Abstract
Within chromatin, the core histone tail domains play critical roles in regulating the structure and accessibility of nucleosomal DNA within the chromatin fiber. Thus, many nuclear processes are facilitated by concomitant posttranslational modification of these domains. However, elucidation of the mechanisms by which the tails mediate such processes awaits definition of tail interactions within chromatin. In this study we have investigated the primary DNA target of the majority of the tails in mononucleosomes. The results clearly show that the tails bind preferentially to “linker” DNA, outside of the DNA encompassed by the nucleosome core. These results have important implications for models of tail function within the chromatin fiber and for in vitro structural and functional studies using nucleosome core particles.
Keywords: nucleosomes, chromatin
The eukaryotic genome is assembled with histones and other nuclear proteins into a large macromolecular complex known as chromatin. Clearly, structural elements of this complex such as the basic repeating unit known as the nucleosome and the folding of arrays of nucleosomes into a chromatin fiber impinge on the utilization of DNA for virtually all nuclear processes (1, 2). Indeed, this facet has been functionally incorporated into the mechanisms that control gene expression (2). Importantly, the core histone tail domains provide a nexus for transduction of intracellular signals associated with preparing functional states of chromatin appropriate for nuclear processes such as transcription or replication. For example, thyroid hormone receptor mediates repression of target genes in part by directing histone deacetylases to promoters, whereas steroid-mediated activation of transcription includes directed histone acetylation (3, 4). These activator- or repressor-mediated posttranslational modifications are likely to alter the charge, structure, and/or interactions of the core histone tails or to serve as targets for the binding of ancillary proteins or other enzymatic functions (5–7). Indeed, it was recently shown that the binding of accessory factors facilitates the essential role of the tails in mitotic chromosome condensation (8). Changes in core histone tail structures and/or interactions in turn are thought to modulate the stability of the chromatin fiber and the accessibility of DNA within individual nucleosomes (9, 10).
Unfortunately, little is known about the exact nature and locations of interactions made by the core histone tail domains in the nucleosome and in various conformations of the chromatin fiber (9). Recent circular dichroism studies indicate that perhaps 20–40% of the tail residues are involved in α-helical structure, perhaps mostly in the H3/H4 tails (11, 12). Interestingly, this α-helical content appears to be substantially increased upon acetylation (12). However, the identity of residues involved in α-helical structures or other defined conformations is not clear, and how the changes caused by acetylation affect tail interactions in chromatin is unknown. Thus, the mechanisms by which posttranslational modifications of the histone tail domains modulate DNA accessibility in the chromatin fiber remain undefined. Crosslinking experiments with nucleosomes and oligonucleosomes (oligosomes) suggest that in physiological salts, the tails are in contact with DNA at defined locations about the nucleosome (13–17). Moreover, quantitative hydrodynamic and gel electrophoresis experiments suggest that the tail domains take part in defined interactions with both DNA and protein within the condensed chromatin fiber (9, 18).
In many experiments nucleosome core particles or similar complexes are used as model systems to study the structural or functional consequences of the core histone tail domains. However, recent studies suggest that most canonical core histone tail–DNA interactions cannot be realized in the core particle and require more physiologically relevant chromatin structures. For example, a recent x-ray crystal structure of the nucleosome core particle indicates that, at elevated salt concentrations (>40 mM MnCl2, ≈35 mM KCl) and in the absence of linker DNA, the tails do not bind nucleosome core DNA in unique and defined conformations (19). Indeed, crosslinking experiments suggest that substantially more tail–DNA interactions occur in structures containing linker DNA—i.e., structures larger than the core particle (14). The C-terminal tail of H2A contacts nucleosomal DNA near the center of the nucleosome core but “shifts” to contact DNA near the edge of the core region in nucleosomes and oligonucleosomal structures containing linker DNA (16, 20). Thus the DNA contacts made by this tail in the nucleosome core particle are unlikely to be representative of contacts in native chromatin. Here we demonstrate that the majority of core histone tail interactions with DNA occur outside of the central 146 bp of DNA within the nucleosome core. These results indicate that extranucleosomal linker DNA is the primary target for a majority of the core histone tail domains in chromatin.
Materials and Methods
Preparation of Linker Histone-Depleted Oligosomes.
Oligosomes were prepared from micrococcal nuclease-digested chicken erythrocyte nuclei as previously described (21). Linker histones H1 and H5 and nonhistone proteins were removed from the soluble chromatin by centrifugation through a 5–20% sucrose gradient containing 0.65 M NaCl. Fractions corresponding to the mononucleosome peak were collected and dialyzed against Tris⋅HCl, pH 7.5/0.25 mM EDTA buffer containing 1, 100, or 120 mM NaCl and 0.5 mM MgCl2 as stated in the figure legends. Nucleosome particles were prepared by digestion of linker histone-depleted oligosomes with micrococcal nuclease (14).
Reconstitution of Defined-Sequence Nucleosomes for UV LaserInduced Crosslinking.
Recombinant Xenopus laevis full-length and tailless domain histone proteins were made in bacteria and purified to homogeneity as described by Luger et al. (19, 22). DNA fragments 147 bp and 207 bp long were prepared and contained the Xenopus borealis somatic 5S RNA gene nucleosome-positioning element. The 147- and 207-bp DNAs were generated by PCR amplification of the pXP-10 plasmid (22) using primers TTCGAGCTCGCCCGGGGATCC and CCAGGCCCGACCCTGCTTGGC, or GAACCGCTCGAGCTCTGTCCTTTTACGAATTCGAGCTCGCC and GCTAGATCTAGACTCTCTCCTTGTACTAACCAGGCCCGACCC, respectively. The primers were chosen so that the predicted dyad axis of the reconstituted nucleosomes would be located at the center of the fragments.
These fragments were reconstituted into nucleosomes as follows. Precisely stoichiometric amounts of the four histones (determined spectrophotometrically and checked by SDS/PAGE) in 10 mM HCl were combined and dialyzed overnight at 4°C against 2 M NaCl/50 mM Tris⋅HCl, pH 7.8/1 mM EDTA/5 mM 2-mercaptoethanol. The next morning, the 32P-end-labeled fragments were added to the dialysis tubing. The ratio of the added DNA to the core histone octamer was 1:0.8. Next, the salt concentration was lowered by successive dialysis against the same buffer as above, but containing decreasing amounts of NaCl (23). The extent of reconstitution and the integrity of the nucleosomes were checked by nucleoprotein gels and DNase I footprinting.
UV Laser Irradiation and Protein-DNA Crosslinking Quantification.
Typically, 250 μl of nucleosome samples at a concentration of 50 μg/ml was irradiated in 10-μl aliquots by a single 5-nsec UV pulse at 266 nm of a Surelite II (Continuum, Santa Clara, CA) neodymium/yttrium/aluminum-garnet laser. The irradiation was carried out in silane-treated 0.65-ml Eppendorf tubes. A calibrated pyroelectrical detector (Ophir Optronics) was used to measure the pulse energy of radiation. The irradiation dose (the pulse energy divided by the beam surface) did not exceed 0.15 J/cm2. These irradiation conditions typically result in a covalent crosslinking of 6–8% of the nucleosomal DNA to the histones in native nucleosomes (17).
Samples (250 μl) of both irradiated and nonirradiated nucleosome were concentrated to 50 μl by using a centrifugal filter device (Amicon), and the samples were run on a native 5% polyacrylamide gel. After electrophoresis, the gel was stained with ethidium bromide and the band corresponding to the nucleosome was cut perpendicular to the direction of migration in 1-mm successive slices. The protein–DNA complexes were electroeluted and radioactively end-labeled with polynucleotide kinase and [γ-32P]ATP. After removal of the free [γ-32P]ATP by filtration through a mini Sephadex G50 column, the amount of crosslinked histone–DNA complexes isolated from each gel slice was determined by a phenol-extraction assay (17, 24). The relative yield of covalently crosslinked histone–DNA complexes was determined as the ratio of the phenol cpm to phenol plus aqueous cpm after subtraction of the background cpm from the nonirradiated controls. An aliquot of each electroeluted complex was treated with proteinase K and extracted with phenol to remove polypeptide fragments, and the DNA was run on a native 5% polyacrylamide gel with appropriate DNA size markers to determine the length of DNAs in each sample.
Immunodetection of Histones Crosslinked to DNA.
The covalently crosslinked histone–DNA complexes in the nucleosome samples were separated from noncrosslinked histones in preformed CsCl gradients centrifuged in an SW 41 Beckman rotor for 36 h at 15°C (25). The gradient was fractionated and the absorbance at 260 nm of the fractions was measured. The DNA-containing fractions were pooled and used directly for immunochemical experiments.
Antisera against core histones were raised in rabbits as described elsewhere (25). Immunospecific antibodies were purified from sera by affinity chromatography with antigen linked to CNBr-activated Sepharose. The antibodies prepared in this way were highly specific and did not detectably crossreact with other proteins (for details see ref. 14). The efficiency of crosslinking of individual histones to DNA was determined by an immunoslot assay. Briefly, 3–5 μg of the CsCl-purified crosslinked material (measured as DNA) was loaded onto nitrocellulose filters, and the presence of individual histones was detected with the appropriate immunopurified antibodies as described (25).
Site-Specific Photochemical Crosslinking in Defined Nucleosomes.
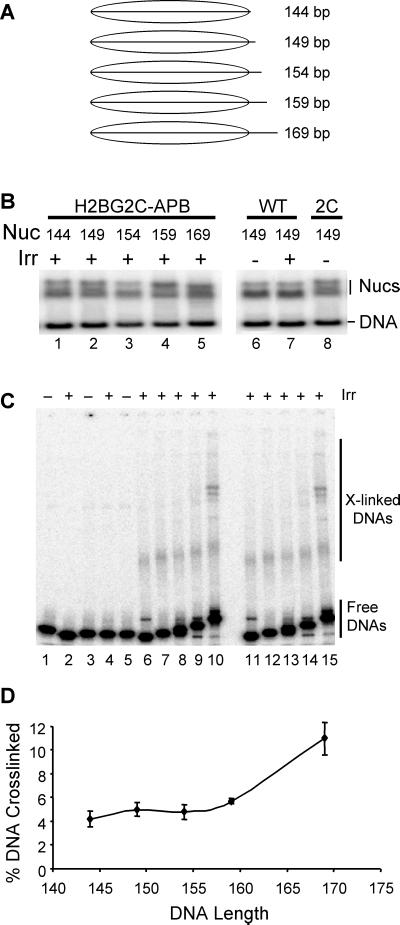
A set of DNA fragments based on the Xenopus borealis somatic 5S rRNA nucleosome-positioning sequence was created by standard PCR methods. These DNAs range in size from 144 to 169 bp (see Fig. 5A) and differ because of progressive 5-bp extensions of the downstream portion of the fragment (26). An H2B coding sequence containing a cysteine at the second codon was constructed by adding the codons for Val-Cys between the Met and the second codon in the wild-type H2B sequence obtained from Xenopus laevis to create H2B2C. The coding sequence for the mutant and wild-type proteins were inserted into the pET3d vector (Novagen) and over-expressed in bacterial cells as described (23). The proteins were purified and dimerized with wild-type H2A, and the Cys residue was modified with azidophenacyl bromide (APB) as described (23). Nucleosomes were reconstituted with either wild-type H2B or the APB-modified H2B2C and the DNA fragments by salt-dialysis described above. The reconstituted nucleosomes and unassembled (naked) DNA fragments were irradiated then separated on 0.7% agarose/0.5× TBE nucleoprotein gels (1× TBE is 90 mM Tris/90 mM boric acid/2.5 mM EDTA, pH 8.3). The wet gels were autoradiographed, and naked DNAs and nucleosomes were extracted from the gel, denatured with SDS, then loaded onto SDS/PAGE “crosslinking” gels to separate crosslinked from uncrosslinked DNA, as described (23). The extent of covalent crosslink formation in each sample was determined by PhosphorImager analysis of the dried gels.
Figure 5.
Efficiency of targeted photochemically induced crosslinking depends on nucleosomal DNA length. Nucleosomes were reconstituted with a recombinant H2B containing a single cysteine at position two in the N-terminal tail (H2B2C) and modified with APB. (A) DNA templates used for reconstitution. Nucleosomes were reconstituted with specific DNA templates ranging in length from 144 to 169 bp. (B) Nucleosomes assembled on various DNA templates. Reconstituted mixtures containing nucleosomes and unassembled (naked) DNAs were separated on a nucleoprotein gel and visualized by autoradiography of the wet gel. Lanes 1–5 contain irradiated nucleosomes reconstituted with 144-, 149-, 154-, 159-, and 169-bp templates, respectively. Lanes 6 and 7 contain nucleosomes reconstituted with completely wild-type histones that were either unirradiated or irradiated before separation on the gel, respectively. Lane 8 contains nucleosomes reconstituted with H2B2C that was not further modified with the crosslinking reagent. (C) Products of site-specific photochemical crosslinking reactions. Nucleosomes and naked DNAs were eluted from the preparative nucleoprotein gel, denatured with SDS, then subjected to SDS/PAGE to visualize protein–DNA crosslinked products. Lanes 1 and 2 contain naked 149-bp DNA either unirradiated or UV irradiated, respectively. Lanes 3 and 4 contain nucleosomes reconstituted with wild-type histones and either unirradiated or UV-irradiated, respectively. Lane 5 contains nucleosomes reconstituted with H2A2C-APB and 149-bp DNA but not irradiated. Lanes 6–10 contain UV-irradiated nucleosomes reconstituted with H2A2C-APB and the 144-, 149-, 154-, 159-, or 169-bp templates, respectively. Lanes 11–15 contain samples identical to those in lanes 6–10 but from a separate experiment. (D) Summary plot of percent DNA crosslinked vs. nucleosomal DNA length (in bp). The fraction of radiolabeled DNA fragment present in crosslinked species (see C) was determined and plotted vs. nucleosomal DNA length.
Results
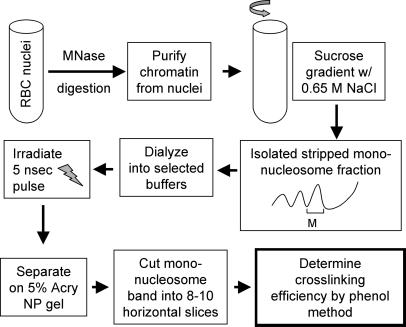
We wished to determine the relative extent to which the core histone tail domains interact with nucleosome core DNA compared with linker DNA. To address this question, we used a UV laser-induced protein–DNA crosslinking technique. Because UV laser light is a “zero-length” crosslinking agent and the crosslinking occurs virtually only between the histone tail domains and DNA, the crosslinking efficiency directly reflects the extent of histone tail–DNA interactions (14, 17). To investigate tail binding to nucleosome core versus linker DNA, mononucleosomes (monosomes) containing a range of DNA sizes were obtained from native chromatin, from nucleosome “core particles” containing 147 bp of DNA and essentially no linker DNA, to nucleosomes containing 210 bp of DNA and up to 63 bp of linker DNA (see Materials and Methods).
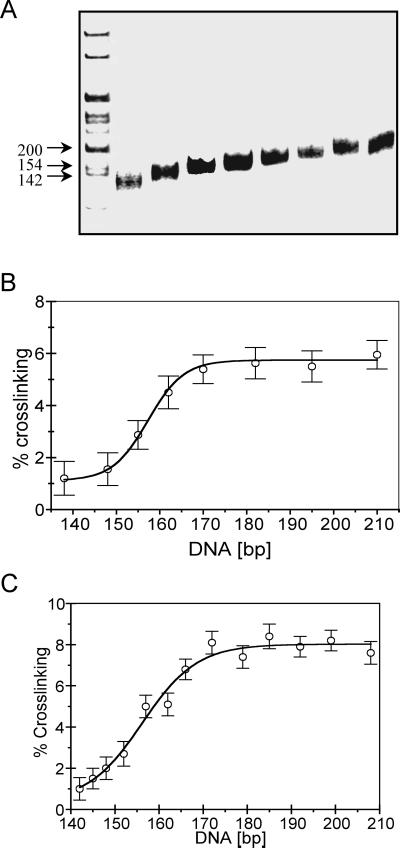
The nucleosomes were irradiated to induce histone tail–DNA crosslinking and then fractionated on high-resolution nucleoprotein gels according to DNA length. Pools of nucleosomes with similar DNA lengths were isolated and the extent of UV laser-induced protein–DNA crosslinking was determined for each fraction (see Fig. 1). We find that the extent of crosslinking is clearly dependent on the length of DNA within the nucleosome (Fig. 2). As the DNA size increases from that of the nucleosome core particle (147 bp) to longer fragments, the efficiency of crosslinking increases significantly, up to an efficiency nearly equivalent to that observed in irradiated native chromatin (Fig. 2B; refs. 14 and 17). Interestingly, the largest increase in efficiency occurs as the DNA size increases from ≈147 bp to ≈170 bp. Further increases in DNA length within the nucleosomes do not lead to significant increases in crosslinking efficiency. Thus, the extent of total tail–DNA interactions depends on the presence of linker DNA outside of the nucleosome core region. Furthermore, these contacts appear to be concentrated within the first ≈25 bp of linker DNA. To eliminate any possibility of differential labeling of the irradiated DNA after the gel isolation procedure, we repeated the experiment with nucleosomes bulk-labeled before the irradiation. As before, we find that the association of the tail domains is exquisitely DNA length-dependent, especially in the range 145–170 bp (Fig. 2C).
Figure 1.
Schematic of the sample preparation and experimental protocol for studying the effect of the length of nucleosomal DNA on the crosslinking efficiency of the core histone tail domains. MNase, micrococcal nuclease; 5% Acry NP gel, native 5% polyacrylamide nucleoprotein gel.
Figure 2.
The efficiency of histone–DNA crosslinking within nucleosomes depends on DNA length. Native H1-depleted oligosomes in buffer containing 80 mM NaCl, 0.25 mM EDTA, and 10 mM Tris⋅HCl, pH 7.5, were irradiated with a single UV laser pulse, then nucleosomes were separated on a 5% polyacrylamide gel. The nucleosome band was cut into several successive slices, the nucleoprotein complexes were electroeluted, and the percentage of crosslinked histone–DNA complexes was determined. (A) DNA length determination for the histone-DNA complexes isolated from the mononucleosome slices. The histone–DNA complexes were extensively digested with Pronase, and DNA was isolated and run on a native 5% polyacrylamide gel. Left lane 1, 1-kb DNA ladder. (B) Plot of the percent of total DNA crosslinked vs. nucleosomal DNA length. (C) As in B except that the nucleosomal DNAs were radioactively end-labeled before the irradiation step (see text). The data represent average values and standard deviations from four different experiments.
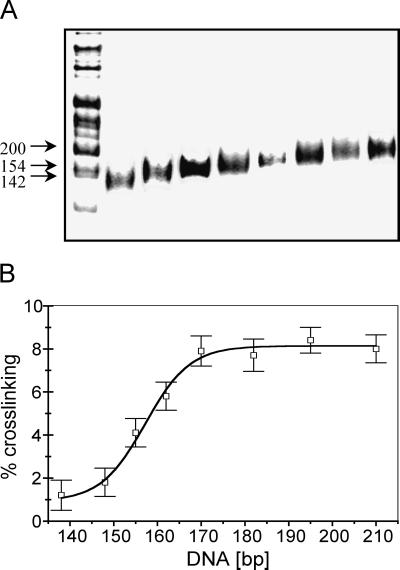
Initially we used buffers containing 80–100 mM NaCl during the crosslinking step to approximate the ionic strength of the intranuclear environment. To see whether the observed DNA length-dependent crosslinking of the tail domains was affected at all by these moderate levels of salt we repeated the experiment under conditions of low ionic strength (Fig. 3). Under such conditions, the highly basic tails are expected to exhibit maximally stable interactions with the DNA. As expected, we find that the tails are clearly associated with the DNA at this ionic strength (27–29). Moreover, we find that the degree of association is still strongly dependent on the length of the nucleosomal DNA even in low-salt solutions, in a manner identical to that found in higher-salt conditions (Fig. 3). To determine whether crosslinking efficiencies are altered in even higher (and perhaps more physiological) ionic strengths than used in Fig. 2, we repeated the crosslinking experiment in 120 mM NaCl/0.5 mM MgCl2. We find that the efficiency and DNA length-dependence of UV laser-induced crosslinking in this buffer is identical to that in 80–100 mM NaCl (results not shown).
Figure 3.
Linker-DNA length-dependent crosslinking of the core histone tails is observed in low ionic strength buffers. Nucleosomes were prepared and irradiated as described in for Fig. 2 except that the buffer contained 1 mM NaCl. (A) DNA length determination for the histone–DNA complexes isolated from the mononucleosome slices. (B) Plot of the percent of total DNA crosslinked vs. nucleosomal DNA length.
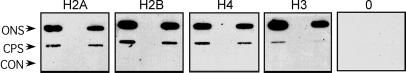
We next determined whether each of the individual core histones exhibits the same DNA-length-dependent crosslinking as the bulk of the core histones. Nucleosome cores and nucleosomes containing linker DNA were irradiated, then individual core histone–DNA crosslinked species were isolated by an SDS/cesium chloride gradient procedure (see Materials and Methods). The crosslinked species were then identified by a slot-immunoblotting technique (17). Equivalent amounts of nucleosomal DNA were blotted onto the membrane and the amount of individual crosslinked histones was determined with immunopurified antibody preparations specific for each of the core histone proteins (17). We find that all four core histones are crosslinked with significantly higher efficiency to DNA in oligonucleosomal structures containing linker DNA than in nucleosome core particles, which lack linker DNA (Fig. 4). Interestingly, we find that the N-terminal tail of H3 exhibits the greatest increase in efficiency of crosslinking in this assay.
Figure 4.
Immunoslot analysis of the reaction of antibodies to the individual core histones H2A, H2B, H3, and H4 and preimmune IgG (0) with covalent histone complexes. H1-depleted oligosomes (essentially monosomes, containing small amount of di- and trisomes, see Fig. 1) and core particles were UV laser irradiated with identical doses, and the covalent complexes were purified on CsCl gradients. A series of 5 μg and 2.5 μg of the complexes (measured as DNA) as well as of the control nonirradiated (CON) CsCl-purified samples were loaded on nitrocellulose filters, and the crosslinked histones were detected with immunopurified antibodies. The slots loaded with H1-depleted oligonucleosomes (ONS) or core particles (CPS) are indicated.
To confirm the above results by another method, we performed site-directed crosslinking experiments with nucleosomes containing a single core histone protein with a photochemical crosslinking probe specifically incorporated in the N-terminal tail of H2B. An H2B containing a single cysteine as the second amino acid residue within the N-terminal tail of this protein was prepared. This protein was modified with APB, introducing a photoactivatable crosslinking probe attached to the cysteine residue. Nucleosomes were reconstituted with the modified protein and native histones H2A, H3, and H4. The DNA templates used for reconstitution were increasingly longer, ranging from 144 to 169 bp (Fig. 5A). Reconstituted nucleosomes and naked DNAs were separated on nucleoprotein gels, the gels were irradiated, and crosslinked and uncrosslinked DNAs were extracted from the nucleosome and naked DNA bands. The extent of crosslinking within each nucleosome and naked DNA control was then determined by separation of irradiated samples by SDS/PAGE (15, 23). We found that maximal crosslinking occurred within nucleosomes assembled with the 169-bp template (n = 3) (Fig. 5 B and C). Minimal crosslinking occurred with the shortest templates, although the majority of the increase in crosslinking occurred as the template was extended from 160 to 169 bp. Previous work has established that the efficiency of the phenylazide-based crosslinking reactivity is not dependent on the location of DNA within the nucleosome core (16). These data suggest that the extension of DNA from 160 to 169 bp provides a more appropriate binding site for the N-terminal tail of H2B.
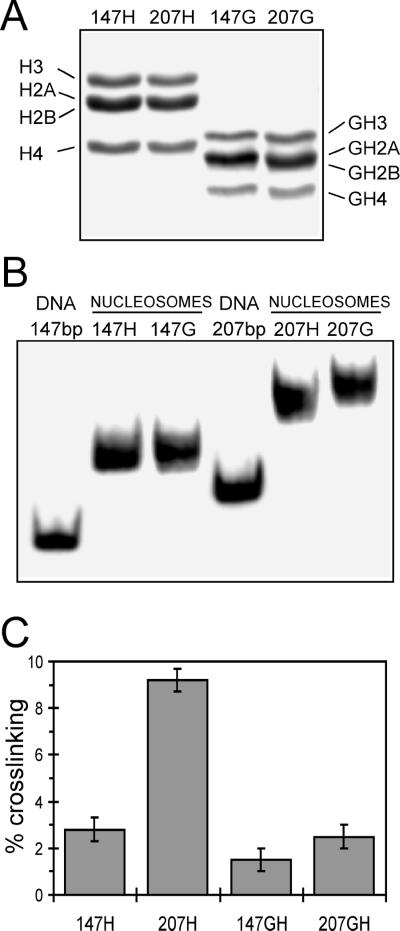
To assess the relationship between the site-specific phenylazide-based crosslinking and the UV laser approach, we prepared nucleosomes containing recombinant histones and defined-length DNA fragments based on the 5S nucleosome-positioning sequence used for the experiments shown in Fig. 5. Histones either containing or lacking tail domains were prepared and reconstituted with 5S DNA fragments of 147 or 207 bp to produce a model nucleosome core particle or a nucleosome containing ≈60 bp of linker DNA (Fig. 6 A and B) (22). Crosslinking was induced within these complexes by means of UV laser irradiation as described for native complexes, and the extent of crosslinking was determined. We found that crosslinking increased from about 3% of total DNA to about 9% as the DNA size associated with the nucleosome increased from 147 to 207 bp. Moreover, experiments with nucleosomes lacking core histone tail domains showed that this increase in crosslinking was entirely dependent on the presence of these domains (Fig. 6). Interestingly, we find that removal of the tails has little or no effect on crosslinking efficiency with the core particle but drastically reduces crosslinking with the nucleosome reconstituted with the 207-bp DNA fragment to levels observed in the absence of linker DNA (Fig. 6C). These results are in agreement with earlier work indicating that a majority of UV laser-induced protein–DNA crosslinking in chromatin occurs with the histone tail domains (14), and they strongly suggest that the linker DNA-dependent increase in crosslinking we observe is due to tail–DNA interactions.
Figure 6.
UV laser-induced crosslinking within a defined sequence nucleosome and nucleosome core particle. Either a 147- or a 207-bp 5S DNA fragment was reconstituted with either full-length or tailless recombinant core histones, then the nucleosomes were purified by sucrose gradient sedimentation. Proteins in mononucleosome fractions were analyzed by SDS/PAGE (A) or nucleosomes were analyzed on polyacrylamide nucleoprotein gels (B). (A) Full-length histones (H) or tailless histones (G) associated with the 147- or 207-bp templates are shown in lanes 1 and 3 or 2 and 4, respectively. (B) Nucleosomes reconstituted with full-length (H) or tailless core histones (G) are shown. (C) Extent of crosslinking in nucleosomes containing full-length (H) or tailless (G) core histones for nucleosomes reconstituted with either the 147- or the 207-bp templates, as indicated. The data represent average values and standard deviations from four different experiments.
Discussion
An understanding of the interactions and binding sites of the core histone tail domains is critical to the elucidation of how these domains regulate the structure and participation of the chromatin fiber in various nuclear processes such as transcription and replication. The experiments presented here suggest that the majority of the core histone tails prefer to bind linker DNA rather than intranucleosome core DNA. We find that the efficiency of UV laser-induced and site-specific arylazide-based tail–DNA crosslinking increases dramatically as the DNA is extended beyond the periphery of the core particle. The dependence of crosslinking efficiency on DNA length is independent of ionic strength over the range 10 mM NaCl to 120 mM NaCl, in the presence of 0.5 mM MgCl2. Previous work has demonstrated that the efficiency of UV laser-induced crosslinking in mononucleosomes retaining substantial linker DNA is equivalent to that found within native chromatin (14, 17). Moreover, crosslinking of native and H1-depeleted chromatin indicates that bulk tail–DNA interactions are not greatly affected by binding of linker histones (14). Thus, the majority of these domains are likely to be directed toward adjacent linker DNA within the native chromatin fiber. Of the 10 core histone tails within each nucleosome, 8 are clearly of sufficient length to contact the linker DNA adjacent to an individual nucleosome core, regardless of the conformation of the linker DNA in the chromatin fiber (1, 10). Moreover, in some models of the chromatin fiber the linker DNA is predicted to follow a superhelical path similar to that within the nucleosome core (1, 2), providing sufficient opportunity for all of the tails to interact with linker DNA.
The increase in crosslinking efficiency as a function of DNA length is interpreted as directly reflective of an increase in interaction of the histone tail domains with DNA. Previous experiments have demonstrated that UV laser-induced protein–DNA crosslinking depends on close approach (binding) of crosslinkable proteins to DNA (14). Moreover, as demonstrated previously and again in Fig. 6C, the majority of histone–DNA crosslinking occurs with the histone tail domains. Although it is formally possible that the increased crosslinking we observe is reflective of greater reactivity of linker vs. core DNA, we believe this is unlikely for the following reasons: (i) UV laser crosslinking reactivity appears to depend only on the presence of the relatively flexible tail domains; (ii) nucleosomal DNA appears to exhibit degrees of freedom and amplitudes of motion nearly identical to those of naked DNA (19); and (iii) both crosslinking techniques used detect increased interactions with linker DNA, and it has been demonstrated that core DNA is not less reactive than linker to nonspecific histone-directed arylazide crosslinking (16).
Our results suggest that the nucleosome core particle may be considered a relatively nonphysiological entity with regard to the proper binding of the tail domains. Indeed, the view of the nucleosome core particle as the most basic repeating unit or building block of eukaryotic chromatin has recently been called into question (30). This particle contains ≈1¾ turns of DNA and only the four core histones and is defined as a relatively kinetically stable intermediate produced from digestion of chromatin with micrococcal nuclease. However, histone octamer–DNA complexes containing anywhere from ≈100 to 180 bp of DNA have been shown to be stable, and the number of superhelical wraps of DNA observed around the octamer varies from ≈1 to 2, depending on the conditions of observation (1, 31). These products are likely to reflect a balance between tail–DNA interactions and the affinity of micrococcal nuclease for nucleosomal DNA and further suggest that the core histone tails require a complete nucleosome subunit for appropriate binding interactions.
Linker histones appear to be an integral component of the nucleosome and require >165 bp of DNA within the nucleosome for stable association (1). Nucleosomal repeats <165 bp have been only rarely observed in eukaryotes (32). This length of DNA approximately corresponds to the length of DNA at which crosslinking peaks in our experiments. Thus, the fundamental repeating unit of chromatin is likely to be a chromatosome—i.e., a nucleosome containing the core histone octamer, 165 bp of DNA, and one linker histone (1, 2, 33). Interestingly, particles containing DNA of chromatosomal length can be produced by micrococcal nuclease digestion of H1-lacking chromatin in buffers containing high concentrations of salt (31). Furthermore, the wrapping of DNA around the histone core is stabilized at the periphery of the nucleosome by the core histone tail domains (34). Our results provide a physical basis for these observations.
Our primary experimental protocol does not require any prior modification to the core histone tails that might alter their interactions with DNA. Irradiation with the UV laser is carried out in such a way as to maximize crosslinking dependent on biphotonic absorption while minimizing monophotonic absorption events, which primarily lead to DNA damage (ref. 14 and references therein). Moreover, crosslinking occurs very rapidly. Indeed, we used a laser pulse of 5 nsec, and crosslinking occurs on a time scale much faster than most potential molecular rearrangements. Thus, the crosslinking can be thought of as a snapshot of the relative population of states of the tails within the various complexes.
Recent results with a human DNA-processing enzyme highlight the potential biological importance of our current findings. Human DNA ligase I can seal a nick located at the center of a 218-bp nucleosome (35). However, the removal of linker DNA, producing a 150-bp core particle, results in a drastic inhibition of the enzyme. This inhibition can be completely reversed by removal of the core histone tail domains. Interestingly, we find that tail removal or addition of linker DNA to the core does not alter the probability of spontaneous DNA site exposure within the nucleosome (35). The most likely explanation for these results is that constraining the core histone tails to bind (apparently nonphysiological) sites within the nucleosome core blocks ligase from accessing nucleosome DNA. The presence of available linker DNA provides an alternative and probably more physiological location for the tail domains, allowing ligase to seal the nick within the nucleosome (35). The present results clearly show that the tails actually prefer to bind the linker DNA vs. the intracore DNA, thus providing an explanation for the large effect of linker DNA on ligase activity.
A myriad of posttranslational modifications within the core histone tails are likely to regulate the structure and functional state of the chromatin fiber (9, 10). One potential effect of these modifications is the alteration of specific structures or interactions of the tails. For example, recent spectroscopic evidence indicates that lysine acetylation substantially increases the extent of α-helical conformation with the tail domains (12). Interestingly, many modifications are targeted to individual tails or subsets of tails within the nucleosome (7), and recent biophysical evidence suggests that alteration of a small subset of tail interactions is sufficient to modulate fiber conformation or stability (36). Thus, in future work, it will be interesting to determine the exact location of individual tail domain–linker DNA interactions within model or natural chromatin fibers.
Acknowledgments
We are grateful to Dr. T. Richmond for the histone expression vectors and to Dr. K. Nightingale for advice regarding recombinant histone purification. We also appreciate the support of Dr. Jean-Jacques Lawrence throughout the course of this work. This work was supported by grants from the Centre National de la Recherche Scientifique and the Institut National de la Santé et de la Recherche Médicale (to S.D.), National Institutes of Health Grant GM52426 and American Cancer Society Grant RPG-00-080-01-GMC (to J.J.H.), and National Science Foundation (Bulgaria) Grant K902 and North Atlantic Treaty Organization Grant CLG 976174 (to D.A.). J.M.V. was supported by National Institutes of Health Grant T32 DE07202. V.M. acknowledges a Centre National de la Recherche Scientifique Poste Rouge Grant.
Abbreviation
- APB
azidophenacyl bromide
References
- 1.van Holde K E. Chromatin. New York: Springer; 1989. [Google Scholar]
- 2.Wolffe A P. Chromatin Structure and Function. San Diego: Academic; 1998. [Google Scholar]
- 3.Struhl K. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 4.Freedman L P. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 5.Edmonson D G, Smith M M, Roth S Y. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 6.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 7.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 8.De la Barre A-E, Gerson V, Gout S, Creaven M, Allis C D, Dimitrov S. EMBO J. 2000;19:379–391. doi: 10.1093/emboj/19.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen J C, Tse C, Wolffe A P. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 10.Wolffe A P, Hayes J J. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baneres J L, Essalouh L, Jariel-Encontre I, Mesnier D, Garrod S, Parello J. J Mol Biol. 1994;243:48–59. doi: 10.1006/jmbi.1994.1629. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Moore S C, Laszckzack M, Ausió J. J Biol Chem. 2000;275:35013–35020. doi: 10.1074/jbc.M004998200. [DOI] [PubMed] [Google Scholar]
- 13.Nacheva G A, Guschin D Y, Preobrazhenskaya O V, Karpov V L, Ebralidse K K, Mirzabekov A D. Cell. 1989;58:27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- 14.Stefanovsky V, Yu., Dimitrov S I, Russanova V R, Angelov D, Pashev I G. Nucleic Acids Res. 1989;23:10069–10081. doi: 10.1093/nar/17.23.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K-M, Hayes J J. Proc Natl Acad Sci USA. 1997;94:8959–8964. doi: 10.1073/pnas.94.17.8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K-M, Hayes J J. Biochemistry. 1998;37:8622–8628. doi: 10.1021/bi980499y. [DOI] [PubMed] [Google Scholar]
- 17.Mutskov V, Gerber D, Angelov D, Ausió J, Workman J, Dimitrov S. Mol Cell Biol. 1998;18:6293–6304. doi: 10.1128/mcb.18.11.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fletcher T M, Hansen J C. J Biol Chem. 1995;270:25359–25362. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 19.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 20.Usachenko S I, Bavykin S G, Gavin I M, Bradbury E M. Proc Natl Acad Sci USA. 1994;91:6845–6849. doi: 10.1073/pnas.91.15.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimitrov S I, Russanova V, Pashev I. EMBO J. 1987;6:2387–2392. doi: 10.1002/j.1460-2075.1987.tb02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luger K, Rechsteiner T J, Richmond T J. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 23.Lee K-M, Chafin D R, Hayes J J. Methods Enzymol. 1999;304:231–251. doi: 10.1016/s0076-6879(99)04014-8. [DOI] [PubMed] [Google Scholar]
- 24.Moss T, Dimitrov S I, Houde D. Methods. 1997;11:225–234. doi: 10.1006/meth.1996.0409. [DOI] [PubMed] [Google Scholar]
- 25.Angelov D, Stefanovsky V, Dimitrov S I, Russanova V, Pashev I. Nucleic Acids Res. 1988;16:4525–4538. doi: 10.1093/nar/16.10.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thiriet C, Hayes J J. J Biol Chem. 1998;273:21352–21358. doi: 10.1074/jbc.273.33.21352. [DOI] [PubMed] [Google Scholar]
- 27.Cary P D, Crane-Robinson C, Bradbury E M, Dixon G H. Eur J Biochem. 1982;127:137–143. doi: 10.1111/j.1432-1033.1982.tb06847.x. [DOI] [PubMed] [Google Scholar]
- 28.Walker I O. Biochemistry. 1984;23:5622–5628. doi: 10.1021/bi00318a037. [DOI] [PubMed] [Google Scholar]
- 29.Smith R M, Rill R L. J Biol Chem. 1989;264:10574–10581. [PubMed] [Google Scholar]
- 30.Van Holde K E, Zlatanova J. BioEssays. 1999;21:776–780. doi: 10.1002/(SICI)1521-1878(199909)21:9<776::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 31.Weischet W O, Allen J R, Riedel G, van Holde K E. Nucleic Acids Res. 1980;6:1843–1862. doi: 10.1093/nar/6.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Godde J, Widom J. J Mol Biol. 1992;226:1009–1025. doi: 10.1016/0022-2836(92)91049-u. [DOI] [PubMed] [Google Scholar]
- 33.Simpson R T. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 34.Ausió J, Dong F, van Holde K E. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 35.Chafin D R, Vitolo J M, Henricksen L A, Bambara R A, Hayes J J. EMBO J. 2000;19:5492–5501. doi: 10.1093/emboj/19.20.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tse C, Sera T, Wolffe A P, Hansen J C. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]