Abstract
Fine-tuning of inflammatory responses by microRNAs (miRNAs) is complex, as they can both enhance and repress expression of pro-inflammatory mediators. In this study, we investigate inflammatory responses following global miRNA depletion, to better define the overall contribution of miRNAs to inflammation. We demonstrate that miRNAs positively regulate Toll-like receptor signaling using inducible Dicer1 deletion and global miRNA depletion. We establish an important contribution of miR-19b in this effect, which potentiates nuclear factor-κB (NF-κB) activity in human and mouse cells. Positive regulation of NF-κB signaling by miR-19b involves the coordinated suppression of a regulon of negative regulators of NF-κB signaling (including A20/Tnfaip3, Rnf11, Fbxl11/Kdm2a and Zbtb16). Transfection of miR-19b mimics exacerbated the inflammatory activation of rheumatoid arthritis primary fibroblast-like synoviocytes, demonstrating its physiological importance in the pathology of this disease. This study constitutes, to our knowledge, the first description of a miR-19 regulon that controls NF-κB signaling, and suggests that targeting this miRNA and linked family members could regulate the activity of NF-κB signaling in inflammation.
INTRODUCTION
MicroRNAs (miRNAs) are critical regulators of gene expression, and exert their activity through the modulation of target mRNA stability or translation efficiency. These small RNAs (∼22 nt) result from the processing of longer transcripts with hairpin-like secondary structure by two successive endonucleases, Drosha and Dicer (1). A miRNA selective activity on a given target mRNA relies on the complementarity of the 5′ ‘seed region’ of the miRNA (2) to a cognate mRNA. Because such a seed region is only 6–8 nt long, seed motifs are frequently repeated in the genome and one miRNA can regulate hundreds of target mRNAs simultaneously (3). However, the miRNA effect on individual protein levels is generally relatively modest (3). Nevertheless, the concurrent regulation of several genes in a given pathway by a single miRNA can have a cumulative effect on the biological process emanating from this pathway (4).
The nuclear factor-κB (NF-κB) family of transcription factors is critical to the rapid response to pathogen infection and pro-inflammatory stimuli, and is sequestered in the cytoplasm by inhibitor κB (IκB) proteins under homeostatic conditions. In the canonical NF-κB pathway, degradation of IκB proteins results in the nuclear translocation of NF-κB subunits and induction of target genes. A complex regulation of signal transduction subsequently operates to ensure the return to homeostasis. In the case of TNF-α signaling, this can be achieved by the termination of signal transduction from the TNF receptor 1 (TNFR1) through the inducible degradation of receptor-interacting protein 1 (RIP1), which is an essential adaptor of TNFR1-dependent NF-κB activation. This inducible degradation of RIP1 relies on the dual activity of the A20/Tnfaip3-ubiquitin editing complex (Tnfaip3/Itch/Rnf11/Tax1bp1/Ccdc50/Tnip1), which replaces the K63-linked polyubiquitin chains of RIP1 with K48-linked polyubiquitin chains, to induce its proteosomal degradation (5). This tight control of NF-κB activity is critical in homeostasis and aberrant NF-κB activation can result in tumor initiation and progression through the production of pro-inflammatory cytokines (e.g. TNF-α, IL-6 and IL-8) (6). Not too surprisingly given its critical role in the termination of NF-κB signal transduction, mutations in A20 are associated with aberrant NF-κB signaling and cancer development (5,7). For example, A20 is frequently inactivated by somatic mutations and/or deletions in human B-cell lymphomas (7).
It is now well accepted that NF-κB signaling is under tight miRNA regulation. For instance, miR-9 is canonically induced by NF-κB following Toll-like receptor (TLR) 4 activation in human neutrophils and monocytes, and acts to repress NF-κB signaling through direct targeting of p50 mRNA (8). Similarly, miR-146a induction by NF-κB negatively represses TLR signaling through its effect on TRAF6, IRAK1 and IRAK2 (9). Regulation is not restricted to canonical NF-κB signaling, as shown with the repression of NF-κB-inducing kinase, which is critical to the non-canonical NF-κB signaling, by miR-31 (10). Moreover, the effect of miRNAs on NF-κB is not limited to the repression of inflammation. Another NF-κB-induced miRNA, miR-301, was shown to positively contribute to NF-κB signaling through its repressive effect on the NF-κB-repressing factor gene (11). Similarly, NF-κB-dependent repression of let-7 processing results in stabilized IL-6 mRNA and increased cytokine production (12). Although evidence points clearly to an important role for miRNAs in the modulation of NF-κB signaling, the opposing effects of these miRNAs led us to investigate the net contribution of miRNAs in the regulation of NF-κB signaling and inflammation.
We show that global miRNA depletion results in the decreased production of inflammatory mediators, and have uncovered a novel role for miR-19b in the potentiation of inflammation through its coordinated activity on repressors of NF-κB signaling, including members of the A20/Tnfaip3-ubiquitin editing complex.
MATERIALS AND METHODS
Ethics statement
The use of human tissues, animals and experimental procedures was approved by the Monash Medical Centre Ethics Committee (references MMCA/2008/26/BC and MMCA 2007/07) and the Human Research Ethics Committee. Rheumatoid arthritis (RA) patients fulfilled the American College of Rheumatology (ACR) criteria for the classification of RA (13). RA primary fibroblast-like synoviocytes (FLS) were obtained from surgical specimens of synovial tissue and cultured as previously described (14).
Cell culture
FLS cells were passaged until P8 in RPMI 1640+l-glutamine medium (Invitrogen Corporation) complemented with 1× antimycotic and 10% FBS (ICPBio Ltd; referred to as complete RPMI). Dicer1flox/floxxCre/Esr1 mice and SV40-mouse embryonic fibroblasts (MEFs) from these mice were previously reported (15). Bone marrow was isolated from the femurs of Dicer1flox/floxxCre/Esr1 mice, and primary bone marrow-derived macrophages (BMDMs) were differentiated in 20% L929-cell-conditioned medium. BMDMs, Dicer1flox/floxxCre/Esr1 and Dicer1wt/floxxCre/Esr1 SV40-MEFs, HEK293T, HEK293-TLR3 and HeLa cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen Corporation, Carlsbad, CA, USA) supplemented with 10% sterile FBS (ICPBio Ltd, Auckland, New Zealand) and 1× antibiotic/antimycotic (Invitrogen; referred to as complete DMEM). Pam3CSK4 (TLR2/1 agonist, Invivogen, San Diego, CA, USA), synthetic monophosphoryl lipid A (Lipid A, Invivogen), LPS from E. coli Serotype O111:B4 (TLR4 agonist, Enzo Life Sciences, Farmingdale, NY, USA), Poly(I:C) (TLR3 agonist, Roche, Nutley, NJ, USA) and ODN1826 (mouse TLR9 agonist, Invivogen) were used at indicated concentrations.
4-hydroxy tamoxifen treatment of the cells
4-Hydroxy tamoxifen (OHT; Sigma Aldrich, St. Louis, MO, USA) was used as previously reported (15). For OHT treatment of SV40-MEFs, the cells were plated at low density (20 000 cells per well of a 6-well plate) and treated with 500 nM OHT for 16 h (day 0). On day 1, the cells were rinsed with fresh complete DMEM, and left to expand further. The cells were collected from each well on day 3 with 300 µl TrypLE™ Express Stable Trypsin (Invitrogen Corporation) and sub-plated into 96 wells. Synthetic miRNA mimics were reverse-transfected at this stage in relevant experiments (Figure 3C). On day 4, the cells were rinsed with 150 µl complete DMEM, and stimulated with 1.5 µl of diluted Lipid A per well, to obtain a final concentration of 200 ng/ml Lipid A. For OHT stimulation of BMDMs, the cells were washed on day 3 with fresh medium supplemented with 20% L929-cell-conditioned medium and 500 nM OHT. The cells were rinsed to remove OHT on day 5, and collected on day 7 before plating in 96-well plates—20% L929-cell-conditioned medium. The BMDMs were subsequently stimulated on day 8 (i.e. 5 days following OHT treatment).
Figure 3.
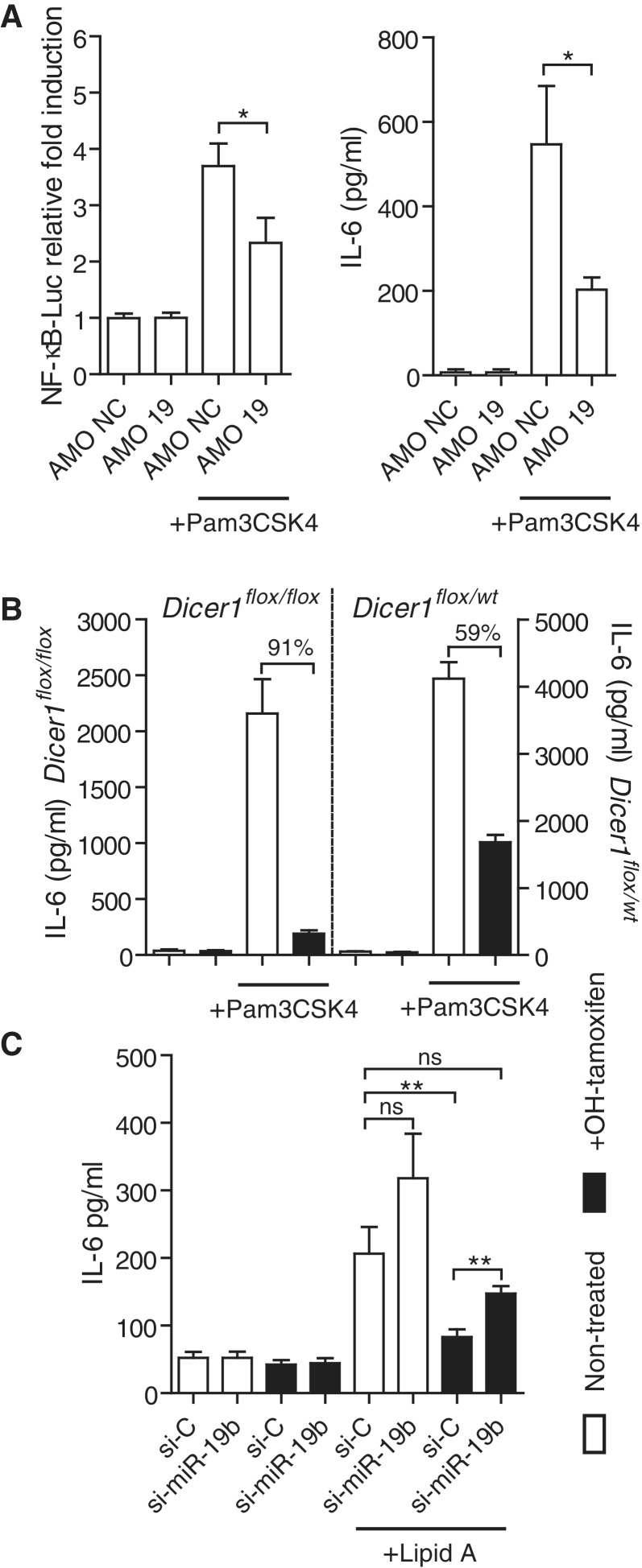
miR-19b is a critical component of the positive regulation of NF-κB signaling by miRNAs. (A) SV40-MEFs were transfected with an NF-κB-luciferase reporter, and 10 nM of the indicated AMO (AMO NC: non-target control). The cells were treated with 10 ng/ml Pam3CSK4 for 6 h. Cell lysates and supernatants were analyzed for NF-κB-luciferase activity (left) and IL-6 production (right), respectively. Ratios of firefly luciferase/Renilla luciferase levels in lysates were reported to ratios of non-treated condition for each AMO (left). The data are averaged from two independent experiments in biological triplicate. Two-tailed t-tests are shown (*P < 0.05). (B, C) Dicer1flox/floxxCre/Esr1+ and Dicer1flox/wtxCre/Esr1+ (B only) SV40-MEFs were treated (black bars) or untreated (white bars) for 16 h with 500 nM OHT, and expanded for a total of 4 days before stimulation with 10 ng/ml Pam3CSK4 (see ‘Materials and Methods’ section). IL-6 levels were measured 6 h after stimulation by ELISA. (B) Data shown are averaged from three independent experiments in biological triplicate, and fold repression of IL-6 production upon OHT treatment is indicated for each cell line. (C) Cells were transfected with 10 nM of indicated si-miRNA (si-C is siRNA control) on day 3 post-OHT treatment, and left for 16 h before Lipid A stimulation at 200 ng/ml. IL-6 levels were measured 6 h after stimulation by ELISA. The data are averaged from three independent experiments in biological triplicate with non-parametric Mann–Whitney U tests shown (*P < 0.05 and **P < 0.01). (A–C) SEM is shown.
Synthetic RNAs
AMO, small interfering RNA (siRNA) and si-miRNA duplexes were synthesized as single-stranded RNAs by Integrated DNA Technologies (IDT) with HPLC purification, and resuspended in duplex buffer (100 mM potassium acetate, 30 mM HEPES, pH 7.5, DNase–RNase free H2O). Annealing of siRNAs and si-miRNAs was performed as previously reported (15). si-miR-19b and si-miR-17 were synthesized as Dicer product duplexes, with a mutated sense strand perfectly matching to the miRNA guide strand:
si-miR-19b-S: 5′AGUUUUGCAUGGAUUUGCACAUU;
si-miR-19b-AS: 5′UGUGCAAAUCCAUGCAAAACUGA.
si-miR-17-S: 5′ACCUGCACUGUAAGCACUUUGUU;
si-miR-17-AS: 5′CAAAGUGCUUACAGUGCAGGUAG.
si-C is a 19 + 2 bp control siRNA duplex targeting human Lamin A/C (NM_170707):
si-C-S: 5′CUGGACUUCCAGAAGAACAdTdT;
si-C-AS: 5′UGUUCUUCUGGAAGUCCAGdTdT.
siRNF11 was synthesized as a Dicer substrate [based on (16)]:
siRNF11-FWD: 5′UAGGAUAGCUCAAAGAAUAGGUCdTdT;
siRNF11-REV: 5′AAGACCUAUUCUUUGAGCUAUCCUAAU.
siEGFPN, a Dicer substrate siRNA targeting EGFP (15) was used as a control for siRNF11. AMOs were designed and synthesized as reverse complements to target miRNAs, with 2′-O-methyl RNA (2′OMe) groups and phosphodiester linkages and a non-nucleotide naphthyl-azo group chemical modifier (dubbed ‘ZEN’) (17).
AMOs are based on the following sequences:
non-targeting control, AMO NC: 5′GzCGUAUUAUAGCCGAUUAACGzA;
AMO 18(a): 5′CzUAUCUGCACUAGAUGCACCUUzA;
AMO 19(b): 5′UzCAGUUUUGCAUGGAUUUGCACzA;
AMO 17(5p): 5′CzUACCUGCACUGUAAGCACUUUzG;
AMO 92(a): 5′CzAGGCCGGGACAAGUGCAAUzA; where ‘z’ denotes the insertion of the ZEN modification between adjacent 2′OMe residues. Noteworthy, because of their close sequence homology, it is anticipated that these AMOs can affect all close family members (for instance, miR-19b AMO would also affect miR-19a activity).
Transfection of small RNAs/AMOs
For Figures 2B and C, 3A and C and 4B–F and Supplementary Figures S3 and S4, 1.35 µl of Lipofectamine 2000 (Invitrogen Corporation) was diluted in 150 µl of Opti-MEM and 1.5 µl of si/mi-RNA/AMO molecules (diluted to 4 µM in duplex buffer) was added. Following 20 min incubation, 50 µl of the Lipofectamine/RNA mix was added per well of a 96-well plate (three biological replicates per condition). A total of 150 µl of cells in suspension in DMEM + 10% FCS without antibiotics was added on the top of the Lipofectamine/RNA mix, giving a final 200 µl per well with 10 nM oligonucleotide. For Figure 5, 8 × 103 RA FLS cells were plated in 96 wells, 24 h before transfection. The cells were rinsed with 150 µl RPMI + 10% FCS without antibiotics per well at time of transfection. One microlitre of Lipofectamine RNAiMax (Invitrogen Corporation) diluted to 75 µl of Opti-MEM was combined to a mix of 75 µl Opti-MEM with 1.5 µl si/mi-RNA molecules (at 4 µM), and incubated 15 min. A total of 50 µl of the resulting mix was added to each well (resulting in a final 200 µl at 10 nM per well). For Figure 4F, 5.4 µl of Lipofectamine 2000 was diluted in 100 µl of Opti-MEM and 0.6 µl of siRNF11 or siEGFPN molecules (at 40 µM) was added. Following 20 min incubation, the 100 µl of resulting mix was added to 400000 HEK293T cells freshly plated in 2 ml DMEM + 10% FCS without antibiotics, in a 6-well plate. After 24 h, the cells were reverse transfected with 10 nM of AMO as indicated above.
Figure 2.
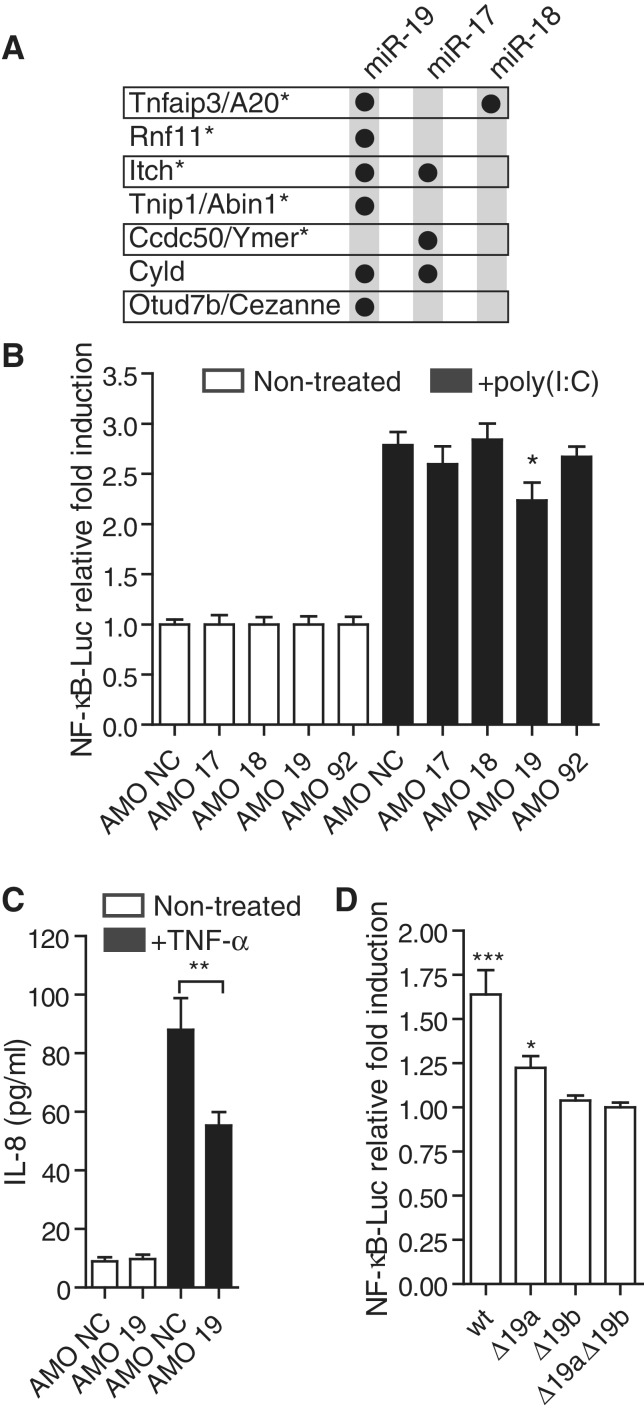
miR-19b is involved in the regulation of NF-κB signaling. (A) Targetscan (v6) (22) predictions of miR-17/20, miR-19 and miR-18 conserved targeting of known negative regulators of NF-κB signaling (5). The star denotes proteins proposed to belong to the A20 ubiquitin-editing complex (5). (B) HEK293 cells stably expressing TLR3 were transfected with an NF-κB-luciferase reporter, and 10 nM of the indicated antagomiR (AMO; AMO NC: non-target control). Cells were stimulated for 6 h with 10 μg/ml of poly(I:C) (black bars). Ratios of firefly luciferase/Renilla luciferase levels in lysates were reported to ratios of non-treated condition for each AMO. The data are averaged from three independent experiments in biological triplicate. (C) HEK293T cells were stimulated for 6 h with 10 ng/ml TNF-α (black bars) following transfection with 10 nM of the indicated AMO. Supernatants were collected and IL-8 levels measured by ELISA. The data are averaged from two independent experiments in biological triplicate. (B, C) Non-parametric Mann–Whitney U tests comparing AMO NC to AMO 19 condition are shown (*P < 0.05 and **P < 0.01). (D) HeLa cells were co-transfected with pN2-miR-17–19 b vectors and an NF-κB-/Renilla luciferase vector and cultured for 48 h. Ratios of firefly luciferase/Renilla luciferase levels in lysates were reported to ratios of Δ19aΔ19b vector condition (see ‘Materials and Methods’ section). Data shown are averaged from three independent experiments in biological triplicate with non-parametric Mann–Whitney U tests compared to Δ19aΔ19b vector condition (*P < 0.05 and ***P < 0.001). (B–D) SEM is shown.
Figure 4.
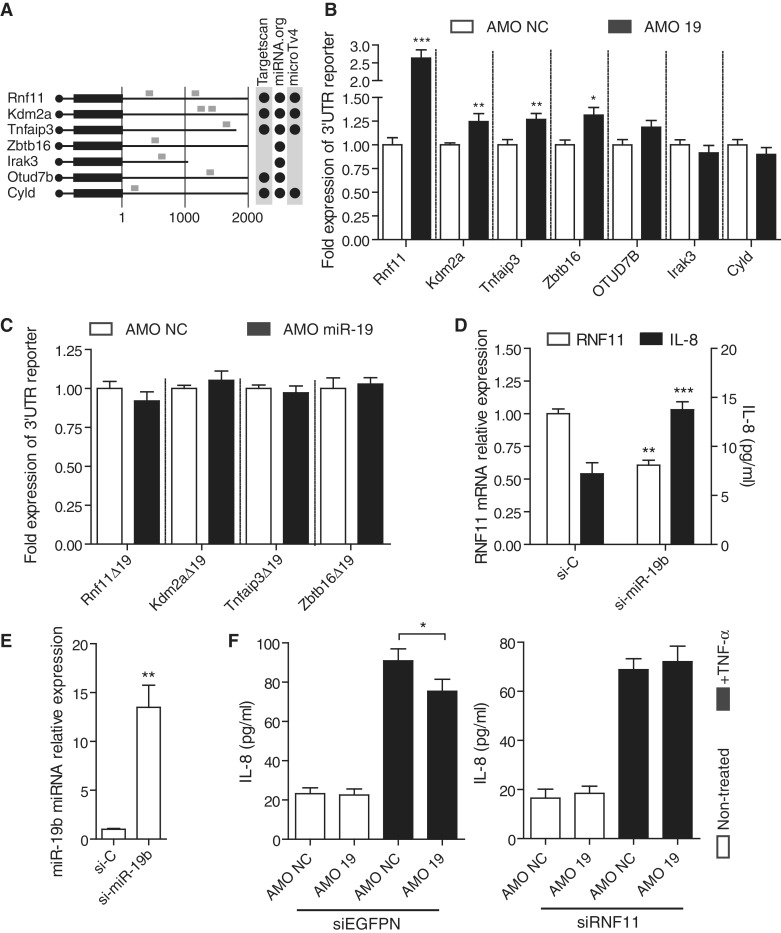
miR-19b controls several negative regulators of NF-κB signaling. (A) Schematic representation of the first 2000 nt of the 3′UTR from seven known regulators of NF-κB signaling (5,27–29). miR-19 predicted conserved target sites are indicated in grey along the murine 3′UTRs, using several prediction algorithms (22,30,31). (B, C) HEK293T cells transiently expressing each individual pRL-3′UTR Renilla reporter (B) or pRL-3′UTR-Δ19 mutated variants (C) were transfected with 10 nM of AMO NC or AMO 19 for 20 h. Ratios of Renilla luciferase/firefly luciferase levels in lysates were reported to ratios of AMO condition for each 3′UTR reporter. Data are averaged from three independent experiments in biological triplicate with non-parametric Mann–Whitney U tests compared to AMO conditions (*P < 0.05, **P < 0.01 and ***P < 0.001). (D, E) HeLa cells were transfected with 10 nM of si-miR-19b mimic or control siRNA (si-C) and incubated for 48 h. Total RNA was extracted from lysates and RNF11 mRNA (D) and miR-19b (E) levels measured by RT-qPCR, while supernatants were collected and IL-8 levels measured (D). Expression was normalized to GAPDH mRNA (D) and U6 RNA (E) and is shown relative to si-C condition. Data are averaged from three independent experiments in biological duplicate (RNF11 mRNA and miR-19b levels) or triplicate (IL-8 levels) with non-parametric Mann–Whitney U tests compared to si-C conditions (**P < 0.01 and ***P < 0.001). (F) HEK293T transfected with 10 nM of siEGFPN (left) or siRNF11 (right) for 24 h were transfected a second time with 10 nM of the indicated AMO for another 14 h. Supernatants were subsequently collected after 6 h of TNF-α stimulation at 20 ng/ml (black bars). Data are averaged from three independent experiments in biological triplicate and two-tailed t-test is shown (*P < 0.05). (B–F) SEM is shown.
Figure 5.
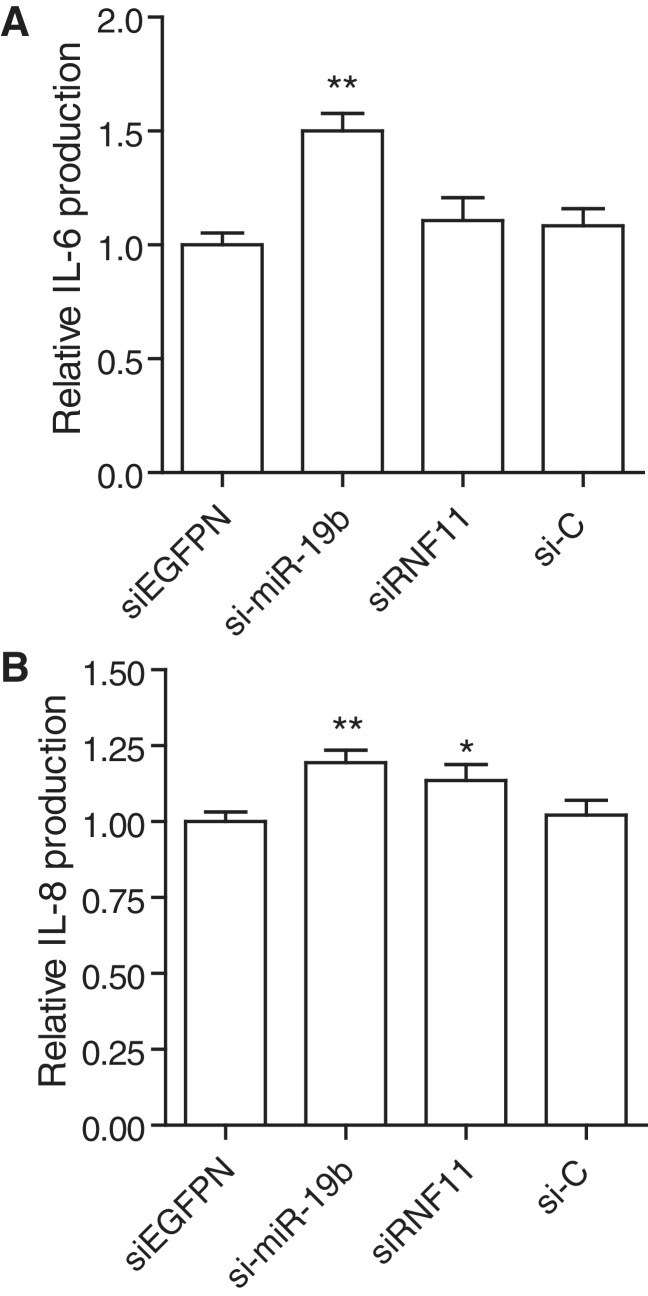
miR-19b transfection increases inflammation in primary human synovial fibroblasts. RA primary FLS were transfected with 10 nM of indicated RNA duplex (siEGFPN is used as control for siRNF11, both being Dicer substrates; see ‘Materials and Methods’ section), and incubated for 15 h. Supernatants were collected and assayed for IL-6 (A) and IL-8 (B) levels. Cytokine levels were normalized to that of siEGFPN condition to remove variations between independent experiments. The data are averaged from two independent experiments in biological triplicate, with Mann–Whitney U tests compared to siEGFPN condition (*P < 0.05 and **P < 0.01). (A–B) SEM is shown.
Detection of cytokines
Human IL-6 and IL-8, and mouse IL-6, IL-12 and TNF-α were measured using BD OptEIA ELISA sets according to the manufacturer’s instructions (BD Biosciences, San Jose, CA, USA). Tetramethylbenzidine substrate (Sigma Aldrich, St. Louis, MO, USA) was used for quantification of the cytokines on a Fluostar OPTIMA (BMG LABTECH, Offenburg, Germany) plate-reader.
Luciferase assays
3′UTRs from indicated genes were cloned into pRL-Spacer, a modified pRL-Control vector (18), expressing Renilla luciferase under the control of a CMV promoter. Cells were co-transfected with individual pRL-3′UTR vectors together with a firefly luciferase control vector. For NF-κB luciferase assays, pIL-8-Luc expressing firefly luciferase under the control of the human IL-8 promoter was co-transfected with pRL-Control.
Cloning of pN2-miR17∼19b vectors
pN2-miR17∼19b were created by sub-cloning the miR-17∼19b cassette (NR_027350, from position 1066 to 1721) from pMXs-17-19b puro (19) into pEGFP-N2 (Clontech, Mountain View, CA, USA)—both vectors being digested by BamHI and NotI. pN2-miR-17∼19b was modified by two successive rounds of site-directed mutagenesis using PfuTurbo polymerase (Agilent Technologies, Santa Clara, CA, USA) and DpnI digestion (New England Biolabs, Ipswich, MA, USA), to mutate the seed regions of miR-19a/b (GTGCAA becomes GacgAA; Supplementary Table S2, for primers used), resulting in pN2-miR17∼19bΔ19a, Δ19b or Δ19aΔ19b. pRL-Spacer used for the cloning of the 3′UTR was generated by inserting a synthetic short multi-cloning site region in between XbaI and NotI of pRL-Control (18) (Supplementary Methods and Supplementary Table S2, for primers).
Statistical analyses
Statistical analyses were carried out using Prism 5 (GraphPad Software Inc.). All statistical analyses rely on a minimum of six biological values (from a minimum of two independent experiments).
RESULTS
Global depletion of miRNAs is associated with decreased TLR signaling in bone marrow-derived macrophages
To investigate the overall impact of miRNAs in the modulation of inflammation, BMDMs were isolated from Dicer1flox/floxxCre/Esr1+ mice, in which the majority of the second RNase III domain of Dicer1 is deleted with OHT treatment of the cells (15). With the exceptions of miR-451 (20), miR-1225 and miR-1228 (21), all mammalian miRNAs require Dicer1 for their final maturation. Treatment of Dicer1flox/floxxCre/Esr1+ cells with OHT, therefore, results in gradual global depletion of miRNAs, and prevents de novo production of inducible miRNAs (15). Strikingly, a marked decreased production of IL-6, TNF-α and IL-12 was observed when these cells were stimulated with various TLR agonists (for TLR1/2, TLR4 and TLR9; Figure 1A). This defect in pro-inflammatory cytokine production was not seen in control BMDMs from Dicer1wt/wtxCre/Esr1− mice treated with OHT, suggesting the specific involvement of Dicer1 and miRNAs in this effect (Supplementary Figure S1). In accordance with our previous results, due to variable decay rates between miRNAs (15), Dicer1flox/floxxCre/Esr1+ BMDMs exhibited a 35–70% depletion of miRNAs 5 days after OHT-mediated Dicer1 deletion in more than 99% of the cells, as demonstrated with let-7i and miR-19b miRNAs, and Dicer1 mRNA levels (Figure 1B and C). These results strongly suggest that the overall effect of miRNAs is to potentiate pro-inflammatory cytokine production in response to TLR agonists.
Figure 1.
Global depletion of miRNAs results in decreased pro-inflammatory cytokine production. (A–C) BMDMs from Dicer1flox/floxxCre/Esr1+ mice were treated (black bars) or untreated (white bars) for 48 h with 500 nM OHT as described in the ‘Materials and Methods’. (A) The cells were treated with 10 ng/ml Pam3CSK4 (TLR1/2 agonist), 10 ng/ml LPS (TLR4 agonist) or 1 μM CpG oligonucleotides (TLR9 agonist) 5 days after initial OHT treatment. IL-6, TNF-α and IL-12 levels were measured in supernatants collected 6 h after TLR stimulation. Data are averaged from two (Pam3CSK4 and CpG conditions) and three (LPS condition) independent experiments in biological triplicate. Two-tailed t-tests are shown (*P < 0.05, **P < 0.01 and ***P < 0.001). (B, C) Total RNA was extracted from non-stimulated cells and miR-19 b/let-7i (B) and Dicer1 mRNA levels (C) were measured by RT-qPCR. Expression was normalized to U6 RNA (B) and Gapdh mRNA (C) and is shown relative to non-OHT treated condition. Data are averaged from three independent experiments. (A–C) SEM is shown.
miR-19b positively regulates NF-κB signaling
The finding that global miRNA depletion inhibited production of cytokines controlled by NF-κB suggested that relief of miRNA control of repressors of NF-κB signaling could be at play. In silico analyses of several known critical regulators of NF-κB signaling, including A20 (Tnfaip3), Cyld, and Cezanne (Otud7b) (5), revealed enriched targeting of miRNAs from the miR-17∼92 cluster (including miR-17/20, miR-19 and miR-18 sites; Figure 2A). Moreover, several members of the Tnfaip3-ubiquitin editing complex (Tnfaip3/Itch/Tnip1/Rnf11) (5) were predicted targets for miR-19 across species (Figure 2A).
To assess whether the miR-17∼92 cluster of miRNAs could be involved in the regulation of NF-κB signaling, we measured the responsiveness of an NF-κB luciferase reporter following AMO (miRNA antagonist)-mediated depletion of each family member of this miRNA cluster, in HEK293 TLR3 cells stimulated with poly(I:C). HEK293 cells have been shown previously to express the miR-17∼92 miRNA cluster in relatively high abundance (23). Although no significant effects were seen with miR-17, miR-18 and miR-92 AMOs, blocking of miR-19 resulted in a significant decrease in NF-κB luciferase induction following TLR3 activation (Figure 2B). Similarly, blocking of miR-19 significantly reduced production of IL-8 following TNF-α stimulation of HEK293T cells (Figure 2C). To delineate further the relative contribution of miR-19a and miR-19b in the regulation of NF-κB signaling, HeLa cells were transfected with over-expression vectors encoding a miR-17∼19b cassette (19), or mutated variants for miR-19a (Δ19a), miR-19b (Δ19b) or both (Δ19aΔ19b; see ‘Materials and Methods’ section). Basal NF-κB activity following miR-19b mutation was indistinguishable from that of the double mutant Δ19aΔ19b, while both wild-type (WT) and miR-19a mutant vectors showed a significant increase in basal NF-κB activity (Figure 2D). Although mutation of miR-19a did result in a decrease in NF-κB activity compared to the WT vector, these results suggested that miR-19b accounts for most of the effect of miR-17∼19b on NF-κB signaling. Altogether, these results indicate a role for miR-19b in the positive regulation of NF-κB signaling.
miR-19b is a critical component of the miRNA regulation of NF-κB signaling
The findings accumulated thus far suggested that the effect of global miRNA depletion on the production of pro-inflammatory cytokines seen in BMDMs would involve miR-19b. TaqMan® reverse transcription quantitative real-time PCR (RT-qPCR) of 335 miRNAs in BMDMs confirmed that miR-19b was one of the five most abundant miRNAs expressed at basal levels in these cells, and could therefore be implicated (Supplementary Figure S2). Inhibition of miR-19 in SV40 large T-antigen immortalized MEFs resulted in decreased NF-κB luciferase activity and IL-6 production with TLR1/2 stimulation (Figure 3A), confirming that the effect of miR-19b on NF-κB signaling is not restricted to human cells. Because miR-19b is the most abundant miRNA expressed in these SV40 MEFs (Supplementary Figure S2) (15), we sought to repeat the BMDM results and analyzed the responsiveness of Dicer1flox/floxxCre/Esr1+ and Dicer1flox/wtxCre/Esr1+ SV40-MEFs following OHT-mediated miRNA depletion. OHT-mediated miR-19b decrease (∼30%) in 99.5% of the cells (Supplementary Figure S2) was concordant with a 91% decrease in TLR1/2-induced IL-6 production in Dicer1flox/floxxCre/Esr1+ cells (Figure 3B). Conversely, the effects of OHT were limited to a 59% decrease in TLR1/2-induced IL-6 production in Dicer1flox/wtxCre/Esr1+, where miR-19b levels were not decreased (Figure 3B and Supplementary Figure S2). Similarly, OHT-mediated miR-19b decrease resulted in a ∼60% decrease in TLR4-induced IL-6 production in Dicer1flox/floxxCre/Esr1+ MEFs (Figure 3C, compare conditions ‘si-C’ with Lipid A stimulation, with and without OHT treatment). Further implicating miR-19b in this effect, rescue of miR-19b levels by transfection of a synthetic double-stranded miRNA mimic for miR-19b (si-miR-19b) restored IL-6 levels in OHT-treated Dicer1flox/floxxCre/Esr1+ MEFs, following TLR4 stimulation, to levels comparable to non-OHT treated cells (Figure 3C, compare conditions ‘si-C Non-treated’ and ‘si-miR-19b +OHT’, with Lipid A stimulation). Collectively, these results indicate that miR-19b has a critical contribution to the miRNA regulation of NF-κB signaling and pro-inflammatory cytokine production.
miR-19b controls several negative regulators of NF-κB
The findings that AMO-mediated blocking of miR-19b decreased both TLR- and TNF-α-driven NF-κB signaling suggested the involvement of the A20-ubiquitin editing complex, which is heavily targeted by miR-19 (Figure 2A). Interestingly, miR-19 is known to regulate expression of suppressor of cytokine signaling 1 (SOCS1) (24), which decreases NF-κB activity through blocking of TLR2/4 signaling (25,26). We therefore searched for other putative targets of miR-19 that could also be at play in the negative regulation of NF-κB signaling. In silico analyses of miRNA targeting sites identified miR-19 target sites in 13 genes negatively affecting NF-κB signaling at different levels of signal transduction (Supplementary Table S1). We selected a sample of seven of these genes (Tnfaip3, Rnf11, Kdm2a, Otud7b, Cyld, Irak3 and Zbtb16) (5,27–29) (Figure 4A), and analyzed the impact of their 3′UTR on mRNA stability/translation efficiency of Renilla luciferase reporters following AMO blocking of miR-19 (Figure 4B). Expression of 3′UTR reporters of four of these genes (Tnfaip3, Rnf11, Kdm2a and Zbtb16) were significantly affected by miR-19 depletion, and this specifically related to the presence of miR-19 target sites in these 3′UTRs as this effect was not seen in 3′UTR reporters with mutated miR-19 target sites (Figure 4C). Interestingly, while miR-19 inhibition exerted a modest effect on the increased expression of Tnfaip3, Kdm2a and Zbtb16 3′UTR reporters, expression of Rnf11 3′UTR reporter was about 10-fold more than that of the other reporters, suggesting a critical role for Rnf11 in the mediation of miR-19 effects on NF-κB signaling. The converse experiment relying on the transfection of synthetic si-miR-19b confirmed the strong effect of miR-19b on Rnf11 3′UTR reporter (Supplementary Figure S3). Transfection of si-miR-19b in HeLa cells strongly increased miR-19b intracellular levels (13-fold) and significantly decreased endogenous Rnf11 mRNA levels (Figure 4D and E). This was coordinated with a significant increase in basal IL-8 production by the cells (Figure 4D). Conversely, inhibition of basal miR-19b resulted in a significant increase in endogenous RNF11 mRNA levels (Supplementary Figure S3). In addition, RNA interference (RNAi)-based down-regulation of RNF11 prior to AMO 19 treatment of the cells suppressed the inhibitory effect of AMO 19 on TNF-α induced IL-8 by HEK293T cells (Figure 4F and Supplementary Figure S3). Altogether, these findings demonstrate the direct involvement of miR-19b in the modulation of expression of several known negative regulators of NF-κB signaling, and indicate an important role for Rnf11 in the effect of miR-19b on NF-κB signaling.
miR-19b increases basal inflammation in primary human fibroblast-like synoviocytes
To characterize further the physiological relevance of miR-19b on NF-κB signaling and inflammation, we investigated the impact of si-miR-19b transfection on basal cytokine production by RA primary FLS. FLS are an important contributor to the inflammatory pathology of RA synovitis, and spontaneously produce large amounts of pro-inflammatory cytokines such as IL-6 and IL-8 without further stimulation (32). Given that production of such cytokines can be further induced through activation of NF-κB signaling (33), we investigated the possibility that increased miR-19b could enhance cytokine production by these primary cells. si-miR-19 transfection significantly increased basal IL-6 and IL-8 secretion by FLS, by ∼50 and ∼20%, respectively (Figure 5). Noteworthy, while the down-regulation of RNF11 mRNA by RNAi (siRNF11 condition and Supplementary Figure S3) did not affect IL-6 levels, it induced a modest increase in IL-8 secretion, which suggests a possible contribution of RNF11 to the effects of miR-19 in these cells. Collectively, these results demonstrate that miR-19b can exacerbate the inflammatory activation of cells crucial to the pathology of an auto-inflammatory disease.
DISCUSSION
miRNAs are critical regulators of cellular homeostasis. As such, imbalanced expression of select miRNAs resulting from chromosomal duplication or inefficient miRNA processing is an important component of tumor cell development (34,35). The miR-17∼92 cluster of miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92-1) was originally described to be over-expressed in human B-cell lymphoma samples, as a result of chromosome 13q31–q32 amplification (35). Critically, over-expression of miR-17∼92 in a Myc-driven mouse model of B-cell lymphoma accelerated tumor development (35), thereby highlighting the direct contribution of miR-17∼92 miRNAs in tumor development. Over 200 publications relating to miR-17∼92 or family members have been published to date, establishing the crucial role of this cluster of miRNAs (also known as oncomiR-1) in oncogenesis. Genetic dissection of the relative contribution of the individual miRNAs of this cluster has demonstrated that miR-19 recapitulated on its own the oncogenic effects of the full cluster (36,37). miR-19 has been shown to exerts its oncogenic effects through its repression of the tumor suppressors Pten and Pp2a (36–38), the pro-apoptotic molecule Bim and the AMP-activated kinase Prkaa1 (38). Crucially, these genes belong to the phosphatidylinositol-3-OH kinase-related survival signals pathway, and illustrate how the combined regulation of several genes by miR-19 participates in its overall oncogenic function.
In this study, we sought to characterize the overall contribution of miRNAs in the control of NF-κB-driven inflammation. While inducible miRNAs such as miR-146a or miR-301 are used to fine-tune stimulus-driven inflammation (9,11), we hypothesized that other miRNAs basally expressed in cells might be involved in the repression of innate immune genes in steady state. This was supported by our prediction that less than 20% of innate immune genes under miRNA repression would be regulated by known inducible miRNAs (39). We investigated the production of pro-inflammatory cytokines following the inducible deletion of Dicer1 and the ensuing global miRNA depletion (15). While anticipating an increased production of TNF-α and IL-6, the levels of which are repressed by miR-19 and let-7 (12,40), respectively, we found that levels of these two cytokines, together with that of IL-12, were markedly decreased following Dicer1 deletion in primary BMDMs (Figure 1). This was independent of the TLR pathway recruited, and was later confirmed for IL-6 in SV40-MEFs stimulated with TLR1/2 and TLR4 agonists (Figure 3). Similar to the positive effect of miR-301 on NF-κB signaling mediated by its repressive action on the NF-κB-repressing factor gene, we suspected the involvement of negative regulators of NF-κB in these phenomena. In silico analyses suggested an important role for the miR-17∼92 cluster of miRNAs in the regulation of several known regulators of NF-κB signaling, including five out of six members of the A20-ubiquitin editing complex (Tnfaip3, Itch, Tnip1, Ccdc50 and Rnf11). In accordance with a putative interplay between NF-κB signaling and the miR-17∼92 cluster, the promoter region of the cluster was previously found to be regulated by the NF-κB p65 subunit (41). In addition, data mining of a miRNA expression atlas suggests an increased expression of the miR-17∼92 cluster following TLR4 activation of B-cells (23).
AMO-mediated blocking of the individual miRNAs from the cluster denoted a possible role for miR-19a/b in potentiation of NF-κB signaling in both TLR and TNF pathways in human HEK293 cells (Figure 2), and this was later confirmed in SV40-MEFs (Figure 3). miR-19b was the prevalent contributor to this positive regulation of NF-κB signaling, as determined in mutagenic studies of the cluster (Figure 2). Critically, transfection of synthetic miR-19b mimic in SV40-MEFs depleted of miR-19b restored ∼70% of the IL-6 levels seen in non-treated cells after TLR4 stimulation (Figure 3). The effects of miR-19b on NF-κB signaling were not limited to induced states, and increased miR-19b levels had a potent effect on the basal NF-κB activity, as seen with increased basal IL-8 levels (Figures 2 and 4). Finally, transfection of synthetic miR-19b in RA primary FLS resulted in the significant exacerbation of inflammation, as measured through IL-6 and IL-8 spontaneous production by these cells. Collectively, these results establish a critical role for miR-19b in the positive regulation of NF-κB and pro-inflammatory cytokine production. As shown with the example of miR-301, positive regulation of NF-κB signaling is however not exclusive to miR-19b. It is most likely that factors other than miR-19b impact on cytokine production in Dicer1-depleted cells. Indeed, Dicer1flox/wtxCre/Esr1+ cells exhibited a 59% decrease of IL-6 production with TLR1/2 stimulation, while miR-19b levels were not decreased (Figure 3 and Supplementary Figure S2).
Our data establish that the positive effect of miR-19b on NF-κB signaling is concurrent with its direct targeting of several known negative regulators of NF-κB signaling: Rnf11, Tnfaip3, Kdm2a, Zbtb16 and Socs1 (Figure 4) (24). Noteworthy, expression of both Tnfaip3 and Rnf11 mRNAs has previously been shown to be repressed upon miR-19 over-expression (38). While Rnf11/Tnfaip3 are involved in the termination of signal transduction (5), Kdm2a/Fbxl11 and Zbtb16 exert their repressive effect through the modulation of transcriptional activity of NF-κB subunits (28,29). This underlines the effect of miR-19b at different levels of NF-κB signaling. In Supplementary Table S1, many other important negative regulators of NF-κB signaling are predicted targets of miR-19b, and it is likely that ∼50% of these predictions are accurate (3). Because these regulators have variable expression levels among cell types, it is improbable that miR-19b relies on each one of these genes to exert its regulatory activity. Rather, the widespread targeting of miR-19b among negative regulators of NF-κB signaling would ensure its regulatory effect in many cell types. This is supported by a recent report describing the repressive effect of endogenous miR-19 on CYLD levels in T-cell leukemia cell lines (Jurkat and Molt-4 cells), but not in HEK293T cells (42). This is in accordance with our results with the murine Cyld 3′UTR reporter (Figure 4). Although modestly regulating Tnfaip3, Zbtb16 and Kdm2a 3′UTR reporters, miR-19b strongly affected Rnf11 3′UTR reporter and endogenous mRNA levels. This observation is in agreement with rescue experiments of miR-19 expression in miR-17∼92-deficient B-cell lymphomas, where Rnf11 was one of the most differentially expressed genes (36). As such, although exerting a regulatory effect on a wide panel of negative regulators of NF-κB, Rnf11 seems to be a critical component of the effect of miR-19b. This is supported by our observation that RNF11 down-regulation by siRNAs suppressed the inhibitory effect of AMO 19 (Figure 4) and mildly potentiated IL-8 production by RA FLS cells (Figure 5). The lack of an effect of RNF11 down-regulation on IL-6 production by RA FLS cells suggests differential downstream effects of RNF11 on IL-6 and IL-8 signaling, although it could also relate to the fact that the efficacy of RNAi in these cells was relatively low (∼60%). Given the potency of the RNF11 siRNA (see the data in HEK293T cells; Supplementary Figure S3), this probably relates to a transfection efficiency of the cells, and differences in IL-6 and IL-8 production could originate from differences in paracrine effects of non-transfected cells.
It has recently been proposed that TLR2 is a direct target for miR-19, and TNF-α has been shown to be destabilized by miR-19, suggesting a repressive effect of miR-19 on inflammation (40,43). Given the numerous targets of miR-19b, it is not surprising that its effects on the modulation of NF-κB signaling are not limited to that of the modulation of negative regulators of NF-κB. Importantly however, we found that in Dicer1 depleted cells TLR2 signaling was strongly decreased, which suggests that although possibly having a repressive effect on TLR2 levels, the net effect of miRNAs on TLR2 signaling in mice is positive regulation. Our findings suggest that the entire miR-17∼92 cluster, not only miR-19, is implicated in the modulation of NF-κB-driven inflammation. As such, we found that several negative regulators of NF-κB were targeted by multiple members of this cluster (Figure 2). This indicates a possible additive effect on the destabilization of their targets. Moreover, we showed that Tnfaip3 3′UTR was directly regulated by both miR-19 and miR-18, the co-depletion of which by AMO resulted in a further increase in Tnfaip3 3′UTR reporter expression (Supplementary Figure S4). This indicates a pro-inflammatory effect of miR-18, which is consistent with a reported role for miR-18 in the facilitation of IL-6 signal transduction (44). Nevertheless, the effects of various members of the miR-17∼92 cluster on the production of select pro-inflammatory factors are finely balanced and sometimes opposed, as seen with the opposite effects of miR-19 and miR-17 on IL-8 levels—IL-8 is a confirmed target of miR-17 (Supplementary Figure S4) (45). We propose that strategies perturbing the balance between miR-19 and miR-17 effects could present novel avenues for the treatment of auto-inflammatory disorders where IL-8 is involved. For instance, we found that AMO-blocking of miR-19 together with transfection of miR-17 mimics resulted in potentiation of the inhibition of IL-8 production upon TNF-α stimulation (Supplementary Figure S4).
The NF-κB pathway is critical in the pathogenesis of inflammatory diseases such as RA, wherein the initiation and perpetuation of inflammation are dependent on cytokines whose expression is regulated by NF-κB. In RA, activation of NF-κB is suggested to be a key link between inflammation and proliferation of FLS, the latter being an essential factor in the hyperplasia of the synovium in this disease (46). Our findings suggest that endogenous miR-19b may have a key regulatory role in constraining the production of pro-inflammatory cytokines and chemokines by FLS, and hence be a contributor to the pathology of this disease. Further work using animals lacking miR-17∼92 in specific immune cell types, should help define further the direct contribution of this cluster of miRNAs in the pathology of RA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4, Supplementary Methods and Supplementary References [47–49].
FUNDING
The Australian National Health and Medical Research Council [1022144] (to M.P.G.), [1006590] (to B.R.G.W.), and [606425] (to B.R.G.W. and D.X.); Victorian Government’s Operational Infrastructure Support Program. C.E.M. is supported by a grant from the Health Research Board Ireland. Funding for open access charge: Monash Institute of Medical Research.
Conflict of interest statement. Mark A. Behlke is employed by Integrated DNA Technologies Inc. (IDT), which offers oligonucleotides for sale similar to some of the compounds described in the manuscript. IDT is however not a publicly traded company, and he does not own any shares or hold equity in IDT.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Aneta Strzelecki (Monash Institute of Medical Research) for her help with maintenance of the mouse colony, and Scott Rose (Integrated DNA Technologies Inc.) for his help in the production of the RNA oligonucleotides.
REFERENCES
- 1.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc. Natl Acad. Sci. USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 4.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 5.Harhaj EW, Dixit VM. Deubiquitinases in the regulation of NF-kappaB signaling. Cell Res. 2011;21:22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 7.Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 8.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl Acad. Sci. USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamagishi M, Nakano K, Miyake A, Yamochi T, Kagami Y, Tsutsumi A, Matsuda Y, Sato-Otsubo A, Muto S, Utsunomiya A, et al. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-kappaB pathway in adult T cell leukemia and other cancers. Cancer Cell. 2012;21:121–135. doi: 10.1016/j.ccr.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Lu Z, Li Y, Takwi A, Li B, Zhang J, Conklin DJ, Young KH, Martin R. miR-301a as an NF-kappaB activator in pancreatic cancer cells. EMBO J. 2011;30:57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 14.Leech M, Metz C, Hall P, Hutchinson P, Gianis K, Smith M, Weedon H, Holdsworth SR, Bucala R, Morand EF. Macrophage migration inhibitory factor in rheumatoid arthritis: evidence of proinflammatory function and regulation by glucocorticoids. Arthritis Rheum. 1999;42:1601–1608. doi: 10.1002/1529-0131(199908)42:8<1601::AID-ANR6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 15.Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011;39:5692–5703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J. 2009;28:513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N, Nir T, Lennox KA, Behlke MA, Dor Y, et al. miRNAs control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. EMBO J. 2011;30:835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 19.Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 20.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl Acad. Sci. USA. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, Nicholson SE, Hilton DJ, O'Neill LA, Hertzog PJ. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat. Immunol. 2006;7:148–155. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 26.Strebovsky J, Walker P, Lang R, Dalpke AH. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. FASEB J. 2011;25:863–874. doi: 10.1096/fj.10-170597. [DOI] [PubMed] [Google Scholar]
- 27.Su J, Zhang T, Tyson J, Li L. The interleukin-1 receptor-associated kinase M selectively inhibits the alternative, instead of the classical NFkappaB pathway. J. Innate. Immun. 2009;1:164–174. doi: 10.1159/000158541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl Acad. Sci. USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu D, Suliman B, Sadler AJ, Williams BRG. Negative regulation of TLR-mediated NF-kappa B activation by PLZF. Cytokine. 2010;52:36. [Google Scholar]
- 30.Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, Kourtis K, Koziris N, Dalamagas T, Hatzigeorgiou AG. DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res. 2011;39:W145–W148. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol. Rev. 2010;233:233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang SY, Kim JY, Kim KW, Park MK, Moon Y, Kim WU, Kim HY. IL-17 induces production of IL-6 and IL-8 in rheumatoid arthritis synovial fibroblasts via NF-kappaB- and PI3-kinase/Akt-dependent pathways. Arthritis Res. Ther. 2004;6:R120–R128. doi: 10.1186/ar1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gantier MP. New perspectives in MicroRNA regulation of innate immunity. J. Interferon Cytokine Res. 2010;30:283–289. doi: 10.1089/jir.2010.0037. [DOI] [PubMed] [Google Scholar]
- 40.Liu M, Wang Z, Yang S, Zhang W, He S, Hu C, Zhu H, Quan L, Bai J, Xu N. TNF-alpha is a novel target of miR-19a. Int. J. Oncol. 2011;38:1013–1022. doi: 10.3892/ijo.2011.924. [DOI] [PubMed] [Google Scholar]
- 41.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye H, Liu X, Lv M, Wu Y, Kuang S, Gong J, Yuan P, Zhong Z, Li Q, Jia H, et al. MicroRNA and transcription factor co-regulatory network analysis reveals miR-19 inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2012;40:5201–5214. doi: 10.1093/nar/gks175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Philippe L, Alsaleh G, Suffert G, Meyer A, Georgel P, Sibilia J, Wachsmann D, Pfeffer S. TLR2 expression is regulated by microRNA miR-19 in rheumatoid fibroblast-like synoviocytes. J. Immunol. 2012;188:454–461. doi: 10.4049/jimmunol.1102348. [DOI] [PubMed] [Google Scholar]
- 44.Brock M, Trenkmann M, Gay RE, Gay S, Speich R, Huber LC. MicroRNA-18a enhances the interleukin-6-mediated production of the acute-phase proteins fibrinogen and haptoglobin in human hepatocytes. J. Biol. Chem. 2011;286:40142–40150. doi: 10.1074/jbc.M111.251793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, McCue PA, Quong AA, Lisanti MP, Pestell RG. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc. Natl Acad. Sci. USA. 2010;107:8231–8236. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miagkov AV, Kovalenko DV, Brown CE, Didsbury JR, Cogswell JP, Stimpson SA, Baldwin AS, Makarov SS. NF-kappaB activation provides the potential link between inflammation and hyperplasia in the arthritic joint. Proc. Natl Acad. Sci. USA. 1998;95:13859–13864. doi: 10.1073/pnas.95.23.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green MR, Gandhi MK, Courtney MJ, Marlton P, Griffiths L. Relative abundance of full-length and truncated FOXP1 isoforms is associated with differential NFkappaB activity in Follicular Lymphoma. Leuk. Res. 2009;33:1699–1702. doi: 10.1016/j.leukres.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Frobose H, Ronn SG, Heding PE, Mendoza H, Cohen P, Mandrup-Poulsen T, Billestrup N. Suppressor of cytokine Signaling-3 inhibits interleukin-1 signaling by targeting the TRAF-6/TAK1 complex. Mol. Endocrinol. 2006;20:1587–1596. doi: 10.1210/me.2005-0301. [DOI] [PubMed] [Google Scholar]
- 49.Ikeda O, Sekine Y, Mizushima A, Oritani K, Yasui T, Fujimuro M, Muromoto R, Nanbo A, Matsuda T. BS69 negatively regulates the canonical NF-kappaB activation induced by Epstein-Barr virus-derived LMP1. FEBS Lett. 2009;583:1567–1574. doi: 10.1016/j.febslet.2009.04.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.