Abstract
In human mitochondria, 10 mRNAs species are generated from a long polycistronic precursor that is transcribed from the heavy chain of mitochondrial DNA, in theory yielding equal copy numbers of mRNA molecules. However, the steady-state levels of these mRNAs differ substantially. Through absolute quantification of mRNAs in HeLa cells, we show that the copy numbers of all mitochondrial mRNA species range from 6000 to 51 000 molecules per cell, indicating that mitochondria actively regulate mRNA metabolism. In addition, the copy numbers of mitochondrial mRNAs correlated with their cellular half-life. Previously, mRNAs with longer half-lives were shown to be stabilized by the LRPPRC/SLIRP complex, which we find that cotranscriptionally binds to coding sequences of mRNAs. We observed that the LRPPRC/SLIRP complex suppressed 3′ exonucleolytic mRNA degradation mediated by PNPase and SUV3. Moreover, LRPPRC promoted the polyadenylation of mRNAs mediated by mitochondrial poly(A) polymerase (MTPAP) in vitro. These findings provide a framework for understanding the molecular mechanism of mRNA metabolism in human mitochondria.
INTRODUCTION
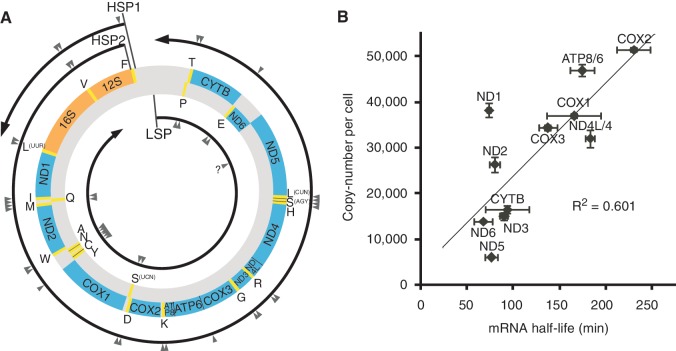
Human mitochondria contain circular, double-stranded DNAs (mtDNA) of 16.6 kb, which encode 37 genes in both the H- and L-strands: 13 of these encode the essential subunits of the respiratory complexes I, III, IV and V; 22 encode tRNAs and 2 encode rRNAs (1) (Figure 1A). To translate the 13 genes that encode proteins, mitochondria have a specific protein synthesis machinery in which all tRNAs and rRNAs are supplied from mtDNA. A long polycistronic precursor RNA of the H-strand is transcribed from the H-strand promoter 2 (HSP2) (Figure 1A) and is then processed to yield 10 mRNAs for 12 genes (ND4L/4 and ATP8/6 are bicistronic), 2 rRNAs and 14 tRNAs (2). Only the mRNA for ND6 is transcribed from the L-strand of mtDNA, together with eight tRNAs (Figure 1A). Hence, in theory, equal copy numbers of 10 mRNA species are generated stoichiometrically from the single polycistronic transcript of the H-strand. However, the steady-state levels of these 10 mRNAs have been reported to differ substantially (3,4). In addition, our group previously determined that the half-life of each mitochondrial mRNA in HeLa cells ranged from 68 to 231 min (5). These studies implied the existence of a post-transcriptional regulatory mechanism that controls the stability and metabolism of mRNAs in mitochondria.
Figure 1.
Variable steady-state levels and half-lives of human mitochondrial mRNAs. (A) Schematic representation of human mtDNA with its gene organization and transcriptional units. The outer and inner circles represent the H- and L-strands of mtDNA, respectively. Protein (blue) and ribosomal RNA (orange) genes are interspersed with 22 tRNA genes (yellow, with single-letter amino acid codes). The L- and H-strand transcripts from the promoters LSP, HSP1 and HSP2 are indicated by circular arrows showing the direction of transcription. The sites for RNA processing are indicated by arrowheads. The processing site for ND6 is shown as a question mark because its 3′-end has not yet been defined (4,18). (B) Copy numbers of mRNAs (means ± SD, n = 3) in HeLa cells plotted against their half-lives in HeLa cells, as previously reported (5). The correlation factor (R2) of the plot is 0.601.
Leigh Syndrome French Canadian variant (LSFC) is an autosomal, neurodegenerative disease from which patients die of fulminant metabolic acidosis (6). LSFC is characterized by a tissue-specific deficiency in complex IV (cytochrome c oxidase) activity, which particularly affects the brain and liver (7). The cause of LSFC was identified as a C-to-T point mutation at nucleotide position 1119 of the leucine-rich pentatricopeptide repeat (PPR) motif-containing protein (LRPPRC) gene (8). This mutation changes the alanine at position 354 to valine (A354V), and the cellular level of mutant LRPPRC is reduced significantly (9). Moreover, the steady-state levels of mitochondrial mRNAs are also significantly decreased (9,10). LRPPRC is predominantly localized to mitochondria (11). LRPPRC contains 16 PPR motifs (12), which are predicted to bind to single-stranded nucleic acids (13). In fact, LRPPRC can bind to a single-stranded RNA in vitro and is found crosslinked to poly(A)+ RNA in mitochondrial fractions isolated from ultraviolet-irradiated cells (11). LRPPRC physically forms a stable complex with the SRA stem-loop-interacting RNA-binding protein (SLIRP) (9), which is a small protein bearing a single RNA recognition motif (RRM) that is localized primarily to mitochondria (14). Knockdown of either LRPPRC (15) or SLIRP (16) results in similar decreases in mRNA levels but does not affect the levels of tRNAs or rRNAs, indicating that the LRPPRC/SLIRP complex plays a specific role in mRNA maturation or stabilization after transcription in mitochondria (9). In addition, a LRPPRC knockout mouse is embryonic lethal and deficient in mRNA polyadenylation (17). Moreover, an aberrant pattern of mitochondrial translation was observed in a LRPPRC knockout mouse, demonstrating that LRPPRC is necessary for regulated translation in mammalian mitochondria.
Human mitochondrial mRNAs have short (∼50 nt) poly(A) tails (18), whose lengths are regulated by mitochondria-specific poly(A) polymerase (MTPAP) (19,20) and polynucleotide phosphorylase (PNPase) (19). The role of the poly(A) tail in mRNA stabilization or destabilization remains elusive. When MTPAP was knocked down by RNAi, the poly(A) tail of each mRNA was shortened and the steady-state levels of several mRNAs, including COX1 and COX2, were reduced, while mRNA levels for ND1, ND2, ND3 and CYTB were unchanged or increased (5,19,20). When artificial deadenylation of mt mRNAs was induced by targeting cytosolic deadenylase (PARN) to mitochondria (21) or by overexpressing PDE12 (22), the steady-state level of COX1 and COX2 mRNAs decreased, while mRNA levels of ND1, ND2, ND3 and CYTB increased.
The entity of the mRNA degradation machinery in human mitochondria has remained elusive (23). PNPase is one of the major 3′–5′ exonucleases in bacteria (24). In human mitochondria, the involvement of PNPase in homeostasis of the poly(A) tail has been suggested. Downregulation of PNPase by RNAi resulted in the elongation of mRNA poly(A) tails for COX1, COX2, COX3, ATP8/6 and ND3 (19), although the steady-state levels of these mRNAs and proteins were unaffected (19,25). This indicated that PNPase participates in the deadenylation of the poly(A) tail of a subset of mRNAs. However, PNPase mainly localizes to the intermembrane space (IMS) of mitochondria where mRNAs are absent (25). In addition, PNPase is involved in the transport of the RNA component for RNaseMRP into mitochondria (26) and participates in the degradation of c-myc mRNA (27) and miR-221 in human melanoma cells (28). These facts further complicate the issue of whether PNPase acts as a 3′–5′ exonuclease of mRNAs in mitochondria.
The mitochondrial RNA degradosome (mtEXO) in Saccharomyces cerevisiae consists of Dss1p, which functions as a 3′–5′ exonuclease (RNase II-like), and Suv3p, which acts as a DExH/D RNA helicase (29). Recombinant protein of the human homolog of SUV3 forms a stable complex with PNPase, which degrades RNA in the 3′–5′ direction in vitro (30). Overexpression of the dominant negative mutant of SUV3 (or SUPV3L1) results in the elongation of the poly(A) tail of mRNAs and the accumulation of aberrant transcripts from the antisense strand, indicating the involvement of SUV3 in mitochondrial mRNA degradation (31).
In this article, we provide evidence that the LRPPRC/SLIRP complex suppresses mRNA degradation mediated by PNPase and SUV3 and promotes polyadenylation of mRNA mediated by mitochondrial poly(A) polymerase MTPAP in vitro.
MATERIALS AND METHODS
Cell culture
HeLa cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum at 37°C under a humidified atmosphere with 5% CO2.
Quantitative reverse-transcription real-time polymerase chain reaction
Total RNA was isolated from cells using TRI Pure (Roche), according to the manufacturer’s instructions. RNA was treated with DNase I (Promega) for 30 min at 37°C. cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Random N6 primers or gene-specific reverse primers [quantitative polymerase chain reaction (qPCR) reverse primers] were used for cDNA synthesis. For mitochondrial tRNAs, qPCR reverse primers were used for the first strand cDNA synthesis. Amplification of cDNA was monitored with the LightCycler 480 SYBR Green I Master (Roche) on a LightCycler 480 (Roche), according to the manufacturer’s instructions. The sequences of the qPCR primers are listed in Supplementary Table S1.
Absolute quantification of human mitochondrial mRNAs in HeLa cells
For absolute quantification of mRNAs, in vitro transcribed mRNAs were prepared as external standards for qRT-PCR. The cDNAs for all mitochondrial mRNAs were amplified by RT-PCR from the total RNA of HeLa cells using the primers listed in Supplementary Table S1, preceded by a T7 class III promoter sequence (32). All 11 mitochondrial mRNAs were transcribed by T7 RNA polymerase in vitro (32) and purified by denaturing polyacrylamide gel electrophoresis. The isolated mitochondrial mRNAs were quantified by measuring the optical density at 260 nm. HeLa cells grown in five dishes (4.86 ± 0.26 × 106 cells/dish) were washed with phosphate-buffered saline (PBS) and lysed by adding TRI Pure (Roche) directly to the dishes. Twenty micrograms of total RNA from Echerichia coli was added to this mixture as a doped marker to estimate the recovery rate of total RNA. Total RNA was isolated from the cells according to the manufacturer’s instructions and dissolved in 100 µl of water. The recovery rate of total RNA from HeLa cells was calculated by analyzing the loss of E. coli gapA mRNA in the doped marker by qRT-PCR. The total RNA (5 µg) from each of the five dishes was treated with DNase I (Promega), and a portion of the total RNA was subjected to cDNA synthesis using gene-specific reverse primers for all mitochondrial mRNAs (qPCR reverse primers listed in Supplementary Table S1). qRT-PCR was conducted as described above. The copy number of each mitochondrial mRNA in the total RNA from one HeLa cell was determined using the calibration line of each mRNA, which was generated by quantifying the amount of in vitro transcribed mRNAs by qRT-PCR. The recovery rate of RNA extraction was also taken into account to determine the copy numbers.
RNAi
siRNAs targeted to LRPPRC, SLIRP, PNPase, PDE12, SUV3 and luciferase were designed using the siRNA design algorithm ‘siExplorer’ (33). The siRNAs used here are listed in Supplementary Table S1. HeLa cells (2.5 × 105) were transfected with 60 pmol of siRNA (5 nM, final concentration) using Lipofectamine RNAi Max (Invitrogen).
Inhibition of mitochondrial transcription
Mitochondrial transcription was inhibited, as previously described (5). Cell culture medium was replaced with DMEM supplemented with 10% fetal bovine serum and 500 ng·ml−1 of ethidium bromide, and total RNA was collected at 0, 1, 2, 4 or 6 h after the initiation of mitochondrial transcription inhibition.
Western blotting
Cells were washed with PBS and collected with a rubber scraper. After centrifugation at 1000g for 2 min, cell pellets were resuspended in 100 µl of lysis buffer [100 mM NaCl, 10 mM Tris–HCl (pH 7.4), 2.5 mM MgCl2, 0.2% sodium dodecyl sulfate (SDS) and protease inhibitor cocktail (Roche)] and sonicated with a Sonifier 450D (Branson) for 5 s at power 3. Lysates were clarified at 20 000 g for 10 min, and the samples were then resolved by SDS polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a nitrocellulose membrane using an iBlot transfer apparatus (Invitrogen). Membranes were blocked with 10% Sea Block blocking buffer (Pierce) in Tris-buffered saline and probed with primary antibodies. Primary antibodies for LRPPRC (Santa Cruz), SLIRP (Santa Cruz) and β-actin (Sigma) were used at a dilution factor of 1:200, 1:100 and 1:5000, respectively. Horseradish peroxidase-conjugated anti-mouse (Dako) and anti-rabbit (Jackson ImmunoResearch Laboratories) secondary antibodies were used at 1:2000 and 1:15 000, respectively. Proteins were illuminated using ECL Plus (GE Healthcare) and visualized with an ImageQuant LAS4000 mini (GE Healthcare).
Immunoprecipitation
Approximately 5 × 106 HeLa cells were washed by PBS and crosslinked by incubation in PBS containing 0.15% formaldehyde for 10 min at room temperature (34). The crosslinking reaction was quenched with 0.25 M glycine (final concentration). Cells were washed with PBS and collected by a rubber scraper, followed by centrifugation at 1000g for 2 min. The cell pellet was resuspended in 1 ml of buffer A [100 mM NaCl, 10 mM Tris–HCl (pH 7.4), 2.5 mM MgCl2, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100 and 1 mM dithiothreitol (DTT)] containing Complete protease inhibitor cocktail (Roche) and SUPERaseIN (Ambion) and passed through a 25-gauge needle 10 times. Lysates were centrifuged at 20 000 g for 25 min and the supernatant was incubated with Protein G agarose for 1 h. The samples were then centrifuged and the supernatant was incubated with LRPPRC antibody (Santa Cruz)-bound Protein G beads for 2 h. The antibody beads were washed five times with buffer A. After immunoprecipitation, the cell lysate and precipitants were decrosslinked by incubating the precipitants in 100 µl of crosslink reversal buffer [50 mM Tris–HCl (pH 7.4), 5 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 1% SDS and 10 mM DTT] at 70°C for 30 min. For the experiment in Figure 3B, immunoprecipitation was conducted in the same way as described above, except that cells were transfected with the LRPPRC-Flag vector (as described below) and anti-Flag M2 agarose (Sigma) was used instead of LRPPRC antibody-bound beads.
Figure 3.
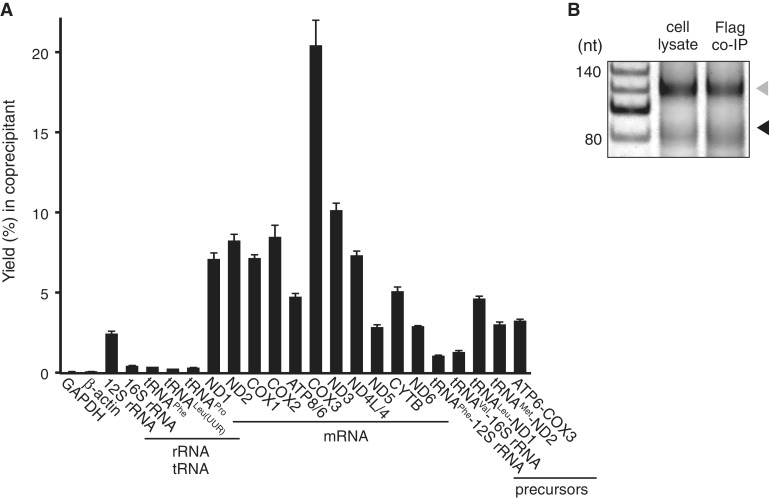
LRPPRC/SLIRP complex binds to mitochondrial mRNA coding sequences. (A) Yields of various RNAs and their precursors (%) co-immunoprecipitated with endogenous LRPPRC and quantified by qRT-PCR (means ± SD, n = 3). (B) A comparison of the poly(A) profiles of ND2 mRNA in the cell lysate and in the precipitate obtained by pull-down of transiently expressed LRPPRC-Flag using the poly(A) tail length assay. The RT-PCR products derived from polyadenylated and oligoadenylated RNAs are indicated by the gray and black triangles, respectively. A 20-nt ladder is shown in the first lane.
Analysis of RNA fragments bound to the LRPPRC/SLIRP complex
The Human Gateway entry clone (35) and DEST12.2 Flag plasmid were used in this study. The LRPPRC expression vector was generated by LR reaction of pDEST12.2 Flag and Entry clone FLJ43793AAAN using LR Clonase (Invitrogen). The C-terminally Flag-tagged LRPPRC vector was generated by changing the stop codon of the LRPPRC expression vector to Tyr, using the primers 5′-actcttcttatgacccagctttcttg-3′ and 5′-caagaaagctgggtcataagaagagt-3′. Approximately, 2 × 107 HeLa cells were transfected with the LRPPRC-Flag expression vector using FuGENE HD (Roche). At 40 h post-transfection, the cells were crosslinked, collected, lysed and precleared and immunoprecipitated (as described above) using anti-Flag M2 affinity beads. Subsequently, the antibody beads were washed twice with buffer B [100 mM NaCl, 10 mM Tris–HCl (pH 7.4), 0.1 mM EDTA, 1 mM DTT], followed by fragmentation of RNAs by incubation in 100 µl of buffer B containing 5 units/µl RNase T1 (Epicentre) and 10 µg/ml RNase A (Ambion) for 1 h at 37°C. The antibody beads were then washed four times with buffer A. 100 µl of crosslink reversal buffer and 10 µl of Proteinase K (Roche) were added, and the beads were incubated at 55°C for 30 min and then at 70°C for 20 min. One milliliter of TRI Pure (Roche) was added, and the RNA fragments were extracted according to the manufacturer’s instructions, except that ethanol precipitation was performed overnight at −80°C. The RNA precipitate was dephosphorylated by bacterial alkaline phosphatase A19 (Takara) and then extracted by phenol–chloroform–isoamyl alcohol (PCI) (Nacalai) and precipitated with ethanol. The adaptor DNA (5′-p-atgtgagatcatgcacagtcata-NH2-3′) was ligated at the 3′ ends of isolated RNA fragments, as described in the poly(A) tail length assay. Next, 3′ adaptor-ligated RNA fragments were extracted by PCI and precipitated with ethanol. The fragments were resolved by denaturing PAGE and stained with SYBR Gold (Invitrogen). Fragments larger than 20 nt were excised, and the RNA fragments were eluted from the gel and ethanol precipitated. The 5′ ends of 3′ adaptor-ligated RNA fragments were phosphorylated by T4 polynucleotide kinase (Toyobo), and the RNA solution was put through a NAP5 column (GE Healthcare) to remove excess ATP and then precipitated with ethanol. The 5′ ends of the RNA fragments were ligated to 5′ adaptor DNA (5′-ctcctggcaaaaggtcagag-3′) using T4 RNA ligase, as described in the mt poly(A) length assay, and were then extracted by PCI and ethanol precipitated. The 5′ and 3′ adaptor-ligated RNAs were resolved by denaturing PAGE and stained with SYBR Gold. Fragments larger than 50 nt were excised from the gel, eluted and recovered by ethanol precipitation. Adaptor-ligated RNA fragments were reverse-transcribed by Transcriptor reverse transcriptase (Roche). cDNAs were amplified by Taq polymerase (Toyobo) with the primers 5′-ctcctggcaaaaggtcagag-3′ and 5′-gactgtgcatgatctcacat-3′. The PCR product was resolved by native PAGE and stained with Mupid Blue (Takara). PCR products larger than 50 bp were excised from the gel, eluted, collected by isopropanol precipitation and cloned into plasmids using the TA cloning kit (Invitrogen) for DNA sequencing.
Poly(A) tail length assay
The assay is based on and altered from the method described by Temperley et al. (36). An adaptor DNA oligonucleotide was ligated to the 3′ termini of total RNA (5 μg) by T4 RNA ligase (New England Biolabs) at 37°C for 3 h, according to the manufacturer’s instructions. The ligated RNA was isolated by TRI Pure (Roche) and isopropanol precipitation and then reverse-transcribed using the First Strand cDNA Synthesis Kit (Roche) with the anti-adaptor primer. A first round of PCR (27 cycles) was carried out using a gene-specific upper primer and the anti-adaptor primer, followed by a second round of 10-cycle PCR using an inner anti-adaptor primer and a gene-specific lower primer. Finally, the PCR product was resolved by 10% polyacrylamide gel electrophoresis in TBE buffer, dyed with SYBR Gold (Invitrogen) and visualized with an FLA-7000 (Fujifilm). All oligo-DNAs are listed in Supplementary Table S1.
Quantification of human LRPPRC protein in HeLa cells
HeLa cells grown in three dishes (2.76 ± 0.20 × 106 cells/dish) were washed with PBS and lysed by direct addition of lysis buffer [100 mM NaCl, 10 mM Tris–HCl (pH 7.4), 2.5 mM MgCl2, 0.2 % SDS and protease inhibitor cocktail (Roche)] to the dishes. Cell lysates were collected from dishes with a rubber scraper and sonicated with a Sonifier 450D (Branson) for 24 s.
For LRPPRC protein quantification, recombinant His-LRPPRC was prepared as an external standard for western blotting. The cDNA for LRPPRC without the mitochondria-targeting signal (amino acid residues 60-1394) was amplified by RT-PCR of the total RNA from HeLa cells using the primers 5′-caacgtgctagcgccattgctgccaaagaaa-3′ and 5′-tgcgtacctaggttaagaagagttttccctcaattttctt-3′, which contain NheI and BamHI sites, respectively. The PCR product was cloned into the corresponding sites of the pET-28 a(+) vector (Novagen) to obtain the expression vector pET-LRPPRC, which produces N-terminal His-tag-fused LRPPRC (His-LRPPRC). The E. coli rosetta (DE3) strain was transformed with pET-LRPPRC and cultured in LB media containing 50 µg/ml kanamycin and 30 µg/ml chloramphenicol. Protein expression was induced by adding lactose to a final concentration of 2% and the cells were grown at 25°C for 20 h. Cells were harvested and suspended in buffer C [50 mM HEPES–KOH (pH 7.6), 100 mM KCl, 10 mM MgCl2 and 7 mM β-mercaptoethanol], followed by sonication on ice. Cell lysates were cleared by ultracentrifugation at 100 000g for 60 min. The supernatant was loaded onto a nickel-charged HiTrap chelating column (GE Healthcare). After washing off unbound proteins with buffer C, recombinant proteins were eluted with a 50 ml linear gradient from 0 to 500 mM imidazole in buffer C. Fractions containing the recombinant proteins, which were visualized by SDS–PAGE followed by CBB staining, were pooled and dialyzed overnight against buffer D [20 mM Tris–HCl (pH 6.8), 100 mM NaCl, 4 mM MgCl2 and 1 mM DTT]. The His-LRPPRC was then loaded onto a Mono Q 5/50 column (GE Healthcare). After washing off unbound proteins with buffer E [20 mM Tris–HCl (pH 6.8), 100 mM NaCl, 4 mM MgCl2, 7 mM β-mercaptoethanol], His-LRPPRC was eluted with a 25 ml linear gradient from buffer E to buffer F [20 mM Tris–HCl (pH 6.8), 500 mM NaCl, 4 mM MgCl2 and 7 mM β-mercaptoethanol]. Eluted fractions were analyzed by SDS–PAGE followed by CBB staining. Glycerol was added to the pooled protein fractions to a final concentration of 10%. The concentration of purified His-LRPPRC was determined by the BCA protein assay (Thermo Scientific) using BSA as a standard. HeLa cell total lysates and the purified His-LRPPRC were subjected to SDS–PAGE and analyzed by western blotting using anti-LRPPRC antibody. The copy number of LRPPRC in one HeLa cell was determined by the signal intensity, using the calibration line of the external standards generated by the purified His-LRPPRC.
Relative quantification of mtDNA in HeLa cells
HeLa cells transfected with Luciferase or PNPase siRNAs and grown in 10 cm dishes were trypsinized and collected. Cell pellets were lysed by 1 ml of buffer G [10 mM Tris–HCl (pH 8.0), 1 mM EDTA (pH 8.0), 0.6% SDS and RNase A]. Cell lysates were incubated at 37°C for 1 h. 5 µl of Proteinase K (Roche) was added, and cell lysates were incubated at 55°C for 2 h. Cell lysates were sonicated by Sonifier 450D (Branson) at power 3 for 24 s. Proteins were removed by addition of PCI (Nacalai) followed by 10 min of vortexing and centrifugation. Total DNA was collected by ethanol precipitation. 0.75 µg of total DNA was used for quantitative real-time PCR of mtDNA or 18 S rDNA, using the LightCycler 480 SYBR Green I Master (Roche) on a LightCycler 480 (Roche) according to the manufacturer’s instructions. The sequences of the qPCR primers are listed in Supplementary Table S1.
In vitro polyadenylation assay
ΔN39 MTPAP-His expression vector and recombinant protein were obtained as previously described (19). Recombinant LRPPRC protein was obtained as described above. COX2 and COX3 mRNA in Figure 5B were made as described above. The DNA templates used for the in vitro T7 RNA transcription of the substrate RNAs shown in Figure 5C and D were made by PCR amplification of sections of the ND1 and ND4L/4 genes. Double-stranded RNAs in Figure 5C were made by annealing the transcripts, and the double-stranded RNAs were then purified by polyacrylamide gel electrophoresis. Polyadenylation reaction was carried out at 37°C for 30 min. Each reaction contained substrate RNA and recombinant MTPAP protein in 20 µl of reaction buffer, which was composed of 50 mM Tris–HCl (pH 8.0), 40 mM KCl, 0.5 mM MgCl2, 0.1 mM MnCl2, 1 mM DTT, 167 nM α-32P ATP and 0.1 mM cold ATP. In Figure 5B, D and E, LRPPRC protein was added to the reaction and incubated at 37°C for 10 min, before the addition of α-32P ATP and MTPAP protein. RNA was purified using TRI Pure (Roche) and ethanol precipitation and was resolved by denaturing PAGE. The RNA was visualized by exposing the gel to FLA-7000 (Fujifilm). All oligo-DNAs are listed in Supplementary Table S1.
Figure 5.
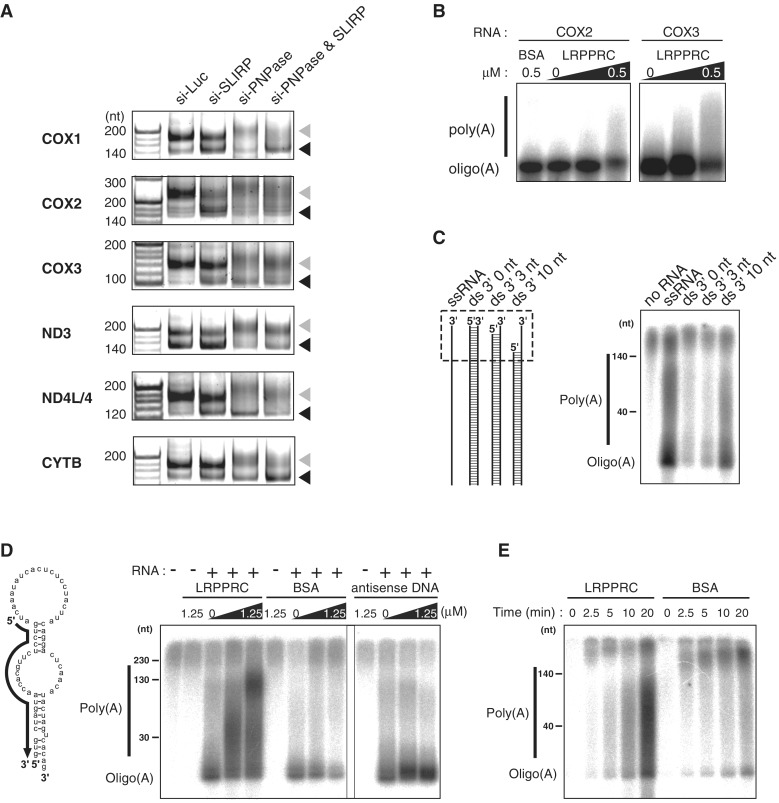
LRPPRC promotes MTPAP-mediated polyadenylation of mRNAs. (A) Polyadenylation profile of mitochondrial mRNAs upon knockdown of luciferase, SLIRP and PNPase or double knockdown of SLIRP and PNPase. The 3′ termini of the mRNAs for COX1, COX2, COX3, ND3, ND4L/4 and CYTB in HeLa cells transfected with siRNAs were analyzed by the poly(A) tail length assay. RT-PCR products derived from polyadenylated and oligoadenylated RNAs are indicated by gray and black triangles, respectively. The first lanes show 20-nt ladders. (B) In vitro polyadenylation of COX2 or COX3 mRNA transcript catalyzed by recombinant MTPAP in the presence of [α-32P] ATP and different amounts of recombinant LRPPRC (0, 0.1 or 0.5 μM) or BSA (0.5 μM). The products were resolved by polyacrylamide gel electrophoresis and the radioactivity was visualized by using an imaging analyzer (B–E). (C) In vitro polyadenylation of single-stranded (ss) or double-stranded (ds) RNA substrates, catalyzed by recombinant MTPAP in the presence of [α-32P] ATP. Schematic depiction of the substrates is shown on the left. The regions containing 3′ ss overhang of each substrate are boxed in the dotted line. (D) In vitro polyadenylation of the structured RNA (shown on the left) catalyzed by recombinant MTPAP in the presence of [α-32P] ATP and different amounts of recombinant LRPPRC (0, 0.25 or 1.25 μM), BSA (0, 0.25 or 1.25 μM) or antisense DNA (0, 0.25 or 1.25 μM). The complementary region of the antisense oligo, shown as a solid arrow, is depicted on the structured RNA substrate. (E) The time course representation of the in vitro polyadenylation of the ssRNA in the presence of LRPPRC or BSA. 1.25 μM of each protein was added to the reaction mixture.
RESULTS
Absolute quantification of human mitochondrial mRNAs in HeLa cells
In human mitochondria, 10 species of mRNA are generated from a single long precursor H-strand RNA. Differences in the steady-state levels of these 10 mRNAs were analyzed by qRT-PCR (3), by measuring the cDNA numbers of polyA(+) RNAs (4) and by deep sequencing (37). These studies reported only the relative and non-absolute values of mRNAs in human mitochondria. Precise analysis of the concentration or number of each species of mRNA in one cell is needed to obtain a more accurate representation of mRNA metabolism in human mitochondria. Thus, we decided to determine the exact copy numbers of all mitochondrial mRNAs in human HeLa cells. For absolute quantification of each mRNA, in vitro transcribed mRNAs were prepared as external standards for qRT-PCR. In addition, the extraction efficiency of mRNAs from HeLa cells was measured precisely by estimating the recovery rate of doped markers added before cell lysis (see Materials and Methods section). As shown in Table 1, the copy numbers of mRNAs ranged from 6000 (ND5) to 51 000 (COX2). The copy number of each mRNA was then plotted against its half-life (5) (Figure 1B). There is a clear correlation (R2 = 0.601) between the copy number and half-life of each mRNA. Abundant mRNAs have a tendency toward longer half-lives, whereas less abundant mRNAs have shorter half-lives. These data strongly suggest that the steady-state levels of mRNAs in mitochondria are largely determined by their half-lives. Notable exceptions are the mRNAs for ND1 and ND2, which show relatively high abundance despite having shorter half-lives (74 and 80 min, respectively).
Table 1.
Copy numbers of human mitochondrial mRNAs in HeLa cells
| mRNA | Copy numbers/cell | Half-life (min) (5) |
|---|---|---|
| ND1 | 38 000 ± 1600 | 74 ± 2 |
| ND2 | 26 000 ± 1600 | 80 ± 6 |
| COX1 | 37 000 ± 490 | 166 ± 29 |
| COX2 | 51 000 ± 480 | 231 ± 18 |
| ATP8/6 | 47 000 ± 1300 | 175 ± 13 |
| COX3 | 34 000 ± 600 | 138 ± 10 |
| ND3 | 15 000 ± 850 | 91 ± 4 |
| ND4L/4 | 32 000 ± 1900 | 183 ± 5 |
| ND5 | 6000 ± 73 | 77 ± 7 |
| CYTB | 16 000 ± 840 | 94 ± 24 |
| ND6 | 14 000 ± 290 | 68 ± 10 |
Mitochondrial mRNAs with longer half-lives are stabilized by the LRPPRC/SLIRP complex in HeLa cells
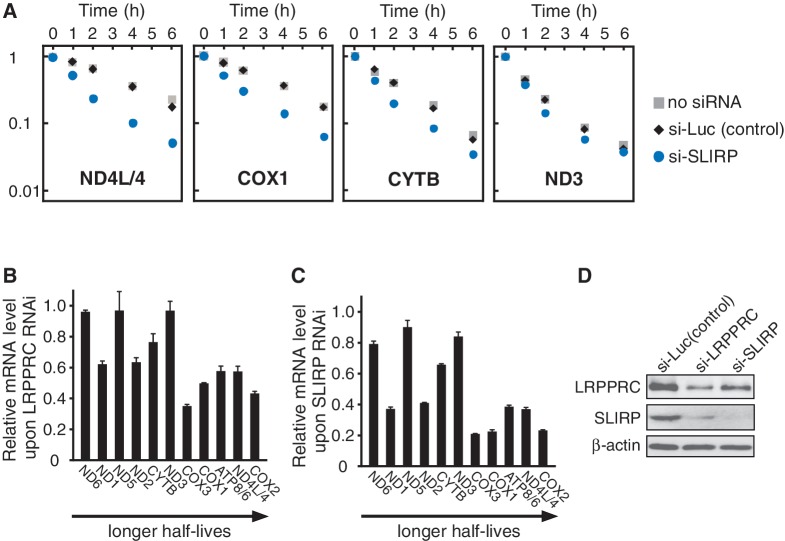
The LRPPRC/SLIRP complex has been assumed to play a regulatory role in post-transcriptional gene expression in human mitochondria (9). To clarify whether the LRPPRC/SLIRP complex is directly involved in the stabilization of mitochondrial mRNA, SLIRP mRNA was knocked down in HeLa cells. As shown in Figure 2A, mitochondrial transcription was specifically inhibited, and mRNA decay was observed. Upon knockdown of SLIRP, accelerated decay of ND4L/4, COX1 and CYTB mRNAs was clearly observed, as compared to the control cells, while the decay of ND3 mRNA was only modestly affected. This result provides direct evidence that SLIRP stabilizes mitochondrial mRNAs post-transcriptionally. Next, the steady-state level of each mRNA was measured after the knockdown of LRPPRC or SLIRP mRNA (Figure 2B and C). The mRNAs with longer half-lives tended to have a greater drop in their steady-state levels in the absence of SLIRP or LRPPRC than mRNAs with shorter half-lives. In addition, as previously mentioned (15), the patterns of relative change for each mRNA after either LRPPRC knockdown or SLIRP knockdown were markedly similar (Figure 2B and C, Supplementary Figure S1). This observation can be explained by the mutual stabilization of LRPPRC and SLIRP at the protein level (Figure 2D). After the knockdown of SLIRP mRNA, the endogenous levels of LRPPRC protein were also decreased. Similarly, knockdown of LRPPRC mRNA caused a specific reduction in endogenous SLIRP at the protein level. The mutual stabilization of these two proteins explains the finding that steady-state levels of SLIRP are significantly decreased in LSFC cells that express unstable LRPPRC with the A354V pathogenic point mutation (9). In addition, the patterns of the decreases in mRNA steady-state levels upon LRPPRC or SLIRP knockdown in HeLa cells (Figure 2B and C) were similar to those reported previously (15,16) in a human fibroblast cell line (MCH58). Thus, our observation is not confined to HeLa cells but is likely to be a general feature of mitochondrial mRNA metabolism in human cells.
Figure 2.
mRNAs with longer half-lives are stabilized by the LRPPRC/SLIRP complex. (A) Decay of ND4L/4, COX1, CYTB and ND3 mRNAs in HeLa cells after siRNA-mediated knockdown of SLIRP (circles) or luciferase (diamonds) or without siRNA (squares). At 0, 1, 2, 4 or 6 h after the inhibition of mitochondrial transcription, total RNA was collected and each mRNA was quantified by qRT-PCR. All data were normalized to GAPDH mRNA. The vertical axis represents the relative mRNA level in logarithmic scale. The steady-state level of SLIRP mRNA decreased to 3% of that in the control cells upon siRNA-mediated knockdown. (B and C) Relative change in each mRNA level upon LRPPRC knockdown (B) or SLIRP knockdown (C) versus luciferase knockdown control cells, aligned in order of the mRNA’s half-life (5). Mean values with SD were obtained from three independent experiments. The steady-state level of LRPPRC (B) and SLIRP (C) mRNAs dropped to 41% (B) and 1.2% (C), respectively, of those in the control cells. (D) Steady-state levels of endogenous LRPPRC and SLIRP after siRNA-mediated knockdown. Four days after siRNA transfection, endogenous LRPPRC and SLIRP in whole cell lysates were detected by western blotting.
The LRPPRC/SLIRP complex binds to mitochondrial mRNAs and their precursors
As both LRPPRC and SLIRP have RNA-binding domains (PPR and RRM, respectively), the LRPPRC/SLIRP complex was predicted to bind directly to RNAs in mitochondria. In fact, several mRNAs had been reported to coprecipitate with LRPPRC (9,17). However, whether the LRPPRC/SLIRP complex binds to other RNAs is unknown. To address this issue, HeLa cells were treated with formaldehyde to crosslink interacting RNAs and proteins, and endogenous LRPPRC was immunoprecipitated from the cell lysate. RNA recovery in the precipitant was analyzed by qRT-PCR (Figure 3A). All 11 mitochondrial mRNA species showed a high recovery ratio compared to cytoplasmic mRNAs and mitochondrial tRNAs, showing that the LRPPRC/SLIRP complex binds to all mRNA species in mitochondria. To elucidate whether the LRPPRC/SLIRP complex cotranscriptionally binds to precursor mRNAs, several sets of primers were designed to detect unprocessed bicistronic regions in precursor RNA: tRNALeu(UUR)-ND1 mRNA, tRNAMet-ND2 mRNA, ATP8/6 mRNA-COX3 mRNA, tRNAPhe-12 S rRNA and tRNAVal-16 S rRNA. The three mRNA precursors were immunoprecipitated with the LRPPRC/SLIRP complex as efficiently as mature mRNAs (Figure 3A), while the two rRNA precursors were not (Figure 3A). These results showed that the LRPPRC/SLIRP complex was associated with the precursor mRNAs before processing, indicating that the LRPPRC/SLIRP binds to the mitochondrial transcripts cotranscriptionally.
The LRPPRC/SLIRP complex recognizes mRNA coding sequences
Most human mitochondrial mRNAs have little or no 5′ and 3′ untranslated regions (UTRs), and nine mRNAs (other than ND5 and ND6) have poly(A) tails of ∼50 nt in length (18). The LRPPRC/SLIRP complex can bind to the coding sequences and/or to the poly(A) tails of mRNAs. As we found that endogenous LRPPRC was coprecipitated with ND5 and ND6 mRNAs that have short and no poly(A) tails, respectively (4,18) (Figure 3A), the LRPPRC/SLIRP complex is not a poly(A)-specific RNA-binding protein. To characterize the mRNA-binding properties of the LRPPRC/SLIRP complex, the poly(A) tail length of ND2 mRNA coprecipitated with LRPPRC-Flag was analyzed, because ND2 mRNA has no 5′- or 3′-UTRs but has both poly(A) and oligoadenylated tails. If binding of the LRPPRC/SLIRP complex is poly(A)-specific, the population of mRNA that accumulated in the precipitant would only have poly(A) tails. However, no difference was observed in the population of mRNAs with poly(A) versus mRNAs with oligoadenylated tails (Figure 3B). To directly analyze the mRNA sequence that the LRPPRC/SLIRP complex binds to, RNA fragments of mRNAs that coprecipitated with the LRPPRC/SLIRP complex from HeLa cells were cloned and sequenced (Supplementary Table S2). Of the 114 clones that were mapped to human transcripts, 112 clones were mapped to mtDNA, while 2 clones were from 28 S rRNA, confirming the quality of the immunoprecipitation. Of the 112 clones mapped to mtDNA, 97 clones mapped to the coding sequences of mitochondrial mRNAs. Notably, none of these 97 clones or other clones contained poly(A) or oligoadenylated tails at their 3′ ends. Therefore, it appears that the LRPPRC/SLIRP complex preferentially binds to the coding sequences of mitochondrial mRNAs and not to their poly(A) tails. In addition, one clone originating from the precursor mRNA containing tRNALeu(UUR) and ND1 was also obtained (Supplementary Table S2). This result supports our observation that the LRPPRC/SLIRP complex binds to precursor mRNAs (Figure 3A).
The LRPPRC/SLIRP complex is required to maintain poly(A) tails of mitochondrial mRNAs
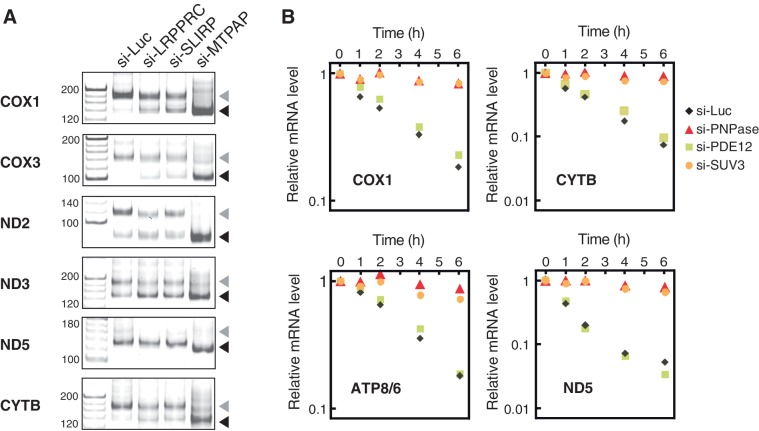
mRNA metabolism in mitochondria was examined to gain mechanistic insight into mRNA stabilization mediated by the LRPPRC/SLIRP complex. Maintenance of the poly(A) tail is required for the stability of several mRNAs, including COX1, COX2, COX3 and ATP8/6 (5,19). Therefore, changes in the poly(A) tail length of several mRNAs were measured when the LRPPRC/SLIRP complex was inactivated by RNAi. Under control conditions (si-Luc), both poly- and oligoadenylated mRNAs were observed (Figure 4A). When MTPAP was knocked down as a positive control experiment, a decrease in the number of polyadenylated mRNAs and the accumulation of no (or oligo-) adenylated mRNAs was observed (Figure 4A). After the knockdown of LRPPRC or SLIRP, oligoadenylated mRNAs of COX1, COX3 and CYTB clearly accumulated, in conjunction with a reduction in the population of polyadenylated mRNAs (Figure 4A). For ND2 and ND3, a slight increase in oligoadenylated mRNAs was observed, but the effect was limited. As ND5 mRNA has a short poly(A) tail (18), little change in the mRNA profile occurred after the knockdown of LRPPRC or SLIRP (Figure 4A). These results indicate that the LRPPRC/SLIRP complex is required for the maintenance of polyadenylated mRNAs. It should be noted that mRNAs (COX1 and COX3) that are strongly stabilized by the LRPPRC/SLIRP complex tend to have a larger poly(A)/oligo(A) ratio. This suggests that the LRPPRC/SLIRP complex might stabilize mRNAs by suppressing 3′deadenylation and/or 3′exonucleolytic degradation.
Figure 4.
PNPase and SUV3 are primarily involved in degradation of human mitochondrial mRNAs. (A) Polyadenylation profile of mitochondrial mRNAs upon LRPPRC or SLIRP knockdown. The 3′ termini of the mRNAs for COX1, COX3, ND2, ND3, ND5 and CYTB in HeLa cells transfected with siRNAs for luciferase, LRPPRC, SLIRP or MTPAP were analyzed by the poly(A) tail length assay. RT-PCR products derived from polyadenylated and oligoadenylated RNAs are indicated by gray and black triangles, respectively. The first lanes show the 20-nt ladders. (B) Decay of COX1, CYTB, ATP8/6 and ND5 mRNAs in HeLa cells after knockdown with siRNA against luciferase (diamonds), PDE12 (squares), PNPase (triangles) or SUV3 helicase (circles). At 0, 1, 2, 4 or 6 h after the inhibition of mitochondrial transcription, total RNA was collected and each mRNA was quantified by qRT-PCR. All data were normalized to GAPDH mRNA. The vertical axis represents the relative mRNA level in logarithmic scale. The steady-state level of PNPase, SUV3 and PDE12 mRNAs dropped to 11.1%, 11.9% and 22%, respectively, of those in the control cells.
PNPase and SUV3 are involved in mRNA degradation in human mitochondria
In human mitochondria, mRNA turnover associated with 3′ terminal metabolism has been extensively studied (23). However, the major component of the RNA degradation machinery has yet to be identified. Our group previously reported that PNPase is involved in 3′ deadenylation of several mRNAs (19). In addition, recombinant PNPase and SUV3 helicase cooperatively degrade RNA in vitro (30). However, whether PNPase is a bona fide 3′ exonuclease in mitochondria has been a controversial issue (23), because PNPase has multivalent functions, including the RNA import of RNase MRP RNA into the mitochondria (26) and the degradation of cytoplasmic mRNA (27) and miRNAs (28). Recently, 2′-phosphodiesterase (PDE12) was found to localize to mitochondria and to possess 3′–5′ exonucleolytic activity for RNAs in vitro, and PDE12 overexpression promoted deadenylation in cultured cells (22,38).
To identify the primary component of the RNA degradation machinery, PNPase or PDE12 was knocked down by RNAi, mitochondrial transcription was specifically inhibited and mRNA decay was monitored (Figure 4B). In control cells, the level of each mRNA decreased in accordance with its specific half-life. Upon knockdown of PNPase, mRNA decay almost completely halted (Figure 4B). On the other hand, when PDE12 was knocked down, the steady-state levels of mRNAs (COX1, CYTB, ATP8/6 and ND5) increased slightly (∼1.3-fold; Supplementary Figure S2), as previously reported (38). However, the rate of mRNA decay was only slightly affected by the inactivation of PDE12 (Figure 4B). These results, together with previous studies (19,30), clearly demonstrate that PNPase is the primary 3′–5′ exonuclease responsible for the degradation of mRNAs in human mitochondria. In addition, PNPase functions not only as a deadenylase but also as an exonuclease, because the inactivation of PNPase also inhibited the degradation of ND5 mRNA (Figure 4B), which is rarely polyadenylated (18).
The recombinant form of the human homolog of yeast Suv3p (SUV3) forms a complex with PNPase, which degrades RNA in the 3′–5′ direction in vitro (30), and SUV3 is involved in mitochondrial mRNA degradation in the cell (31). To investigate whether SUV3 helicase is absolutely necessary for the degradation of mitochondrial mRNAs, mRNA decay was monitored after knockdown of SUV3 expression. Similar to the effect of the knockdown of PNPase, the decay of each mRNA was halted (Figure 4B). Therefore, both SUV3 helicase and PNPase are essential for mRNA degradation in human mitochondria.
Taken together with the accelerated decay of several mRNAs upon SLIRP knock down (Figure 2A), the results suggest that the LRPPRC/SLIRP complex stabilizes a set of mRNAs by suppressing 3′–5′ exonuclease activity performed by PNPase and SUV3.
LRPPRC promotes MTPAP-mediated polyadenylation of mRNAs
Next, we examined whether the LRPPRC/SLIRP complex has a function to promote polyadenylation of mRNAs. In six species of mRNAs (Figure 5A), oligoadenylated forms clearly accumulated when SLIRP was inactivated (as observed in Figure 4A), and elongated poly(A) tails were observed upon knockdown of PNPase, as previously reported (19). Upon the simultaneous knockdown of SLIRP and PNPase (Figure 5A), the oligoadenylated forms of COX1, COX2, ND3 and CYTB mRNAs accumulated to slightly higher levels than those observed after single knockdown of PNPase, suggesting that the LRPPRC/SLIRP complex promotes the polyadenylation of some mRNAs. Although we cannot rule out completely the possibility that insufficient PNPase knockdown led to the persistence of weak deadenylation activity under these conditions, we speculate that mitochondrial poly(A) polymerase (MTPAP) cannot efficiently synthesize poly(A) tails of mRNAs in the absence of the LRPPRC/SLIRP complex.
To demonstrate whether the LRPPRC/SLIRP complex plays a functional role in promoting polyadenylation of mRNAs, we conducted an in vitro polyadenylation experiment using recombinant proteins. Either COX2 or COX3 mRNA transcript was oligoadenylated with the recombinant MTPAP in the presence of [α-32P] ATP (Figure 5B). Polyadenylation of these mRNAs was stimulated in a dose-dependent manner after the addition of recombinant LRPPRC (Figure 5B), indicating that LRPPRC directly promoted MTPAP-mediated polyadenylation of mRNAs in vitro. Only a small effect of recombinant SLIRP on polyadenylation was observed (data not shown), indicating that LRPPRC plays the dominant role in promoting polyadenylation, while SLIRP plays a supportive role in stabilizing LRPPRC in the cell (Figure 2D).
To gain mechanistic insights into the functional role of LRPPRC in MTPAP-mediated polyadenylation, we characterized the substrate specificity of MTPAP. Considering that LRPPRC has PPR motifs that bind to single-stranded RNAs (11,13), we speculated that the LRPPRC/SLIRP complex binds to mRNAs and prevents their 3′-terminal regions forming secondary/tertiary structures, facilitating the polyadenylation by MTPAP. In fact, a plant PPR motif protein (PPR10) was shown to unwind a hairpin RNA structure and recognize a single-stranded target sequence (39). This observation inspired us to examine whether MTPAP preferentially adenylates the single-stranded termini of mRNAs. As a model substrate, we used a 62-nt-long single-stranded (ss)RNA (with an mfold prediction folding ΔG = −1.4 kcal·mol−1), corresponding to the 3′ terminal region of ND1 mRNA (Figure 5C). This ssRNA was oligoadenylated efficiently and was polyadenylated to some extent (Figure 5C). When the ssRNA was hybridized with complementary RNAs to prepare double-stranded (ds)RNA substrates with 0 - and 3-nt 3′ overhangs, the dsRNAs were hardly adenylated (Figure 5C). In contrast, the dsRNA with a 10-nt 3′ overhang was adenylated by MTPAP (Figure 5C). These results clearly demonstrate that MTPAP specifically adenylates RNAs with a single-stranded 3′-end. ND5 and ND6 mRNAs have short and no poly(A) tails, respectively (4,18). In addition, ND5 and ND6 mRNAs are the only mitochondrial mRNAs with long 3′ UTRs (4,18). The 3′ UTR of ND5 is complementary to the ND6 coding sequence; conversely, the ND6 3′ UTR is complementary to the ND5 coding sequence (Figure 1A). Some fractions of ND5 and ND6 mRNAs can hybridize with each other to form a duplex structure. This would prevent MTPAP from performing polyadenylatation, because MTPAP preferentially polyadenylates RNAs with single-stranded 3′ ends (Figure 5C). In addition, deep sequence analysis detected abundant antisense transcripts that were complementary to ND5 and ND6 mRNAs in human mitochondria (40). These transcripts might be involved in the suppression of polyadenylation. Alternatively, 3′ UTRs of ND5 and ND6 mRNAs may form stable intramolecular tertiary structures that prevent polyadenylation by MTPAP.
Mitochondrial mRNAs, in general, form complex secondary structures (41), which might prevent polyadenylation by MTPAP. We next designed a structured RNA substrate with a duplex 3′-end (mfold folding ΔG = −12.8 kcal·mol−1) (Figure 5D) that mimics the 3′ terminal region of mitochondrial mRNAs. When the substrate was used for in vitro polyadenylation in the absence of LRPPRC, a small portion of this transcript was oligoadenylated (Figure 5D), whereas in the presence of LRPPRC, polyadenylation occurred in a dose-dependent manner (Figure 5D). A poly(A) tail of up to about 150 nt was observed under this condition. Addition of BSA instead of LRPPRC did not show any effect on polyadenylation (Figure 5D), showing that the increased poly(A) synthesis by LRPPRC is not caused by non-specific stabilization of MTPAP by increased protein concentration. Next, we replaced LRPPRC with a DNA probe that is complementary to the 5′ half of the substrate used for in vitro polyadenylation. The DNA probe hybridizes to the structured RNA substrate, liberating its 3′-end as a single strand. Surprisingly, addition of the DNA probe only resulted in the accumulation of oligoadenylated RNA (Figure 5D) with no clear enhancement of polyadenylation (Figure 5D). In addition, when the ssRNA was employed as a substrate for in vitro polyadenylation, efficient polyadenylation was observed in the presence of LRPPRC (Figure 5E). These data demonstrated that the single-stranded 3′-end of RNA is not sufficient for polyadenylation and that MTPAP requires LRPPRC for efficient elongation of polyadenylated chains.
DISCUSSION
In this report, we describe the first absolute quantification of mitochondrial mRNAs in a human cell. The copy numbers of mitochondrial mRNAs ranged from 6000 (ND5) to 51 000 (COX2) per cell. Thus, an up to 8.5-fold difference in the steady-state levels of mRNAs from the long H-strand transcript was observed. We also noted good correlation between the copy number and the half-life of each mRNA (R2 = 0.601). Abundant mRNAs, such as COX1, COX2, COX3, ATP8/6 and ND4L/4, have longer half-lives, whereas less abundant mRNAs, such as ND3, ND5 and CYTB, have shorter half-lives. These data indicate that the steady-state levels of mitochondrial mRNAs in human HeLa cells are basically determined by their half-lives. Notable exceptions are ND1 and ND2 mRNAs, which were relatively abundant despite having short half-lives (74 and 80 min, respectively). According to the gene organization of mtDNA (Figure 1A), ND1 and ND2 are localized at the beginning of the long H-strand transcript. Recently, it was reported that TEFM (transcription elongation factor of mitochondria), which is a partner protein of mitochondrial RNA polymerase (POLRMT), is required for synthesizing promoter-distal transcripts from both strands (42). Limited availability of TEFM in rapidly growing cells might result in an increase of promoter-proximal transcripts. Alternatively, as mtDNAs are damaged by reactive oxygen species and aldehydes, H-strand transcription by POLRMT might be arrested by damaged bases in mtDNA (43), which produces shorter H-strand transcripts.
PPR motif-containing proteins constitute a large family of RNA-binding proteins that are primarily found in higher plants (44,45). In general, most proteins bearing PPR motifs bind to specific RNA sequences (44). This sequence-specific recognition is accomplished by the alignment of PPR motifs. Although LRPPRC contains 16 PPR motifs (12), the LRPPRC/SLIRP complex does not show strong sequence specificity for mRNA binding. In fact, all mRNA species were coprecipitated with the LRPPRC/SLIRP complex (Figure 3A). Sequence analysis of the bound RNA fragments revealed that the LRPPRC/SLIRP complex bound to mRNA coding regions and not to poly(A) tails (Supplementary Table S2). This result is in good agreement with an in vitro study that showed LRPPRC does not bind to poly(A) RNA immobilized on beads (11). In addition, ND5 and ND6 mRNAs, which have little or no poly(A) tails, were also coprecipitated with the LRPPRC/SLIRP complex (Figure 3A). Collectively, these data showed that the LRPPRC/SLIRP complex directly binds to mRNA coding sequences without strong sequence specificity. Nonetheless, considering the different degrees of mRNA stabilization by the LRPPRC/SLIRP complex (Figure 2B and C), there might still exist some specific sequences in these mRNAs that are preferentially recognized by the LRPPRC/SLIRP complex.
The ND6 mRNA is only weakly coprecipitated with LRPPRC-Flag in the absence of crosslinking (17). We performed immunoprecipitation of the RNP complex with endogenous LRPPRC from the cells that were crosslinked with formaldehyde (Figure 3A). The data showed that ND6 mRNA was coprecipitated with LRPPRC, when compared to control tRNAs and cytoplasmic RNAs, but the efficiency of coprecipitation was relatively low (Figure 3A). This indicates that the LRPPRC/SLIRP complex is weakly associated with ND6 mRNAs in mitochondria. In addition to mitochondrial mRNAs, a small amount of 12 S rRNA was observed in the LRPPRC immunoprecipitate (Figure 3A). The 28S small subunit of mammalian mitochondrial ribosome binds to mitochondrial mRNA even in the absence of auxiliary factors, whereas the large subunit does not (46). Thus, under crosslinking conditions, the 12 S rRNA in the 28 S subunit might have been coprecipitated with the LRPPRC/SLIRP complex through its tethering to mRNAs.
According to proteomic data (47), LRPPRC and SLIRP are abundant in mitochondria. We roughly estimated the copy numbers of LRPPRC in HeLa cells by western blotting, using recombinant LRPPRC as a reference (see ‘Materials and Methods’ section), and found approximately 2.0 × 106 LRPPRC molecules per cell. Given that there are 316 000 mitochondrial mRNA molecules in each HeLa cell (Table 1), the average number of LRPPRC to each mRNA would be 6, or 1 LRPPRC per 170 nt of mitochondrial mRNA. Such a high abundance of LRPPRC in mitochondria may facilitate cotranscriptional binding of the LRPPRC/SLIRP complex to many locations on the mRNA coding sequence, which could potentially affect mRNA secondary/tertiary structure.
How does the LRPPRC/SLIRP complex specifically discriminate mRNA regions from tRNAs and rRNAs in the precursor RNA? As the LRPPRC/SLIRP complex binds to mRNA coding regions without much sequence specificity, once transcription begins, the LRPPRC/SLIRP complex is loaded onto the nascent chain (Figure 6). By contrast, tRNAs spontaneously fold into secondary and tertiary structures that are recognized by various RNA-modifying enzymes and processing endonucleases, including RNaseP (MRPP1,2,3) (48) and tRNase Z (ELAC2) (49), which separate tRNAs, mRNAs and rRNAs from polycistronic transcripts. During this process, the LRPPRC/SLIRP complex would probably be unable to access tRNA regions, because PPR motifs generally bind to single-stranded RNAs (13). Moreover, the precursor RNAs, including both 12 S and 16 S rRNAs, also fold into programmed secondary and tertiary structures, which result in the recruitment of dozens of ribosomal proteins and non-ribosomal factors for their assembly into small and large ribosomal particles. Little is known about the mitoribosome assembly mechanism. However, as the assembly of bacterial ribosomes takes place cotranscriptionally (50), mitochondrial rRNAs and ribosomal proteins are also likely to assemble cotranscriptionally; thus, the ribosomal proteins are likely to bind to rRNAs with more specificity and affinity than the LRPPRC/SLIRP complex. This speculation would explain our observation that the LRPPRC/SLIRP complex was not efficiently coprecipitated with the precursor transcripts composed of tRNA and rRNA (Figure 3A). Therefore, the mRNA specificity of the LRPPRC/SLIRP complex might be established by a passive mechanism in which tRNA and rRNA regions are excluded from binding to the LRPPRC/SLIRP complex.
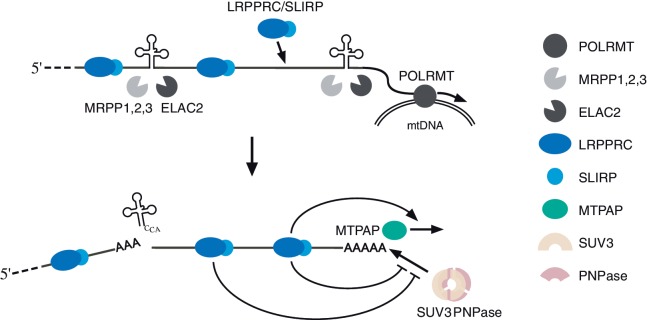
Figure 6.
Schematic depiction of biogenesis and the metabolism of mRNAs in human mitochondria. As mitochondrial RNA polymerase (POLRMT) transcribes mtDNA, the LRPPRC/SLIRP complex binds to the mRNA coding sequences of the long polycistronic precursor. tRNA 5′ and 3′ processing conducted by MRPP1,2,3 (48) and ELAC2 (49) generate mRNAs that are bound to the LRPPRC/SLIRP complex. The LRPPRC/SLIRP complex promotes mRNA polyadenylation by MTPAP. The LRPPRC/SLIRP complex protects mRNAs from 3′ exonucleolytic degradation mediated by PNPase and SUV3 helicase.
We have shown that PNPase and SUV3 are essential factors for mRNA degradation in human mitochondria. As knockdown of either PNPase or SUV3 alone inhibited mitochondrial mRNA decay (Figure 4B), PNPase and SUV3 helicase are likely to cooperatively degrade mitochondrial mRNAs in the cell. This speculation was also supported by the fact that recombinant PNPase and SUV3 form a heteropentamer which degrades dsRNA in vitro (30). However, in HeLa cell lines in which PNPase was stably silenced by RNAi (51), ND3 and ND5 poly(A) tails were elongated, while the COX3 poly(A) tail was unaffected and COX1 lost its poly(A) tail, indicating that PNPase is indirectly involved in poly(A) metabolism. This result conflicts with our observation that transient knockdown of PNPase caused poly(A) elongation in most of transcripts, including COX1 and COX3 (Figure 5) (19). In fact, in their article (51), transient PNPase silencing was shown to cause elongation of the COX1 mRNA poly(A) tail, which was clearly self-contradictory. PNPase has multivalent functions, including the transport of the RNA component for RNaseMRP into mitochondria (26) and the degradation of cytoplasmic mRNA (27) and miRNA (28). Thus, long-term, stable PNPase silencing (51) may have caused various secondary effects. Also, some revertants to complement the reduced PNPase function might have arisen.
Although we have strong evidence that PNPase is directly involved in deadenylation and degradation of mRNAs in human mitochondria, the steady-state levels of several mRNAs and proteins were unchanged upon knockdown of PNPase (19,25). If PNPase functions only in mRNA degradation, downregulation of PNPase should have resulted in the accumulation of mitochondrial mRNAs and proteins to some extent. PNPase is also involved in the mitochondrial import of the RNA component for RNaseMRP (26), which is an essential factor for generating the RNA primers required for the initiation of mtDNA replication (52). This fact prompted us to speculate that PNPase might be involved in the maintenance of mtDNA steady-state level. To investigate this possibility, we quantified the mtDNA level when PNPase was knocked down. As expected, PNPase knockdown caused a decrease in the mtDNA level (Supplementary Figure S3A), which could have decreased the transcriptional level of mRNAs, thereby counteracting the accumulation of mitochondrial mRNAs caused by loss of PNPase exonucleolytic activity. Four days after PNPase knockdown, the mtDNA level was 15% of the control level. However, mRNA degradation had almost completely stopped at this time (Figure 4B). When PNPase was knocked down for 2 days and the mtDNA level decreased to about 50% (Supplementary Figure S3A), a clear halt in mRNA degradation was still observed (Supplementary Figure S3B). As the measurement of mRNA degradation is independent of changes in mtDNA copy number, the results in this study cannot be easily attributed to a secondary effect of a decrease in the mtDNA level upon PNPase knockdown.
The LRPPRC/SLIRP complex was found to stabilize mRNAs by suppressing 3′ exonucleolytic degradation catalyzed by PNPase and SUV3. The mechanism by which the LRPPRC/SLIRP complex suppresses mRNA degradation awaits further elucidation. As the LRPPRC/SLIRP complex bound to mRNA coding sequences and not to poly(A) tails (Figure 3B and Supplementary Table S2), the LRPPRC/SLIRP complex could not directly impede deadenylation by PNPase and SUV3 by covering the poly(A) tail. Considering that PNPase and SUV3 preferentially degrade dsRNAs with a 3′ overhang (30), we speculate that the binding of the LRPPRC/SLIRP complex to the coding regions of mRNAs inhibits the folding of their 3′ termini into secondary/tertiary structures, thereby preventing mRNAs from becoming good substrates for degradation. Otherwise, if the poly(A) tail hybridizes with the uridine-rich region in the coding sequence to form intramolecular dsRNA, the LRPPRC/SLIRP complex might be involved in releasing the poly(A) tail from the coding region, which would make it a poor substrate for 3′exonucleolytic degradation by PNPase and SUV3.
A loss of poly(A) tails was found in mitochondrial mRNAs from a LRPPRC null mouse (17). Similarly, loss of poly(A) tails and accumulation of oligo(A) tails were observed in mitochondrial mRNAs from the RNAi knockdown flies of bicoid stability factor, which is a homolog of LRPPRC in Drosophila (53). These reports demonstrated that the LRPPRC/SLIRP complex is necessary to maintain mitochondrial mRNA poly(A) tails. In accordance with these reports, knockdown of LRPPRC or SLIRP in HeLa cells caused the accumulation of oligoadenylated mRNAs with a reduced population of polyadenylated mRNAs (Figure 4A). We have shown that the LRPPRC/SLIRP complex promotes mRNA poly(A) synthesis (Figure 5A and B). More precisely, the LRPPRC/SLIRP complex is likely to be involved in the unwinding the 3′ terminus in the initiation step of adenylation (oligoadenylation), because MTPAP preferentially adenylates RNA substrates bearing a single-stranded 3′ terminus (Figure 5C). In fact, artificial unwinding of the structured RNA substrate by the antisense DNA enhanced its oligoadenylation by MTPAP (Figure 5D). The most important finding in this experiment was that LRPPRC also promoted the elongation step of polyadenylation (Figure 5D and E). Further studies will be necessary to reveal how LRPPRC promotes poly(A) elongation. Our observation would explain why the mitochondrial mRNA poly(A) tail was lost and the oligo(A) tail was retained in a LRPPRC null mouse (17).
In conclusion, we report the different steady-state levels of human mitochondrial mRNAs through the accurate determination of their copy numbers. We also show a clear correlation between copy number and the half-life of each mRNA. mRNAs with longer half-lives are stabilized by the LRPPRC/SLIRP complex, which cotranscriptionally binds to mRNA coding sequences, promotes mRNA poly(A) elongation and suppresses 3′exonucleolytic degradation mediated by PNPase and SUV3.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online: Supplementary Figures 1–3 and Supplementary Tables 1 and 2.
FUNDING
Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (to T.S.); New Energy and Industrial Technology Development Organization (NEDO) (to T.S.). Funding for open access charge: Japan Ministry of Education, Science, Sports and Culture.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the members of the Suzuki laboratory for fruitful discussions and suggestions. We also thank Drs. Masayuki Sakurai (University of Pennsylvania), Shinichi Nakagawa (RIKEN) and Yuko Hasegawa (RIKEN) for technical support.
REFERENCES
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 3.Duborjal H, Beugnot R, Mousson de Camaret B, Issartel JP. Large functional range of steady-state levels of nuclear and mitochondrial transcripts coding for the subunits of the human mitochondrial OXPHOS system. Genome Res. 2002;12:1901–1909. doi: 10.1101/gr.194102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol. Cell. Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagao A, Hino-Shigi N, Suzuki T. Measuring mRNA decay in human mitochondria. Methods Enzymol. 2008;447:489–499. doi: 10.1016/S0076-6879(08)02223-4. [DOI] [PubMed] [Google Scholar]
- 6.Morin C, Mitchell G, Larochelle J, Lambert M, Ogier H, Robinson BH, De Braekeleer M. Clinical, metabolic, and genetic aspects of cytochrome C oxidase deficiency in Saguenay-Lac-Saint-Jean. Am. J. Hum. Genet. 1993;53:488–496. [PMC free article] [PubMed] [Google Scholar]
- 7.Merante F, Petrova-Benedict R, MacKay N, Mitchell G, Lambert M, Morin C, De Braekeleer M, Laframboise R, Gagne R, Robinson BH. A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-Lac-Saint-Jean region of Quebec. Am. J. Hum. Genet. 1993;53:481–487. [PMC free article] [PubMed] [Google Scholar]
- 8.Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci. USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mili S, Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sterky FH, Ruzzenente B, Gustafsson CM, Samuelsson T, Larsson NG. LRPPRC is a mitochondrial matrix protein that is conserved in metazoans. Biochem. Biophys. Res. Commun. 2010;398:759–764. doi: 10.1016/j.bbrc.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Small ID, Peeters N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 14.Hatchell EC, Colley SM, Beveridge DJ, Epis MR, Stuart LM, Giles KM, Redfern AD, Miles LE, Barker A, MacDonald LM, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol. Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z, Mootha VK. A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet. 2009;5:e1000590. doi: 10.1371/journal.pgen.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Camara Y, Milenkovic D, Zickermann V, Wibom R, Hultenby K, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs—like members of all families, similar but different. Biochim. Biophys. Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 20.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wydro M, Bobrowicz A, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Targeting of the cytosolic poly(A) binding protein PABPC1 to mitochondria causes mitochondrial translation inhibition. Nucleic Acids Res. 2010;38:3732–3742. doi: 10.1093/nar/gkq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rorbach J, Nicholls TJ, Minczuk M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res. 2011;39:7750–7763. doi: 10.1093/nar/gkr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borowski LS, Szczesny RJ, Brzezniak LK, Stepien PP. RNA turnover in human mitochondria: more questions than answers? Biochim. Biophys. Acta. 2010;1797:1066–1070. doi: 10.1016/j.bbabio.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 24.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell. Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar D, Leszczyniecka M, Kang DC, Lebedeva IV, Valerie K, Dhar S, Pandita TK, Fisher PB. Down-regulation of Myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPaseold-35) in human melanoma cells. J. Biol. Chem. 2003;278:24542–24551. doi: 10.1074/jbc.M302421200. [DOI] [PubMed] [Google Scholar]
- 28.Das SK, Sokhi UK, Bhutia SK, Azab B, Su ZZ, Sarkar D, Fisher PB. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc. Natl Acad. Sci. USA. 2010;107:11948–11953. doi: 10.1073/pnas.0914143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 30.Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J. Biol. Chem. 2009;284:20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh T, Suzuki T. Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res. 2007;35:e27. doi: 10.1093/nar/gkl1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parrott AM, Walsh MR, Mathews MB. Analysis of RNA:protein interactions in vivo: identification of RNA-binding partners of nuclear factor 90. Methods Enzymol. 2007;429:243–260. doi: 10.1016/S0076-6879(07)29012-3. [DOI] [PubMed] [Google Scholar]
- 35.Goshima N, Kawamura Y, Fukumoto A, Miura A, Honma R, Satoh R, Wakamatsu A, Yamamoto J, Kimura K, Nishikawa T, et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods. 2008;5:1011–1017. doi: 10.1038/nmeth.1273. [DOI] [PubMed] [Google Scholar]
- 36.Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, Chrzanowska-Lightowlers ZM. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum. Mol. Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- 37.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poulsen JB, Andersen KR, Kjaer KH, Durand F, Faou P, Vestergaard AL, Talbo GH, Hoogenraad N, Brodersen DE, Justesen J, et al. Human 2′-phosphodiesterase localizes to the mitochondrial matrix with a putative function in mitochondrial RNA turnover. Nucleic Acids Res. 2011;39:3754–3770. doi: 10.1093/nar/gkq1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rackham O, Shearwood AM, Mercer TR, Davies SM, Mattick JS, Filipovska A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA. 2011;17:2085–2093. doi: 10.1261/rna.029405.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denslow ND, Michaels GS, Montoya J, Attardi G, O’Brien TW. Mechanism of mRNA binding to bovine mitochondrial ribosomes. J. Biol. Chem. 1989;264:8328–8338. [PubMed] [Google Scholar]
- 42.Minczuk M, He J, Duch AM, Ettema TJ, Chlebowski A, Dzionek K, Nijtmans LG, Huynen MA, Holt IJ. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cline SD, Lodeiro MF, Marnett LJ, Cameron CE, Arnold JJ. Arrest of human mitochondrial RNA polymerase transcription by the biological aldehyde adduct of DNA, M1dG. Nucleic Acids Res. 2010;38:7546–7557. doi: 10.1093/nar/gkq656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 45.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Liao HX, Spremulli LL. Interaction of bovine mitochondrial ribosomes with messenger RNA. J. Biol. Chem. 1989;264:7518–7522. [PubMed] [Google Scholar]
- 47.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 49.Brzezniak LK, Bijata M, Szczesny RJ, Stepien PP. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 2011;8:616–626. doi: 10.4161/rna.8.4.15393. [DOI] [PubMed] [Google Scholar]
- 50.de Narvaez CC, Schaup HW. In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J. Mol. Biol. 1979;134:1–22. doi: 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- 51.Slomovic S, Schuster G. Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA. 2008;14:310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl Acad. Sci. USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bratic A, Wredenberg A, Gronke S, Stewart JB, Mourier A, Ruzzenente B, Kukat C, Wibom R, Habermann B, Partridge L, et al. The bicoid stability factor controls polyadenylation and expression of specific mitochondrial mRNAs in Drosophila melanogaster. PLoS Genet. 2011;7:e1002324. doi: 10.1371/journal.pgen.1002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.