Abstract
Transcripts of the human tumor susceptibility gene 101 (TSG101) are aberrantly spliced in many cancers. A major aberrant splicing event on the TSG101 pre-mRNA involves joining of distant alternative 5′ and 3′ splice sites within exon 2 and exon 9, respectively, resulting in the extensive elimination of the mRNA. The estimated strengths of the alternative splice sites are much lower than those of authentic splice sites. We observed that the equivalent aberrant mRNA could be generated from an intron-less TSG101 gene expressed ectopically in breast cancer cells. Remarkably, we identified a pathway-specific endogenous lariat RNA consisting solely of exonic sequences, predicted to be generated by a re-splicing between exon 2 and exon 9 on the spliced mRNA. Our results provide evidence for a two-step splicing pathway in which the initial constitutive splicing removes all 14 authentic splice sites, thereby bringing the weak alternative splice sites into close proximity. We also demonstrate that aberrant multiple-exon skipping of the fragile histidine triad (FHIT) pre-mRNA in cancer cells occurs via re-splicing of spliced FHIT mRNA. The re-splicing of mature mRNA can potentially generate mutation-independent diversity in cancer transcriptomes. Conversely, a mechanism may exist in normal cells to prevent potentially deleterious mRNA re-splicing events.
INTRODUCTION
Pre-mRNA splicing takes place precisely and efficiently in a spatially and temporally regulated manner to generate mRNAs for translation into active protein products. Defects in the splicing process are closely associated with the formation and development of cancers [reviewed in (1–3)].
Tumor susceptibility gene 101 (TSG101) was originally identified with a screen for potential tumor suppressors using a transformation assay in mouse NIH3T3 cells (4), but its role as a tumor suppressor gene remained controversial. As a component of the endosomal sorting complex required for transport I (ESCRT-I), the TSG101 protein mediates a variety of endosomal and non-endosomal functions, such as protein sorting, the biogenesis of multivesicular bodies, virus budding, stimulation of the cell cycle, regulation of transcription and the maintenance of epithelial cell polarity [reviewed in (5,6)]. The fragile histidine triad (FHIT) gene has also been identified as encoding a putative tumor suppressor, diadenosine triphosphate/tetraphosphate (Ap3A/Ap4A) hydrolase involved in the regulation of signaling pathways (7,8). Because ApnA is a signaling molecule that responds to cellular stress and affects several cellular processes, FHIT protein plays an important role in the promotion of apoptosis and the control of the cell cycle in response to oxidative and replicative stress [reviewed in (9–11)].
TSG101 and FHIT pre-mRNAs are aberrantly spliced in many cancers, although no mutations have been found in the authentic splice sites of these pre-mRNAs in most cases [reviewed in (1,2)]. Some types of transcript variants have also been detected in normal tissues, but the overall complexity and frequency of their aberrant splicing are clearly increased in cancer tissues, suggesting a progressive loss of splicing fidelity during malignant transformation (7,12–24). Most of the aberrant mRNAs result from activation of distant weak splice sites accompanied by elimination of multiple exons. Considering that this aberrant splicing occurs via conventional one-step pathway, we do not yet know why the splicing machinery ignores many strong bona fide splice sites that exist between the activated 5′ and 3′ splice sites, which are conspicuously weak. By analyzing the typical cancer-specific aberrant splicing of TSG101 and FHIT pre-mRNAs, we have obtained considerable evidence that normal constitutive splicing precedes aberrant splicing, which clarifies the above issue. Our discovery of re-splicing of the mRNAs in cancer cells may reflect a general mechanism for alternative splicing between extremely distant 5′ and 3′ splice sites, which must be well controlled in normal cells.
MATERIALS AND METHODS
Full descriptions, including the detailed experimental conditions, are provided in Supplementary Materials and Methods.
Splice sites scoring methods
We calculated the 5′ and 3′ splice site scores using nine computer programs (Table 1), which are available online (the URLs are provided in Supplementary Materials and Methods). Detailed descriptions and the original references for the algorithms used have been given previously (25–27).
Table 1.
Strengths of the authentic and alternative splice sites in TSG101 pre-mRNA estimated with different scoring methods
| 5′ splice site | S&Sa | ΔGb | H-bondc | NNd | MAXENTe | MMf | MDDg | WMMh | SDi | 3′ splice site | S&Sa | NNd | MAXENTe | MMf | WMMh |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intron 1 | 89.63 | −8.5 | 15.7 | 0.99 | 9.16 | 9.62 | 13.98 | 10.39 | −2.09 | Intron 1 | 90.89 | 0.90 | 7.85 | 7.41 | 8.19 |
| Alternative | 67.27 | −0.6 | 10.7 | 0.78 | 6.33 | 6.24 | 9.48 | 3.26 | −4.05 | Intron 2 | 91.04 | 0.99 | 11.58 | 11.86 | 13.87 |
| Intron 2 | 86.25 | −7.2 | 16.1 | 0.98 | 10.13 | 9.22 | 14.08 | 9.29 | −2.17 | Intron 3 | 98.93 | 0.98 | 11.22 | 12.00 | 14.07 |
| Intron 3 | 54.14 | −5.5 | 14.9 | 0.92 | 8.39 | 7.31 | 12.38 | 8.42 | −2.73 | Intron 4 | 87.15 | 0.96 | 7.63 | 8.98 | 11.08 |
| Intron 4 | 86.77 | −4.3 | 19.1 | 0.99 | 10.16 | 9.72 | 15.28 | 10.22 | −2.99 | Intron 5 | 70.07 | 0.03 | 3.15 | 3.40 | −2.44 |
| Intron 5 | 79.30 | −4.2 | 15.7 | 0.95 | 8.65 | 6.49 | 13.18 | 7.50 | −2.96 | Intron 6 | 81.26 | 0.97 | 8.90 | 11.43 | 12.26 |
| Intron 6 | 82.79 | −6.0 | 15.8 | 0.95 | 9.43 | 7.98 | 13.48 | 8.23 | −2.43 | Intron 7 | 94.05 | 0.99 | 13.05 | 13.81 | 14.67 |
| Intron 7 | 87.40 | −7.3 | 18.7 | 1.00 | 10.47 | 9.09 | 15.08 | 9.00 | −2.27 | Intron 8 | 95.99 | 0.99 | 9.62 | 10.51 | 9.43 |
| Intron 8 | 76.53 | −4.2 | 14.0 | 0.98 | 8.01 | 5.57 | 12.58 | 6.66 | −3.31 | Alternative | 80.74 | 0.31 | 6.67 | 6.41 | 5.76 |
| Intron 9 | 77.74 | −4.1 | 12.3 | 0.92 | 7.64 | 6.06 | 10.28 | 5.86 | −2.66 | Intron 9 | 82.22 | 0.89 | 6.82 | 8.48 | 11.68 |
aS&S: Shapiro and Senapathy Score, a position-weight matrix.
bΔG: Free energy of the 5′ splice site–U1 snRNA duplex.
cH-Bond: An algorithm based on the hydrogen bonding of the 5′ splice site–U1 base pairing.
dNN: Neural network, a machine learning approach.
eMAXENT: Maximum entropy model, an algorithm that considers dependencies between positions.
fMM: First-order Markov model, an algorithm that considers dependencies between adjacent positions.
gMDD: Maximum dependence deposition, a decision-tree approach.
hWMM: Weight matrix model, a quantification of the relative likelihood of the candidate splice site sequence.
iSD: SD score, a common logarithm of the frequency of the 5′ splice sites used in the human genome.
Construction of plasmids
The expression plasmids for the TSG101-EGFP fusion proteins (TSG101-EGFP, TSG101-EGFP[+] and TSG101-EGFP[−]; Figure 2C) were constructed by subcloning the corresponding polymerase chain reaction (PCR) fragments from the TSG101 gene together with PCR fragments from the pBI-EGFP plasmid (Clontech) into the pCXN2 mammalian expression vector (28). The FHIT-EGFP fusion plasmid (FHIT-EGFP; Figure 5C) was constructed with the same procedure. Overlap-extension PCR was performed with Ex Taq HS DNA polymerase (Takara Bio) and the corresponding primers (Invitrogen).
Figure 2.
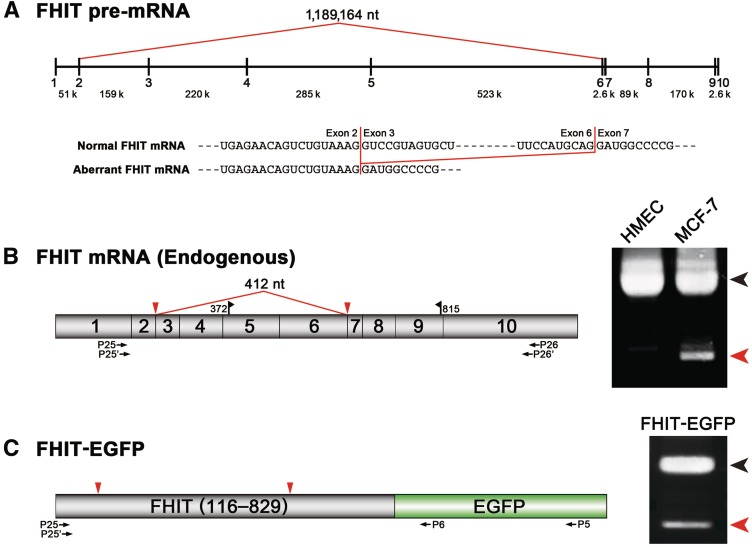
Ectopically expressed intronless TSG101 gene (cDNA) is spliced and generates equivalent product as the endogenous aberrant mRNA in cancer cells. (A) The structure of the TSG101 mRNA. Red line and triangles indicate the postulated mRNA re-splicing via activated alternative 5′ and 3′ splice sites. Blue triangle indicates the PTC (1112–1114) generated by the postulated splicing. Black flags indicate the open reading frame (ORF; 127–1296). The positions of the PCR primers (P1–P4, P7–P10) are shown. (B) Using normal cells (HMEC) and cancer cells (MCF-7), endogenous TSG101 mRNAs were analyzed by RT-PCR (RT) and the genomic DNA was analyzed by PCR (Ge) with primer sets (P1–P4, P7–P10) annealed to the indicated exons. The black and red arrowheads denote the full-length and major aberrant mRNAs, respectively (Figure 1). (C) The structures of three reporter TSG101-EGFP plasmids (see (A) for the red and blue triangles). The postulated splicing-dependent EGFP expression of these plasmids is indicated with (−) and (+). The positions of the PCR primer sets (P1–P5, P3–P6) are shown. (D) Cancer cells (MCF-7) were transfected with the indicated TSG101-EGFP reporter plasmids and the EGFP expression was analyzed by fluorescence microscopy. (E) These ectopically expressed TSG101-EGFP transcripts were analyzed by RT-PCR. The black and red arrowheads indicate the unspliced and spliced products via the alternative splice sites, respectively. Two minor spliced products (indicated with * and **) were generated from different alternative 3′ splice sites, i.e. Δ190–776 (587-nt deletion) and Δ190–1236 (1047-nt deletion), respectively.
Figure 5.
Ectopically expressed intronless FHIT gene (cDNA) is spliced, generating a product equivalent to the endogenous aberrant mRNA in cancer cells. (A) Schematic representation of the FHIT pre-mRNA and the observed aberrant splicing in cancer cells (red line). The aligned sequences of the normal (full-length) and aberrant FHIT mRNAs reveal the skipping of exon 3 to exon 6. (B) Using normal cells (HMEC) and cancer cells (MCF-7), endogenous FHIT mRNAs (schematically shown) were analyzed by RT-PCR with the indicated primer sets (P25–P26, P25′–P26′). The full-length mRNA (black arrowhead) and aberrantly spliced mRNA (red arrowhead) are indicated. Black flags indicate the ORF (372–815) and the red line indicates the postulated mRNA re-splicing that skips the whole exon 3 to exon 6 region. (C) Cancer cells (MCF-7) were transfected with the FHIT-EGFP reporter plasmid (schematically shown) and the spliced products were analyzed by RT-PCR with the indicated primer sets (P25–P5, P25′–P6). The black and red arrowheads indicate the unspliced and spliced products, respectively. Sequence analysis mapped the spliced sites (red triangles) to the 5′ end of exon 3 and the 3′ end of exon 6 (data not shown).
Cell lines, transfection of cells and preparation of total RNA
Most of the indicated cell lines were purchased from Lonza, the Cell Resource Center for Biomedical Research (at the Tohoku University), the Cell Bank of the RIKEN BioResource Center and ATCC.
MCF-7 cells were transiently transfected with indicated expression plasmids using Lipofectamine LTX (Invitrogen), according to the manufacturer’s instructions. At 24 h after transfection, the cells were examined for the expression of the green fluorescent protein (GFP) signal. At 48 h after transfection, their total cellular RNA was extracted with TRIzol reagent (Invitrogen) and digested with DNase I (Takara Bio).
RT-PCR detection of TSG101 and FHIT mRNAs
cDNA was synthesized from the total cellular RNA using PrimeScript reverse transcriptase (Takara Bio) and an oligo(dT) primer (Invitrogen), according to the manufacturers’ instructions. The sequences of all the indicated RT-PCR primers (Invitrogen) are listed in Supplementary Table S1. All the PCR products were analyzed by 2% agarose gel electrophoresis.
The reaction mixture containing cDNA was used for the first PCR amplification (24 and 22 cycles for TSG101 and FHIT mRNAs, respectively) with Ex Taq HS DNA polymerase (Takara Bio) and the indicated primers (Figures 2A, B and 5B). After the amplified products were purified with Nucleospin Extract II (Macherey-Nagel), the second nested PCR was performed as described earlier, with the indicated inner primers (Figures 2A, B and 5B). The PCR was also performed with genomic DNA prepared from the cells, with the indicated primers (Figure 2A and B).
To detect the spliced products from the TSG101-EGFP and FHIT-EGFP-transfected cells (Figures 2E and 5C), 1:100-diluted cDNA solutions, prepared with SuperScript III RNase H− reverse transcriptase (Invitrogen), were used for the first PCR (30 and 22 cycles, respectively) with the indicated primers (Figures 2C and 5C). After purification, as described earlier, 1:100-diluted PCR solutions were used for the second nested PCR, also with the indicated inner primers (Figures 2C and 5C).
Detection/identification of TSG101 and FHIT splicing products
The selective identification of lariat RNA products by RT-PCR of RNase R-digested samples was performed essentially as described previously (29). Total cellular RNA was digested with purified recombinant RNase R (provided by Dr A. Malhotra) at 37°C for 1 h, as described previously (29). All the PCR products were analyzed by 2% agarose gel electrophoresis.
The linear RNA-free sample obtained was reverse transcribed using the PrimeScript reverse transcriptase (Takara Bio) or SuperScript III reverse transcriptase (Invitrogen) with random hexamer primers (Figure 3B) or random hexamer primers with lariat-specific primers (Figures 4A, 6 and 7A). The reaction mixture contained cDNA was used for the first PCR (35 or 36 and 40 cycles for the TSG101 and FHIT products, respectively) with PrimeSTAR Max or Ex Taq HS DNA polymerase (Takara Bio) and the indicated primers (Figures 3B, 4A, 6 and 7A). The amplified products were purified and used for the second nested PCR, as described earlier, with the corresponding inner primers. The isolated DNA fragments were subcloned into the pGEM-T Easy vector (Promega) and the sequences were verified.
Figure 3.
Detection of the lariat RNA consisting of only exons demonstrates the re-splicing of the endogenous TSG101 mRNA. (A) Schematic representation of the two postulated pathways leading to the generation of the aberrant mRNA, i.e. a conventional one-step direct aberrant splicing (right) and the proposed two-step process, including the re-splicing of the constitutively spliced mRNA (left). The pre-mRNA [1] and specific splicing products [2–5] from these two pathways were analyzed by RT-PCR with the indicated primers (arrows). (B) Detection of the specific splicing products [2–5] by RT-PCR using RNase R-digested (+) or RNase R-undigested (−) endogenous total RNA. The detected RT-PCR signals in the RNase R-digested sample (containing no linear RNAs) indicates an RNA species with either a 5′–2′ lariat or a 5′–3′ circular structure. However, the latter case was ruled out by the identification of a 5′–2′ branched structure (Figure 4A). Primers P19, P20, P21 and P22 anneal to introns 6, 7, 7 and 8, respectively. Primers P13/14 and P15/16 anneal to introns 7 and 8, respectively. Primers P11, P12, P17 and P18 anneal to exons 8, 10, 5 and 8, respectively. We used exactly the same PCR cycle numbers to amplify all these RNAs (Supplementary Materials and Methods).
Figure 4.
The lariat RNA containing TSG101 exon 2 to exon 9 was identified by sequencing. (A) Detection of the lariat exons by lariat-specific RT-PCR amplification across the branch point with the indicated primer sets (P23–P24, P23′–P24′). The RT-PCR signal remained in the RNase R-digested (+) RNA sample, revealing a closed lariat structure. (B) Sequencing analysis of the lariat-specific RT-PCR product. Electropherogram of the sequence containing the branch point (arrow), followed by the alternative 5′ splice site (arrow) in the lariat exons. (C) A sequence alignment of the PCR fragment (blue) and the TSG101 gene (black) reveals a 2′–5′ branched connection between the end of the alternative 5′ splice site (GT) and the branch point (A), which is located upstream from the alternative 3′ splice site (AG).
Figure 6.
Detection of a lariat RNA consisting only of exons demonstrates the re-splicing of the endogenous FHIT mRNA. RT-PCR analysis of the specific splicing products with the indicated primer sets (P27–P28, P29–P30, P31–P32) using RNase R-digested (+) or RNase R-undigested (−) endogenous total RNA. The RT-PCR signals in the RNase R-digested (+) sample (containing no linear RNAs) indicate either a lariat or circular structure, but a lariat structure was confirmed by the detection of the branched structure (Figure 7A). We used exactly the same number of PCR cycles to amplify all these RNAs (Supplementary Materials and Methods).
Figure 7.
The lariat RNA containing FHIT exon 3 to exon 6 was identified by sequencing. (A) Detection of the lariat exons by a lariat-specific RT-PCR across the branch point with the indicated primer sets (P33–P34, P33′–P34′). The RT-PCR signal remained in the RNase R-digested (+) RNA sample, revealing a closed lariat structure. (B) The sequence of the RT-PCR product is shown in an electropherogram. The alternative 5′ splice site is indicated. (C) A sequence alignment of the PCR fragment (blue) with the FHIT gene (black) reveals a branched connection between the end of the alternative 5′ splice site (GT) and the branch point (A), which is located upstream from the alternative 3′ splice site (AG). A gap at the branch point (indicated with hyphens) results from skipping that occurred during reverse transcription, as described previously (48).
RESULTS
Alternative splice sites are weaker than the authentic splice sites in TSG101 pre-mRNA
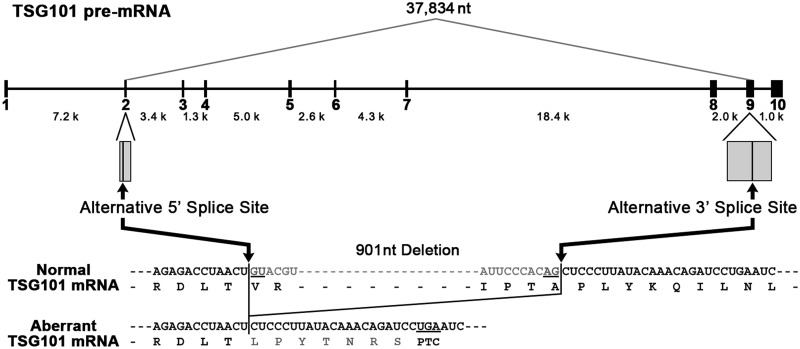
The most frequent aberrant splicing event on TSG101 pre-mRNA in cancers generates an mRNA with a 901-nucleotide (nt) deletion (Δ190–1090, formerly Δ154–1054) (12,13–18). It arises through activation of alternative 5′ and 3′ splice sites in distant exons 2 and 9, respectively, thus eliminating a large 37 834-nt span of sequence present in the pre-mRNA, which includes seven pairs of authentic splice sites (Figure 1) (16). Here, we analyzed the aberrant splicing that occurs in cancer cells, which can be classified as combinatorial alternative splicing, i.e. multi-exon skipping coupled to the selection of distant 5′ and 3′ splice sites (30).
Figure 1.
TSG101 pre-mRNA is aberrantly spliced in cancer cells by the activation of distant weak alternative splice sites. The structure of the TSG101 pre-mRNA and the major aberrant splicing often observed in various cancers are shown schematically. The sequences of the normal and aberrant TSG101 mRNAs are aligned with the encoded amino acids. The premature termination codon (PTC) generated in the aberrant mRNA is indicated.
The sequence-dependent strength of splice sites is a crucial determinant of their utilization (31,32). Therefore, we first statistically estimated the strengths of all the authentic and alternative splice sites on the TSG101 pre-mRNA with nine computer programs, which produced consistent results (Table 1). The scores indicated that the 5′ and 3′ alternative splice sites are significantly weaker than all the authentic constitutive splice sites, with the exception of one authentic 3′ splice site (numbers in bold, Table 1). Weak alternative splice sites activated when the adjacent strong authentic splice sites are destroyed by mutation, but are not otherwise utilized, are termed ‘cryptic’ splice sites (31,32).
The adjacent authentic splice sites were not mutated in the TSG101 gene tested, and the aberrant mRNA was indeed detected in several tissues from fetuses (12). Therefore, the activated splice sites in cancers are not cryptic but alternative. We postulated that a natural mechanism that eliminates the competing authentic splice sites must precede activation of the alternative splice sites. Using the TSG101 pre-mRNA as a model substrate, we tested the hypothesis that the product of the normal constitutive splicing process is re-spliced in cancer cells.
The major aberrant TSG101 mRNA is detected in various cancer cells with different efficiencies
We first confirmed that the endogenous production of a reported major aberrant product with a 901-nt deletion (Figure 2A) was recapitulated in various cancer cell lines with varying efficiencies, whereas it was not detectable in normal cells derived from adult tissues (Figure 2B, lane ‘RT’, and Supplementary Figure S1A). In the series of breast cancer cell lines, a modest correlation was found between the tumor stage and the efficiency of aberrant splicing (Supplementary Figure S1B). These data suggest that this specific aberrant splicing event is under the control of certain trans-acting factor(s), which could be deficient or over-expressed to varying degrees in cancer cells.
To evaluate the relative abundance of this particular aberrant mRNA and normal mRNA in these breast cancer cell lines, we performed single-round RT-PCR (no nested PCR) with specific primer sets that exclusively amplified either the aberrant mRNA or normal mRNA (Supplementary Figure S1C, lower gel picture and map). The semi-quantitative RT-PCR products revealed the substantial production of this aberrant mRNA in these breast cancer cell lines (MCF-7 and HCC series), whereas it was barely detectable in normal cells (HMEC) (Supplementary Figure S1C, upper histogram), consistent with previous reports (13,15,18). Notably, this particular aberrant product was often generated predominantly or even exclusively in the native tissues from patients with malignant or metastatic cancer (13–15,18). We also observed an overall increase in normal TSG101 mRNA production in breast cancer cells (MCF-7 and HCC series) compared with that in normal cells (HMEC), consistent with the finding that TSG101 expression is up-regulated in advanced stages of various human cancers (33–37).
No processed TSG101 pseudogene was detected in the human genome; however, a pseudogene has been identified in mouse, although its expression is very unlikely (38,39). To rule out possible aberrant TSG101 products generated from a processed TSG101 pseudogene, we performed a PCR analysis of human genomic DNA. We detected no processed TSG101 pseudogene with a primer set covering all the exons, whereas we observed the authentic TSG101 gene with primer sets that targeted exon 1 and exon 9 to exon 10 with intron 9 (Figure 2B, lane ‘Ge’). We thus confirmed that the observed aberrant TSG101 transcripts did not arise from a deleted gene or a processed pseudogene.
Ectopically expressed TSG101 cDNA can be spliced in cancer cells
If the aberrantly spliced TSG101 mRNA arises through re-splicing of a mature mRNA as we hypothesized, ectopically expressed TSG101 mRNA in cancer cells must be a substrate for further splicing. Consistent with this assumption, we found that an aberrantly spliced product with a 901-nt deletion that is equivalent to that from the endogenous TSG101 gene could be generated in a breast cancer cell line (MCF-7) transfected with a plasmid expressing an intronless TSG101 gene (cDNA). To detect the aberrant splicing of the TSG101 cDNA transcripts in vivo, we constructed three reporter minigenes fused to an enhanced GFP (EGFP) cDNA, so that the aberrant splicing could be monitored visually (Figure 2C).
MCF-7 cells transfected with the TSG101-EGFP plasmid, which included the full-length (exons 1–10) TSG101 mRNA, expressed GFP properly in the Golgi complex, as previously reported (Figure 2D, TSG101-EGFP) (40). The TSG101-EGFP[+] reporter was designed so that EGFP could not be translated unless the aberrant splicing event occurred. Remarkably, the transfected cells expressed GFP and the fused protein showed a diffuse cellular distribution (Figure 2D, TSG101-EGFP[+]). This demonstrates that in-frame aberrant splicing via the alternative splice sites took place on the transcript. The TSG101-EGFP[−] reporter was a frame-shifted negative control that did not result in the translation of EGFP after the aberrant splicing event. This reporter did not express GFP in the transfected cells (Figure 2D, TSG101-EGFP[−]), confirming that the GFP fluorescence in the TSG101-EGFP[+] cells was not attributable to leaky expression.
The transcripts generated from these reporter minigenes, together with those generated from the endogenous TSG101 gene, were then analyzed by RT-PCR (Figure 2E). The verified sequence of the spliced product from the transfected TSG101-EGFP[+] construct exactly matched the endogenous aberrant TSG101 mRNA (Supplementary Figure S2). These results demonstrate that cancer cells (MCF-7) can potentially splice mature TSG101 mRNA via the alternative sites.
Spliced TSG101 mRNA from the endogenous TSG101 gene is re-spliced in cancer cells
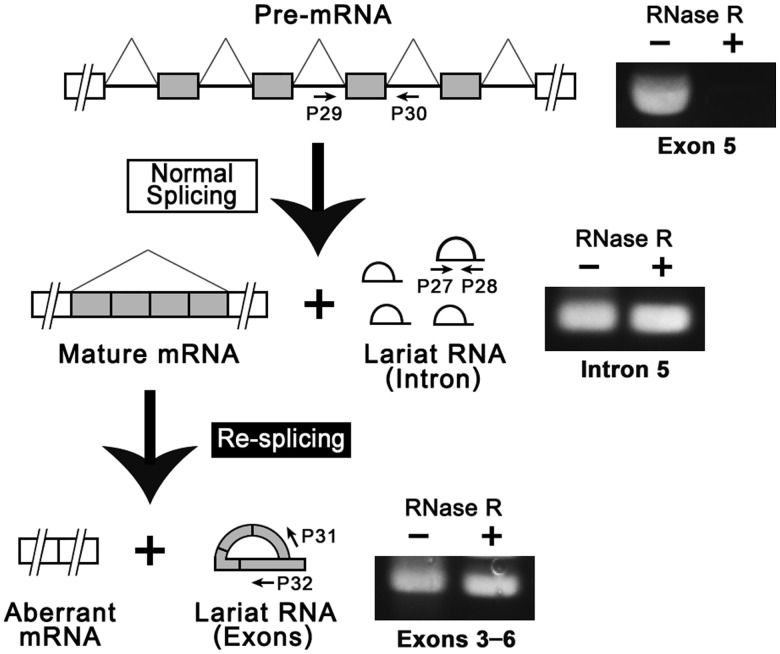
We next analyzed the aberrantly spliced products (with a 901-nt deletion) from the endogenous TSG101 gene, which could have been generated through two possible pathways (Figure 3A): by conventional alternative splicing in one step, or by splicing in two steps with re-splicing of the mature mRNA. In an effort to distinguish between these pathways, we searched for the lariat RNA molecules predicted to be generated in each case. If one-step aberrant splicing occurred on the nascent pre-mRNA, a large lariat product consisting of both exonic and intronic sequences would be predicted to exist, albeit transiently ([5] in Figure 3A). However, if aberrant re-splicing occurs on the endogenous mature mRNA, then a lariat product consisting solely of exonic sequences should be generated ([4] in Figure 3A).
To distinguish between the two pathways, we took advantage of a sensitive method that we previously developed to selectively detect splicing-specific lariat RNAs. The method uses RT-PCR with RNase R (E. coli 3′ to 5′ exoribonuclease)-treated total cellular RNA, which lacks the majority of linear RNA species (29). We demonstrated previously that RNase R thoroughly degrades the abundant linear RNAs, including rRNAs, tRNAs, pre-mRNAs and mRNAs, while preserving the loop portion of lariat RNAs and fully circular RNAs (29). Here, we show that the RT-PCR signal produced by the mature TSG101 mRNA was completely abolished by RNase R digestion before the RT-PCR ([2] in Figure 3A and B), whereas the signals detected from the excised lariat introns produced by the normal constitutive splicing of endogenous TSG101 pre-mRNA were fully resistant to RNase R digestion ([3] in Figure 3A and B). In this RNase R-treated RNA sample, we could not detect any RT-PCR signals corresponding to a large lariat RNA that included exons and flanking introns ([5] in Figure 3A and B). Therefore, the signals in the RNase R-untreated sample could be derived from the pre-mRNA [1]. In contrast, we did detect a lariat RNA consisting of exonic sequences only, because the RT-PCR signal remained after RNase R digestion ([4] in Figure 3A and B). These data provide compelling evidence that the two-step pathway involving a re-splicing event on the spliced TSG101 mRNA does indeed occur (Figure 3A). However, because the large lariat molecule predicted to be generated by the one-step pathway might be more susceptible to breakage, rendering it RNase R-sensitive, we cannot completely exclude the possibility it also occurs.
In theory, the exonic lariat RNA ([4] in Figure 3A) could also be produced from the large lariat RNA [5] through re-splicing of its intron portions. If this were the case, the large lariat RNA [5] must exist, albeit transiently, as the precursor of the exonic lariats [4]. We therefore performed RT-PCR at very high cycle number (35 cycles followed by 35 cycles of nested PCR). As no signal for the large lariat RNA [5] was detected, whereas all other predicted lariat RNAs in the total RNA sample were detected (Figure 3B) (29), we believe it is highly unlikely that this is the pathway through which the exonic lariat is generated. Our finding that the ectopically expressed intronless TSG101 cDNA (Figure 2A) was spliced at exactly the same alternative splice sites in the same cells provides further evidence in support of a re-splicing event on the constitutively spliced TSG101 mRNA (Figure 2E and Supplementary Figure S2).
Exonic lariat was verified as a re-spliced product from TSG101 mRNA
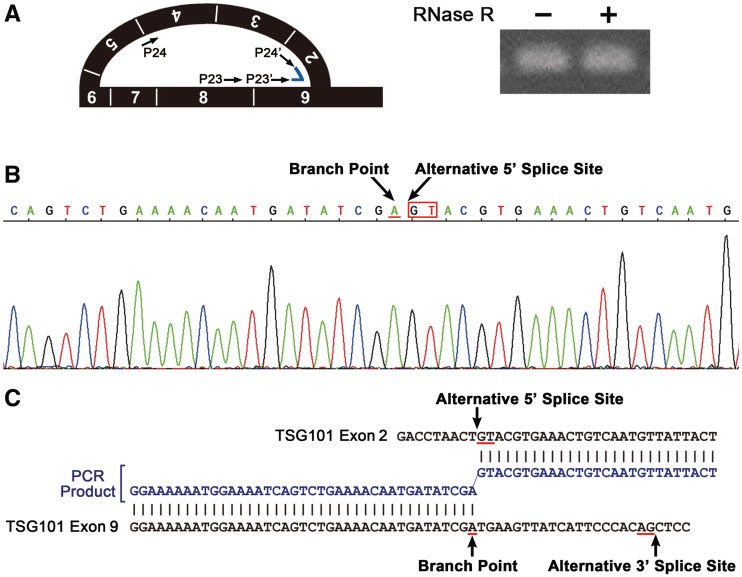
Because RNase R-digested RNA samples might contain not only lariat RNAs but also circular RNAs (29), we further confirmed that the exonic lariat product has the canonical structure through RT-PCR amplification across the branch site, an established technique for identifying trace amounts of splicing-specific lariat molecules (29,41–43).
We performed the assay using a DNA primer set that targeted the branched exon junction (Figure 4A, left panel). We observed a discrete RT-PCR signal of 112 bp, which was resistant to RNase R digestion (Figure 4A, right panel), indicating that it is derived from an intact lariat structure, not a cleaved lariat or Y-shaped structure (29). Sequencing of the PCR product revealed that the fragment contained the junction between the alternative 5′ splice site and the branch point ‘A’ nucleotide located 19-nt upstream from the alternative 3′ splice site (Figure 4B and C), verifying its identity as a bona fide exonic lariat RNA excised by re-splicing.
Altogether, we have provided compelling evidence for an aberrant re-splicing event that occurs on the constitutively spliced TSG101 mRNA in cancer cells.
A second mRNA re-splicing event was identified on the FHIT pre-mRNA
One of the major aberrant splicing events on the FHIT pre-mRNA in cancers generates an mRNA with a 412-nt deletion, extending from exon 3 to exon 6 (Figure 5A and B). This aberrant splicing eliminates a huge 1 189 164-nt span of the pre-mRNA sequence, which includes four exons and five flanking huge introns (Figure 5A) (14,24). By applying the same strategy and logic used in the analysis of TSG101, we show that the aberrant FHIT mRNA is also the product of re-splicing. In this case, the initial constitutive splicing events greatly shorten the distance between the activated 5′ and 3′ splice sites joined to produce the final mRNA (to <1/2800).
We first confirmed that the endogenous generation of this aberrant FHIT mRNA was recapitulated in various cancer cell lines, including MCF-7, CaSki, HepG2 and HEK293 cells, whereas it was not detected in normal cells (HMEC) (Figure 5B, right panel; data not shown). We then showed that breast cancer cells (MCF-7), transfected with an intronless FHIT-EGFP construct, were able to splice the FHIT-EGFP transcripts at the exonic 5′ and 3′ splice sites, exactly at the 5′ end of exon 3 and the 3′ end of exon 6, respectively (Figure 5C). These results indicate that cancer cells can potentially splice mature FHIT mRNA via exonic splice sites, which is consistent with but not sufficient to prove the occurrence of re-splicing on endogenous FHIT mRNA. Therefore, we assayed for two-step versus one-step pathway-specific splicing products from the endogenous FHIT gene by RT-PCR (Figure 6). We were able to detect an RNase R-resistant RT-PCR signal for the excised lariat intron 5 (middle panel), whereas no RNase R-resistant RT-PCR signal for the predicted large lariat RNA containing exon 5 and the flanking introns was observed (top panel). RT-PCR performed under the same conditions did allow detection of a lariat RNA consisting of the entire exon 3 to exon 6 region in the RNase R-treated sample (lower panel). This experiment also confirmed that there are no introns between these exons in the lariat form, because intron 3 (∼220 kb), intron 4 (∼286 kb) and intron 5 (∼523 kb) are huge. It is highly unlikely that a pathway exists that generates exonic lariats from a large lariat containing exons and introns because no such large lariat, which must exist as an intermediate, was detected, even when a large number of PCR cycles was used. Finally, the canonical lariat structure, and the absence of a circular structure, was verified by RT-PCR across the branch site, followed by sequencing (Figure 7). Taken together, these data indicate that a re-splicing event occurs on the spliced FHIT mRNA, while providing no evidence for one-step direct splicing of the FHIT pre-mRNA.
The discovery of a second case of mRNA re-splicing supports the generality of this pathway, and argues against the possibility that mis-splicing is unique to a single gene. An extensive analysis of exonic lariat molecules in the whole human transcriptome is underway.
DISCUSSION
Re-splicing of constitutively spliced mRNA is a novel pathway of multi-step splicing
Previously, two cases of multi-step splicing that occur on particular pre-mRNAs were reported (Figure 8, left column). (i) ‘Recursive splicing’ in the fruit fly Ultrabithorax (Ubx) gene and several other genes, i.e. the stepwise removal of introns by sequential re-splicing at composite 3′/5′ splice sites (0-nt length exons) (42–44); to date, recursive splicing has not been found in human or other vertebrate cells. Interestingly, the alternative use, but not the sequential use, of a composite 3′/5′ splice site, termed dual-specificity splice site, was reported in both human and mouse (45); in contrast to recursive splicing or the events described here, this is not multi-step splicing. (ii) ‘Intrasplicing’ in the human EPB41 gene (encoding the 4.1R protein), i.e. prerequisite internal splicing followed by specific external splicing (joining the far upstream promoter to an alternative 3′ splice site) (46).
Figure 8.
Different types of multi-step pre-mRNA splicing pathways. ‘Recursive splicing’ and ‘intrasplicing’ have been previously identified in Drosophila Ubx pre-mRNA and human EPB41 pre-mRNA, respectively (42,44,46). Novel multi-step splicing pathways were identified in human TSG101 and FHIT pre-mRNAs (right column), which are distinctive from the previously known multi-step splicing (left column). Each pathway of multi-step splicing is represented here with minimal numbers of exons/introns. 5′ and 3′ denote the 5′ splice site and the 3′ splice site, respectively. The black and gray colors indicate the active and inactive splice sites, respectively, in the each process.
In the known instances of pre-mRNA re-splicing, intronic sequences are partially removed to generate intermediates that are eventually spliced to produce the final mature mRNAs (Figure 8, left column). A specific intronic element, i.e. a composite 3′/5′ splice site, is essential for the subsequent splicing step in the case of ‘recursive splicing’. In contrast, we demonstrate that normal constitutive splicing occurs on TSG101 and FHIT pre-mRNAs and that the fully exonic mRNAs produced are substrates for subsequent aberrant re-splicing (Figure 8, right column). The constitutive splicing removes many competitive authentic splice sites, which should be important to allow activation of suboptimal exonic alternative splice sites on the mature mRNAs to produce the final aberrant mRNAs with internal deletions. All of the previously reported instances of pre-mRNA re-splicing take place on particular pre-mRNAs in normal cells, whereas mRNA re-splicing can be induced or activated on any pre-mRNAs under the abnormal or deregulated conditions in cancer cells. Considering the evidence together, we infer that the re-splicing of constitutively spliced mRNA is a distinct pathway of multi-step splicing. A fascinating question that remains to be answered is what causes these mature mRNAs to remain substrates for the splicing machinery, the identity of which is currently under investigation.
Although both involve re-splicing, from the mechanistic viewpoint of splice-site activation, the pathways that generate aberrant TSG101 and FHIT mRNAs differ. In FHIT mRNA re-splicing, the new 5′ and 3′ splice sites are regenerated at the exon–exon junctions by the preceding constitutive splicing event (Figure 8, right column). Therefore, this process is analogous to ‘recursive splicing’ of the Ubx pre-mRNA, in which the first upstream splicing event creates a composite 5′ splice site with the joined upstream exon. In TSG101 mRNA re-splicing, the activated alternative splice sites are not altered by the preceding constitutive splicing event (right column); therefore, this process is similar to ‘intrasplicing,’ in which the distal splice site pair is already present in the pre-mRNA. All these types of multi-step splicing may partly share a common mechanism, which remains a fascinating issue to be explored.
Functional and physiological implications of mRNA re-splicing
The re-spliced TSG101 mRNA contains a premature termination codon (PTC) in exon 9, and the re-spliced FHIT mRNA lacks an initiation codon for the translation. Therefore, these particular aberrant mRNAs do not encode the corresponding proteins. Thus, the functional contribution of these aberrant mRNAs to tumorigenesis, if any, would most likely arise through negative regulation or partial silencing of the genes. Endogenous aberrant TSG101 mRNA, including the PTC, may be subject to nonsense-mediated mRNA decay (NMD), which might account for the low yield of aberrant TSG101 splicing we observed in cultured cells.
The unprecedented pathway we have uncovered, re-splicing after constitutive splicing, might generally account for particular types of functional alternative splicing that occur over very long distances, eliminating multiple exons with their surrounding huge introns (30). The fact that aberrant mRNA re-splicing is induced in cancer cells implies that a mechanism controlling re-splicing operates in normal cells. Indiscriminate re-splicing in cancer cells could involve not only the deregulation of splicing but also deficiencies in NMD and mRNA export, which could allow further splicing of mRNAs containing potential splice sites. This scenario is consistent with the observation that aberrant proteins accumulate globally in cancer cells, irrespective of the corresponding genomic mutations [reviewed in (1,47)].
Our experimental system provides a potentially useful tool to investigate the presumptive mechanism that prevents ‘extra’ splicing of mRNAs, which could be important for the quality control of mRNAs to ensure that they can serve as proper blueprints for proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2, Supplementary Table 1 and Supplementary Materials and Methods.
FUNDING
Grants-in-Aid for Challenging Exploratory Research, Japan Society for the Promotion of Science (JSPS) [21657033 and 23651198 to T.K., 20651051 to A.M.]; Grant-in-Aid for Scientific Research on Innovative Areas ‘Diversity and asymmetry achieved by RNA program’, Japan Ministry of Education, Culture, Sports, Science and Technology (MEXT) [23112722 to A.M.]; Program for the Strategic Research Foundation at Private Universities, MEXT (to A.M., in part). Funding for the open access charge: Grant-in-Aid, MEXT [23112722].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs A. Malhotra and R. Reeves for providing the purified recombinant RNase R and Hs 578T cell line, respectively. We thank Drs G.R. Screaton, J. Wang and M.Q. Zhang for their constructive idea in the initial stages of this study; Drs R.J. Roberts and X. Roca for their critical reading of the manuscript; Drs M. Garcia-Blanco, A.R. Krainer and C.B. Burge for their helpful discussions before publication; and Drs R.J. Roberts and T. Tsukahara for their generous support and encouragement.
REFERENCES
- 1.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 2.Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 3.Pajares MJ, Ezponda T, Catena R, Calvo A, Pio R, Montuenga LM. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8:349–357. doi: 10.1016/S1470-2045(07)70104-3. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Cohen SN. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 5.Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka N, Kyuuma M, Sugamura K. Endosomal sorting complex required for transport proteins in cancer pathogenesis, vesicular transport, and non-endosomal functions. Cancer Sci. 2008;99:1293–1303. doi: 10.1111/j.1349-7006.2008.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 8.Barnes LD, Garrison PN, Siprashvili Z, Guranowski A, Robinson AK, Ingram SW, Croce CM, Ohta M, Huebner K. Fhit, a putative tumor suppressor in humans, is a dinucleoside 5′,5′″-P1,P3-triphosphate hydrolase. Biochemistry. 1996;35:11529–11535. doi: 10.1021/bi961415t. [DOI] [PubMed] [Google Scholar]
- 9.Hassan MI, Naiyer A, Ahmad F. Fragile histidine triad protein: structure, function, and its association with tumorogenesis. J. Cancer Res. Clin. Oncol. 2010;136:333–350. doi: 10.1007/s00432-009-0751-9. [DOI] [PubMed] [Google Scholar]
- 10.Saldivar JC, Shibata H, Huebner K. Pathology and biology associated with the fragile FHIT gene and gene product. J. Cell. Biochem. 2010;109:858–865. doi: 10.1002/jcb.22481. [DOI] [PubMed] [Google Scholar]
- 11.Martin J, St-Pierre MV, Dufour JF. Hit proteins, mitochondria and cancer. Biochim. Biophys. Acta. 2011;1807:626–632. doi: 10.1016/j.bbabio.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Lee MP, Feinberg AP. Aberrant splicing but not mutations of TSG101 in human breast cancer. Cancer Res. 1997;57:3131–3134. [PubMed] [Google Scholar]
- 13.Sun Z, Pan J, Bubley G, Balk SP. Frequent abnormalities of TSG101 transcripts in human prostate cancer. Oncogene. 1997;15:3121–3125. doi: 10.1038/sj.onc.1201521. [DOI] [PubMed] [Google Scholar]
- 14.Gayther SA, Barski P, Batley SJ, Li L, de Foy KA, Cohen SN, Ponder BA, Caldas C. Aberrant splicing of the TSG101 and FHIT genes occurs frequently in multiple malignancies and in normal tissues and mimics alterations previously described in tumours. Oncogene. 1997;15:2119–2126. doi: 10.1038/sj.onc.1201591. [DOI] [PubMed] [Google Scholar]
- 15.Oh Y, Proctor ML, Fan YH, Su LK, Hong WK, Fong KM, Sekido YS, Gazdar AF, Minna JD, Mao L. TSG101 is not mutated in lung cancer but a shortened transcript is frequently expressed in small cell lung cancer. Oncogene. 1998;17:1141–1148. doi: 10.1038/sj.onc.1202029. [DOI] [PubMed] [Google Scholar]
- 16.Wagner KU, Dierisseau P, Rucker EB, III, Robinson GW, Hennighausen L. Genomic architecture and transcriptional activation of the mouse and human tumor susceptibility gene TSG101: common types of shorter transcripts are true alternative splice variants. Oncogene. 1998;17:2761–2770. doi: 10.1038/sj.onc.1202529. [DOI] [PubMed] [Google Scholar]
- 17.Ferrer M, Lopez-Borges S, Lazo PA. Expression of a new isoform of the tumor susceptibility TSG101 protein lacking a leucine zipper domain in Burkitt lymphoma cell lines. Oncogene. 1999;18:2253–2259. doi: 10.1038/sj.onc.1202551. [DOI] [PubMed] [Google Scholar]
- 18.Klaes R, Kloor M, Willeke F, Melsheimer P, von Knebel Doeberitz M, Ridder R. Significant increase of a specific variant TSG101 transcript during the progression of cervical neoplasia. Eur. J. Cancer. 1999;35:733–737. doi: 10.1016/s0959-8049(99)00016-7. [DOI] [PubMed] [Google Scholar]
- 19.Lin SF, Lin PM, Liu TC, Chang JG, Sue YC, Chen TP. Clinical implications of aberrant TSG101 transcripts in acute myeloblastic leukemia. Leuk. Lymphoma. 2000;36:463–466. doi: 10.3109/10428190009148393. [DOI] [PubMed] [Google Scholar]
- 20.McIver B, Grebe SK, Wang L, Hay ID, Yokomizo A, Liu W, Goellner JR, Grant CS, Smith DI, Eberhardt NL. FHIT and TSG101 in thyroid tumours: aberrant transcripts reflect rare abnormal RNA processing events of uncertain pathogenetic or clinical significance. Clin. Endocrinol. 2000;52:749–757. [PubMed] [Google Scholar]
- 21.Wang NM, Chang JG, Liu TC, Lin SF, Peng CT, Tsai FJ, Tsai CH. Aberrant transcripts of FHIT, TSG101 and PTEN/MMAC1 genes in normal peripheral mononuclear cells. Int. J. Oncol. 2000;16:75–80. [PubMed] [Google Scholar]
- 22.Lee SH, Kim CJ, Park HK, Koh JW, Cho MH, Baek MJ, Lee MS. Characterization of aberrant FHIT transcripts in gastric adenocarcinomas. Exp. Mol. Med. 2001;33:124–130. doi: 10.1038/emm.2001.22. [DOI] [PubMed] [Google Scholar]
- 23.Matthews CP, Shera K, Kiviat N, McDougall JK. Expression of truncated FHIT transcripts in cervical cancers and in normal human cells. Oncogene. 2001;20:4665–4675. doi: 10.1038/sj.onc.1204622. [DOI] [PubMed] [Google Scholar]
- 24.Kraggerud SM, Aman P, Holm R, Stenwig AE, Fossa SD, Nesland JM, Lothe RA. Alterations of the fragile histidine triad gene, FHIT, and its encoded products contribute to testicular germ cell tumorigenesis. Cancer Res. 2002;62:512–517. [PubMed] [Google Scholar]
- 25.Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahashi K, Masuda A, Matsuura T, Shinmi J, Zhang Z, Takeshima Y, Matsuo M, Sobue G, Ohno K. In vitro and in silico analysis reveals an efficient algorithm to predict the splicing consequences of mutations at the 5′ splice sites. Nucleic Acids Res. 2007;35:5995–6003. doi: 10.1093/nar/gkm647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartmann L, Theiss S, Niederacher D, Schaal H. Diagnostics of pathogenic splicing mutations: does bioinformatics cover all bases? Front. Biosci. 2008;13:3252–3272. doi: 10.2741/2924. [DOI] [PubMed] [Google Scholar]
- 28.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng CL, Kwon YS, Li HR, Zhang K, Coutinho-Mansfield G, Yang C, Nair TM, Gribskov M, Fu XD. MAASE: an alternative splicing database designed for supporting splicing microarray applications. RNA. 2005;11:1767–1776. doi: 10.1261/rna.2650905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roca X, Sachidanandam R, Krainer AR. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003;31:6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vorechovsky I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu RT, Huang CC, You HL, Chou FF, Hu CC, Chao FP, Chen CM, Cheng JT. Overexpression of tumor susceptibility gene TSG101 in human papillary thyroid carcinomas. Oncogene. 2002;21:4830–4837. doi: 10.1038/sj.onc.1205612. [DOI] [PubMed] [Google Scholar]
- 34.Young TW, Rosen DG, Mei FC, Li N, Liu J, Wang XF, Cheng X. Up-regulation of tumor susceptibility gene 101 conveys poor prognosis through suppression of p21 expression in ovarian cancer. Clin. Cancer Res. 2007;13:3848–3854. doi: 10.1158/1078-0432.CCR-07-0337. [DOI] [PubMed] [Google Scholar]
- 35.Oh KB, Stanton MJ, West WW, Todd GL, Wagner KU. Tsg101 is upregulated in a subset of invasive human breast cancers and its targeted overexpression in transgenic mice reveals weak oncogenic properties for mammary cancer initiation. Oncogene. 2007;26:5950–5959. doi: 10.1038/sj.onc.1210401. [DOI] [PubMed] [Google Scholar]
- 36.Ma XR, Edmund Sim UH, Pauline B, Patricia L, Rahman J. Overexpression of WNT2 and TSG101 genes in colorectal carcinoma. Trop. Biomed. 2008;25:46–57. [PubMed] [Google Scholar]
- 37.Liu DC, Yang ZL, Jiang S. Identification of PEG10 and TSG101 as carcinogenesis, progression, and poor-prognosis related biomarkers for gallbladder adenocarcinoma. Pathol. Oncol. Res. 2011;17:859–866. doi: 10.1007/s12253-011-9394-7. [DOI] [PubMed] [Google Scholar]
- 38.Wagner KU, Dierisseau P, Rucker EB, III, Robinson GW, Hennighausen L. Genomic architecture and transcriptional activation of the mouse and human tumor susceptibility gene TSG101: common types of shorter transcripts are true alternative splice variants. Oncogene. 1998;17:2761–2770. doi: 10.1038/sj.onc.1202529. [DOI] [PubMed] [Google Scholar]
- 39.Wagner KU, Dierisseau P, Hennighausen L. Assignment of the murine tumor susceptibility gene 101 (tsg101) and a processed tsg101 pseudogene (tsg101-ps1) to mouse chromosome 7 band B5 and chromosome 15 band D1 by in situ hybridization. Cytogenet. Cell Genet. 1999;84:87–88. doi: 10.1159/000015221. [DOI] [PubMed] [Google Scholar]
- 40.Xie W, Li L, Cohen SN. Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc. Natl Acad. Sci. USA. 1998;95:1595–1600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel J, Hess WR, Borner T. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170:661–674. doi: 10.1534/genetics.104.039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conklin JF, Goldman A, Lopez AJ. Stabilization and analysis of intron lariats in vivo. Methods. 2005;37:368–375. doi: 10.1016/j.ymeth.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Hatton AR, Subramaniam V, Lopez AJ. Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon–exon junctions. Mol. Cell. 1998;2:787–796. doi: 10.1016/s1097-2765(00)80293-2. [DOI] [PubMed] [Google Scholar]
- 45.Zhang C, Hastings ML, Krainer AR, Zhang MQ. Dual-specificity splice sites function alternatively as 5′ and 3′ splice sites. Proc. Natl Acad. Sci. USA. 2007;104:15028–15033. doi: 10.1073/pnas.0703773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parra MK, Tan JS, Mohandas N, Conboy JG. Intrasplicing coordinates alternative first exons with alternative splicing in the protein 4.1R gene. EMBO J. 2008;27:122–131. doi: 10.1038/sj.emboj.7601957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J. Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 48.Gao K, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 2008;36:2257–2267. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.