Abstract
The functional centers of the ribosome in all organisms contain ribosomal RNA (rRNA) modifications, which are introduced by specialized enzymes and come at an energy cost for the cell. Surprisingly, none of the modifications tested so far was essential for growth and hence the functional role of modifications is largely unknown. Here, we show that the methyl groups of nucleosides m2G966 and m5C967 of 16S rRNA in Escherichia coli are important for bacterial fitness. In vitro analysis of all phases of translation suggests that the m2G966/m5C967 modifications are dispensable for elongation, termination and ribosome recycling. Rather, the modifications modulate the early stages of initiation by stabilizing the binding of fMet-tRNAfMet to the 30S pre-initiation complex prior to start-codon recognition. We propose that the m2G966 and m5C967 modifications help shaping the bacterial proteome, most likely by fine-tuning the rates that determine the fate of a given messenger RNA (mRNA) at early checkpoints of mRNA selection.
INTRODUCTION
Ribosomes from all organisms contain modified nucleosides that are mostly located at the functional centers of ribosomal RNA (rRNA), clustering at the binding sites for messenger RNA (mRNA) and transfer RNAs (tRNAs) in the A and P sites, in the peptidyl transferase center, the peptide exit tunnel and at the intersubunit bridges (for recent review, see (1)). In bacteria, specialized enzymes have evolved for each group of modifications, and the reactions proceed at a substantial cost of energy-rich substrates, suggesting that introducing modifications must be important for the cell. In some cases, the presence of the modification is even more conserved than the nucleotide identity: for instance, nucleoside at position 966 in Escherichia coli 16S rRNA is m2G in bacteria, acp3U in archaea and m1acp3Ψ in eukaryotes (2,3), implying an important role of rRNA modification at this position. Surprisingly, none of modified nucleosides whose modification enzymes are known appeared to be essential for cell growth at laboratory conditions (4–9), suggesting that rRNA modifications are not obligatory for the core reactions of the translation cycle. rRNA modifications may regulate ribosome assembly (10,11), stabilize ribosome structure (12,13) or modulate interactions with ligands (14,15). However, an exact function could not be attributed to the majority of modified rRNA residues.
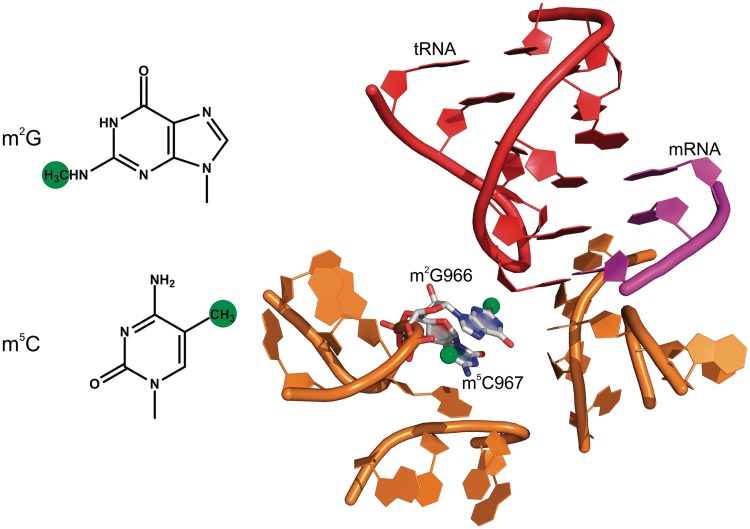
Many modified nucleosides are found in the P site where they seem to monitor the codon–anticodon complex between mRNA and tRNA (16–18). The mRNA is in contact with nucleotides m4Cm1402 and m3U1498 of 16S rRNA (16,18,19) which are engaged in a hydrophobic contact with one another (16); m3U1498 is close to the ribose of the second position of the P-site codon, while m4Cm1402 interacts with the phosphate bridging the second and third positions of the mRNA codon. The mutation m3U1498G and the transversion of the adjacent G1401-C1501 base pair affect the formation of the first peptide bond (20–23). From the anticodon side, the ribose of nucleotide 34 in tRNA forms stacking interactions with m2G966 in helix 31 of 16S rRNA, whereas the other side of m2G966 is stacked with m5C967 (Figure 1) (18,24).
Figure 1.
Modification of m2G966 and m5C967 in the ribosome. Left, chemical structures of the methylated nucleosides. Methyl groups are shown in green. Right, the 30S decoding center surrounding the anticodon of the P site-bound tRNA. The tRNA is shown in red, the initiation codon in magenta and the rRNA in orange (PDB 3I9D) (24).
A growing body of data suggests that modifications of 16S rRNA modulate translation initiation. Recognition of one characteristic feature of fMet-tRNAfMet, i.e. the three consecutive GC pairs in the anticodon stem, is impaired in strains lacking the m62A1518, m62A1519 and m2G966 methylations (25). Moreover, the mutation in the folD gene which leads to a reduction of the intracellular level of S-adenosyl-l-methionine, the methyl donor for the rRNA methylation enzymes, caused a relaxation in initiator tRNA selection in a genetic screen using as initiator tRNA fGln-tRNAfMet with anticodon CUA reading an UAG start codon (25). In the genetic background of diminished overall methylation in the folD122 strain, modifications of m2G1207 and m3U1498 appeared to have a dramatic effect on the stringency of fMet-tRNAfMet selection (25,26). Similarly, the knockout of rsmH, the gene coding for the methyltransferase of N4-C1402, resulted in an increased usage of AUU as initiation codon (16). Thus, modified nucleotides in the P site of the 30S subunit appear to influence the efficiency and fidelity of translation initiation; however, the mechanism by which methylations affect initiation is not understood.
Although genetic screens suggested the role of m2G966 and m5C967 in the selection of the correct initiator tRNA and the start codon during translation initiation (25), earlier studies reported that mutations of nucleotides G966/C967 do not cause major phenotypic changes, except for the deletion of C967, which was lethal (27). Later experiments utilizing a specialized translation system, which allows to specifically monitor the effects of mutations independent of bulk translation, suggested that various G966 substitutions lead to a decrease in translation activity (28). The inhibition caused by the m2G966A mutation could be rescued by the G1338A mutation, which increased the binding of the initiator tRNA, fMet-tRNAfMet, to the P site in the presence of excess of initiation factor (IF) 3 (29). In contrast, exhaustive mutagenesis of helix 31 of 16S rRNA performed in a different specialized ribosome system revealed that mutations of nucleotides at positions 966 and 967 produced hyperactive ribosomes which could be down-regulated to wild-type (wt) expression levels by IF3 overexpression, whereas overexpression of IF2 inhibited the activity of the mutant ribosomes (30). Understanding the exact roles of the m2G966 and m5C967 methylations—in contrast to base substitutions—requires knockout strains that lacked the modification enzymes, the methyltransferases RsmB and RsmD, which are responsible for the methylation of C967 and G966, respectively (5,31,32). RsmB recognizes free 16S rRNA through a 16-nucleotide-long stem-loop structure containing C967 (nt 960–975 of 16S rRNA) and apparently does not require extensive secondary or tertiary structure in the RNA for recognition (31,32). In contrast, RsmD acts late in the assembly process and is able to modify a completely assembled 30S subunit (5,33,34). Both enzymes are very specific for the respective nucleotide position in 16S rRNA and do not act on 23S rRNA; as the activity toward small RNAs was not tested, a dual specificity cannot be ruled out but seems unlikely given the sequence specificity (RsmB) or 30S assembly state specificity (RsmD) of the two methytransferases. Here, we investigated the functional role of methylations at positions m2G966/m5C967 of 16S rRNA using a combination of genetic screens and biophysical techniques. We used strains in which the genes coding for the methyltransferases RsmB and RsmD were knocked out and studied the effects in vivo. Furthermore, we isolated ribosomes that lacked the m2G966/m5C967 modifications and studied in vitro their performance in all phases of translation, initiation, elongation, termination and ribosome recycling.
MATERIALS AND METHODS
Bacterial strains and growth rates
Escherichia coli strains BW25113 and JW3250, carrying a kanR cassette inserted into the rsmB gene, and JW3430, carrying a kanR cassette inserted into the rsmD gene, were from the Keio collection and were kindly provided by Dr H. Mori (National Institute of Genetics, Japan) (35). The double-knockout ΔrsmB/ΔrsmD was constructed from the JW3250 strain by disrupting the rsmD gene with a gene coding for the chloramphenicol acetyl transferase. The gene knockout was performed with the help of the phage λ Red recombinase system (36). The knockouts were verified by polymerase chain reaction using genomic DNA as template. The lack of methylations in the ΔrsmB, ΔrsmD and double knockout strains was verified by reverse transcription as follows. Because m2G966 modification results in the stop of reverse transcription (5), the lack of methylation at position G966 was monitored by the disappearance of the m2G966-specific band. In contrast, m5C967 does not cause a stop of reverse transcription. To detect the m5C967 modification, cytosine deamination by bisulfite treatment was performed followed by reverse transcription. Upon bisulfite treatment, unmodified cytosine is converted to uracil, while methylated m5C967 is not. After elimination of all unmethylated cytosines, the methylated cytosine can be detected as a stop of reverse transcription in the presence of ddGTP in 1:10 ratio to dGTP. Reaction mixtures containing rRNA (50 μg), NaHSO3 (2 mM) and hydroquinone (0.7 mM) were incubated at 55°C for 3 h, rRNA precipitated with 3 vol of ethanol and 1/10 vol of 3 M sodium acetate at pH 5.5. Reverse transcription was performed as described (5). In vitro modification of ribosomes purified from ΔrsmD strain using radioactive S-adenosyl-methionine and recombinant RsmD protein led to stoichiometric labeling of 16S rRNA; modification of wt ribosomes resulted in only 10–20% modification suggesting that 80–90% of wt ribosomes are modified at position 966 (data not shown). Likewise, 16S rRNA isolated from native ribosomes, that is rRNA containing its normal complement of modified bases, was not modified by RsmB, suggesting that most of wt ribosomes are modified at position 967 (32).
Growth rates at conditions where strains were grown alone or in a mixture with the wt were measured as described (7). The proportion of mutant and wt cells in the mixture was determined by colony counting after plating the cells on the agar plates with and without the appropriate antibiotics. Cold sensitivity of cells was tested by growing the cells on LB (Luria-Bertrani medium) agar broth at 18°C for 3 days. Overexpression of azurine was studied after transformation of the ΔrsmB/ΔrsmD and the isogenic BW25113 strains with pASK-IBA4 vector carrying azurine gene under the control of the tetracycline promoter (IBA, Goettingen). Cells were grown in M9 media with 50 µg/ml ampicillin. Protein expression was induced at OD590 = 0.01 by adding 2 or 200 µg/ml anhydrotetracycline (AHTC).
Growth rates of wt and ΔrsmB/ΔrsmD strains expressing plasmid-encoded IF1 or IF3 were measured using the automated station Janus (Perkin Elmer). Optical densities of the cells were measured using the Victor X5 detector (Perkin Elmer). Wt and ΔrsmB/ΔrsmD strains were transformed with plasmids pQE30IF1 and pQE30IF3 carrying an ampicillin resistance marker and coding for IF1 and IF3, respectively (37). Fresh LB media with ampicillin (50 μg/ml) were inoculated with a single colony of the respective strain, containing either of the two plasmids or no plasmid. After incubation at 37°C for 12 h, 5 µl of culture was transferred to 200 μl of fresh LB medium with ampicillin (50 µg/ml) and with isopropyl β-D-1-thiogalactopyranoside at concentrations 0, 0.1, 0.5 or 1 mM and incubated at 18°C with moderate shaking. OD600 was measured every 120 min.
Sucrose density centrifugation
Cells were grown at 37°C to OD600 = 0.2, then split into two portions and grown for 1.5 h at 37°C or 12 h at 18°C to OD600 = 0.4–0.8, and cultures were rapidly cooled on ice. Cells were collected by centrifugation (6000 rpm at 4°C for 10 min; rotor JA-14, Beckman), washed twice with 20 ml of ice-cold buffer A (20 mM Hepes-KOH at pH 7.5, 200 mM ammonium acetate and 6 mM 2-mercaptoethanol) containing 1 or 12 mM magnesium acetate, resuspended in the same buffer and lysed by ultrasonication. Cell lysates were cleared by centrifugation (15 000 rpm at 4°C for 40 min; rotor JA-20, Beckman). Sucrose gradient sedimentation was performed essentially as described (38) with the exception that buffer A with 1 or 12 mM magnesium acetate as indicated in figure legends was used for sucrose gradients. To monitor the potential biogenesis effects, equal amounts (10 A260) of lysates were loaded on 10–30% (wt/vol) gradients of sucrose in buffer A containing 1 mM magnesium acetate and centrifuged in the SW28 rotor (Beckman) at 22 000 rpm for 16.5 h. To monitor subunit association, equal amounts (10 A260) of lysates were loaded on 5–40% (wt/vol) gradients of sucrose in buffer A containing 12 mM magnesium acetate and centrifuged in the SW41 rotor (Beckman) at 35 000 rpm for 2.5 h. Gradient analysis was done using a flow-through spectrophotometer UV-900 (Amersham Biosciences).
Biochemical methods
All biochemical experiments were performed in buffer B (50 mM Hepes-KOH at pH 7.5, 70 mM NH4Cl, 30 mM KCl and 7 mM MgCl2) unless stated otherwise. Ribosomal subunits were prepared from purified 70S ribosomes by sucrose gradient centrifugation in a zonal rotor (Ti 15, Beckman) (39,40). 30S subunits were reactivated in buffer B containing 20 mM MgCl2 for 30 min at 37°C. fMet-tRNAfMet was purified by high-pressure (or high-performance) liquid chromatography (40) and was 95% aminoacylated and formylated. mRNA was 93 nucleotides long and was prepared by in vitro T7 RNA-polymerase transcription (nucleotide sequence: 5′-GGGAAUUCAAAAAUUUAAAAGUUAACAGGUAUACAUACUAUGUUUACGAUUACUACGAUCUUCUUCACUUAAUGCGUCUGCAGGCAUGCAAGC-3′ (start codon underlined)). Single-cysteine mutants of IF1 and IF3 were prepared and labeled with maleimide derivatives of Alexa 488 (Invitrogen, Germany) or Atto565 (Atto-tec, Germany) fluorophores as described (40); for some experiments, the non-fluorescent resonance energy transfer acceptor QSY35 (Invitrogen) was used. The functional activity of fluorescence labeled components ((fMet-tRNAfMet(QSY35), IF3(Alx488) and IF1(Atto565)) was tested as described and was close to that of wt components (41,42).
To form 70S initiation complex (IC), 30S subunits (0.3 μM), 50S subunits (0.3 μM), f[3H]Met-tRNAfMet (0.6 μM), IF1 (0.6 μM), IF2 (0.6 μM), IF3 (0.6 μM), mRNA (2 μM) and GTP (1 mM) were incubated in buffer B for 60 min at 37°C. The efficiency of 70S IC formation was measured by nitrocellulose filtration. In vitro translation of HemK mRNA was performed as described (43) in buffer C (50 mM Hepes-KOH at pH 7.5, 70 mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 0.5 mM spermidine, 8 mM putrescine and 2 mM DTT) using BodipyFL-Met-tRNAfMet for visualization of the synthesized peptides. Translation was performed at 18 and 37°C; reactions were quenched with 2 M NaOH and the length of the translated peptide was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorescence imaging. HemK mRNA was prepared by in vitro transcription using T7 RNA-polymerase.
Kinetic experiments
Fluorescence stopped-flow measurements were performed in a SX-20MV stopped-flow apparatus (Applied Photophysics, Leatherhead, UK). Experiments were performed by rapidly mixing equal volumes (60 μl each) of reactants at 20 or 37°C, as indicated. In a single experiment, 1000 data points were acquired in the logarithmic sampling mode. Excitation wavelength was 470 nm for Alx488. Fluorescence changes and FRET were measured after passing KV500 (for Alx488) or KV590 (for Atto565) cutoff filters (Schott). To measure light scattering, the excitation wavelength was set to 430 nm, and the scattered light was measured at an angle of 90° to the incident beam without a filter. Data were evaluated by two-exponential fitting using Prism (Graphpad Software). Standard deviations of all values were calculated from four to eight time courses.
The IF2-dependent binding of fMet-tRNAfMet(QSY35) to the pre-formed 30S pre-IC (PIC) containing IF1, IF2, IF3(Alx488) and mRNA was performed as described (40) using 30S subunits (0.05 μM), mRNA (0.5 μM) where indicated, IF1, IF2 and IF3(Alx488) (0.1 µM each), GTP (0.25 mM) and different concentrations (0.075–0.5 µM) of fMet-tRNAfMet(QSY35) (final concentrations after mixing). To measure the dissociation of fMet-tRNAfMet from the 30S PIC in the absence of mRNA, the 30S PIC was formed by incubating 30S subunits (0.2 µM), IF1 (0.4 µM), IF2 (0.4 µM), IF3(Alx488) (0.24 µM) and fMet-tRNAfMet(QSY35) (0.4 µM) in buffer B for 30 min at 37°C. Next, the 30S PIC (final concentration after mixing 0.1 µM) containing IF3(Alx488) and fMet-tRNAfMet(QSY35) was mixed with unlabeled fMet-tRNAfMet (2 μM, a 10-fold excess over fMet-tRNAfMet(QSY35)); the FRET decrease due to fMet-tRNAfMet(QSY35) dissociation was monitored. To determine the rate of fMet-tRNAfMet dissociation from the 30S IC containing mRNA, 30S IC (0.1 µM) containing radioactively labeled [3H]fMet-tRNAfMet was mixed with a 10-fold excess unlabeled fMet-tRNAfMet, and the dissociation of [3H]fMet-tRNAfMet was monitored by nitrocellulose filtration.
Dissociation of IF3 from the 30S IC was measured as described (42) using IF1(Atto565) and IF3(Alx488). To form the 30S IC, 30S subunits (0.2 µM), IF1(Atto565) (0.4 µM), IF2 (0.4 µM), IF3(Alx488) (0.24 µM), mRNA (1 µM) and fMet-tRNAfMet were incubated in buffer B for 30 min at 37°C. Then, 30S IC (final concentration after mixing 0.1 µM) containing IF3(Alx488) and IF1(Atto565) was mixed with unlabeled IF3 (1 μM, an 8.3-fold excess over IF3(Alx488)), and the FRET decrease due to IF3(Alx488) dissociation was monitored.
RF2-dependent fMet release was measured at 18°C as described (44). The rate of ribosome recycling was measured by light scattering as described (41,45). 70S ribosomes were formed incubating 30S and 50S subunits in buffer B containing 20 mM MgCl2 for 2 min at 37°C. 70S ribosomes (0.1 µM) were mixed in the stopped-flow apparatus in buffer B at 37°C with ribosome recycling factor (RRF) (2.5 µM), elongation factor (EF)-G (0.8 µM), IF3 (0.3 µM) and IF1 (0.5 µM).
RESULTS
Fitness costs and cold sensitivity
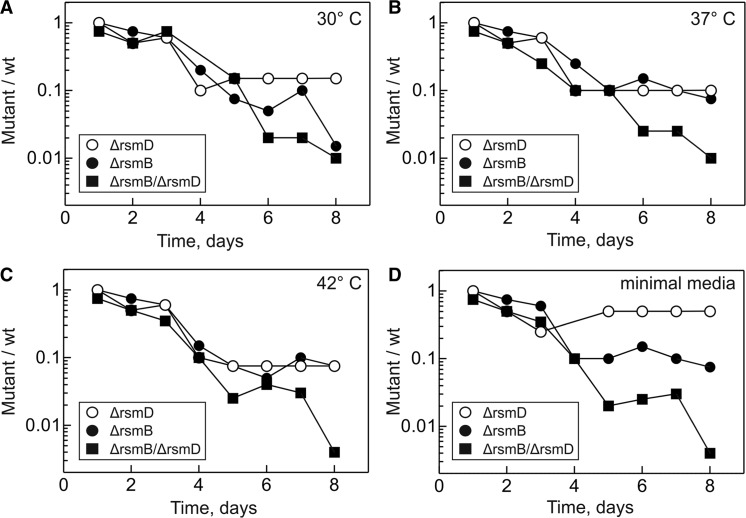
The effects of m2G966/m5C967 methylations were tested using single- and double-knockout strains lacking methyltransferase RsmD (ΔrsmD), RsmB (ΔrsmB) or both (ΔrsmB/ΔrsmD). The absence of the modifications was verified by primer extension (Materials and Methods). The doubling times of the mutant strains were slightly increased (39 ± 2, 40 ± 2 and 38 ± 2 min for ΔrsmD, ΔrsmB and ΔrsmB/ΔrsmD, respectively) compared with the wt strain (33 ± 1 min). As the growth rate measurements have limited sensitivity and allow to compare cells only during exponential growth phase, we also measured the relative fitness of mutant and wt strains when cultivated together at different temperatures and medium conditions (Figure 2). In the absence of methylations, the viability of cells was compromised which resulted in counter-selection against mutant strains after several growth cycles in mixed culture. The fitness costs of the single ΔrsmD and ΔrsmB mutations were similar, whereas the double-knockout ΔrsmB/ΔrsmD had more pronounced defects. In most of the further experiments, we chose to work with the ΔrsmB/ΔrsmD double-knockout strain to analyse the cumulative functional role of m2G966/m5C967 modifications.
Figure 2.
Competitive fitness of mutant strains compared with isogenic wt strain at 30°C (A), 37°C (B), 42°C (C) or in minimal M9 medium at 37°C (D).
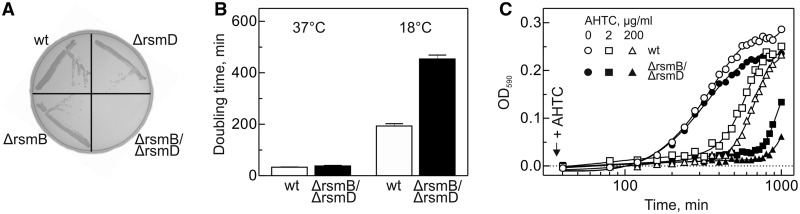
To further scan for the potential effects on bacterial physiology, we studied the response of the mutant strains to cold shock. Inactivation of both RsmB and RsmD genes caused an extreme slow-growth phenotype at 18°C (Figure 3A and B). Because cold sensitivity is often associated with defects of ribosome biogenesis (11,46–49), we screened for defects in ribosome assembly and subunit association in the ΔrsmB/ΔrsmD strain by sucrose gradient sedimentation analysis (Supplementary Figure S1). The relative amounts of ribosomal subunits and 70S ribosomes were not significantly different in the ΔrsmB/ΔrsmD and wt strains both at optimal temperature and at cold shock conditions. No additional peaks were found which could be attributed to assembly intermediates, indicating that the cold sensitivity of the ΔrsmB/ΔrsmD strain is probably not caused by a defect in the biogenesis or association of ribosomal subunits.
Figure 3.
Effects of m2G966/m5C967 on cell growth at conditions of stress. (A) Cold sensitivity (18°C). (B) Effect of cold shock (18°C) on the growth rate. (C) Influence of protein overexpression on cell growth (37°C). Overexpression of azurin was induced by addition of AHTC at time zero. Growth curves of wt (open symbols) and ΔrsmB/ΔrsmD strain (closed symbols) upon induction with different amounts of AHTC: no addition (circles), 2 µg/ml (squares) or 200 µg/ml (triangles).
During the cold shock response, the ribosome selectively translates ∼25 cold shock proteins (for review, see (50)). IFs play an important role in cold adaptation. IF3 selectively promotes the translation of cold shock mRNAs (50), whereas IF1 stimulates translation preferentially at low temperatures without affecting mRNA selectivity (51) and may also play a non-ribosome-associated role as an RNA-chaperone (52,53). Furthermore, an increased ratio of IFs to ribosomes during cold shock may be required to compensate for the higher affinity between the ribosomal subunits at low temperatures to maintain the proper levels of free ribosomal subunits (50). To investigate whether the cold-sensitive phenotype of the ΔrsmB/ΔrsmD strain was related to defects in the interaction between ribosomes and IF1/IF3, we tested whether cell growth of the mutants at 18°C could be rescued by overexpression of IF1 or IF3 (Supplementary Figure S2). Changing either IF1 or IF3 concentration in the ΔrsmB/ΔrsmD strain had no effect on the growth rate, suggesting that the cold-sensitive phenotype does not result from IF1/IF3-related defects. Effects of increasing IF2 concentration were not tested, because overexpression of IF2 is strongly inhibitory to wt cells as well as to mutant strains with m2G966U and m5C967U mutations (30).
Finally, to test effects of the methylations at yet another type of stress conditions, we monitored growth rates during overexpression of a heterologous protein azurin at 37°C. Azurin has been identified in some bacteria, e.g. Pseudomonas aeruginosa, where it functions as a periplasmic redox protein expressed in stress situations. The protein is absent in E. coli and was chosen as a non-toxic soluble protein that is unlikely to specifically interact with E. coli components. A plasmid encoding azurin from P. aeruginosa under the control of Tet promoter was introduced into both wt and ΔrsmB/ΔrsmD strains. Induction of azurin expression inhibited the growth of the ΔrsmB/ΔrsmD strain much stronger than that of the wt cells (Figure 3C) due to an extended lag phase of more than 12 h in the growth of ΔrsmB/ΔrsmD strain, compared with 6 h with wt stain expressing azurin or 2.7 h with un-induced cells of both wt and ΔrsmB/ΔrsmD strains. Taken together, the sensitivity of the ΔrsmB/ΔrsmD strain to cold shock and protein overexpression suggests defects in ribosome function that are particularly clearly manifested at conditions when the translational machinery is tuned toward the production of a limited number of selected proteins.
To identify the stages of protein synthesis which are affected by the lack of the m2G966/m5C967 modifications, we purified ribosomes from the ΔrsmB/ΔrsmD strain (denoted as Δm ribosomes) and investigated the effect of m2G966/m5C967 methylation on the activity of ribosomes at every step of protein synthesis in vitro.
Effect on translation initiation
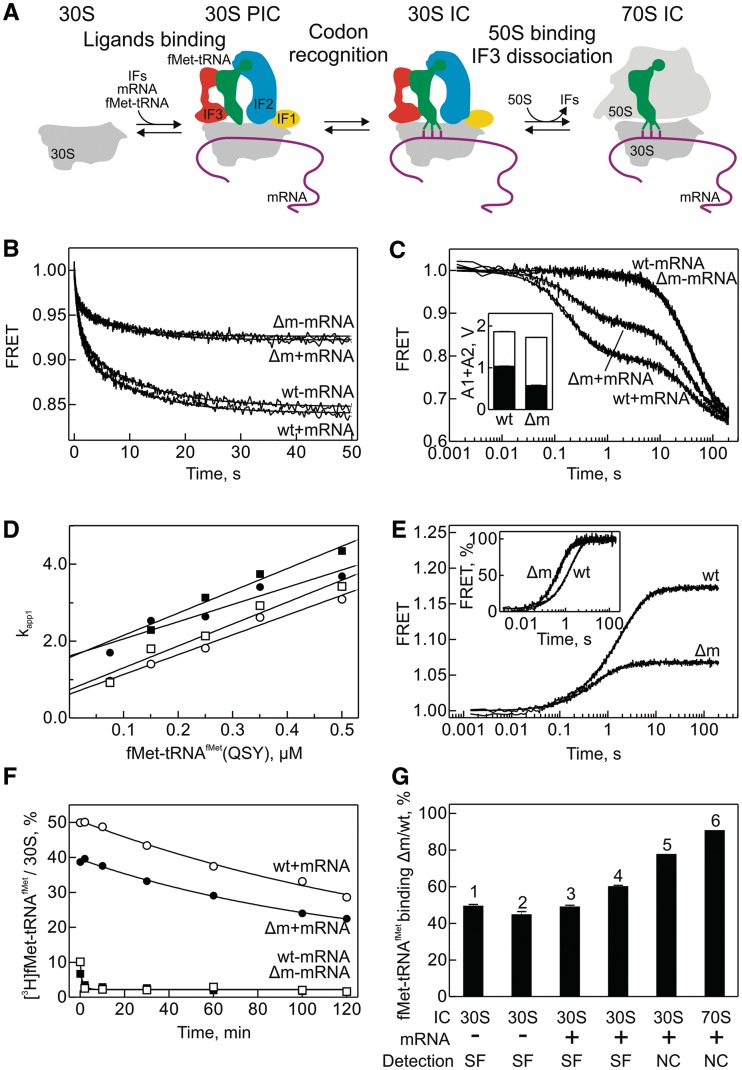
Substitutions of residues G966 and C967 were previously implicated in IF3 binding (30) and the selection of the correct initiator tRNA and initiation codon (25). This prompted us to test the effect of the methylations on initiation. In bacteria, translation initiation comprises three main phases (Figure 4A). Recruitment of the three IFs (IF1, IF2 and IF3), mRNA and fMet-tRNAfMet to the 30S subunit first leads to the assembly of the 30S PIC which upon recognition of the start codon is converted to the mature 30S IC (54–56). Binding of the 50S subunit to the 30S subunit leads to GTP hydrolysis by IF2, dissociation of the IFs and formation of an elongation-competent 70S IC (55,57) (Figure 4A). The formation of initiation intermediates is rapid and completed within a few seconds, which necessitates the use of rapid kinetics approaches for studying the process (41,42,58). First, we studied the binding of fMet-tRNAfMet to 30S complexes by stopped-flow monitoring FRET between IF3 labeled with Alexa 488 (IF3(Alx488)) and fMet-tRNAfMet labeled with QSY35 (fMet-tRNAfMet(QSY35)). 30S complexes were prepared with the Δm or wt 30S subunits by adding the three IFs (of which IF3 is fluorescent) in the presence or absence of mRNA. The apparent rates of fMet-tRNAfMet binding to the 30S were very similar with the Δm and wt ribosomes and independent of the presence or absence of mRNA (Figure 4B); the latter observation is consistent with the notion that the recruitment of fMet-tRNAfMet to the 30S PIC is mediated by IF2 and does not require mRNA (41,59). However, the amplitude of the FRET change was 2-fold lower with Δm compared with wt 30S subunits (Figure 4B). Because the amplitude of the FRET change is sensitive to the extent of binding, this difference could arise from impaired occupancy of Δm ribosomes by either fMet-tRNAfMet(QSY35) or IF3(Alx488). To distinguish between these alternatives, we monitored the dissociation of IF3(Alx488) from the 30S PIC or 30S IC containing IF1 labeled with the FRET acceptor Atto565 (IF1(Atto565)) upon addition of excess unlabeled IF3 (Figure 4C). The overall amplitude of the FRET change was essentially the same for the Δm and wt 30S subunits irrespective of the presence of mRNA (Figure 4C), suggesting that identical amounts of labeled IF3 were initially bound to any type of complexes. In the absence of mRNA, IF3 dissociates from the 30S PIC at the same slow rate from the Δm and wt 30S subunits, which indicates that binding of IF3 to the 30S PIC was not affected by the lack of m2G966/m5C967 methylations. IF3 dissociation from the 30S IC which formed upon recognition of the start codon by fMet-tRNAfMet was more complex: the reaction was biphasic, with the rapid phase corresponding to the dissociation of IF3 from the 30S IC containing all components (4 s−1) and the slow phase (0.025 s−1) reflecting the dissociation from the complexes that lack either mRNA or fMet-tRNAfMet (42). The rates of the fast and slow phases were not affected by the methylations; however, the ratio between rapid and slow phases was changed in favor of the slow phase on Δm ribosomes (Figure 4C, inset). These results suggest that a larger proportion of ribosomes lacking the m2G966/m5C967 methylations remains in the 30S PIC conformation, which—given the observed decrease in the amplitude of fMet-tRNAfMet binding (Figure 4B)—can be attributed to a defect in fMet-tRNAfMet recruitment.
Figure 4.
Effect of m2G966/m5C967 methylations on the initiation of protein synthesis in vitro. (A) Schematic of initiation (57). (B) IF2-dependent recruitment of fMet-tRNAfMet to the 30S PIC containing IF1, IF2 and IF3 in the presence and absence of mRNA. FRET between IF3(Alx488) and fMet-tRNAfMet(QSY35) was monitored. Δm denotes ribosomes from the ΔrsmB/ΔrsmD strain. (C) Time courses of IF3 dissociation from the 30S IC upon addition of excess unlabeled IF3. FRET between IF3(Alx488) and IF1(Atto565) was monitored. The bar graph (inset) represents the distribution of amplitudes of the rapid (black bar) and slow (white bar) phases of IF3(Alx488) dissociation. (D) Concentration dependence of the apparent rate constant of fMet-tRNAfMet binding to the 30S PIC as in (B). Δm (closed symbols) and wt (opened symbols) 30S subunits in the presence (circles) or absence of mRNA (squares). FRET between IF3(Alx488) and fMet-tRNAfMet(QSY35) was monitored. (E) Time courses of fMet-tRNAfMet(QSY35) dissociation from the 30S PIC in the absence of mRNA upon addition of excess of unlabeled fMet-tRNAfMet. Dissociation was monitored by the change of FRET as in (B). For clearer visualization of the rate differences, normalized traces are presented in the inset. (F) Time courses of [3H]fMet-tRNAfMet dissociation from the 30S IC in the presence of mRNA measured by nitrocellulose filtration. Δm (closed symbols) and wt (open symbols) 30S subunits were studied in the presence (circles) or absence of mRNA (squares). (G) Summary of data on fMet-tRNAfMet occupancy in different ICs. The extent of fMet-tRNAfMet binding to Δm relative to wt 30S subunits or 70S ribosomes is shown. 1, IF2-dependent binding of fMet-tRNAfMet to the 30S PIC in the absence of mRNA (from B); 2, fMet-tRNAfMet(QSY35) dissociation from the 30S PIC in the absence of mRNA (from D); 3, the amplitude of IF2-dependent binding of fMet-tRNAfMet(QSY35) to the 30S PIC in the presence of mRNA (from B); 4, proportion of the 30S IC from which IF3 can rapidly dissociate (from C); 5, [3H]fMet-tRNAfMet dissociation from 30S IC in the presence of mRNA (from F); 6, 70S IC formation after incubation of 30S and 50S subunits with mRNA, [3H]fMet-tRNAfMet, IF1, IF2, IF3 and GTP. SF, measured by stopped-flow; NC, measured by nitrocellulose filtration.
To further verify the effect of methylations on fMet-tRNAfMet binding to the 30S subunit, we measured the concentration dependence of the apparent association rate constants and followed the dissociation of fMet-tRNAfMet from the 30S PIC and 30S IC using the IF3(Alx488):fMet-tRNAfMet(QSY35) FRET pair. The association rate constants (obtained from the slope of the linear concentration dependence of the apparent rate constants) were almost identical, about 5 µM−1 s−1 for fMet-tRNAfMet binding to the Δm and wt 30S subunits both in the presence and absence of mRNA (Figure 4D), consistent with values reported previously (41). The dissociation of fMet-tRNAfMet from the 30S PIC in the absence of mRNA was studied by stopped-flow, because the reaction was rapid and the complex was not sufficiently stable to be isolated by nitrocellulose filtration. The dissociation was about two times faster, and the dissociation amplitude was reduced by 50%, for Δm compared with wt ribosomes (Figure 4E). The latter observation was consistent with the amplitude difference of fMet-tRNAfMet binding for Δm and wt ribosomes (Figure 4B). The small, but reproducible and significant 2-fold increase in the dissociation rate of fMet-tRNAfMet from the Δm 30S PIC explains the observed effect on both binding and dissociation amplitude of fMet-tRNAfMet and suggests that Δm ribosomes are less efficient in 30S PIC formation.
The dissociation of fMet-tRNAfMet from the 30S IC after start codon recognition could be followed by nitrocellulose filtration, because it is very slow (Figure 4F) (41). In this case, a similar rate of fMet-tRNAfMet dissociation was observed and the difference in initial [3H]fMet-tRNAfMet binding to Δm and wt 30S subunit was only 20%, suggesting that the effect of the methylations was at least partially alleviated by start-codon selection. An even smaller difference in fMet-tRNAfMet binding to Δm and wt ribosomes was observed after the addition of 50S subunits to the 30S IC, i.e. after formation of the 70S IC, as monitored by nitrocellulose filtration (Figure 4G, bar 6). The relative efficiencies of 30S PIC, 30S IC and 70S IC formation are summarized in Figure 4G. The data suggest that the lack of methylations at positions m2G966/m5C967 affects fMet-tRNAfMet binding preferentially at an early step of initiation, i.e. 30S PIC formation, and do not affect the stability of IF3 binding on the 30S PIC and 30S IC.
Translation elongation, termination and ribosome recycling
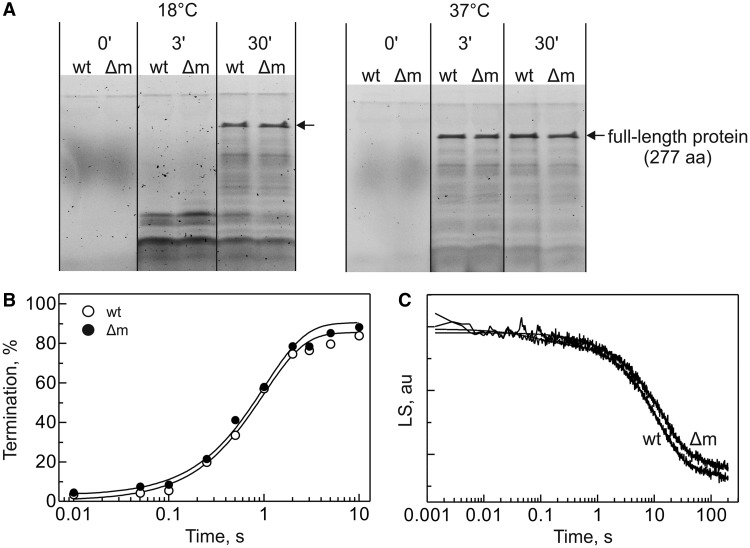
To test whether translation elongation is affected, we studied the synthesis of the 277-amino acid protein HemK in vitro. Because the ΔrsmB/ΔrsmD strain exhibited a cold-sensitive phenotype, the experiments were performed at 18 and 37°C. Translation was initiated by the addition of EF-Tu, EF-G and a mixture of all aminoacyl-tRNAs to the pre-formed 70S IC containing fluorescence-labeled BodipyFL-Met-tRNAfMet, such that only the effects on a single round of translation elongation were monitored. Translation products were resolved by SDS-PAGE and visualized using fluorescence imaging (Figure 5A). Δm ribosomes were as efficient as the wt in peptide elongation at both temperatures.
Figure 5.
Translation elongation, termination and ribosome recycling. (A) SDS-PAGE of the products of HemK mRNA translation at 18°C (left) and 37°C (right). (B) Time courses of RF2-catalyzed peptide release from the wt (opened symbols) or the Δm (closed symbols) ribosomes. (C) Time courses of 70S ribosome dissociation into subunits upon addition of RRF, EF-G, IF3 and IF1 measured by changes in light scattering (LS).
Methylations at positions 1518 and 1519 of 16S rRNA near the interface with the 50S subunit in the vicinity of the IF3 binding site affect the efficiency of RRF-mediated ribosome recycling (26). This prompted us to test the effects of the m2G966/m5C967 modifications on the two remaining steps of protein synthesis, i.e. termination by RF2 and ribosome recycling by RRF and EF-G in the presence of IF3. The efficiency of RF2-catalyzed peptide release in vitro was measured using a model system with ribosomes containing fMet-tRNAfMet bound to an AUG codon in the P site followed by an UAA stop codon in the A site (44). Both rates and final levels of fMet release from wt and Δm 70S ribosomes were identical (Figure 5B). Ribosome recycling was measured by the change in light scattering after mixing 70S ribosomes with RRF, EF-G, IF1 and IF3 in a stopped-flow apparatus (45). The time courses of dissociation of the wt and Δm ribosomes were identical (Figure 5C). Thus, the lack of m2G966/m5C967 modifications has no effect on translation elongation, termination or ribosome recycling.
DISCUSSION
Here, we present a comprehensive analysis of the functional role of the two modified residues m2G966 and m5C967 of 16S rRNA in the P site of the 30S subunit. Consistent with previous notions, suggesting that the lack of these methylations is not lethal, the lack of m2G966 and m5C967 modifications in the strain where both rsmB and rsmD methyltransferase genes were deleted did not cause severe growth defects, but rather resulted in compromised fitness of bacteria compared with the isogenic wt strain. The ΔrsmB/ΔrsmD strain was cold sensitive and sensitive to protein overexpression. The cold sensitivity was not due to the defects in ribosome assembly or impaired association of the ribosomal subunits. Furthermore, the cold sensitivity phenotype was not alleviated by overexpression of IF1 or IF3, which are important factors for cold adaptation (50). Together, the results of the in vivo experiments suggest that the lack of m2G966/m5C967 methylations affects the cells in a complex way, which is particularly strongly manifested upon stress.
The in vitro analysis suggested that the stress sensitivity may be related to an impaired translation initiation, consistent with previous observations that nucleotides at positions 966 and 967 are important for the initiation of protein synthesis (25,30). Our detailed kinetic analysis suggests that the primary role of the two modifications is to stabilize fMet-tRNAfMet binding to the 30S PIC, resulting in a longer residence time of the fMet-tRNAfMet in the complex, whereas binding of IF3 is not affected. The effect on the fMet-tRNAfMet binding stability agrees well with the crystal structures that indicate contacts of m2G966/m5C967 with the anticodon of fMet-tRNAfMet (18). The observed 2-fold change in the rate constant of fMet-tRNAfMet dissociation was significant and highly reproducible. In addition, the observed change in FRET amplitudes may reflect an altered orientation of fMet-tRNAfMet on the ribosomes lacking the methylations, compared with the wt ribosomes. Either effect—the increased dissociation of fMet-tRNAfMet from the complexes or its altered positioning—results in an impaired transition from the 30S PIC to 30S IC, as manifested by a larger portion of 30S complexes that retain tightly bound IF3 despite the presence of mRNA in the complex, i.e. ribosomes escaping the affinity switch of IF3 binding (42).
In addition to their role in the stabilization of fMet-tRNAfMet binding to the 30S PIC, the m2G966/m5C967 modifications may take part in shaping the cellular proteome by altering the intricate kinetic balance between reactions early in initiation. The transition from the 30S PIC to the mature 30S IC constitutes an important checkpoint for both mRNA and fMet-tRNAfMet selection (54,55). The underlying principle of the selection is kinetic partitioning: the fraction of mRNA selected for translation is determined by the relative kinetics of collective steps of mRNA and tRNA docking to the 30S subunit and the start codon recognition versus the rates of mRNA and fMet-tRNAfMet dissociation, which are rather high at this phase (57). Kinetic studies indicated that an mRNA with strong secondary structure in the initiation region, which requires a considerable time to unfold, is more likely to be rejected from the 30S PIC than a non-structured mRNA, even if the intrinsic dissociation rates are the same (54). Upon start codon recognition, the 30S PIC is converted to the 30S IC (60), which is accompanied by a major rearrangement in the complex, resulting in the stabilization of fMet-tRNAfMet and mRNA binding to the 30S subunit and the destabilization of IF3 binding (42). A longer residence time of fMet-tRNAfMet in the 30S PIC may increase the probability that an mRNA which transiently exposes a start codon will be selected for translation due to the recognition of the start codon by fMet-tRNAfMet. For the bulk of mRNAs, a 2-fold decrease in the residence time of fMet-tRNAfMet in the 30S PIC may not result in any significant disadvantage at normal conditions, consistent with the notion that at optimal growth conditions, protein synthesis in the ΔrsmB/ΔrsmD strain is not significantly affected. However, the observed reduced fitness of the mutant in competition with the wt strain suggests that some changes in the proteome composition must occur. For instance, it is possible that changes in the selection stringency at the 30S PIC checkpoint are non-uniform for different mRNAs and particularly affect those mRNAs that are intrinsically slow in the transition from the 30S PIC to the 30S IC (56). At normal growth conditions, such imbalance in the otherwise fine-tuned mRNA selection machinery apparently is tolerable for the cell; however, at conditions of competition with wt cells or during conditions of stress, the bias in mRNA selection due to the lack of the methylations leads to impaired cell growth. Thus, although the primary role of m2G966/m5C967 modifications in 16S rRNA is to stabilize fMet-tRNAfMet binding to the 30S subunit, the modifications may indirectly affect the balance of kinetic parameters that govern mRNA selection, thereby altering the translation efficiency of individual mRNAs and contributing to the response and adaptation to environmental changes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Max Planck Society (to M.V.R., D.E.B. and P.M.); Deutsche Forschungsgemeinschaft (to M.V.R.); Russian Foundation for Basic Research [10-04-01345-a to P.V.S., 11-04-01314-a to O.A.D., 11-04-01018-a to A.A.B., 11-04-12060-ofi to P.V.S.]; Russian Ministry of Science [16.512.11.2108 to O.A.D.]; Federal Agency for Science and Innovations [02.740.11.0706 to O.A.D.]; Moscow University Development Program [PNR 5.13 to O.A.D]. Funding for open access charge: Max Planck Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wolfgang Wintermeyer for critically reading this article; Jörg Mittelstaet for the help with the translation system; Christina Kothe, Olaf Geintzer, Sandra Kappler, Tanja Wiles, Theresia Uhlendorf and Michael Zimmermann for expert technical assistance and H. Mori and T. Ueda for strains and plasmids.
REFERENCES
- 1.Sergiev PV, Golovina AY, Prokhorova IV, Sergeeva OV, Osterman IA, Nesterchuk MV, Burakovskii DE, Bogdanov AA, Dontsova OA. Modifications of ribosomal RNA: from enzymes to function. In: Rodnina MV, Wintermeyer W, Green R, editors. Ribosomes: Structure, Function, and Dynamics. New York: Springer; 2011. pp. 97–110. [Google Scholar]
- 2.Youvan DC, Hearst JE. A sequence from Drosophila melanogaster 18S rRNA bearing the conserved hypermodified nucleoside am psi: analysis by reverse transcription and high-performance liquid chromatography. Nucleic Acids Res. 1981;9:1723–1741. doi: 10.1093/nar/9.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kowalak JA, Bruenger E, Crain PF, McCloskey JA. Identities and phylogenetic comparisons of posttranscriptional modifications in 16 S ribosomal RNA from Haloferax volcanii. J. Biol. Chem. 2000;275:24484–24489. doi: 10.1074/jbc.M002153200. [DOI] [PubMed] [Google Scholar]
- 4.Sergiev P, Golovina A, Prokhorova I, Sergeeva O, Osterman I, Nesterchuk M, Burakovsky D, Bogdanov A, Dontsova O, editors. Modifications of Ribosomal RNA: From Enzymes to Function. New York: Springer-Verlag; 2011. [Google Scholar]
- 5.Lesnyak DV, Osipiuk J, Skarina T, Sergiev PV, Bogdanov AA, Edwards A, Savchenko A, Joachimiak A, Dontsova OA. Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure. J. Biol. Chem. 2007;282:5880–5887. doi: 10.1074/jbc.M608214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sergiev PV, Golovina AY, Sergeeva OV, Osterman IA, Nesterchuk MV, Bogdanov AA, Dontsova OA. How much can we learn about the function of bacterial rRNA modification by mining large-scale experimental datasets? Nucleic Acids Res. 2012;40:5694–5705. doi: 10.1093/nar/gks219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sergiev PV, Lesnyak DV, Bogdanov AA, Dontsova OA. Identification of Escherichia coli m2G methyltransferases: II. The ygjO gene encodes a methyltransferase specific for G1835 of the 23 S rRNA. J. Mol. Biol. 2006;364:26–31. doi: 10.1016/j.jmb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Sergiev PV, Serebryakova MV, Bogdanov AA, Dontsova OA. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23 S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J. Mol. Biol. 2008;375:291–300. doi: 10.1016/j.jmb.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 9.Lesnyak DV, Sergiev PV, Bogdanov AA, Dontsova OA. Identification of Escherichia coli m2G methyltransferases: I. the ycbY gene encodes a methyltransferase specific for G2445 of the 23 S rRNA. J. Mol. Biol. 2006;364:20–25. doi: 10.1016/j.jmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Hager J, Staker BL, Bugl H, Jakob U. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 2002;277:41978–41986. doi: 10.1074/jbc.M205423200. [DOI] [PubMed] [Google Scholar]
- 11.Connolly K, Rife JP, Culver G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 2008;70:1062–1075. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desaulniers JP, Chang YC, Aduri R, Abeysirigunawardena SC, SantaLucia J, Jr, Chow CS. Pseudouridines in rRNA helix 69 play a role in loop stacking interactions. Org. Biomol. Chem. 2008;6:3892–3895. doi: 10.1039/b812731j. [DOI] [PubMed] [Google Scholar]
- 13.Demirci H, Murphy F, 4th, Belardinelli R, Kelley AC, Ramakrishnan V, Gregory ST, Dahlberg AE, Jogl G. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010;16:2319–2324. doi: 10.1261/rna.2357210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ejby M, Sorensen MA, Pedersen S. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc. Natl Acad. Sci. USA. 2007;104:19410–19415. doi: 10.1073/pnas.0706558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Connor M, Gregory ST. Inactivation of the RluD pseudouridine synthase has minimal effects on growth and ribosome function in wild-type Escherichia coli and Salmonella enterica. J. Bacteriol. 2011;193:154–162. doi: 10.1128/JB.00970-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010;38:1341–1352. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brimacombe R, Mitchell P, Osswald M, Stade K, Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993;7:161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- 18.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 19.Basturea GN, Rudd KE, Deutscher MP. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA. 2006;12:426–434. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ringquist S, Cunningham P, Weitzmann C, Formenoy L, Pleij C, Ofengand J, Gold L. Translation initiation complex formation with 30 S ribosomal particles mutated at conserved positions in the 3′-minor domain of 16 S RNA. J. Mol. Biol. 1993;234:14–27. doi: 10.1006/jmbi.1993.1560. [DOI] [PubMed] [Google Scholar]
- 21.Cunningham PR, Nurse K, Bakin A, Weitzmann CJ, Pflumm M, Ofengand J. Interaction between the two conserved single-stranded regions at the decoding site of small subunit ribosomal RNA is essential for ribosome function. Biochemistry. 1992;31:12012–12022. doi: 10.1021/bi00163a008. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham PR, Nurse K, Weitzmann CJ, Negre D, Ofengand J. G1401: a keystone nucleotide at the decoding site of Escherichia coli 30S ribosomes. Biochemistry. 1992;31:7629–7637. doi: 10.1021/bi00148a026. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham PR, Nurse K, Weitzmann CJ, Ofengand J. Functional effects of base changes which further define the decoding center of Escherichia coli 16S ribosomal RNA: mutation of C1404, G1405, C1496, G1497, and U1498. Biochemistry. 1993;32:7172–7180. doi: 10.1021/bi00079a014. [DOI] [PubMed] [Google Scholar]
- 24.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 25.Das G, Thotala DK, Kapoor S, Karunanithi S, Thakur SS, Singh NS, Varshney U. Role of 16S ribosomal RNA methylations in translation initiation in Escherichia coli. EMBO J. 2008;27:840–851. doi: 10.1038/emboj.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seshadri A, Dubey B, Weber MH, Varshney U. Impact of rRNA methylations on ribosome recycling and fidelity of initiation in Escherichia coli. Mol. Microbiol. 2009;72:795–808. doi: 10.1111/j.1365-2958.2009.06685.x. [DOI] [PubMed] [Google Scholar]
- 27.Jemiolo DK, Taurence JS, Giese S. Mutations in 16S rRNA in Escherichia coli at methyl-modified sites: G966, C967, and G1207. Nucleic Acids Res. 1991;19:4259–4265. doi: 10.1093/nar/19.15.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdi NM, Fredrick K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli. RNA. 2005;11:1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Saraiya AA, Lamichhane TN, Chow CS, SantaLucia J, Jr, Cunningham PR. Identification and role of functionally important motifs in the 970 loop of Escherichia coli 16S ribosomal RNA. J. Mol. Biol. 2008;376:645–657. doi: 10.1016/j.jmb.2007.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu XR, Gustafsson C, Ku J, Yu M, Santi DV. Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:4053–4057. doi: 10.1021/bi982364y. [DOI] [PubMed] [Google Scholar]
- 32.Tscherne JS, Nurse K, Popienick P, Michel H, Sochacki M, Ofengand J. Purification, cloning, and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:1884–1892. doi: 10.1021/bi981880l. [DOI] [PubMed] [Google Scholar]
- 33.Sergeeva OV, Prokhorova IV, Ordabaev Y, Tsvetkov PO, Sergiev PV, Bogdanov AA, Makarov AA, Dontsova OA. Properties of small rRNA methyltransferase RsmD: mutational and kinetic study. RNA. 2012;18:1178–1185. doi: 10.1261/rna.032763.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weitzmann C, Tumminia SJ, Boublik M, Ofengand J. A paradigm for local conformational control of function in the ribosome: binding of ribosomal protein S19 to Escherichia coli 16S rRNA in the presence of S7 is required for methylation of m2G966 and blocks methylation of m5C967 by their respective methyltransferases. Nucleic Acids Res. 1991;19:7089–7095. doi: 10.1093/nar/19.25.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 38.Culver GM, Noller HF. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 2000;318:446–460. doi: 10.1016/s0076-6879(00)18069-3. [DOI] [PubMed] [Google Scholar]
- 39.Rodnina MV, Wintermeyer W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl Acad. Sci. USA. 1995;92:1945–1949. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milon P, Konevega AL, Peske F, Fabbretti A, Gualerzi CO, Rodnina MV. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007;430:1–30. doi: 10.1016/S0076-6879(07)30001-3. [DOI] [PubMed] [Google Scholar]
- 41.Milon P, Carotti M, Konevega AL, Wintermeyer W, Rodnina MV, Gualerzi CO. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 2010;11:312–316. doi: 10.1038/embor.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol. Cell. 2008;30:712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burakovsky DE, Sergiev PV, Steblyanko MA, Kubarenko AV, Konevega AL, Bogdanov AA, Rodnina MV, Dontsova OA. Mutations at the accommodation gate of the ribosome impair RF2-dependent translation termination. RNA. 2010;16:1848–1853. doi: 10.1261/rna.2185710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15:772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dammel CS, Noller HF. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- 47.Awano N, Xu C, Ke H, Inoue K, Inouye M, Phadtare S. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J. Bacteriol. 2007;189:5808–5815. doi: 10.1128/JB.00655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue K, Chen J, Tan Q, Inouye M. Era and RbfA have overlapping function in ribosome biogenesis in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2006;11:41–52. doi: 10.1159/000092818. [DOI] [PubMed] [Google Scholar]
- 49.Roy-Chaudhuri B, Kirthi N, Kelley T, Culver GM. Suppression of a cold-sensitive mutation in ribosomal protein S5 reveals a role for RimJ in ribosome biogenesis. Mol. Microbiol. 2008;68:1547–1559. doi: 10.1111/j.1365-2958.2008.06252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gualerzi CO, Giuliodori AM, Pon CL. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 2003;331:527–539. doi: 10.1016/s0022-2836(03)00732-0. [DOI] [PubMed] [Google Scholar]
- 51.Giuliodori AM, Brandi A, Gualerzi CO, Pon CL. Preferential translation of cold-shock mRNAs during cold adaptation. RNA. 2004;10:265–276. doi: 10.1261/rna.5164904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber MH, Beckering CL, Marahiel MA. Complementation of cold shock proteins by translation initiation factor IF1 in vivo. J. Bacteriol. 2001;183:7381–7386. doi: 10.1128/JB.183.24.7381-7386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phadtare S, Severinov K. Comparative analysis of changes in gene expression due to RNA melting activities of translation initiation factor IF1 and a cold shock protein of the CspA family. Genes Cells. 2009;14:1227–1239. doi: 10.1111/j.1365-2443.2009.01346.x. [DOI] [PubMed] [Google Scholar]
- 54.Studer SM, Joseph S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell. 2006;22:105–115. doi: 10.1016/j.molcel.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 55.Gualerzi CO, Brandi L, Caserta E, Garofalo C, Lammi M, La Teana A, Petrelli D, Spurio R, Tomsic J, Pon CL. Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harb. Symp. Quant. Biol. 2001;66:363–376. doi: 10.1101/sqb.2001.66.363. [DOI] [PubMed] [Google Scholar]
- 56.Rodnina MV. Quality control of mRNA decoding on the bacterial ribosome. Adv. Protein Chem. Struct. Biol. 2012;86:95–128. doi: 10.1016/B978-0-12-386497-0.00003-7. [DOI] [PubMed] [Google Scholar]
- 57.Milon P, Rodnina MV. Kinetic control of translation initiation in bacteria. Crit. Rev. Biochem. Mol. Biol. 2012;47:334–348. doi: 10.3109/10409238.2012.678284. [DOI] [PubMed] [Google Scholar]
- 58.Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat. Struct. Mol. Biol. 2012;19:609–615. doi: 10.1038/nsmb.2285. [DOI] [PubMed] [Google Scholar]
- 59.Gualerzi CO, Brandi L, Caserta E, Garofalo C, Lammi M, La Teana A, Petrelli D, Spurio R, Tomsic J, Pon CL. Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harb. Symp. Quant. Biol. 2001;66:363–376. doi: 10.1101/sqb.2001.66.363. [DOI] [PubMed] [Google Scholar]
- 60.Calogero RA, Pon CL, Canonaco MA, Gualerzi CO. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USA. 1988;85:6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.