation (52–61), gamma-aminobutyric acid B receptor 1 in depression (62) and latrophilin in synapse exocyotsis and schizophrenia (63). Therefore, there is a group of critical neuronal transcripts containing similarly localized and at least two copies of hnRNP K target motifs, which are sufficient to confer forskolin/PKA- or hnRNP K-regulation of splicing in heterologous genes (Figures 1–4).
Figure 5.
Functional categories of transcripts containing similar hnRNP K target motifs within U2AF65 binding sites and an example for the essential role of hnRNP K in forskolin-regulated splicing of an endogenous neuronal gene. (A) Percentage of the 46 hnRNP K target transcripts/genes in each significant category of biological functions and diseases. These transcripts, containing similar hnRNP K target motifs within U2AF65 binding sites, were identified from the alternative splicing database ASAPII. The categories are as follows: 1, cell morphology; 2, cell cycle; 3, neurological diseases; 4, drug metabolism; 5, endocrine system; 6, inflammatory response and 7, lipid metabolism. (B) Diagram of the upstream 3′ splice site sequences (upper), splicing pattern (middle) and protein domains (lower) of the vertebrate Runx1 (also called acute myeloid leukemia (AML)-1B, GenBank accession #: NM_001754) gene. The relative position of the splice site, exon, encoded peptide and its location in the protein domains are linked with dotted lines. The black dot denotes the 3′ splice site with TCCCT motifs. The 192 nt exon 6-encoded peptide (red) contains the critical sequence (underlined) required for binding by the Sin3A transcriptional corepressor. Runt: Runt homology DNA-binding domain (brown), AD: activation domain (green). Relative locations of the AD domain and Sin3A binding site/activity are based on published data by Lutterbach, with the expected variant transcriptional activities in brackets at the bottom. (C) Sequences of wild type or mutant RunX1 RNA probe for UV cross-linking experiments. Note that all cytidines (underlined) in the hnRNP K target motifs are mutated to guanines. (D) HnRNP K and U2AF65 binding to the upstream 3′ splice site of RunX1 exon 6 in UV cross-linking with HeLa nuclear extracts, carried out similarly as in Figure 3F. On the right (lanes 6 and 7) is a higher contrast image of the IP samples in lanes 4 and 5. a: protein bands also abolished by the mutations, at sizes similar to PTB doublets. b: a protein only in the mutant sample, likely preferring the G tracts, at a size similar to hnRNP F/H. (E) An agarose gel of the Runx1 transcripts in PC12 cells with or without expressing the shRNA against hnRNP K (upper) and a bar graph of the relative reduction of exon 6 inclusion by forskolin (lower, average ± SD, n ≥ 3). **P < 0.01, compared with controls. Arrowheads in B and C demarcate intron–exon junctions.
HnRNP K binds the same polypyrimidine tracts as U2AF65 within the Runx1 transcript and is essential for its regulated alternative splicing by forskolin
To demonstrate that similarly localized hnRNP K target motifs of neuronal genes also mediate splicing regulation by forskolin or hnRNP K, we examined several exons of transcripts that are involved in neurological diseases and found that the Runx1 exon 6 was both alternatively spliced and regulated by forskolin in PC12 cells (Figures 5B–E). The exon skipping loses the corepressor-binding domain of this transcription factor (Figure 5B), whose repressor/activator activity changes specify neuronal lineages (53,64).
To determine whether hnRNP K and U2AF65 also bind directly to the same polypyrimidine tracts at the 3′ splice site, we carried out similar UV cross-linking-IP experiment as in Figure 3F. The upstream 3′ splice site of this exon contains 4–6 copies of TCCCT in mammals and less or none in lower species examined (Figure 5B). UV cross-linking-IP indicates specific direct binding of both hnRNP K and U2AF65 to the polypyrimidine tracts at the Runx1 3′ splice site (Figures 5C and D), as to the Snap25 5a RNA. Interestingly, here the bound hnRNP K signal is almost at the same amount as U2AF65, instead of being half of it as for the Snap25 exon 5a (Figure 3F). This higher hnRNP K/U2AF65 ratio is consistent with the multiple hnRNP K consensus motifs within the Runx1 3′ splice site and hnRNP K competition with U2AF65.
To determine whether hnRNP K is essential for forskolin-regulation of Runx1, we examined the hnRNP K knockdown samples as in Figure 4A. The inclusion level of exon 6 was reduced by about 20% on forskolin addition in shLuc-expressing PC12 cells (Figure 5E, lanes 1 and 2). In contrast, in the hnRNP K knockdown cells, the forskolin-induced reduction was abolished (Figure 5E, lanes 3 and 4, compared with lanes 1 and 2). Thus, hnRNP K is essential for forskolin-induced repression of the endogenous exon 6 of Runx1.
Together these data indicate that hnRNP K, binding the same polypyrimidine tracts at the upstream 3′ splice site as U2AF65, is essential for forskolin-regulated alternative splicing of Runx1 during neuronal differentiation.
HnRNP K is essential for forskolin-induced neurite growth
The enrichment of hnRNP K target transcripts involved in neurological diseases, the critical splicing switch from Snap25 5a to 5b during neuronal differentiation (45,46) and the presence/absence of the Sin3A corepressor binding site in the Runx1 variants (Figure 5B) suggest that hnRNP K plays a critical role in neurons through its competition with U2AF65. One important feature of the forskolin-treated PC12 cells is that they differentiate and grow neurites (Figure 1), as we have also shown in a previous study (37). We thus examined the loss-of-hnRNP K effect on the forskolin-induced neurite growth.
We transduced the PC12 cells with lentivirus expressing shRNA against either luciferase (shLuc) or hnRNP K (shK), as in the earlier experiments (Figures 3D and 5E), and then treated them with forskolin to induce neurite growth. Cells expressing the shLuc still grow neurites abundantly (Figure 6, left). In contrast, cells expressing the shK exhibited much shorter and less neurites (middle). On average, the neurite density in the shK samples is only about half of that in the shLuc control samples (0.54 ± 0.03, n = 5, right). Therefore, knocking down hnRNP K severely disrupts forskolin-induced neurite growth, supporting that hnRNP K plays an essential role for forskolin-induced neuronal differentiation. Considering the critical role of Snap25 splicing switch, Runx1 and other target genes in neurological diseases (Figure 5A and Supplementary Table SII), hnRNP K-regulated splicing of their transcripts through competition with U2AF65 is likely a critical part of its control of neuronal differentiation.
Figure 6.
Effect of knocking down hnRNP K on forskolin-induced neurite growth. Shown are bright field images of PC12 cells expressing shRNA against luciferase (shLuc, control) or hnRNP K (shK) and treated with forskolin (10 µM) for 18 h as indicated. To the right of the images is a bar graph of the levels of neurite densities measured in the two groups (***P < 0.005). Images representative of three samples treated separately.
Taken together, this work demonstrates that forskolin, a stimulus of neuronal differentiation, targets the critical spliceosome component U2AF65 through hnRNP K to control the alternative splicing of critical neuronal genes in this important process (Figure 7).
Figure 7.
Diagram of our proposed model for the observed forskolin effect on exon skipping through direct hnRNP K competition with U2AF65 at the 3′ splice site during neuronal differentiation of PC12 cells. Briefly, on stimulation of PC12 cells with the external stimulus forskolin and activation of the PKA or other pathways, hnRNP K binding to the KARRE motifs at the upstream 3′ splice sites of the exons is enhanced inside the nucleus (gray oval), competing with U2AF65 binding (detailed in the enlarged box with Snap25 and Runx1 3′ splice sites as examples). The hnRNP K target consensus KARRE motifs are in red and underlined. For Snap25, hnRNP K likely acts together with an unknown factor (blue oval with a question mark) for the forskolin repression of exon 5a. For Runx1, hnRNP K alone is sufficient to mediate the forskolin repression. The two hnRNP K proteins above the polypyrimidine tract of Runx1 are to reflect the multiple binding motifs available for the protein to compete with U2AF65. The regulation leads to exon skipping and neurite growth in the presence of the resulting splice variants (such as reduced Snap25 a/b ratio or increased Runx1-E16). Without hnRNP K and this regulation (upper right corner), the splicing switches/changes of a group of neuronal splice variants (Supplementary Tables SI and SII), including Snap25a/b and the Runx1 + E6, are altered contributing to disrupted neurite growth. Black dot: potential branch point of Runx1. The upstream branch point of Snap25 exon 5a is further upstream according to our experiment data.
DISCUSSION
Little has been known about how external stimuli target the 3′ splice sites to regulate splicing. The identification of hnRNP K as a direct competitor of U2AF65 for the 3′ polypyrimidine tract of forskolin-controlled neuronal genes provides a straightforward way for cell signal regulation of 3′ splice sites. This also provides a molecular target for the stimulus to signal the splicing machinery to control critical alternative splicing events during neuronal differentiation.
One important remaining question is how hnRNP K binding to the KARRE is enhanced by forskolin for competition with U2AF65. The total protein level of hnRNP K is not increased on forskolin addition (Figure 4A, lanes 1 and 2); the mainly nuclear localization of hnRNP K is not changed either (Supplementary Figure S1). However, interestingly, hnRNP K is phosphorylated in cells (42) and is also directly phosphorylated by PKA in vitro (Figure 3C), consistent with the idea that hnRNP K phosphorylation regulates its binding to the polypyrimidine tracts (Figure 3B). Moreover, hnRNP K contains three RNA binding domains. How these domains interact with the two or more copies of target motifs at the 3′ splice sites (Figure 7), to compete with the dynamic interaction of the multidomain U2AF65 (65) and how the interaction is influenced by the phosphorylation would be important questions for future investigations.
As one of the hnRNP K target neuronal exons/genes, Runx1 is critical for neuronal differentiation; particularly, its repressor/activator changes are important for specifying neuron lineages during development (53). Interestingly, based on mutagenesis and transcriptional assays (64,66), the inclusion or exclusion of its exon 6 determines whether this transcription factor contains the binding site of the corepressor Sin3A (Figure 5B). Together with its regulation by forskolin, the alternative splicing provides a novel mechanism to explain the activity switch (53). It would also be interesting to examine the exon 6 changes seen in genetic diseases particularly acute myeloid leukemia (66–71).
A striking finding about the biological functions of the hnRNP K target genes is that almost half of them including Runx1 are involved in neurological diseases (Figure 5A and Supplementary Tables SI and SII). Consistent with the importance of these genes and the role of Snap25 in neurons, hnRNP K is essential for forskolin-induced neurite growth in PC12 cells (Figure 6). HnRNP K also plays a crucial role in axonogenesis during neuronal differentiation in Xenopus (72,73). Taken together with the critical role of alternative splicing in neuronal differentiation and diseases (5,6,74,75), it is possible that a coordinated program of critical splicing changes by hnRNP K plays a crucial role for the forskolin-induced neuronal differentiation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1 and 2.
FUNDING
Manitoba Health Research Council (MHRC) and Canadian Institutes of Health Research (CIHR) [FRN 106608 to J.X.]; MHRC [postdoctoral fellowship (2007–09) to W.G.C.]. Funding for open access charge: CIHR Operating Grant [FRN 106608].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Ze’ev Ronai and Dr Hasem Habelhah for the HA-hnRNP K, Dr Arzu Osturk for the His-hnRNP K, Hongzhao Li for the 175NK, Guodong Liu for the shRNA plasmid against hnRNP K, Dr Sam Kung’s laboratory for help with lentivirus preparations and the Xie laboratory members for helpful discussion. We are grateful to Benoit Chabot for critical comments during preparation of the manuscript.
REFERENCES
- 1.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger rna splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim. Biophys. Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 9.Heyd F, Lynch KW. Degrade, move, regroup: signaling control of splicing proteins. Trends Biochem. Sci. 2011;36:397–404. doi: 10.1016/j.tibs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 11.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell. Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 12.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore MJ. Intron recognition comes of AGe. Nat. Struct. Biol. 2000;7:14–16. doi: 10.1038/71207. [DOI] [PubMed] [Google Scholar]
- 15.Tronchere H, Wang J, Fu X-D. A protein related to splicing factor U2AF-35 that interacts with U2AF-65 and SR proteins in splicing of pre-mRNA. Nature (London) 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- 16.Singh R, Banerjee H, Green MR. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA. 2000;6:901–911. doi: 10.1017/s1355838200000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 18.Valcarcel J, Singh R, Zamore PD, Green MR. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 19.Shepard J, Reick M, Olson S, Graveley BR. Characterization of U2AF26, a splicing factor related to U2AF35. Mol. Cell. Biol. 2002;22:221–230. doi: 10.1128/MCB.22.1.221-230.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcarcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 21.Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcarcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3' splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 22.Heiner M, Hui J, Schreiner S, Hung LH, Bindereif A. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol. 2010;7:56–64. doi: 10.4161/rna.7.1.10402. [DOI] [PubMed] [Google Scholar]
- 23.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl Acad. Sci. USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thisted T, Lyakhov DL, Liebhaber SA. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest Distinct modes of RNA recognition. J. Biol. Chem. 2001;276:17484–17496. doi: 10.1074/jbc.M010594200. [DOI] [PubMed] [Google Scholar]
- 25.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 27.Revil T, Pelletier J, Toutant J, Cloutier A, Chabot B. Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J. Biol. Chem. 2009;284:21458–21467. doi: 10.1074/jbc.M109.019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Expert-Bezancon A, Le Caer JP, Marie J. Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken beta-tropomyosin pre-mRNA. J. Biol. Chem. 2002;277:16614–16623. doi: 10.1074/jbc.M201083200. [DOI] [PubMed] [Google Scholar]
- 29.Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol. Cell. Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Hai Y, Liu G, Fang T, Kung SK, Xie J. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca++/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J. Biol. Chem. 2009;284:1505–1513. doi: 10.1074/jbc.M805113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Liu G, Yu J, Cao W, Lobo VG, Xie J. In vivo selection of kinase-responsive RNA elements controlling alternative splicing. J. Biol. Chem. 2009;284:16191–16201. doi: 10.1074/jbc.M900393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie J. Control of alternative pre-mRNA splicing by Ca(++) signals. Biochim. Biophys. Acta. 2008;1779:438–452. doi: 10.1016/j.bbagrm.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hai Y, Cao W, Liu G, Hong SP, Elela SA, Klinck R, Chu J, Xie J. A G-tract element in apoptotic agents-induced alternative splicing. Nucleic Acids Res. 2008;36:3320–3331. doi: 10.1093/nar/gkn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao WG, Sohail M, Liu G, Koumbadinga GA, Lobo VG, Xie JY. Differential effects of PKA-controlled CaMKK2 variants on neuronal differentiation. RNA Biol. 2011;86:1061–1072. doi: 10.4161/rna.8.6.16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma S, Liu G, Sun Y, Xie J. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim. Biophys. Acta. 2007;1773:912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Benderska N, Becker K, Girault JA, Becker CM, Andreadis A, Stamm S. DARPP-32 binds to tra2-beta1 and influences alternative splicing. Biochim. Biophys. Acta. 2010;1799:448–453. doi: 10.1016/j.bbagrm.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kvissel AK, Orstavik S, Eikvar S, Brede G, Jahnsen T, Collas P, Akusjarvi G, Skalhegg BS. Involvement of the catalytic subunit of protein kinase A and of HA95 in pre-mRNA splicing. Exp. Cell. Res. 2007;313:2795–2809. doi: 10.1016/j.yexcr.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Shi J, Qian W, Yin X, Iqbal K, Grundke-Iqbal I, Gu X, Ding F, Gong CX, Liu F. Cyclic AMP-dependent Protein Kinase Regulates the Alternative Splicing of Tau Exon 10: a mechanism involved in tau pathology of Alzheimer disease. J. Biol. Chem. 2011;286:14639–14648. doi: 10.1074/jbc.M110.204453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarnaess E, Stokka AJ, Kvissel AK, Skalhegg BS, Torgersen KM, Scott JD, Carlson CR, Tasken K. Splicing factor arginine/serine-rich 17A (SFRS17A) is an A-kinase anchoring protein that targets protein kinase A to splicing factor compartments. J. Biol. Chem. 2009;284:35154–35164. doi: 10.1074/jbc.M109.056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, Hentze MW, Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell. Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 43.Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3'-UTR. RNA. 2007;13:1103–1115. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl Acad. Sci. USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bark C, Bellinger FP, Kaushal A, Mathews JR, Partridge LD, Wilson MC. Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission. J. Neurosci. 2004;24:8796–8805. doi: 10.1523/JNEUROSCI.1940-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johansson JU, Ericsson J, Janson J, Beraki S, Stanic D, Mandic SA, Wikstrom MA, Hokfelt T, Ogren SO, Rozell B, et al. An ancient duplication of exon 5 in the Snap25 gene is required for complex neuronal development/function. PLoS Genet. 2008;4:e1000278. doi: 10.1371/journal.pgen.1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hepp R, Dupont JL, Aunis D, Langley K, Grant NJ. NGF enhances depolarization effects on SNAP-25 expression: induction of SNAP-25b isoform. Neuroreport. 2001;12:673–677. doi: 10.1097/00001756-200103260-00011. [DOI] [PubMed] [Google Scholar]
- 48.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 1990;265:5267–5272. [PubMed] [Google Scholar]
- 49.Xie J, McCobb DP. Control of alternative splicing of potassium channels by stress hormones. Science (Washington, DC) 1998;280:443–446. doi: 10.1126/science.280.5362.443. [DOI] [PubMed] [Google Scholar]
- 50.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Kim N, Alekseyenko AV, Roy M, Lee C. The ASAP II database: analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007;35:D93–8. doi: 10.1093/nar/gkl884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marmigere F, Montelius A, Wegner M, Groner Y, Reichardt LF, Ernfors P. The Runx1/AML1 transcription factor selectively regulates development and survival of TrkA nociceptive sensory neurons. Nat. Neurosci. 2006;9:180–7. doi: 10.1038/nn1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marmigere F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat. Rev. Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- 54.Theriault FM, Nuthall HN, Dong Z, Lo R, Barnabe-Heider F, Miller FD, Stifani S. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J. Neurosci. 2005;25:2050–2061. doi: 10.1523/JNEUROSCI.5108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kramer I, Sigrist M, de Nooij JC, Taniuchi I, Jessell TM, Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi A, Senzaki K, Ozaki S, Yoshikawa M, Shiga T. Runx1 promotes neuronal differentiation in dorsal root ganglion. Mol. Cell. Neurosci. 2011;49:23–31. doi: 10.1016/j.mcn.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 57.Stifani N, Freitas AR, Liakhovitskaia A, Medvinsky A, Kania A, Stifani S. Suppression of interneuron programs and maintenance of selected spinal motor neuron fates by the transcription factor AML1/Runx1. Proc. Natl Acad. Sci. USA. 2008;105:6451–6456. doi: 10.1073/pnas.0711299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vilardell M, Rasche A, Thormann A, Maschke-Dutz E, Perez-Jurado LA, Lehrach H, Herwig R. Meta-analysis of heterogeneous Down Syndrome data reveals consistent genome-wide dosage effects related to neurological processes. BMC Genomics. 2011;12:229. doi: 10.1186/1471-2164-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katzaki E, Morin G, Pollazzon M, Papa FT, Buoni S, Hayek J, Andrieux J, Lecerf L, Popovici C, Receveur A, et al. Syndromic mental retardation with thrombocytopenia due to 21q22.11q22.12 deletion: report of three patients. Am. J. Med. Genet. A. 2010;152A:1711–1717. doi: 10.1002/ajmg.a.33478. [DOI] [PubMed] [Google Scholar]
- 60.Lindstrand A, Malmgren H, Sahlen S, Schoumans J, Nordgren A, Ergander U, Holm E, Anderlid BM, Blennow E. Detailed molecular and clinical characterization of three patients with 21q deletions. Clin. Genet. 2010;77:145–154. doi: 10.1111/j.1399-0004.2009.01289.x. [DOI] [PubMed] [Google Scholar]
- 61.Beri-Dexheimer M, Latger-Cannard V, Philippe C, Bonnet C, Chambon P, Roth V, Gregoire MJ, Bordigoni P, Lecompte T, Leheup B, et al. Clinical phenotype of germline RUNX1 haploinsufficiency: from point mutations to large genomic deletions. Eur. J. Hum. Genet. 2008;16:1014–1018. doi: 10.1038/ejhg.2008.89. [DOI] [PubMed] [Google Scholar]
- 62.Cryan JF, Slattery DA. GABAB receptors and depression. Current status. Adv. Pharmacol. 2010;58:427–451. doi: 10.1016/S1054-3589(10)58016-5. [DOI] [PubMed] [Google Scholar]
- 63.Silva JP, Ushkaryov YA. The latrophilins, “split-personality” receptors. Adv. Exp. Med. Biol. 2010;706:59–75. doi: 10.1007/978-1-4419-7913-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lutterbach B, Westendorf JJ, Linggi B, Isaac S, Seto E, Hiebert SW. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- 65.Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature. 2011;475:408–411. doi: 10.1038/nature10171. [DOI] [PubMed] [Google Scholar]
- 66.Mikhail FM, Sinha KK, Saunthararajah Y, Nucifora G. Normal and transforming functions of RUNX1: a perspective. J. Cell. Physiol. 2006;207:582–593. doi: 10.1002/jcp.20538. [DOI] [PubMed] [Google Scholar]
- 67.Giguere A, Hebert J. CLCA2, a novel RUNX1 partner gene in a therapy-related leukemia with t(1;21)(p22;q22) Cancer Genet. Cytogenet. 202:94–100. doi: 10.1016/j.cancergencyto.2010.07.116. [DOI] [PubMed] [Google Scholar]
- 68.Mikhail FM, Coignet L, Hatem N, Mourad ZI, Farawela HM, El Kaffash DM, Farahat N, Nucifora G. A novel gene, FGA7, is fused to RUNX1/AML1 in a t(4;21)(q28;q22) in a patient with T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2004;39:110–118. doi: 10.1002/gcc.10302. [DOI] [PubMed] [Google Scholar]
- 69.Nucifora G, Begy CR, Kobayashi H, Roulston D, Claxton D, Pedersen-Bjergaard J, Parganas E, Ihle JN, Rowley JD. Consistent intergenic splicing and production of multiple transcripts between AML1 at 21q22 and unrelated genes at 3q26 in (3;21)(q26;q22) translocations. Proc. Natl Acad. Sci. USA. 1994;91:4004–4008. doi: 10.1073/pnas.91.9.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Emmanuel N, Kamboj G, Chen J, Shurafa M, Van Dyke DL, Wiktor A, Rowley JD. PRDX4, a member of the peroxiredoxin family, is fused to AML1 (RUNX1) in an acute myeloid leukemia patient with a t(X;21)(p22;q22) Genes Chromosomes Cancer. 2004;40:365–370. doi: 10.1002/gcc.20050. [DOI] [PubMed] [Google Scholar]
- 71.Sacchi N, Nisson PE, Watkins PC, Faustinella F, Wijsman J, Hagemeijer A. AML1 fusion transcripts in t(3;21) positive leukemia: evidence of molecular heterogeneity and usage of splicing sites frequently involved in the generation of normal AML1 transcripts. Genes Chromosomes Cancer. 1994;11:226–236. doi: 10.1002/gcc.2870110405. [DOI] [PubMed] [Google Scholar]
- 72.Liu Y, Gervasi C, Szaro BG. A crucial role for hnRNP K in axon development in Xenopus laevis. Development. 2008;135:3125–3135. doi: 10.1242/dev.022236. [DOI] [PubMed] [Google Scholar]
- 73.Liu Y, Szaro BG. hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development. 2011;138:3079–3090. doi: 10.1242/dev.066993. [DOI] [PubMed] [Google Scholar]
- 74.Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 75.Raj B, O'Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol. Cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.