Abstract
IκB kinase α (IKKα) is part of the cytoplasmic IKK complex regulating nuclear factor-{kappa}B (NF-κB) release and translocation into the nucleus in response to pro-inflammatory signals. IKKα can also be recruited directly to the promoter of NF-κB-dependent genes by NF-κB where it phosphorylates histone H3 at serine 10, triggering recruitment of the bromodomain-containing protein 4 and the positive transcription elongation factor b. Herein, we report that IKKα travels with the elongating form of ribonucleic acid polymerase II together with heterochromatin protein 1 gamma (HP1γ) at NF-κB-dependent genes in activated macrophages. IKKα binds to and phosphorylates HP1γ, which in turn controls IKKα binding to chromatin and phosphorylation of the histone variant H3.3 at serine 31 within transcribing regions. Downstream of transcription end sites, IKKα accumulates with its inhibitor the CUE-domain containing protein 2, suggesting a link between IKKα inactivation and transcription termination.
INTRODUCTION
The nuclear factor-{kappa}B (NF-κB) family of transcription factors is involved in a variety of physiological and pathological processes such as development, immunity, tissue homeostasis and inflammation with abundant epidemiological data showing a strong correlation between inflammation and cancer incidence (1). In unstimulated cells, NF-κB is sequestered in the cytoplasm bound to a family of proteins known as IκB. On stimulation, IκB is phosphorylated by the IκB kinase (IKK) complex, allowing NF-κB to translocate into the nucleus and to stimulate transcription of its target genes. The IKK complex includes two catalytic subunits, IKKα, IKKβ and a regulatory subunit IKKγ/Nemo. In the nucleus, NF-κB interacts with several chromatin modifiers including the histone H3K4 methyltransferase Set7/9 (2), the lysine acetyltransferase CBP/p300 (3) and IKKα for which specific chromatin modifier activity has been described (4). Some NF-κB-dependent genes, which function as rapidly induced primary response genes, produce a low level of non-processed primary transcripts in macrophages even before activation (5). On induction, these NF-κB-dependent genes recruit positive transcription elongation factor b (p-TEFb), which controls both the release of the promoter-proximal paused ribonucleic acid (RNA) polymerase II (RNAPII) and productive transcription elongation, to generate high levels of functional messenger RNA (mRNA) (5,6). This recruitment of the p-TEFb complex by bromodomain-containing protein 4 (Brd4) is triggered by a cascade of events initiated by histone H3 Serine 10 (H3S10) phosphorylation and leading to histone H4 acetylation (7). H3S10 phosphorylation is directed by IKKα at NF-κB-dependent genes (8,9). p-TEFb activates elongation and RNA processing by phosphorylating the C-terminal domain (CTD) of RNAPII at Serine 2 (RNAPII S2p). This CTD is the largest subunit of RNAPII (10); it comprises up to 52 tandemly repeated heptapeptides in mammals and serves as a scaffold for a large range of nuclear factors (11). Interestingly, the state of transcribing chromatin is intimately linked to the phosphorylation state of the RNAPII CTD (12,13). For example, the Set2 histone methyltransferase is recruited during transcription through CTD S5 and S2 phosphorylation and Set2 methylation of histone H3 lysine 36 (H3K36me3) increases towards the 3′-end of genes where the S2/S5 double phosphorylation mark is the highest (11,14). Interactions between the RNAPII CTD and nuclear factors are not restricted to chromatin modifier enzymes because heterochromatin protein 1 gamma (HP1γ) also interacts with the RNAPII CTD domain when it is phosphorylated on S2 or S5 and recruits the facilitates chromatin transcription (FACT) complex to RNAPII (15,16). HP1γ is part of the HP1 family of proteins together with HP1α and HP1β in vertebrates. HP1 proteins are commonly associated with heterochromatin formation, as they are recruited to methylated lysine 9 of histone H3. They can in turn recruit methyltransferase to propagate silencing marks along chromatin (17,18). In contrast to HP1α and HP1β, HP1γ is also located in euchromatin (19) where it can be found phosphorylated at serine 93 (20). This post-translational modification abrogates the transcriptionally repressive function of HP1γ (21).
Because IKKα is recruited to transcribing regions (22), we have studied the dynamics of IKKα recruitment to NF-κB-dependent genes in macrophages after treatment with lipopolysaccharide (LPS). We show that IKKα phosphorylates not only histone H3S10 but also H3.3S31 and HP1γS93. IKKα binds to RNAPII S2p, and its association with chromatin, mainly at the 3′-end of genes seen in both cell lines and primary macrophages, is transcription elongation dependent. Interestingly, we show that IKKα does not bind to chromatin in absence of HP1γ, the latter being needed for phosphorylation of histone H3.3S31. In vitro, HP1γ directly interacts with IKKα, accelerates IKKα-mediated phosphorylation of H3.3S31 and inversely, decreases the rate of H3S10 phosphorylation. In addition, the presence of adenosine triphosphate (ATP) favours the interaction between IKKα and HP1γ over the interaction between IKKα and RNAPII S2p, suggesting a model in which both HP1γ and IKKα are recruited to the polymerase and then loaded onto chromatin after IKKα−dependent phosphorylation of HP1γ. Finally, we show that IKKα co-localizes at the 3′-end of tumour necrosis factor (TNF) with its repressor CUE-domain containing protein 2 (CUEDC2) (23). These data indicate that IKKα chromatin modifier activity is playing a role in transcription elongation beyond the p-TEFb recruitment step and is controlled by HP1γ.
MATERIALS AND METHODS
Cell culture
RAW264.7 cells were grown in Dulbecco’s modified Eagle’s medium with 10% foetal calf serum and penicillin–streptomycin. Mouse primary macrophages were obtained from bone marrow by culturing in Iscove‘s modified dulbecco‘s medium containing 10% foetal calf serum and penicillin–streptomycin and 10% L cell conditioned medium containing Macrophage colony-stimulating factor (24) for 7 days. Where indicated, cells were treated with 1 µg/ml LPS (Sigma) and 200 µM 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (Alexis). HP1γ knockdown was achieved by transient transfection of pMSCV-mir30-based shRNAs, designed against mouse HP1γ and firefly luciferase (25), using Fugene HD (Promega). After transfection, RAW264.7 cells were incubated for 24 h and treated with LPS for an additional hour.
Reverse transcription polymerase chain reaction
Total RNA was prepared and reverse transcription polymerase chain reaction (PCR) performed as described previously (26). Relative expression was calculated as a ratio of specific transcript to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (for primer sequences see Supplementary Table S1).
Chromatin immunoprecipitation assays and real-time PCR analysis
Chromatin immunoprecipitation (ChIP) was performed as described previously (22) using dynabeads protein G (Invitrogen) with 2.4 µg per 10 µl beads of anti-p65 (Santa Cruz sc-372X), anti-histone H3K9acS10p (Abcam ab12181), anti-H3S10p (Millipore, 06-570), anti-H3 (Abcam ab1791), anti-H3.3S31p (Abcam ab92628), anti-HP1γ (Millipore 05-690), anti-HP1γS93p (Abcam ab45270), anti-IKKα (Santa Cruz sc-7606X), anti-cdk9 (Santa Cruz sc-8338X), anti-Brd4 (Abcam ab46199), anti-RNAPII S2p (Abcam ab5095), anti-RNAPII S5p (Abcam ab5131), anti-RNAPII CTD (Abcam, ab817), anti-RNAPII (Santa-Cruz sc-900X) and anti-CUEDC2 (Sigma PRS4839). Micrococcal Nuclease (MNase) experiments have been conducted as described previously (27). Depletion experiments were performed after a first immunoprecipitation (IP) with anti-RNAPII S2p with chromatin prepared from 4 × 105 cells/IP, after which the unbound fraction (supernatant collected after 2 h incubation chromatin/beads/antibody) was divided into two fractions to be incubated with anti-HP1γ or anti-IKKα. Re-ChIP was performed after a first immunoprecipitation with anti-RNAPII S2p with chromatin prepared from 107 cells/IP. After 2-h incubation, the beads were washed and then incubated with 25 µl of 10 mM dithiothreitol (DTT) for 30 min at RT. This eluted fraction was separated from the beads diluted in 325 µl of IP buffer, and 100 µl was incubated with anti-p65, anti-HP1γ or anti-IKKα (22). Quantitative Real-time PCR (qPCR) was performed as described previously (28) with data normalized versus input and then versus the average of control regions representing enrichment base line (for primer sequences see Supplementary Table S1).
Protein extraction
For whole cell lysate preparation, RAW264.7 cells were washed in phosphate-buffered saline (PBS) and lysed on ice for 20 min in 300 mM NaCl hypertonic cell lysis buffer (HCLB-300) [20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH7.5, 10 mM KCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 0.1% Triton X-100, 10% glycerol, 300 mM NaCl] with phosphatase and protease inhibitors [1 mM b-glycerophosphate, 10 mM NaF, 0.5 mM Na3VO4, 1xproteinase inhibitors cocktail (Roche) and 0.5 mM phenylmethanesulfonylfluoride]. Lysates were centrifuged and supernatant stored on ice for endogenous co-immunoprecipitation (eCoIP). For cytosolic fractioning, cells were washed in PBS and lysed on ice in HCLB-0 for 10 min, spun at 200 g and the supernatant stored on ice for CoIP. Nuclear pellets were then washed in HCLB-0 and resuspended in HCLB-300 for 10 min, spun and supernatant retained on ice for eCoIP. For DRB expression assays, proteins were collected from cell lysed in radio immunoprecipitation assay (RIPA) buffer [10 mM Tris–HCl pH 7.5, 1 mM EDTA, 0.5 mM ethylene glycol tetraacetic acid, 140 mM NaCl, 0.1% sodium dodecyl sulphate (SDS), 1% Triton X100, 0.1% sodium deoxycholate] on ice for 20 min.
Endogenous co-immunoprecipitation
HCLB extracted cell lysates or nuclear extracts were all equilibrated to 150 mM NaCl. Lysates were pre-cleared with 2.4 µg of either mouse or rabbit IgG isotype control (Santa Cruz sc2025 and sc2027) immobilized on 15 µl protein G dynabeads (Invitrogen) for 1 h at 4°C. Supernatants were then incubated with 2.4 µg primary antibody immobilized on 15 µl protein G dynabeads overnight at 4°C. Antibodies used are described earlier (see ‘Chromatin Immunoprecipitation assays and real-time PCR analysis’ section) except for anti-IKKα (abcam ab32041), mouse IgG2b (R+D systems MAB004). Immobilized complexes were washed with HCLB buffer (0, 150 and 300 mM NaCl successively) and eluted in elution buffer (100 mM NaHCO3, 1% SDS) at room temperature for 0.5 h. Samples were run on SDS-polyacrylamide gel electrophoresis (PAGE) and detected with primary antibodies, see Supplementary Table S2.
In vitro CoIP
Recombinant proteins IKKα-GST (glutathione S-transferase) (200 ng, Millipore 14-461), HP1γ-GST (Abnova H00011335-P01) and HP1α-His (Abcam ab92015) were incubated in kinase buffer (KB) (1 mg/ml BSA, 25 mM HEPES pH 7.9, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 10 mM KCl and 10% glycerol) for 1 h in the presence or absence of 200 µM ATP. Immunoprecipitations were then carried out as described for eCoIP.
Kinase assays
Recombinant proteins (100 ng) HP1γ-GST or HP1α-HIS and 1 µg of H3.3 (NEB M2507S), were added to KB. ATP was added where indicated to 200 µM final concentration. Reactions were allowed to warm to 16°C and started by the addition of 50 ng IKKα-GST. Aliquots were removed at indicated time points, mixed with SDS-LB and stored at −20°C for western blot analysis against HP1γS93p, H3S10p and H3.3S31p as described in Supplementary Table S2. Antibodies were first assayed to confirm they did not recognize unphosphorylated forms of the recombinant proteins.
Pull-down assays
Synthetic peptide corresponding to CTD-S2p (Abcam ab1771) was biotinylated with Lightning Link (InnovaBiosciences 7040030); 250 ng of biotinylated peptide per reaction was immobilized on 10 µl of M280 streptavidin dynabeads (Invitrogen 112-09D) as per manufacturer’s instructions. Recombinant proteins (200 ng; IKKα-GST and HP1γ-GST or HP1α-His) were suspended in KB with biotinylated peptides in the presence or absence of 200 µM ATP final concentration at room temperature for 30 min. The immobilized complexes were washed in PBS-T (0.05% Tween), twice in KB and eluted in RIPA buffer. Samples were stored at −20°C for analysis by western blot.
Peptide binding assay
Peptide (600 pmol) was incubated with 2 pmol of IKK-GST recombinant protein in peptide-binding buffer (PBB) (25 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM DTT, 5% glycerol, 0.03% NP-40) for 30 min before 5 µl of magnetic glutathione beads (Pierce 88821) were added for a further hour. Complexes were removed by magnet, washed three times in PBB and eluted with 10 mM glutathione at room temperature. Supernatant was spotted onto nitrocellulose and probed with anti-S5p or anti-S2p (Supplementary Table S2). Densitometry of blots was conducted using Quantity One on a GelDoc XR. Measurements were taken from an area of set size.
Western blots
Western blots were carried out under denaturing conditions with SDS-PAGE, and proteins were transferred to polyvinylidene fluoride membranes (for blocking conditions and primary antibody concentrations see Supplementary Table S2). HRP-conjugated secondary antibodies were used at 1:20 000 (eBioscience).
RESULTS
IKKα accumulates at the 3′-end of TNF after LPS treatment in macrophages
LPS is a potent activator of NF-κB signalling pathways in macrophages. On LPS stimulation, NF-κB binds to specific deoxyribonucleic acid (DNA) sequences to stimulate high-level production of processed transcripts of its target genes by recruitment of the Brd4/p-TEFb complex (5). PIM1-dependent histone H3S10 phosphorylation is necessary for Brd4/p-TEFb recruitment and activation of transcription elongation of the FOSL1 gene (7). As IKKα targets H3S10 at NF-κB-dependent genes (8,9), we expected IKKα chromatin modifier activity to be associated with Brd4/p-TEFb recruitment. However, we have previously shown that IKKα can also be recruited to transcribing regions (22), suggesting an additional role for this kinase in transcription elongation. To tackle questions regarding the impact of IKKα on transcription regulation, we focused on the murine TNF gene in the RAW264.7 macrophage cell line as an experimental model. Global transcriptional activation following pro-inflammatory stimuli is heterogeneous, each gene responding with specific intensities and kinetics, making it difficult to perform global mechanistic studies with multiple genes at the same time. However, the general mechanisms of transcription regulation are conserved between genes and across species; therefore, observations from our TNF model can serve as a paradigm for other genes. TNF is a small gene that is rapidly induced and highly transcribed in response to LPS treatment in macrophages (Figure 1A). Using this system, we performed ChIP experiments to confirm that NF-κB was binding to the TNF promoter after LPS treatment in RAW264.7 cells (Supplementary Figure S1A) and in primary macrophages (Supplementary Figure S2A). We observed recruitment of Brd4 mainly at the promoter of TNF, whereas Cdk9, the catalytic subunit of p-TEFb, was present throughout the coding region of this gene (Supplementary Figure S1B and S1C). These data are in agreement with the described model in which Brd4 recognizes and binds to acetylated histone H3 and H4 (5,7,29) and then recruits p-TEFb, which in turn phosphorylates RNAPII S2 to drive productive transcription elongation and mRNA processing (5,7).
Figure 1.
The response of the TNF locus to LPS treatment. (A) Time course of TNF mRNA levels in RAW264.7 cells in response to LPS treatment. Results are expressed relative to GAPDH expression. Error bars represent standard deviation (SD) from three independent experiments. (B) Positions and names of the amplicons designed within the TNF locus for the ChIP experiments. ChIP performed with RAW264.7 cells treated with LPS for the indicated time points in minutes: (C) anti-H3K9acS10p, (D) anti-IKKα, (E) anti-HP1γ, (F) anti-H3.3S31p and (G) anti-HP1γS93p. Horizontal axis indicates primers used for the real-time PCR (distance in kb from the transcription start site or CCCTC-binding factor CTCF-binding site at the vicinity of the indicated gene, see B). Data are normalized versus input and then versus an average of background control regions. Error bars represent SD from three independent qPCR replicates. (C–G) Data are representative of at least three independent experiments.
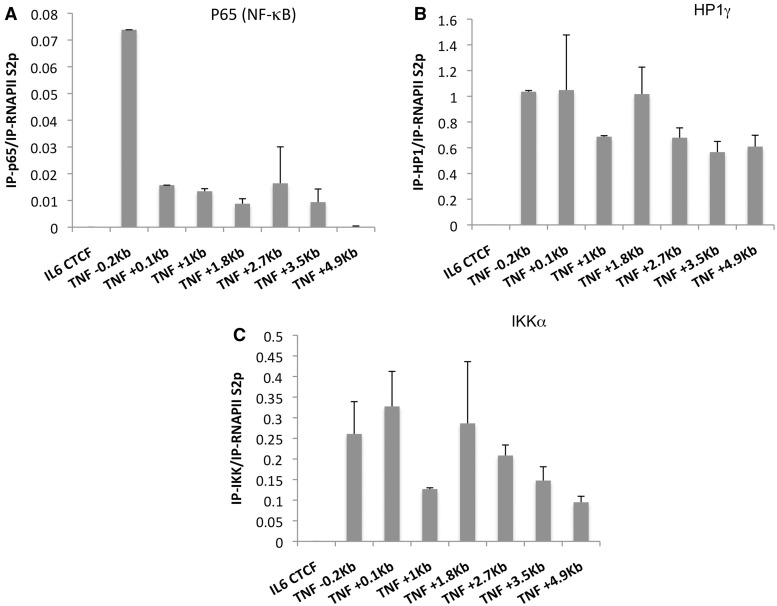
Next, we performed additional ChIP experiments to look for H3S10p enrichment and IKKα recruitment at the TNF locus after LPS treatment. Because many genes undergo global changes in chromatin structure on activation, we first analysed the nucleosome landscape of TNF using anti-histone H3 antibody and observed a rapid decrease in nucleosome content extending beyond TNF (Supplementary Figure S1D), as described previously for heat-shock genes (30). We also detected a rapid and transient increase in the level of histone H3 modified by both lysine 9 acetylation and H3 serine 10 phophorylation, with a peak at 15 min localized 100 bp downstream of the transcription start site at the +1 nucleosome (Figure 1C) in parallel with recruitment of Brd4 and p-TEFb in this region. The same profile was detected by ChIP with antibodies against either the double mark H3K9acS10p or the single mark H3S10p (Supplementary Figure S1E). However, H3S10 phosphorylation appeared to be an early event in TNF transcriptional activation and was not maintained throughout the time course of TNF transcription, in contrast to both Brd4 and Cdk9 recruitments. These data support the view that H3S10p initiates Brd4 recruitment to the promoter but that it is not necessary to maintain this modification after the onset of transcription. When we focused our analysis on the 5′-end of the gene, we also detected the highest IKKα enrichment at the +1 nucleosome correlated with total RNAPII (Supplementary Figure S3A and S3B) suggesting recruitment of IKKα to the region of RNA polymerase pausing. However, a wider view of IKKα distribution along TNF revealed a much more substantial accumulation of IKKα downstream of the transcription end site (TES) of the gene in both the RAW264.7 cell line and primary macrophages (Figure 1D and Supplementary Figure S2B). This 3′-end accumulation of IKKα was not restricted to TNF, as other LPS inducible and NF-κB-dependent genes presented a similar distribution of this protein around coding regions (Ccl3 and Il1β, Supplementary Figures S3C, S3D, S4A and S4E). A DNase I hypersensitive site (DHS) has previously been detected downstream of the human TNFα transcription end site (31) (Figure 1B). This DHS is occupied by NF-κB and interferon regulatory factor 5 in macrophages and dendritic cells after LPS treatment (32,33). However, the possibility that the recruitment of IKKα at the 3′-end of TNF occurs solely by transcription factors at this DHS is very unlikely, because IKKα enrichment (25 times higher than observed at the promoter) covers a large region extending from the TES to at least 600 bp downstream from this DHS. In addition, the +2.7 kb amplicon, in which the mouse conserved sequences for the two described NF-κB-binding sites are included, did not reveal significant binding of NF-κB in this region after LPS treatment (Supplementary Figures S1A and S2A). Besides the role of IKKα as the kinase phosphorylating histone H3S10 at NF-kB-dependent genes, our data argue for additional roles of this enzyme in transcription elongation downstream p-TEFb recruitment.
LPS treatment induces accumulation of HP1γ and H3.3S31p in the coding region of TNF
To unravel the new roles of IKKα in transcription, we first looked for potential substrates catalysed by this enzyme. HP1γ and the histone variant H3.3 are specifically enriched at transcriptionally active genes (15,34) with H3.3 enrichment observed at the TES of transcribed sequences (35). Because HP1γS93p is a marker of transcription elongation (20) and H3.3S31 represents the only amino acid difference between the N-terminal tails of the histone variants H3.1 and H3.3, both serines represent attractive potential chromatin targets for IKKα. Indeed, our ChIP experiments revealed that both HP1γ and H3.3S31p were enriched within the coding region of TNF and Ccl3 genes after LPS treatment (Figure 1E and F, Supplementary Figures S2C, S4C and S4D), and a peak of HP1γS93p was detected at the 3′-end of TNF in a region where IKKα enrichment was the highest (Figure 1G). In addition, In vitro kinase assays using recombinant IKKα incubated with HP1γ and histone H3.3 showed that IKKα phosphorylates not only H3.3S10 but also H3.3S31 and HP1γS93 (Figure 2A and B). In these assays, the presence of HP1γ altered the IKKα kinase specificity, whereas HP1α did not. The presence of HP1γ accelerated the rate of H3.3S31 phosphorylation, whereas the rate and level of H3S10 phosphorylation was decreased (Figure 2B). In addition, eCoIP experiments done with anti-HP1γ antibody and nuclear extracts from LPS-treated cells (untreated cells showing no detectable level of IKKα in the nucleus) or with recombinant proteins and anti-HP1γ or anti-IKKα antibodies showed interaction between IKKα and HP1γ but not between IKKα and HP1α (Figure 2C–E). The reciprocal experiment with anti-IKKα antibody did not co-precipitate HP1γ when incubated with nuclear extracts. We hypothesized that the epitope of this monoclonal antibody may be masked in vivo. In addition, using recombinant proteins, we observed that the interaction between IKKα and HP1γ was direct and stronger in the presence of ATP (Figure 2D). This ATP-dependent direct interaction between IKKα and HP1γ would justify the observed co-localization of IKKα and HP1γS93p on chromatin template. In summary, we identified two new targets for IKKα during transcription and lifted the veil on what appears to be a complex interdependency between HP1γ and IKKα.
Figure 2.
IKKα interacts with HP1γ and histone H3.3. (A) IKKα kinase assay with recombinant HP1γ. Western blot has been performed with anti-IKKα, anti-HP1γS93p and anti-HP1γ. (B) IKKα kinase assay with recombinant IKKα and histone H3.3 in presence or absence of recombinant HP1γ (top) or HP1α (bottom). Western blot has been performed with anti-IKKα, anti-H3S10p, anti-H3.3S31p, anti-HP1γ and anti-HP1α antibodies. (C) eCoIP experiments with nuclear extract from RAW264.7 cells treated with LPS for 1 h. Antibodies used for immunoprecipitation are named at the top of the figure and antibodies used for western blot named on the left. Input was 5% of material used for CoIP. (D) In vitro CoIP experiment performed with recombinant HP1γ–GST or IKKα–GST incubated with or without ATP and immunoprecipitated with anti-HP1γ or anti-IKKα antibodies. Western blot performed with anti-HP1γ or anti-IKKα antibodies. (E) In vitro CoIP experiment performed with recombinant HP1α-HIS incubated with recombinant HP1γ–GST or IKKα–GST and immunoprecipitated with anti-HP1α or anti-IKKα antibodies. Western blot performed with anti-HP1α antibody.
Accumulation of both IKKα and HP1γ to TNF is transcription elongation dependent
IKKα distribution along TNF coding region is correlated with RNAPII S2p enrichment (Supplementary Figures S1F and S2D) suggesting that IKKα may travel with the elongating RNA polymerase. To test the link between the progression of the elongating form of RNAPII through chromatin and the accumulation of IKKα and also HP1γ and H3.3S31p to TNF after LPS treatment, we treated RAW264.7 cells with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB). DRB is a selective inhibitor of transcription elongation by inhibition of Cdk9 kinase activity (36). We treated RAW264.7 cells with LPS for 30 min in the presence or absence of DRB, or alternatively, we added DRB 30 min after the addition of LPS for an additional 30 min. We first confirmed that DRB treatment was blocking TNF expression and RNAPII transcription elongation (Supplementary Figure S5A and S5B) and checked whether this treatment was affecting global levels of IKKα or HP1γ proteins or H3.3S31 phosphorylation (Figure 3A). In this experiment, we observed a severe reduction of total H3.3S31p, implying that this post-translational modification is a transient mark, which is directly dependent on transcription elongation (Figure 3A). We did previously establish that DRB was preventing LPS-dependent recruitment of IKKα to chromatin (22). In agreement with this observation, DRB treatment prevented not only recruitment but also maintenance of IKKα and HP1γ proteins to the TNF locus (Figure 3B and C). In contrast, DRB treatment increased total RNAPII enrichment and NF-κB binding to the TNF promoter, suggesting that initiation complexes remained bound to the chromatin and indicating that DRB did not lead to a non-specific loss of chromatin-associated proteins from the TNF locus (Supplementary Figure S5C and S5D). Next we performed a ChIP experiment with an anti-RNAPII S2p antibody, eluted the complexes and repeated the immunoprecipitation (re-ChIP), with antibodies against IKKα, HP1γ or p65 (NF-κB). NF-κB was only detected together with RNAPII S2p in the promoter region (Figure 4A). In contrast, IKKα and HP1γ associated with RNAPII S2p were both equally spread along the gene (Figure 4B and C). Such co-purification and the fact that DRB prevents, as described previously (22), and even reverses accumulation of both IKKα and HP1γ to the TNF locus, without affecting NF-κB binding, support a model, in which both HP1γ and IKKα interact with the elongating polymerase during transcription elongation.
Figure 3.
Transcription elongation-dependent accumulation of IKKα, HP1γ and H3.3S31 phosphorylation at the TNF locus. (A) Quantification of total IKKα, HP1γ, H3.3S31p, H3.3 and β-actin protein levels by western blot with total cell extract, after RAW264.7 cells incubation with LPS+DRB. RAW264.7 cells treated with LPS in presence or absence of DRB for 30 min LPS (30), 30 min LPS+DRB (30+DRB), 60 min LPS (60) or 30 min LPS following by 30 min with DRB (60+DRB). ChIP performed with (B) anti-IKKα and (C) anti-HP1γ. Horizontal axis indicates primers used for the real-time PCR. Data are normalized versus input and then versus an average of control regions. Data are representative of at least three independent experiments. Error bars represent standard deviation (SD) from three independent qPCR replicates.
Figure 4.
Re-ChIP. Re-ChIP performed in RAW264.7 cells treated with LPS for 1 h, with anti-RNAPII S2p antibody and then with (A) anti-IKKa, (B) anti-HP1g and (C) anti-p65 antibodies. Horizontal axis indicates primers used for the real-time PCR. Data are normalized versus IP performed with anti-RNAPII S2p antibody. Data are representative of two independent experiments. Error bars represent standard deviation (SD) from three independent qPCR replicates.
IKKα can bind to phosphorylated RNAPII CTD in the absence of HP1γ
FACT recruitment to RNAPII has been shown to be dependent of HP1c in drosophila (16). Because both HP1γ and IKKα detection to TNF was transcription elongation dependent, we tested whether IKKα was recruited by HP1γ to the RNA polymerase. For this, we first performed eCoIP using whole cell extracts and showed an interaction between both IKKα and HP1γ and phosphorylated RNAPII CTD, whereas these proteins were inefficient at co-immunoprecipitating the unphosphorylated RNAPII CTD (Figure 5A and Supplementary Figure S6). Then GST pull-down experiments performed with recombinant IKKα and both CTD S2p and CTD S5p peptides revealed that IKKα was binding to phosphorylated RNAPII CTD in absence of HP1γ (Figure 5B and D). In addition, when both peptides were incubated with IKKα, the amount of bound CTD S5p was reduced compared with incubation of IKKα with only CTD S5p (compare bound fractions bottom of Figure 5B; lanes 4 and 5). This was not the case for the same comparison with only CTD S2p (compare bound fractions top of Figure 5B; lanes 3 and 5). This observation was confirmed by densitometric quantification (Figure 5C). Together these results show that IKKα has a stronger affinity for CTD S2p than CTD S5p. Finally, additional pull-down experiments with recombinant proteins incubated with a biotinylated peptide corresponding to the CTD S2p showed that the interaction between IKKα and the CTD S2p peptide was weaker in the presence of both HP1γ and ATP, whereas the interaction between HP1γ and this peptide was unaffected by the presence of ATP (Figure 5D, compare lane 8 with lane 9). In the same experiment, we also observed that HP1γ interaction with the CTD S2p peptide was strongly reduced in the presence of IKKα (Figure 5D, compare lane 4 with lane 8). Taken together, we highlighted a complex and dynamic interaction between phosphorylated RNAPII, IKKα and HP1γ.
Figure 5.
IKKα binds directly to RNAPII CTD S2p. (A) eCoIP performed with whole cell extract from RAW264.7 cells treated with LPS for 1 h. Antibodies used for immunoprecipitation are named at the top of the figure and antibodies used for western blot named on the left. Input was 5% of material used for CoIP. (B) In vitro pulldown followed by dot blot assay. GST-IKKα (2 pmol) was incubated in the absence of peptide (lane 2) or in the presence of 600 pmol CTD-S2p (lane 3) or CTD-S5p (lane 4) peptides or an equimolar mix of the two (lane 5). A GST-control peptide, not interacting with RNAPII, was incubated with the same equimolar mix of the two peptides (lane 1). Pulldown was performed with anti-GST beads. Unbound and bound factions were transferred onto the membrane. Antibodies used for dot blot are named on the left. (C) The competition between peptides for binding to IKKα was quantified using densitometry. Data are expressed as a ratio (bound/unbound)S2p + S5p/(bound/unbound)S2p or S5p. Error bars represent standard deviation (SD) of two independent experiments. (D) In vitro pull-down experiment performed with a biotinylated peptide corresponding to the CTD S2p incubated with recombinant HP1α, HP1γ and IKKα proteins in the presence or absence of ATP.
HP1γ controls IKKα recruitment to chromatin
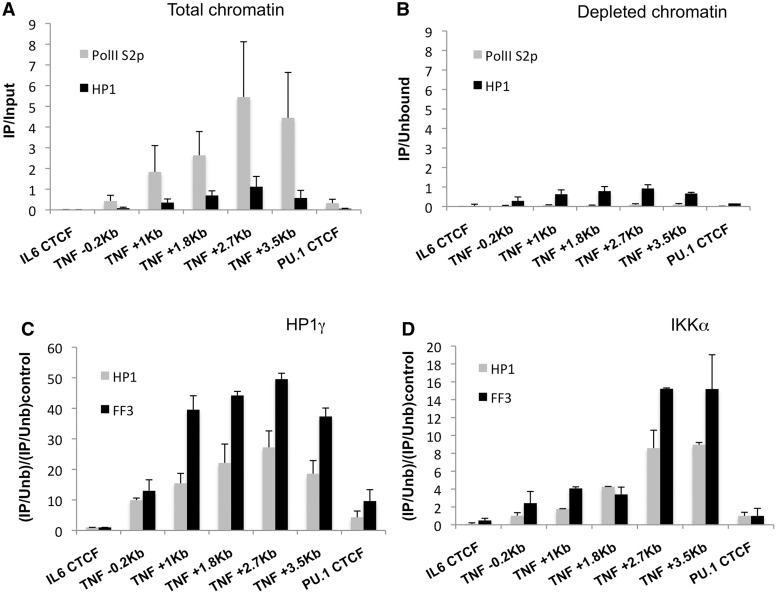
As HP1γ can alter the dynamics of histone H3.3 phosphorylation by IKKα and because HP1γ and H3.3S31p profiles at TNF overlap, we were aiming to further investigate the role of HP1γ in IKKα chromatin modifying activity. Manipulation of IKKα protein level or kinase activity affects NF-κB-dependent gene expression before the p-TEFb recruitment step (4,37–39) making it difficult to properly assess events happening downstream of transcription initiation. Therefore, to further study the relationship between HP1γ and IKKα kinase activity during transcription elongation, we performed HP1γ knockdown and assessed MNase accessibility, TNF mRNA expression and localization of IKKα and RNAPII S2p at this locus (Figure 6). After up to 80% and 50% reduction of HP1γ mRNA and protein levels, respectively (Figure 6A and B), we did not observe any effect on NF-κB recruitment to the promoter (Supplementary Figure S7A) indicating that HP1γ knockdown does not interfere with activation of the NF-κB pathway on LPS activation. However, we detected increased TNF and Ccl3 expression (Figure 6A and Supplementary figure S7B) correlated with an increase in RNAPII S2p recruitment within TNF coding region (Figure 6C). These changes were accompanied by an increased chromatin accessibility of TNF as shown by the reduced qPCR signal obtained after amplification of genomic DNA prepared from chromatin treated with MNase, whereas total histone H3 was unaffected (Figure 6D and Supplementary Figure S7E). Altogether these data are in agreement with a repressive role of HP1γ in transcription. Finally, we also observed a reduction of H3.3S31 phosphorylation suggesting that HP1γ controls H3.3S31 phosphorylation (Figure 6E). However, in these experiments, we did not see any significant effect of HP1γ knockdown on the recruitment of HP1γ and IKKα or the enrichment of HP1γS93p at this locus (Supplementary Figure S7C, S7D and S7F). Because our previous experiments showed an association between HP1γ, IKKα and elongating RNA polymerase and because HP1γ was only partially knocked down, we hypothesized that the decreased association of HP1γ with chromatin at this locus after knock down was masked by an increased amount of ‘travelling’ HP1γ in correlation with the increased level of elongating polymerase detected in the TNF locus. Therefore, to distinguish between ‘travelling’ and chromatin-associated HP1γ and IKKα, we performed ChIP on RNAPII S2p ‘depleted’ chromatin. As a proof of principle, the unbound fraction of a first RNAPII S2p ChIP (Figure 7A) was used for a second round of ChIP with anti-RNAPII S2p or anti-HP1γ antibodies (Figure 7B). This control experiment showed that anti-RNAPII S2p antibody precipitated 50 times less DNA from this ‘unbound’ fraction compared with the first round of ChIP, whereas HP1γ ChIP was not significantly affected, confirming a specific and efficient depletion of the elongating polymerase associated with chromatin from the template (Figure 7A and B). Then, we performed a second round ChIP with RNAPII S2p-depleted chromatin and antibodies against NF-κB, HP1γ and IKKα. As expected, we did not see any difference in NF-κB recruitment after HP1γ knockdown but saw a significant reduction of both HP1γ and IKKα accumulation at the TNF locus (Supplementary Figures S8A, S7C and S7D). In this experiment, the ratio of IKKα versus HP1γ enrichment was conserved after HP1γ knockdown indicating a strict correlation between the losses of chromatin-bound HP1γ and chromatin-bound IKKα (Supplementary Figure S8B). Taken together, these results show that IKKα recruitment to chromatin and histone H3.3S31 phosphorylation are HP1γ dependent. It also confirms that chromatin bound IKKα does exist in a form dissociated from RNAPII.
Figure 6.
HP1γ knockdown increases expression of TNF and abolishes H3.3S31 phosphorylation. (A) HP1γ, TNF and IKKα mRNA levels in RAW264.7 cells after HP1γ knockdown using three shRNAs against HP1γ (sh-1, -2 and -3) or firefly luciferase (sh-c) after 1 h LPS treatment. Results are expressed relative to GAPDH expression. Error bars represent standard deviation (SD) from three independent experiments. (B) Quantification of total HP1γ and β-actin protein levels by western blot with total cell extract, after HP1γ knockdown (sh-1) in RAW264.7 cells. (C,E) ChIP performed, after HP1γ knockdown in RAW264.7 cells treated with LPS for 1 h, with (C) anti-RNAPII S2p and (E) anti-H3.3S31p antibodies. Horizontal axis indicates primers used for the real-time PCR. Data are normalized versus input or (E) total histone H3 and then versus an average of background control regions. (D) qPCR quantification of MNase-treated chromatin. (C–E) Data are representative of at least three independent experiments. Error bars represent SD from three independent qPCR replicates.
Figure 7.
HP1γ knockdown reduces chromatin-associated IKKα. HP1γ knockdown in RAW264.7 cells treated with LPS for 1 h. (A) ChIP performed with anti-RNAPII S2p and anti-HP1γ normalized versus input only. The unbound fraction of a first anti-RNAPII S2p ChIP was used as a template for a second ChIP, (B) with anti-RNAPII S2p and anti-HP1γ normalized versus the unbound fraction only, (C) with anti-HP1γ and (D) anti-IKKα. (A–D) Horizontal axis indicates primers used for the real-time PCR. (C,D) Data are normalized versus the unbound fraction (Unb) and then versus an average of control regions. Data are representative of at least five independent experiments. Error bars represent ± standard deviation (SD) from three independent qPCR replicates.
CUEDC2 co-localizes with IKKα at the 3′-end of TNF and Ccl3
We found a correlation between HP1γ and H3.3S31p accumulation within the coding region of TNF. However, if IKKα mediates H3.3S31 phosphorylation, then the presence of H3.3S31p should peak where RNAPII S2p independent IKKα enrichment is at the maximum, i.e. at the 3′-end of the TNF gene (Figure 1D and Supplementary Figure S2B). IKKα activity is regulated by NIK and CUEDC2 (23,40). For this reason, a non-strict correlation between chromatin-associated IKKα and H3.3S31p could be explained by modulations of the IKKα kinase activity during the transcription elongation process. Looking for interactions between IKKα and the CUEDC2/PP1 complex by eCoIP, we showed that IKKα was indeed interacting with this complex in the nucleus and in the cytoplasm (Figure 8A). In addition, ChIP experiments revealed an accumulation of CUEDC2 in the 3′-end of the gene together with IKKα, suggesting that at the 3′-end of TNF, IKKα was inactivated (Figure 8B). We observed the same accumulation at the 3′-end of Ccl3 (Supplementary Figure S4B). These results suggest a close control of IKKα activity during transcription elongation.
Figure 8.
CUEDC2 accumulates at the 3′-end of TNF gene. (A) eCoIP experiment with nuclear and cytoplasmic extracts from RAW264.7 cells treated with LPS for 1 h and precipitated with anti-IKKα antibody followed by western blot with anti-HPRT, anti-PP1 and anti-CUEDC2 antibodies. (B) ChIP with RAW264.7 cells treated with LPS performed with anti-CUEDC2 antibody. Horizontal axis indicates primers used for the real-time PCR. Data are normalized versus input and then versus an average of control regions. Data are representatives of at least three independent experiments. Error bars represent ± standard deviation (SD) from three independent qPCR replicates. (C) Suggested model of IKKα cycling activity during transcription elongation. (Step 1) Both IKKα and HP1γ bind to phosphorylated RNAPII CTD. IKKα phosphorylates HP1γS93. (Step 2) The complex IKKα/HP1γS93p is transferred to chromatin. (Step 3) The presence of HP1γ favours H3.3S31 phosphorylation by IKKα. (Step 4) After the passage of the RNA polymerase, H3.3S31 is phosphorylated, unphosphorylated HP1γS93 is high and IKKα low. (transcription termination) CUEDC2 co-localizes with the IKKα/HP1γS93p complex on chromatin, whereas H3.3S31 is not phosphorylated.
DISCUSSION
Despite extensive studies, the role of IKKα in NF-κB signalling pathways is still controversial, mainly due to its contribution at different stages of the signalling cascades, making functional studies difficult. IKKβ appears to be the main kinase controlling NF-κB release into the nucleus (8,9). However, IKKα increases NF-κB-mediated transcription by removing repressor complexes from targeted promoters and by facilitating p65 binding to DNA for a selected number of NF-κB-dependent genes (37,39). Conversely, IKKα also accelerates both the turnover of the NF-κB subunits and their removal from pro-inflammatory gene promoters, limiting macrophage activation and inflammation (38). Downstream of NF-κB recruitment to DNA, this kinase is also known to phosphorylate H3S10, a histone mark triggering Brd4/p-TEFb recruitment (5,7–9). Herein, we reveal new chromatin targets for IKKα during transcription elongation of NF-κB-dependent genes. We show that IKKα plays a role in transcription even further downstream of the initiation of productive transcription by phosphorylating not only H3S10 but also H3.3S31 and HP1γS93. Absence of H3S10p at the promoter of TNF in response to LPS was previously reported (41). In our hand, H3S10p stays at a relatively low level at the promoter but peaks at the +1 nucleosome of TNF, early after LPS stimulation, in correlation with Brd4/p-TEFb recruitment. H3.3S31p then progressively replaces H3S10p in the gene body in parallel with accumulation of HP1γ. Both events correlate with enrichment of IKKα at the 3′-end of genes, downstream of the TES in a region where HP1γS93 is phosphorylated and RNAPII S2p high. In addition, even if NF-κB recruits IKKα to NF-κB-dependent genes (8,9), the interaction between NF-κB and IKKα on DNA appears to be unstable. This hit-and-run type of interaction seems to be independent of IKKα loading onto the RNA polymerase, as DRB treatment does not induce any accumulation of this enzyme at the promoter.
Our work reveals a central role for HP1γ in controlling IKKα chromatin-modifying activity and a role for IKKα in controlling HP1γ (see proposed model in Figure 8C). We show that IKKα directly binds to HP1γ· Both proteins interact with phosphorylated RNAPII CTD and their accumulation, and maintenance at the TNF locus is strictly dependent on RNAPII S2p. This interaction between HP1γ and phosphorylated RNAPII CTD is in agreement with previous publications (15,16). Moreover, we identified two populations of HP1γ and IKKα proteins during transcription: one travelling with the polymerase and the other associated with chromatin. HP1γ affinity for RNAPII S2p decreases in the presence of IKKα, and the presence of ATP favours IKKα interaction with HP1γ over RNAPII S2p, suggesting a mechanism in which both proteins are recruited to the RNAPII CTD and loaded onto chromatin after HP1γ phosphorylation driven by IKKα. The correlation between IKKα and HP1γS93p enrichment at the 3′-end of TNF and the fact that no change in Hp1γS93p enrichment level is observed after HP1γ knockdown, establishing that travelling HP1γS93p compensates for the loss of chromatin-associated HP1γS93p, argue in support of such a mechanism. In addition, our ChIP experiments performed with chromatin ‘depleted’ for elongating polymerase show that HP1γ allows association of IKKα with chromatin and subsequent phosphorylation of H3.3S31. Similarly in Drosophila, the expression of heat-shock genes is dependent on recruitment of FACT to heat-shock loci by HP1c (16). However, in contrast with what we observe in our system, FACT cannot bind to the active forms of RNAPII in the absence of HP1c.
Transcription termination occurs after dismantling of the elongation complex from DNA by the polyadenylation cleavage factor Pcf11 (42,43), and therefore, RNA polymerase does not pass through nucleosomes downstream of the TES, providing clues to determining the order of events which may occur when RNAPII reaches a nucleosome (Figure 8C). First, the IKKα/HP1γS93p complex could be loaded from the phosphorylated RNAPII CTD onto the chromatin. In the body of the actively transcribing TNF, IKKα enrichment is low compared with the 3′-end, HP1γS93 is unphosphorylated and H3.3S31 phosphorylated. Again, the IKKα enrichment profile at TNF suggests a hit-and-run mechanism for this enzyme in the body of the gene. Consequently, we concluded that the passage of the polymerase through the nucleosome would be associated with H3.3S31 phosphorylation by IKKα (hit), IKKα eviction from the chromatin (run) and HP1γ dephosphorylation. Interestingly, at the 3′-end of TNF, we found a significant amount of chromatin-bound IKKα not associated with RNAPII S2p. In this region, IKKα and its repressor CUEDC2 co-localize, suggesting that, in parallel with transcription termination, CUEDC2 prevents H3.3S31 phosphorylation and IKKα hit-and-run activity.
The exact function of the described sequential post-translational modifications is still not understood. HP1γ is commonly associated with transcribing chromatin (15,16,19,44), where it is found phosphorylated, a post-translational modification impairing HP1γ-repressive activity (20,21). We have determined that HP1γ knockdown correlates with a decrease in chromatin-associated HP1γ and a higher rate of TNF transcription in agreement with a repressive role for this protein during transcription elongation. However, we also revealed a correlation between the elongating polymerase present at TNF locus and HP1γ associated with this polymerase. This correlation suggests a dual role for HP1γ during transcription, a positive role by recruiting FACT to chromatin (16) and a repressive role when bound to chromatin after the passage of the polymerase through nucleosomes. By targeting HP1γ, IKKα would control HP1γ dual activity.
When recruited to chromatin, IKKα may also play a role to facilitate or repress the elongating polymerase to read through nucleosomes during transcription elongation. In this context, the function of H3.3S31p is unclear. Because H3.3S31 is not phosphorylated downstream of TNF where IKKα and CUEDC2 co-localize, we hypothesize that H3.3S31p could facilitate transcription elongation. This histone mark is evidently co-enriched in coding regions with non-phosphorylated HP1γ, the ‘repressive’ form of HP1γ, whereas HP1γS93p is restricted to the 3′-end. However, transcription elongation being a very dynamic process, it may just indicate that H3.3S31p is dephosphorylated a step later than HP1γS93p. In addition, an increased rate of TNF transcription in absence of both H3.3S31p and HP1γ may mean that H3.3S31p specifically antagonizes HP1γ-repressive function and is therefore dispensable in absence of the latter. Histone H3K36 methylation is another histone mark associated with transcription elongation (45). Similar to HP1γ, H3K36me3 is found within coding regions of actively transcribing genes where it negatively influences gene expression including the repression of cryptic promoters (46). H3K36me3 is specifically enriched within exons (47,48) where it may slow the rate of RNAPII elongation to increase inclusion of alternatively spliced exons (49), a role also described for HP1γ (50). H3.3S31p may have a positive action on transcription by antagonizing H3K36me3 by different mechanisms. Similar to H3S10, H3.3S31p may trigger histone acetyl transferase recruitment (7) to antagonize H3K36me3-mediated recruitment of histone deacetylase (51) or it may control H3P30 and H3P38 isomerizations, post-translational modifications preventing H3K36me3 (52).
HP1γ and H3.3 co-enrichment can be restricted to promoters as recently described for HSP70 gene after heat shock (53), suggesting distinct mechanisms preventing or allowing spreading of HP1γ/H3.3 along coding regions. Differences in chromatin structure may account directly or indirectly for such repartition along genes. Although further investigations will be necessary to decipher these mechanisms, we did not establish a link between HP1γ/H3.3 recruitment to coding regions and nucleosome density because the nucleosome landscape across the entire coding region of HSP70 after heat shock (30) is similar to the one we described for TNF on LPS induction.
In conclusion, we have unravelled new functional interactions for the chromatin modifying enzyme IKKα and HP1γ during transcription elongation of NF-κB-dependent genes. Because NF-κB is a complex family of transcription factor activated by multiple stimuli, it will be important to determine whether IKKα is selectively recruited to a subclass of NF-kB-dependent genes or whether this enzyme is widely recruited by the different subunits of this family including non-canonical subunits such as p52 or RelB, the latter regulating transcription elongation (54). In addition, because, neither incorporation of histone H3.3 into chromatin nor Hp1γS93 phosphorylation is restricted to the transcription of NF-κB-dependent genes, we expect the mechanism we are describing herein to be more universal with other kinases fulfilling the role of IKKα for other genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–8.
FUNDING
The Biotechnology and Biological Sciences Research Council UK (BBSRC) [BB/G001391/1]; Medical Research Council UK [G0801215]. Funding for open access charge: BBSRC [BB/G001391/1].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Maarten Hoogenkamp and Peter N. Cockerill (Section of Experimental haematology, LIMM, University of Leeds) for advice and critical comments on the manuscript.
REFERENCES
- 1.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J. Biol. Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 4.Swaminathan S. IKKalpha: a chromatin modifier. Nat. Cell. Biol. 2003;5:503. doi: 10.1038/ncb0603-503. [DOI] [PubMed] [Google Scholar]
- 5.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 7.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 9.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 10.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 11.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends. Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 13.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 14.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes. Dev. 2010;24:2133–2145. doi: 10.1101/gad.1959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 18.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- 20.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 2006;8:407–415. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 21.Koike N, Maita H, Taira T, Ariga H, Iguchi-Ariga SM. Identification of heterochromatin protein 1 (HP1) as a phosphorylation target by Pim-1 kinase and the effect of phosphorylation on the transcriptional repression function of HP1(1) FEBS Lett. 2000;467:17–21. doi: 10.1016/s0014-5793(00)01105-4. [DOI] [PubMed] [Google Scholar]
- 22.Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol. Cell. 2008;32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li HY, Liu H, Wang CH, Zhang JY, Man JH, Gao YF, Zhang PJ, Li WH, Zhao J, Pan X, et al. Deactivation of the kinase IKK by CUEDC2 through recruitment of the phosphatase PP1. Nat. Immunol. 2008;9:533–541. doi: 10.1038/ni.1600. [DOI] [PubMed] [Google Scholar]
- 24.Tagoh H, Himes R, Clarke D, Leenen PJ, Riggs AD, Hume D, Bonifer C. Transcription factor complex formation and chromatin fine structure alterations at the murine c-fms (CSF-1 receptor) locus during maturation of myeloid precursor cells. Genes Dev. 2002;16:1721–1737. doi: 10.1101/gad.222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefevre P, Lacroix C, Tagoh H, Hoogenkamp M, Melnik S, Ingram R, Bonifer C. Differentiation-dependent alterations in histone methylation and chromatin architecture at the inducible chicken lysozyme gene. J. Biol. Chem. 2005;280:27552–27560. doi: 10.1074/jbc.M502422200. [DOI] [PubMed] [Google Scholar]
- 27.Lefevre P, Bonifer C. Analyzing histone modification using crosslinked chromatin treated with micrococcal nuclease. Methods Mol. Biol. 2006;325:315–325. doi: 10.1385/1-59745-005-7:315. [DOI] [PubMed] [Google Scholar]
- 28.Lefevre P, Melnik S, Wilson N, Riggs AD, Bonifer C. Developmentally regulated recruitment of transcription factors and chromatin modification activities to chicken lysozyme cis-regulatory elements in vivo. Mol. Cell. Biol. 2003;23:4386–4400. doi: 10.1128/MCB.23.12.4386-4400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl Acad. Sci. USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsytsykova AV, Rajsbaum R, Falvo JV, Ligeiro F, Neely SR, Goldfeld AE. Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers. Proc. Natl Acad. Sci. USA. 2007;104:16850–16855. doi: 10.1073/pnas.0708210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smallie T, Ricchetti G, Horwood NJ, Feldmann M, Clark AR, Williams LM. IL-10 inhibits transcription elongation of the human TNF gene in primary macrophages. J. Exp. Med. 2010;207:2081–2088. doi: 10.1084/jem.20100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krausgruber T, Saliba D, Ryzhakov G, Lanfrancotti A, Blazek K, Udalova IA. IRF5 is required for late-phase TNF secretion by human dendritic cells. Blood. 2010;115:4421–4430. doi: 10.1182/blood-2010-01-263020. [DOI] [PubMed] [Google Scholar]
- 34.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 35.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoberg JE, Yeung F, Mayo MW. SMRT derepression by the IkappaB kinase alpha: a prerequisite to NF-kappaB transcription and survival. Mol. Cell. 2004;16:245–255. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 39.Gloire G, Horion J, El Mjiyad N, Bex F, Chariot A, Dejardin E, Piette J. Promoter-dependent effect of IKKalpha on NF-kappaB/p65 DNA binding. J. Biol. Chem. 2007;282:21308–21318. doi: 10.1074/jbc.M610728200. [DOI] [PubMed] [Google Scholar]
- 40.Park GY, Wang X, Hu N, Pedchenko TV, Blackwell TS, Christman JW. NIK is involved in nucleosomal regulation by enhancing histone H3 phosphorylation by IKKalpha. J. Biol. Chem. 2006;281:18684–18690. doi: 10.1074/jbc.M600733200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat. Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 42.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mateescu B, Bourachot B, Rachez C, Ogryzko V, Muchardt C. Regulation of an inducible promoter by an HP1beta-HP1gamma switch. EMBO Rep. 2008;9:267–272. doi: 10.1038/embor.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 46.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 48.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat. Struct. Mol. Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 51.Lee JS, Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat. Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 52.Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 53.Kim H, Heo K, Choi J, Kim K, An W. Histone variant H3.3 stimulates HSP70 transcription through cooperation with HP1{gamma} Nucleic Acids Res. 2011;39:8329–8341. doi: 10.1093/nar/gkr529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suhasini M, Pilz RB. Transcriptional elongation of c-myb is regulated by NF-kappaB (p50/RelB) Oncogene. 1999;18:7360–7369. doi: 10.1038/sj.onc.1203158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.