Abstract
Recent advances in RNA sequencing technology (RNA-Seq) enables comprehensive profiling of RNAs by producing millions of short sequence reads from size-fractionated RNA libraries. Although conventional tools for detecting and distinguishing non-coding RNAs (ncRNAs) from reference-genome data can be applied to sequence data, ncRNA detection can be improved by harnessing the full information content provided by this new technology. Here we present NorahDesk, the first unbiased and universally applicable method for small ncRNAs detection from RNA-Seq data. NorahDesk utilizes the coverage-distribution of small RNA sequence data as well as thermodynamic assessments of secondary structure to reliably predict and annotate ncRNA classes. Using publicly available mouse sequence data from brain, skeletal muscle, testis and ovary, we evaluated our method with an emphasis on the performance for microRNAs (miRNAs) and piwi-interacting small RNA (piRNA). We compared our method with Dario and mirDeep2 and found that NorahDesk produces longer transcripts with higher read coverage. This feature makes it the first method particularly suitable for the prediction of both known and novel piRNAs.
INTRODUCTION
The involvement of non-coding RNAs (ncRNAs) in many genetic and epigenetic processes is well documented (1). To date, the best studied class of small ncRNAs is micro RNA(miRNA), which regulates gene expression by repressing messenger RNA (mRNA) translation, but other classes of small ncRNAs, such as piwi-interacting small RNA (piRNA) and small nucleolar RNA (snoRNA), have also been characterized (1). It has been said that the majority of the genome is transcribed at some stage (2), therefore, the exact expression profile is necessary to functionally annotate known and novel ncRNAs.
Progress in the study of the active RNA transcriptome in a genome-wide manner is supported by recent advances in sequencing technology. The initial sequencing protocols developed for capturing large RNAs such as mRNAs, are inadequate to detect small ncRNAs; new protocols have thus been developed to specifically capture small ncRNA (small-RNA-Seq). In these protocols, only the fragments that fall within a predefined size distribution are selected for sequencing. Although conventional ncRNA discovery methods can be applied to analyze assembled sequences derived from this data, programs specifically developed for deep sequencing data yield better performance (3,4).
Most methods developed for deep sequencing data analysis to date are either in-house-only bioinformatic tools to generate a resource dataset (5,6) or report simply the clusters of mapped reads (7), mainly with a focus on miRNA (8–11) or small silencing RNAs (12). Friedländer et al. (3) observed that the biological miRNA maturation process leaves a distinct footprint in the read coverage of deep sequencing data, which can be leveraged to improve the specificity of miRNA prediction [mirDeep2 (13)]. miRNA transcripts (known as pri-miRNAs) are transcribed by the RNA polymerase II from either independent transcripts of intergenic regions (IGRs) or from the introns of protein-coding genes (14). These pri-miRNA transcripts are processed into hairpin shaped precursors (pre-miRNAs) of about 70 nucleotides (nt) in length by the Drosha-DGCR8 complex, which are then cleaved into ∼22 nt long duplexes of mature miRNAs by Dicer (14). Chaing et al. observed miRNA gene expression in high-throughput data has certain characteristics: miRNA tend to have pairs of expressed contigs connected by a sequence able to form a predicted hairpin structure, and regions where one of these proximal pairs is absent are therefore likely to be degradation intermediates rather than miRNA (15). Therefore, the simplest way to reconstruct miRNA transcripts from deep sequence reads is to examine whether proximal reads are able to form pre-miRNA like hairpin structures. This observation can be extended to the prediction of other ncRNA classes (16) such as piRNAs, which also have a characteristic footprint in the read distribution data. piRNAs are short RNAs of 26–31 nt in length with a bias for 5′ uridine that interact with Piwi proteins, a subgroup of Argonaute proteins that are required for germ- and stem-cell development (17–19). In mammals, the main role of piRNAs is to regulate Piwi proteins to repress transposons (20). piRNAs are derived from clusters of genome regions of 20–90 kilobases in length, since all transcripts in one cluster originate from the same strand it has been suggested that they are processed from one long primary transcript (17,18).
Although the origin of the observed uneven and gapped read distribution of ncRNAs is not well understood, Langenberger et al. (16) developed a machine learning-based framework, Dario (21), which firstly reports all annotated ncRNA with non-zero coverage and secondly uses a machine learning-based framework to successfully predict and classify novel ncRNA types from these indicative features in the sequencing data. However, the accuracy of any machine learning based classifier strongly depends on how well the training set represents the features of yet unseen ncRNAs. Dario's training set size is 434 ncRNA of which half are miRNAs. Its ability to generalize is thus likely to be limited and is further restricted to the species the classifier was trained on (currently human, mouse, Caenorhabditis elegans and Dorsophila melanogaster).
Here we present NorahDesk, the first unbiased and universally applicable tool for predicting ncRNA that exploits the read characteristics of ncRNAs in deep sequencing data. We utilize two biological observations: first, small ncRNAs generally possess stable secondary structures with each class of ncRNAs clearly distinguishable by their length and structures (22); second, many small ncRNAs are transcribed from large primal transcripts (23).
To leverage these observations, our method first joins overlapping reads into contigs and then tests whether neighboring contigs can hybridise to form a sound secondary structure. By hybridizing contigs, we reconstruct the full-length of the putative ncRNA transcript, which enables us to evaluate the structural energies before reporting the high-confidence candidates.
To evaluate the performance of our approach in predicting ncRNAs from deep sequencing data, we report the frequency of hyridization-based merging and the size distribution of assembled transcripts in four publicly available datasets. Secondly, we report the fraction of known ncRNA within NorahDesk's predictions. In the ‘Results’ section, we compare the performance of our method to DARIO and mirDeep2. Fourthly, we report NorahDesk's ability to predict miRNA transcripts and piRNAs-clusters, and conclude by surveying the fraction of novel ncRNA predicted by our method.
MATERIALS AND METHODS
Implementation
The functionality of NorahDesk can be separated into two stages: (i) reconstructing small ncRNA transcripts from deep sequencing data; (ii) and annotating these transcripts with known RNAs.
Transcriptome reconstruction
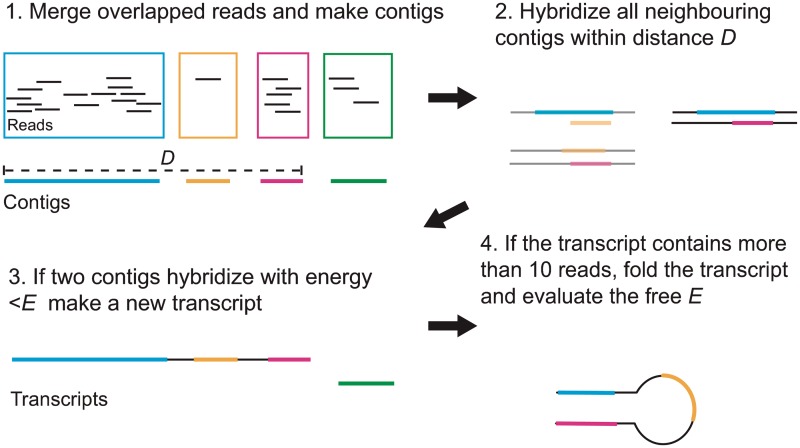
NorahDesk takes a list of deep sequence reads aligned to the reference genome in BAM or BED format produced by sequence read aligners such as BWA (24) and Bowtie (25) as input. Figure 1 shows the overview of the four steps NorahDesk undertakes to reconstruct ncRNA transcripts from deep sequencing data. The ‘first’ step merges all overlapping reads into one longer sequence fragment (a contig). The ‘second’ step hybridizes closely located contigs to each other. To do this, we group all contigs by chromosome and strand, then we hybridize all transcripts within a group that are at most a certain distance (D) apart and screen the pairs of hybridized transcripts that have a free energy below a certain threshold (Eh). We test two different maximum distances, D = {250 nt, 500 nt}, measured by the start position of one read and end position of another read, and three different maximum hybridization free energies, Eh = {−5 kcal/mol, −7.5 kcal/mol, −10 kcal/mol}, computed by using RNAduplex (Vienna RNA package http://www.tbi.univie.ac.at/RNA/). The third step merges overlapping newly created transcripts and original contigs to create the final reconstructed transcripts. The transcripts containing <10 reads are discarded at this stage. In the final step, we compute the folding energies of remaining transcripts and discard the transcripts with free energy Ef >−5 kcal/mol. The structural energies of the transcripts are computed using RNAfold (26). We compute the folding energies for only the transcripts ≤3000 nt in length due to computational limitations; and assign −100 kcal/mol for those >3000 nt, assuming the long transcripts have lower folding energies than −5 kcal/mol.
Figure 1.
Steps to reconstruct small ncRNA transcripts.
Throughout the processes, BEDTools (27) are used to merge reads (‘merge’ program) and obtain the sequences for the merged reads (‘getSequences’ program). The output of this stage is a list of predicted small ncRNA transcripts in BED format. All displayed structures are predicted using RNAfold (26).
The run time of NorahDesk largely depends on the numbers and the length of transcripts predicted by the software. For example, it took from ∼11 min (using a threshold of D = 250 nt and Eh = 5 kcal/mol) to ∼12 min (D = 500 nt and Eh = 10 kcal/mol) to compute brain sample (SRR042477) using a linux cluster with Intel Xeon CPU 2.13 GHz with (restricted to) 8 GB memory. It also took from ∼204 min (D = 250 nt and Eh = 5 kcal/mol) to ∼265 minutes (D = 500 nt and Eh = 10 kcal/mol) to compute testis samples (SRR042485), which contain many long piRNA transcripts (see Results) to compute. For the same dataset, mirDeep2 took from ∼101 min (SRR042485) to ∼233 min (SRR042486).
Transcripts annotation
In the second stage, NorahDesk categorizes predicted transcripts into classes of known ncRNAs and novel ncRNAs. To do this, we construct an annotation file of known ncRNAs using fRNAdb version 3.4 (28), (http://www.ncrna.org/frnadb/download). Since the miRNA entries in fRNAdb are old (miRBase version 9), we supplement the precursor miRNA data using miRBase version 18 (29) (www.mirbase.org/ftp.shtml) and construct a single ncRNA annotation file. To detect the transcripts that overlapped with protein coding genes, we also construct an annotation file for Ensemble genes using ensGene.txt (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/database/). All annotation files are converted into BED format. We use the intersect program in BEDTools (27) to map the locations of our predicted RNA transcripts to known RNAs. We require 90% overlap (90% of the shorter transcripts, either the predicted or known transcripts, have to overlap with the longer transcripts) for fRNAdb and miRBase data, and 10% overlap with Ensemble data. We search predicted transcripts against, first, the ncRNA file, then the Ensemble gene file. We identify transcripts that overlap with only ncRNA files, i.e. miRBase and fRNAdb, as known ncRNAs, and transcripts that do not overlap with any of the above database are classified as potential novel ncRNAs. We also count transcripts that overlap with Ensemble entries but not overlap with ncRNAs as mRNAs; however, we do not supply an annotation file of mRNA transcripts. Additionally, we categorize ncRNAs into classes of: miRNA, piRNA, snoRNA, other small ncRNA (sncRNA), rRNA, tRNA, antisense transcript and other ncRNA. sncRNA class includes small cajal body specific RNA (scaRNA), small nuclear RNA (snRNA) and minor spliceosomal RNA (snRNA_splicing). If a predicted transcript overlaps with multiple classes of ncRNAs, e.g. as miRNA and piRNA, the known ncRNA annotation file contains multiple entries. For counting statistics, we select each transcript as miRNA, piRNA, snoRNA, sncRNA, rRNA, tRNA, antisense transcript and other ncRNA in this order and do not allow multiple counts; The outputs of this program are a list of categorized known ncRNAs, a list of potential novel ncRNAs and a summary (count) file. NorahDesk requires only 4–6 s to annotate each set of data.
Sequence data
Four sets of mouse RNA-Seq reads from Kuchen et al. (30) [brain (SRR042477), skeletal muscle (SRR042483), testis (SRR042485) and ovary (SRR042486)] were downloaded from NCBI Sequence Reads Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra). These data were generated by the protocol optimizing for small RNAs (18–30 nt in length) with the read length of 32 nt and single strand (adaptor in one side) (30). The number of reads in each sample ranges from 3 to 7.5 million. The downloaded reads (in SRA format) are converted into FASTQ files using SRA Toolkits (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=software) and mapped to the mouse genome (mm9) using BWA (24) with the default setting. About 65–89% of reads map to the genome, after discarding the reads that mapped to mitochondrial DNA (Supplementary Table 1),
Shuffled reads
To query the statistical validity of the hybridization step we generate 100 sets of shuffled sequences for each set of contigs using a first order Markov model, which keeps the di-nucleotide distribution intact (‘squid’, ftp://selab.janelia.org/pub/software/squid/). We hybridize these shuffled contigs as mentioned above (the second step of ‘Transcriptome reconstruction’) and construct shuffled transcripts. Then we compare the number of the resulting transcripts between the shuffled and real data (Wilcoxon Rank-sum test).
Coevolution and PeptideAtlas annotation
The secondary structure conservation is obtained by employing tools that investigate thermodynamic stability [RNAz (31)] and covariance [SISSIz (32)] in multiple sequence alignments (multiz30way from UCSC genome browser), as also used in Mercer et al. (33). After filtering for size >30 and species ≥3, there are 800 regions with alignment information. All transcripts exceeding the default score as determined by this approach are reported to have conserved secondary structure.
To investigate whether the transcripts are translated to known peptides, we first generate the amino acid sequence for all open reading frames of the predicted transcripts and then query the resulting peptides against the the online database of PeptideAtlas (34) using a REST protocol. All transcripts overlapping with known peptides are from our ncRNA prediction.
RESULTS
We predict 612 unique transcripts (which contain ∼4.6 million reads) in brain, 729 transcripts (∼6.2 million reads) in muscle, 5397 transcripts (∼2.4 million reads) in testis, and 1284 transcripts (∼2.8 million reads) in ovary. The number of predicted transcripts and transcripts overlapping with known ncRNAs is largely robust to parameter changes (Supplementary Tables S2 and S3). We hence select D = 250 nt and a maximum hybridization free energy Eh = −5 kcal/mol, and describe the results obtained using this criteria for the rest of the document.
Hybridizing proximal reads significantly improves transcript prediction
To establish that the hybridization-based reconstruction of transcripts is contributing significantly to the prediction-outcome, we show that the size distribution of the initial contigs (see the first step of ‘Transcriptome reconstruction’ in ‘Materials and Methods’ section) is substantially different from the size distribution of the predicted transcripts.
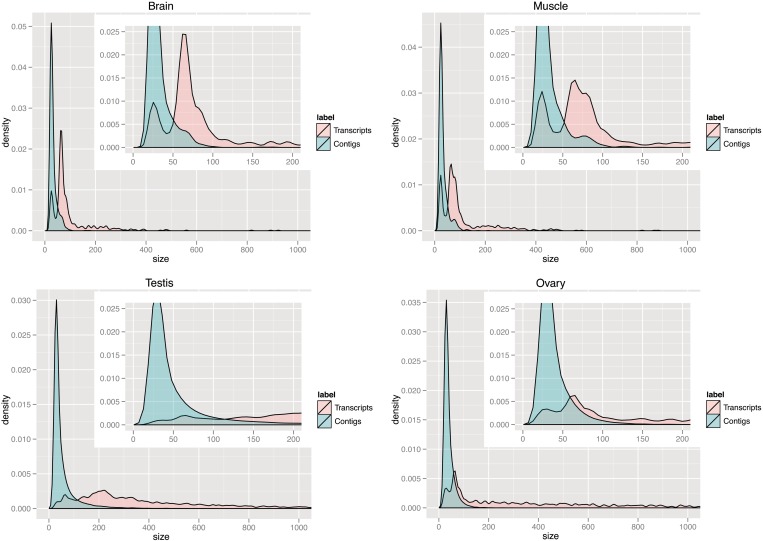
The length distributions are visualized in Figure 2. Starting off with a mean ‘read’-length of ∼21–22 nt in brain, skeletal muscle and ovary, and 26 nt in testis, we observe almost no change in the mean ‘contig’-length (22–31 nt; Supplementary Table 1). When we select contigs that contain ≥10 reads, we observe a small change in length (27–28 nt) in brain and skeletal muscle, and a slightly larger increase in ovary (46 nt) and testis (92 nt) (Figure 2 and Supplementary Table 1). However, the mean length of predicted ncRNA ‘transcripts’ is substantially shifted to 101 nt in brain, 110 nt in muscle, 498 nt in testis and 666 nt in ovary (Figure 2 and Supplementary Table 2), implying that neighboring fragments indeed originated from the same transcript.
Figure 2.
Size distribution of contigs and predicted transcripts. The shift in the size distribution of contigs (blue) and predicted transcripts (red) in brain, muscle, testis and ovary. The x-axis shows the size in number of nucleotides and the y-axis shows the corresponding density as the smoothed and normalized contig- and transcript count, respectively.
Figure 2 is showing that ‘contigs’ are of similar length across the different tissues. In contrast, the length distributions of reconstructed ‘transcripts’ varies greatly, suggesting that our method is better suited for revealing the different ncRNA populations present in each tissue than methods based on contigs alone. For example, the size distribution peak at ∼23 nt in brain, muscle and ovary suggests a large fraction of transcripts are mature miRNAs, whereas the peak at ∼65–75 nt is indicative of pre-miRNA transcripts. We will show in the next section that these transcripts indeed overlap with known miRNAs. The miRNA peaks are less dominant in testis, which has an additional broader peak around ∼200 nt. Both testis and ovary have a general tendency for longer transcripts, i.e. 11% of transcripts in testis and 20% in ovary are ≥1000 nt in length, which imply both testis and ovary contain long piRNA transcripts (explained in the piRNA section). Importantly, the highest peak shown at ∼200 nt in testis indicates that the dominant small ncRNA in testis is piRNA and not miRNA.
Furthermore, to establish that the hybridization-based transcript reconstruction is building upon biologically relevant properties rather than noise, we show that randomly generated data have a different contig-hybridization property. Successful hybridization depends on two determinants: location (distance of neighboring contigs) and sequence (base pairing compatibility). We hypothesize that contigs from the same transcript are more likely to hybridize with each other compared with random sequences or contigs originating from different transcripts.
In brain we observe 1477 hybridization events that satisfy above criteria. After shuffling the sequence of each contig keeping their relative location fixed we observe on average 1332 and at most 1366 hybridization events (repeated 100 times). This shows that real data are significantly more likely to contain neighboring contigs that are able to hybridize (Wilcoxon Rank-sum test; P-value ≤ 0.045). We obtain similar results for the other tissues (data not shown).
In summary, our hybridization-based reconstruction of transcript is a statistically sound approach to greatly extend the length of the predicted transcripts thereby may be able to trace the ncRNA population in different tissues.
Discovery of ncRNAs with known function
In this section we survey the predicted transcripts with respect to known ncRNA classes in the different tissues to further investigate NorahDesk's sensitivity.
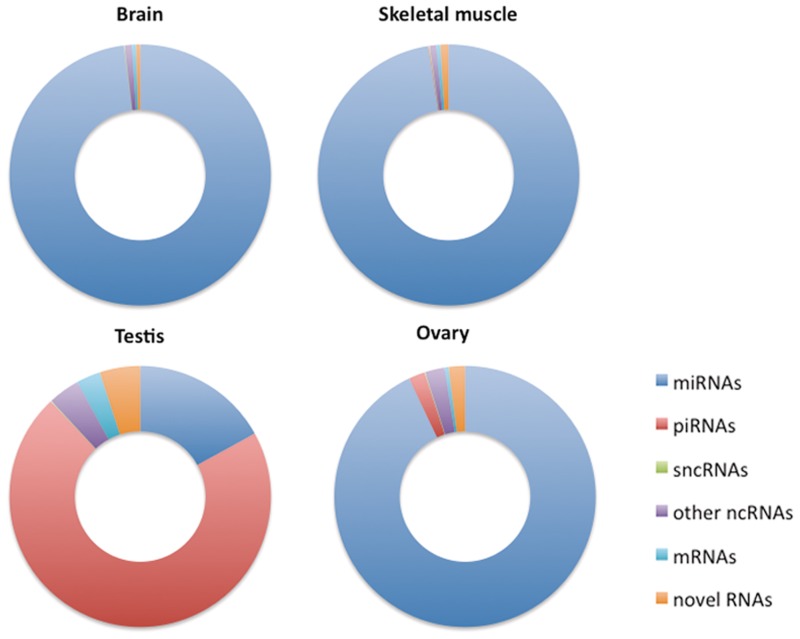
We observe that 48% (291) and 37% (268) of the predicted transcripts in brain and muscle, respectively, overlap with known ncRNA annotations. In testis and ovary this ratio increases to 66% (3557) and 76% (979), respectively. As shown in Figure 3, these transcripts comprise between 92 and 99% of the total reads in these tissues. This means that any ncRNA our method fails to report must have a very low expression level, which attests to the precision of our method.
Figure 3.
Distribution of ncRNA types in different tissues. The figures show the fraction of reads overlap with known and novel classes of RNAs in brain, skeletal muscles, testis and ovary.
As noted in the previous section, the ncRNA population in the four tissues is very different. The highly abundant transcripts predicted by NorahDesk for brain, muscle and ovary are predominantly miRNAs, whereas the predictions in testis are dominated by piRNAs. These observations are in agreement with Kuchen et al. (30) and will be further discussed in the following sections.
Concordance with published ncRNA prediction methods
In this section, we benchmark NorahDesk against the current state-of-the-art in miRNA and ncRNA prediction by comparing our results to the predictions made by mirDeep2 and Dario. We used the same input (mapped sequence) file for all three methods.
We predict 246 miRNA transcripts that overlap with known miRNAs in brain, see Table 1 (the results on other tissues are available in Supplementary Table S4). Although, NorahDesk predicts only ∼60% of the miRNAs predicted by Dario and mirDeep2 (Table 1), our miRNA transcripts comprise a similar number of reads as Dario, which means that on average Dario's predictions are supported by fewer reads. To ensure only high-confidence transcripts are reported, NorahDesk requires the support of at least 10 reads per transcript, where DARIO also includes ncRNAs consisting of a single read in the known ncRNA prediction. In fact, 119 out of 427 (28%) miRNAs predicted by DARIO contain <10 reads.
Table 1.
Comparison with other methods
| Method | miRNAs |
piRNAs |
snoRNAs |
sncRNAs |
tRNAs |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| transcripts | reads | transcripts | reads | transcripts | reads | transcripts | reads | transcripts | reads | |
| NorahDesk | 246 | 4 497 220 | 9 | 1150 | 12 | 1237 | 3 | 103 | 0 | 0 |
| Dario | 427 | 4 811 848 | NA | NA | 282 | 43 075 | NA | NA | 334 | 8486 |
| mirDeep2 | 442 | 8 781 481 | NA | NA | NA | NA | NA | NA | NA | NA |
| mirDeep2 > 0 | 343 | 7 429 586 | NA | NA | NA | NA | NA | NA | NA | NA |
| mirDeep2 > 9 | 208 | 3 712 002 | NA | NA | NA | NA | NA | NA | NA | NA |
Number of transcripts and reads predicted by NORAHDESK, DARIO and MIRDEEP2. >0 and >9 show that the predicted miRNAs with MIRDEEP2 quality scores >0 and 9. The numbers are actual (not normalized) reads.
The 442 miRNAs predicted by mirDeep2 on the other hand, comprise nearly double the number of reads than Dario and NorahDesk (Table 1), which means that the average number of supporting reads is as high as NorahDesk's. This indicates that a single predicted miRNA transcript is assigned to a miRNA (or a miRNA family) that derived from multiple locations. However, mirDeep2 implements a quality criteria to remove low-quality predictions including a miRNA transcript that mapped to multiple known miRNA locations. Due to the length and the sequence composition of miRNA the fraction of non-uniquely mapping reads is natively high; rather than excluding a very large fraction of reads at the mapping stage, NorahDesk reduces the negative effect of multi-mappings by reconstructing miRNA precursors. In this way, we are able to predict each miRNA transcript that derived from a distinct (unique) locus.
Despite the different approaches, all three methods predict a similar set of high-abundant miRNAs (Supplementary Table S5).
Similar to the miRNA prediction performance, NorahDesk predicts fewer snoRNAs, rRNAs and tRNAs compared with Dario. However, of the 282 snoRNAs only 40 snoRNAs contain ≥10 reads, which means the majority of known snoRNAs predicted by Dario would not pass our quality criteria. fRNAdb contains only one tRNA entry; therefore, we do not expect to predict any tRNA; however, some novel ncRNA transcripts NorahDesk predicts are overlapped with tRNAscan-SE (35) predicted tRNAs (see ‘Discovery of novel ncRNAs’ section).
To evaluate how well a method is able to predict unannotated miRNAs, we compare the novel miRNAs predicted by mirDeep2 and NorahDesk using miRBase v16 to the content of miRBase v18. The proportion of novel miRNA that are included in the most recent annotation indicates the power of miRNA prediction. We are unable to compare Dario's results in the same way, as the documentation does not indicate which database version Dario uses. In the brain sample, mirDeep2 predicts 62 novel miRNAs (including six transcripts, which are also predicted as rRNA/tRNA) using miRBase v16. Of these 62 predicted novel miRNAs, three are listed as new miRNAs in v18. NorahDesk predicts 268 novel ncRNA transcripts, including 150 transcripts that are overlapped with predicted tRNAscanSE (see ‘Discovery of novel ncRNAs’ section) using an annotation file that contains miRBase v16. Of these, four transcripts are now listed in v18. MiRBase v18 includes a further three transcripts that were predicted by NorahDesk but previously annotated as piRNA, piRNA/snoRNAs and other ncRNAs in fRNAdb. In total, NorahDesk predicts seven new miRNAs that were shown to be correct in v18; this demonstrates the ability of NorahDesk to correctly predict un-annotated miRNAs transcripts. Note, one miRNA (miR-1994) is deleted from v18 but annotated as snoRNA in fRNAdb.
To summarize, while all three methods are able to detect the highly abundant transcripts, NorahDesk's strict selection criteria applied for all predicted transcripts, both known and novel small ncRNAs, predict fewer but high-quality transcripts supported by more reads.
On data that do not contain many long transcripts such as brain samples, the run time of NorahDesk (∼11 min) is considerably shorter than mirDeep2 (∼157 min); however, NorahDesk requires more computational resource to process data containing many long transcripts (see ‘Transcriptome reconstruction’ section in ‘Materials and Methods’ section). Since Dario is a web server, we are unable to directly compare its run time with other stand-alone programs.
Prediction of miRNA precursors
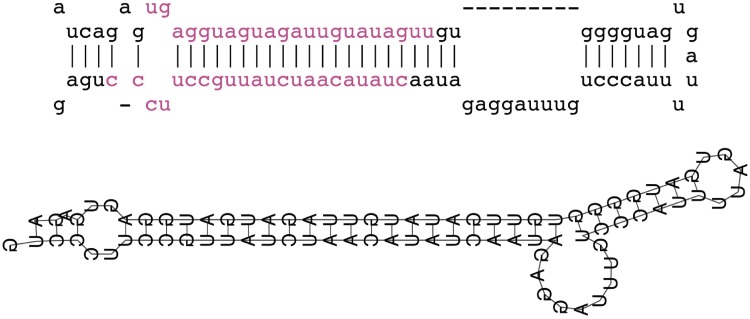
This section explores the properties of miRNA transcripts predicted by NorahDesk. As previously described, our method relies on reconstructing miRNA precursors to predict miRNA transcripts. Being able to accurately predict the secondary structure of the precursor is hence paramount. Figure 4 shows an example of similar folded pre-miRNA transcript predicted by our method is in comparison with the known fold as annotated by miRBase v18.
Figure 4.
Known versus predicted structure of mmu-let-7f1. The top figure shows the known structure of mmu-let-7f1 from miRBase and the bottom shows the predicted structure of reconstructed transcript. The mature miRNA-duplex is shown in purple.
The average length of precursors in miRBase v18 is 84 nt (48 – 133 nt), where we predict transcripts in brain with the average length of 87 nt with 9% of them being >133 nt (4% are <48 nt). Furthermore, we observe that these large transcripts contain multiple known miRNAs, e.g. the largest predicted transcript is 817 nt (Chromosome 14 + strand) and overlaps with 5 known miRNAs (miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92a-1). Rather than being an artifact of an over-agressive contig merging behavior, NorahDesk's predictions reflect biological properties: miRNA clusters indeed exist where several miRNA genes are transcribed in a single pri-miRNA, as discussed in Berezikov (36). Following on from this, we also predict a small number of transcripts, which contain a single miRNA and one or more contigs that are either un-annotated or annotated as ncRNAs (e.g. non-coding transcripts from FANTOM3), indicating yet unknown miRNA clusters (Figure 5).
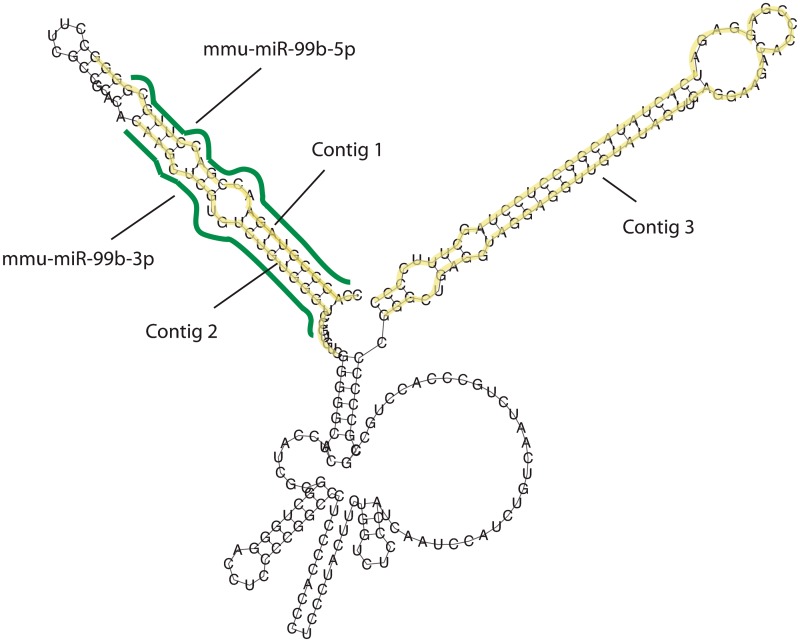
Figure 5.
Example of long miRNA transcript. Predicted miRNA transcript from chr17:17967156-17967398 (+strand) overlaps with miR-99b precursor and contains additional one un-annotated contig.
To summarize, NorahDesk's approach of reconstructing full transcripts by hybridizing neighboring fragments can be successfully applied to miRNA discovery.
Prediction of piRNA sub clusters
In this section, we explore how suitable NorahDesk is for identifying piRNAs; as piRNAs are thought to be derived from long primary transcripts (17), to reconstruct long transcripts (resembling the pre-processed form) by hybridizing neighboring contigs would aid in the discovery of all piRNAs from one transcription unit.
As shown in Figure 3, we predict piRNAs mostly in testis and to a lesser degree in ovary. This is in agreement with what the raw sequence data indicates: the average length of sequence reads in testis (26 nt) is longer than in other tissues (21–22 nt), with the largest frequency of reads at 31 nt, which falls within the range of a typical piRNA (26–31 nt) (Supplementary Figure S6). Indeed, 3099 out of 5397 predicted transcripts (57%) overlap with known piRNAs in testis. As discussed earlier, a group of piRNAs are transcribed from a large cluster, and in many cases, individual piRNAs are located close each other in the cluster. Thus using our method, clustered piRNAs are merged into a single long transcript, each of them contains several known piRNAs. Alrhough the average length of predicted piRNA-cluster transcripts are longer than the ones overlapping with known miRNAs in testis (577 versus 124 nt), they are still shorter than the piRNA-cluster that have been reported to extend 20–90 k nt (18). Our longest predicted transcripts is 6505 nt and the majority (85%) are <1 k nt; therefore, the piRNA-clusters analyzed here seem to be derived from substantially shorter primary transcripts or contain gaps that are longer than our hybridization approach can bridge (>250 nt on this analysis).
Figure 6 shows two predicted transcripts that overlap with known piRNAs. Both transcripts form energetically stable complexes and contain one or more known piRNAs (green), which are overlapped with contigs (yellow), indicating that the piRNAs were indeed derived from the longer transcript. Additionally, these transcripts contain contigs that are not annotated as piRNA, but when individually folded form piRNA like secondary structures (Supplementary Figure S7) and the majority contains 5′ uridine.
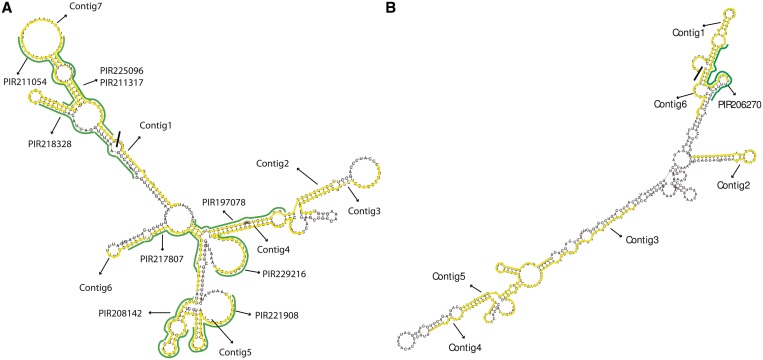
Figure 6.
Examples of predicted piRNA transcripts. (A) Predicted piRNAs transcripts from chr10:18517077-18517536 (-strand); 4 out of 7 contigs overlap with known piRNAs and all contigs have 5′ uridine. (B) Predicted piRNAs transcripts from chr6:128121896-128122316 (-strand); only one out of 6 contigs overlap with a known piRNA and 4 out of 6 contigs have 5′ uridine.
By reconstructing the full—or at least a part of the—extended transcriptional unit, we are able to predict additional or even a complete set of piRNAs transcribed from a single piRNA-cluster. This classification-by-association approach enables us to annotate novel ncRNA with yet unknown function as piRNAs. Indeed, while Kuchen et al. (30) was able to annotate only ∼50% of the reads to be piRNA and classified ∼23% to be novel ncRNAs, we predict that 82% of all reads in testis belong to piRNA.
To summarize, our analysis supports the evidence that piRNAs are transcribed from longer transcripts and processed into mature forms. NorahDesk's hybridization-based approach is particularly suited to the identification of a set of piRNAs from one transcriptional unit, which includes many predicted novel piRNAs.
Discovery of novel ncRNAs
In this section, we investigate whether NorahDesk is able to recover meaningful transcripts in the four publicly available datasets that have not been reported before. We predict 264 novel transcripts in brain, 359 in skeletal muscle, 1182 in testis and 230 in ovary. The average lengths of the novel ncRNA transcripts in brain, muscle and testis are 6–16% shorter than the average length of over all transcripts, and in ovary the transcript-size was reduced by half. Apart from a smaller size, they also occupy only a very small fraction of reads: 0.4% in brain, 1.1% in muscle, 4.6% in testis, 2.2% in ovary. However, as discussed by Clark et al. (37), low-level transcripts may be functional and hence must not be discarded.
To investigate whether the predicted low-abundant transcripts are potential novel miRNAs, we screen transcripts in brain that fall in the pre-miRNA size range (48–133 nt). We randomly select 8 transcripts and manually investigate their secondary structure for the pre-miRNA like hairpin structure. In total, 7 out of 8 randomly selected transcripts overlap with predicted tRNA by tRNAscan-SE (35). The structure of the remaining transcript resembles a snoRNA (Supplementary Figure 8).
Since one structure resembles a snoRNA, we also investigate whether the predicted transcripts in brain may be other types of ncRNA. We therefore first remove all transcripts that overlapped with predicted tRNA structures (tRNAscan-SE), which filters out about half of all transcripts leaving 114 putatively novel ncRNA in brain, 180 in muscle, and 128 in ovary, except in testis where almost all transcripts remain (1107).
Next we measure whether the secondary structure of the predicted RNA transcripts is evolutionary conserved, by using the same methodology as reported in Mercer et al. (33) (see ‘Materials and Methods’ section). Only 4–10% of the predicted transcripts show evolutionary conserved RNA structure (12 brain, 7 in muscle, 8 in ovary and 98 in testis). After excluding the ones overlapping with known peptides from PeptidAtlas (34), we predict 10 novel ncRNA in brain, 7 in muscle, 8 in ovary and 96 in testis. When limiting our analysis to regions expressed in all four tissues, we observe 20 regions, of which two are conserved throughout mammalian evolution and one overlaps with a known peptide.
To summarize, while NorahDesk predicts small numbers of novel ncRNA transcripts in this dataset, these transcripts are likely degradated products of either tRNA or peptide producing genes.
DISCUSSION
The collection of short sequence reads from RNA-Seq reflects the expression level of each transcript as well as the state of the transcripts where RNA synthesis, degradation and interim processes of ncRNA biogenesis have occurred simultaneously. Thus, it is meaningful to reconstruct transcripts from proximal reads found in the same sample, since they may be derived from the same primary transcript. Evaluating whether proximal reads/contigs hybridise and form a stable structure is important to distinguish degraded fragments from functional ncRNA transcripts and reduce the risk of annotating fragmented, miss-mapped and/or multi-mapped reads as novel ncRNAs.
Using these assumptions, we have developed a program, NorahDesk, to predict small ncRNA transcripts from small-RNA-Seq data. We tested our method on publicly available mouse data (brain, muscle, testis and ovary) using different selection criteria. We showed that different criteria produce similar results, suggesting that NorahDesk robustly detect biological signals rather than sequencing artifacts.
Transcripts that overlapped with known ncRNAs including the un-annotated contigs in these transcripts, occupy 92–99% of reads that mapped to the genome. An overwhelming majority of reads belongs to miRNAs in brain, muscle and ovary, and miRNAs and piRNAs in testis.
Compared with Dario and mirDeep2, our method recovers similar highly expressed known miRNAs; however, it produces longer transcripts with higher coverage. Our method is capable of detecting known miRNAs without extensive read mapping strategies.
We detect many known piRNAs in large transcripts, where piRNAs are clustered, indicating that piRNAs indeed are transcribed from a long primary transcripts. Moreover, we found that many un-annotated contigs that resemble piRNA structures are clustered with annotated contigs (known piRNAs) in these long transcripts. NorahDesk is hence the first program to specifically exploit the piRNA biogenesis to predict piRNA transcripts from high-throughput sequencing data.
CONCLUSION
NorahDesk reconstructs full-length putative ncRNA transcripts from short sequence reads by hybridizing contigs. It analyzes not only the distinct read distribution of true ncRNA classes in an unbiased way but also utilizes secondary structures as an independent confirmation source to reliably predict ncRNA from deep sequencing data. NorahDesk and the mouse small ncRNA annotation file in BED format used in this study are available at http://www.bioinformatics.org.au/NorahDesk.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 6–8.
FUNDING
The Zaccari Ph.D. Scholarship in Mental Health Research, University of Queensland (to C.R.); the Australian National Health and Medical Research Council, Grant [#631406 to B.J.M.]. Funding for open access charge: The University of Queensland, Queensland Brain Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Martin Smith for generating the co-evolution data used in ‘Discovery of novel ncRNAs’, Jake Gratten for his input throughout the project, and Marc Friedländer, Sebastian Mackowiak and Nikolaus Rajewsky for making miRDeep2 available prior to the publication.
REFERENCES
- 1.Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information–moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–28. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 4.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief. Bioinform. 2009;10:490–497. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar M, Wyman SK, Fritz BR, Qi JL, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erhard F, Zimmer R. Classification of ncRNAs using position and size information in deep sequencing data. Bioinformatics. 2010;26:i426–i432. doi: 10.1093/bioinformatics/btq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang PJ, Liu YC, Lee CC, Lin WC, Gan RR, Lyu PC, Tang P. DSAP: deep-sequencing small RNA analysis pipeline. Nucleic Acids Res. 2010;38:W385–W391. doi: 10.1093/nar/gkq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronen R, Gan I, Modai S, Sukacheov A, Dror G, Halperin E, Shomron N. miRNAkey: a software for microRNA deep sequencing analysis. Bioinformatics. 2010;26:2615–2616. doi: 10.1093/bioinformatics/btq493. [DOI] [PubMed] [Google Scholar]
- 10.Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, Sun Z, Wu J. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38:W392–W397. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantano L, Estivill X, Marti E. A non-biased framework for the annotation and classification of the non-miRNA small RNA transcriptome. Bioinformatics. 2011;27:3202–3203. doi: 10.1093/bioinformatics/btr527. [DOI] [PubMed] [Google Scholar]
- 13.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 15.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langenberger D, Bermudez-Santana CI, Stadler PF, Hoffmann S. Identification and classification of small RNAs in transcriptome sequence data. Pac. Symp. Biocomput. 2010:80–87. doi: 10.1142/9789814295291_0010. [DOI] [PubMed] [Google Scholar]
- 17.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 18.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 19.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 21.Fasold M, Langenberger D, Binder H, Stadler PF, Hoffmann S. DARIO: a ncRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2011;39:W112–W117. doi: 10.1093/nar/gkr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 23.Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- 27.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K. The functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 2009;37:D89–D92. doi: 10.1093/nar/gkn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruber AR, Findeiss S, Washietl S, Hofacker IL, Stadler PF. Rnaz 2.0: improved noncoding RNA detection. Pac. Symp. Biocomput. 2010;15:69–79. [PubMed] [Google Scholar]
- 32.Gesell T, von Haeseler A. In silico sequence evolution with site-specific interactions along phylogenetic trees. Bioinformatics. 2006;22:716–722. doi: 10.1093/bioinformatics/bti812. [DOI] [PubMed] [Google Scholar]
- 33.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell Proteomics. 2011;10:M110 006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 37.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.