Abstract
Mitochondria lack the ability to repair certain helix-distorting lesions that are induced at high levels in mitochondrial DNA (mtDNA) by important environmental genotoxins and endogenous metabolites. These lesions are irreparable and persistent in the short term, but their long-term fate is unknown. We report that removal of such mtDNA damage is detectable by 48 h in Caenorhabditis elegans, and requires mitochondrial fusion, fission and autophagy, providing genetic evidence for a novel mtDNA damage removal pathway. Furthermore, mutations in genes involved in these processes as well as pharmacological inhibition of autophagy exacerbated mtDNA damage-mediated larval arrest, illustrating the in vivo relevance of removal of persistent mtDNA damage. Mutations in genes in these pathways exist in the human population, demonstrating the potential for important gene–environment interactions affecting mitochondrial health after genotoxin exposure.
INTRODUCTION
Mitochondria serve several critical cellular functions, including energy production via oxidative phosphorylation (OXPHOS) and key roles in apoptosis. The majority of mitochondrial proteins involved in these processes are encoded by nuclear DNA (nDNA), but a small subset, 13 polypeptides that are incorporated into 4 of the 5 complexes of the OXPHOS system plus 2 rRNAs and 22 tRNAs, are encoded by mitochondrial DNA (mtDNA). A large and growing number of inherited mitochondrial diseases result from mutations in or depletion of mtDNA and likely affect >1 in 6000 people (1), emphasizing the importance of mtDNA integrity to human health. Furthermore, there is substantial evidence to support a role of mitochondrial dysfunction and mtDNA mutation in the pathogenesis of more common diseases such as neurodegenerative conditions (2,3), type 2 diabetes mellitus (4) and cancer (5).
Compared with nDNA, mtDNA is particularly susceptible to damage from prevalent environmental contaminants. For instance, depending on exposure conditions, mtDNA is 2–100 times more susceptible to bulky adducts when exposed to the carcinogenic, polycyclic aromatic hydrocarbon (PAH) benzo(a)pyrene (6–11). This susceptibility is partially because of the fact that mitochondria lack nucleotide excision repair (NER), the repair pathway utilized to repair many helix-distorting or ‘bulky’ nDNA damage induced by common environmental agents including PAHs, mycotoxins and ultraviolet C radiation (UVC) (12–15). Thus, helix-distorting mtDNA lesions resulting from exposure to these genotoxins are persistent and reduce mtDNA replication (8,16–18) and transcription (19). The resulting reduction of mtDNA copy number or protein synthesis could result in mitochondrial dysfunction, as suggested by cytochrome c release, reduced mitochondrial membrane potential (MP), decreased respiration and increased lipid peroxidation reported in studies with PAHs and the mycotoxin Aflatoxin B1 (20–22).
Helix-distorting lesions induce nDNA mutations (23,24) and in vitro evidence supports the potential for these lesions to be mutagenic in mtDNA (25,26). However, exposures to benzo(a)pyrene diol epoxide and UVC in cell culture have resulted in little to no increase in mutation frequency above background (7,18,27), prompting the hypothesis that mtDNAs harboring helix-distorting lesions are degraded and mutations resulting from these lesions are rare (28,29). Mitochondrial degradation is carried out by macroautophagy (30,31). Non-selective autophagy of mitochondria as well as other organelles and cytoplasmic materials (31) is triggered by starvation and nutrient deprivation. On the other hand, selective autophagy of mitochondria or ‘mitophagy’ (32) is responsible for clearance of paternal mtDNA in sperm of Caenorhabditis elegans (33,34), of mitochondria during erythrocyte maturation (35) and has emerged as a more specific degradation mechanism for dysfunctional mitochondria in somatic cells (30,32,36–38). Although the detailed mechanism of dysfunctional mitochondrial clearance by mitophagy has yet to be resolved, current research suggests that spontaneous or stress-induced depolarization results in isolation of dysfunctional mitochondria followed by removal by autophagy (30,39–43). The Parkinson’s disease-associated genes encoding Parkin and PINK1 have been shown to selectively accumulate on mitochondria with reduced MP and facilitate mitophagy under certain conditions (44–47). The role of mitophagy in removal of dysfunctional mitochondria with damaged DNA has not been previously addressed but this could play an important role in preventing fixation of mtDNA mutations and mitochondrial-mediated apoptosis or necrosis by removal of unstable mitochondria (48,49).
The goals of this research were to determine the long-term fate of helix-distorting mtDNA damage and investigate the role of autophagy and mitochondrial dynamics in removal of and recovery from this mtDNA damage. Using UVC, we induced helix-distorting mtDNA lesions in adult C. elegans with and without RNAi knockdown of autophagy, fusion and fission genes and measured mtDNA damage over time using quantitative polymerase chain reaction (qPCR). We show that UVC-induced mtDNA lesions are removed slowly over 72 h in adult C. elegans and that this removal is dependent upon genes involved in autophagy and mitochondrial dynamics. To better understand the implications of persistent mtDNA damage removal in vivo, we investigated in C. elegans the effect of UVC-induced mtDNA damage on the frequency and duration of larval arrest, a well-described phenotype mediated by mitochondrial function that serves as an indicator of overall mitochondrial health (50–53). We show that high levels of persistent mtDNA damage induce L3 larval arrest and mitochondrial dysfunction in C. elegans. Furthermore, recovery from larval arrest is dependent on mitochondrial fusion, mtDNA replication and removal of damaged mtDNA via autophagy. Thus, we show for the first time that, although persistent, UVC-induced mtDNA lesions are removed gradually and that mitochondrial dynamics and autophagy are involved in removal of and recovery from persistent mtDNA damage.
MATERIALS AND METHODS
Caenorhabditis elegans strains and culture
Populations of C. elegans were maintained on K agar plates seeded with OP50 bacteria unless otherwise stated. N2 (wild-type), JK1107 glp-1(q224), VC1024 pdr-1(gk448), CB369 unc-51(e369) were obtained from the Caenorhabditis Genetics Center (University of Minnesota). DA631 eat-3(ad426), BC10210 fzo-1(tm1133) and N2 + Ex[myo-3::mls::matrixGFP + rol-6] were provided by Alexander van der Bliek, University of California (Los Angeles, CA, USA). drp-1 (tm1108) was provided by Ding Xue, University of Colorado (Boulder, CO, USA). Guy Caldwell, University of Alabama (Tuscaloosa, AL, USA), provided UA86 pink-1(tm1779); Pdat-1::GFP; Pdat-1::α-syn [baIn11]. Luminescent PE255 (feIs5) were provided by Christina Lagido, University of Aberdeen (Aberdeen, UK). Alicia Melendez, Queens College, City University of New York (Flushing, NY, USA) kindly provided QU1 izEx1[Plgg-1::gfp::lgg-1 + rol-6].
Quantitative PCR
DNA damage analysis was performed using qPCR optimized for whole C. elegans as described in Boyd et al. (54) with some modifications. glp-1 adults were picked at a 1 worm to 15 µl lysis buffer ratio. In RNAi experiments, nuclear and mtDNA copy number for each QPCR sample determined by quantitative, real-time PCR was used to normalize for copy number variation between samples. This assay relies on the ability of DNA damage to block progression of the DNA polymerase used for PCR; thus, the amount of amplification is inversely related to the amount of DNA damage (55,56). In each sample, a long (∼10 kb) nuclear and mitochondrial genomic region is amplified. The long product is used to calculate lesion frequency as compared with control samples.
DNA damage and repair quantification
Synchronized glp-1 L1 larvae were obtained by bleach-sodium hydroxide isolation of eggs and liquid egg hatch in K+ medium (54). Nematodes were grown to young adult stage at 25°C, then transferred to K-agar plates without OP50 and exposed to 50 J/m2 UVC using an ultraviolet lamp (UVLMS-38 EL Series 3UV Lamp, UVP, Upland, CA, USA) with peak emission at 254 nm. Doses were quantified using a UVX digital radiometer. Nematodes were lysed at 0, 24, 48 and 72 h after exposure. Nematodes were incubated at 25°C during recovery periods. Analysis was performed on 7–12 sample tubes (6 nematodes/tube) for each treatment condition generated from at least two independent, time-separated experiments creating an ‘n’ of 7–12. At least two PCR technical replicates were carried out for each sample. Lesion frequency in UVC-treated samples was calculated relative to RNAi clone untreated controls. Global analysis of variance (ANOVA) analysis was used to determine effects and interactions of treatment, recovery and RNAi clone (Statview 5.0.1). When warranted by global ANOVA, Fisher’s PLSD was used to determine the effect of individual variables.
Mitochondrial copy number analysis
Absolute mitochondrial copy number was measured by quantitative, real-time PCR as described in Bratic et al. (57) using the lysed worm samples collected for QPCR as described earlier in the text. For each sample, three technical replicates were averaged from a single real-time PCR run.
RNA interference
RNAi-mediated interference was carried out by feeding. HT115 (DE3) RNase III-deficient Escherichia coli engineered to express dsRNA for eat-3 (D2013.5), drp-1 (T12E12.4), pink-1 (EEED8.9) and pdr-1 (K08E3.7) were obtained from Geneservice (Cambridge, UK) or Open Biosystems (Huntsville, AL, USA). atg-18 (F41E6.13), bec-1 (T19E7.3), fzo-1 (ZK1248.14) and unc-51 (F41E6.13) were retrieved from the Vidal C. elegans RNAi library (58), and pL4440 (empty vector, negative RNAi control) and unc-22 (ZK617.1, positive RNAi control) were retrieved from the Ahringer C. elegans RNAi library (59). All RNAi vectors were sequenced to verify the identity of the insert.
RNAi stocks were selected on LB plates containing 15 µg/ml tetracycline and 100 µg/ml ampicillin (Sigma Aldrich). Bacteria were grown in LB containing 50 µg/ml ampicillin (final concentration) for 8 h and were added to NGM plates containing 50 µg/ml ampicillin and 1 mM isopropyl β-d-1-thiogalactopyranoside (Sigma Aldrich). With the exception of eat-3, fzo-1 and drp-1 RNAi knockdown, L1 glp-1 nematodes were synchronized as described earlier in the text and grown on RNAi-seeded NGM plates for two generations. Nematodes were grown at 15°C throughout the experiment except for a 15-h period between the L3 and L4 stage during which nematodes were transferred to 25°C for sterilization (60). At the adult stage of the second generation, glp-1 nematodes were dosed with 50 J/m2 UVC and lysed immediately and 120 h after the UVC dose for DNA damage analysis. For eat-3, fzo-1 and drp-1 RNAi knockdown, an identical procedure was carried out except over one generation instead of two because of embryonic lethality and larval arrest in F1 progeny of nematodes treated with eat-3, fzo-1 (61) and drp-1 (62) RNAi.
Mitochondrial morphology and autophagy analysis
Synchronized young adult N2 myo-3::matrixGFP and N2 LGG-1::GFP nematodes were maintained and UVC exposed as described earlier in the text except at 20°C instead of 25°C. At recovery periods, 6–10 nematodes were picked onto 10% agar pads with 10 µl of 10 mM levamisole (Sigma Aldrich, dissolved in water). Single-plane images were taken of muscle cell mitochondria and seam cells from five to 10 nematodes per experiment in two, time-separated experiments on a Zeiss 780 confocal microscope at 63× magnification. Approximately 9–15 seam cells and 25–50 muscle cells were analyzed per treatment per time point. LGG-1::GFP foci in each seam cell were counted manually. An overall effect of treatment determined by ANOVA (P < 0.0001) (Statview 5.0.1) and a difference between specific treatments was determined by Fisher’s PLSD. Images of muscle cell mitochondria were analyzed using MetaMorph Premier and methods described in (63,64). Briefly, background subtraction, a top hat morphological filter and a threshold were applied to images. After converting to a binary image, mitochondrial area excluding the area of ‘holes’, perimeter and form factor for each muscle cell was measured using Integrated Morphology Analysis.
Larval arrest protocol
L1 worms were plated in equal amounts on unseeded, non-peptone (to prevent growth of bacterial contamination), K-agar plates with or without ethidium bromide (Sigma Aldrich) for 48 h. L1 nematodes were starved during this period to prevent development. L1 worms were exposed to UVC at 0, 24 and 48 h after plating. Immediately following the 48-h UVC dose, worms were transferred to 24-well, seeded, K-agar plates (1–3 worms/well, 12 wells/treatment, 1 plate/strain) with or without ethidium bromide. For 3-methyladenine (3-MA) exposure following the 48 h UVC dose, worms were transferred to 24-well, seeded, liquid K+ as described earlier in the text with or without 3-MA (Sigma Aldrich, dissolved in dH2O). Every 24 h for 96 h, each worm was staged and the percentage of nematodes that reached L4 was determined based on the presence of the vulval crescent. All mutant strains and/or treatments were screened in a minimum of three time-separated experiments and the % ≤ L3 from each experiment was considered a biological replicate. % ≤ L3 across all time points was compared between strains (if applicable) and treatments by repeated measures ANOVA (Statview 5.0.1). Given a significant interaction in the repeated measures ANOVA, ANOVA was used to determine overall effects and interactions at each time point followed by Fisher’s PLSD, if warranted.
Relative ATP analysis
PE255 nematodes were dosed according to the UVC dosing scheme described earlier in the text (Larval Arrest Protocol), and ATP analysis was conducted immediately after the third UVC dose (0 h) or 24 h after the third dose and placement on food. Each experiment had two biological replicates dosed on separate no-peptone, no-food plates and three time-separated experiments were performed (n = 6). Luminescence and GFP were measured using a FLUOstar Optima (BMG Labtech) on ∼100 nematodes in 100 µl of K-media added to a 96-well plate with two to four technical replicates/biological replicate. GFP was measured initially using 485/520 filter set. Following GFP measurement, 50 µl of luminescence buffer (citrate phosphate buffer pH 6.5, 0.1 mM d-luciferin, 1% DMSO and 0.05% Triton-X, final concentrations) was injected into each sample, and luminescence was measured 3 min after injection using a 590 nm emission filter. Luminescence was normalized to GFP fluorescence intensity to account for differences in number of worms per well. Two-way ANOVA was used to determine if there was an effect of treatment and/or recovery on % control luminescence (Statview 5.0.1). After establishing that there was a treatment by recovery effect (P = 0.0017), the effect of treatment at each time point was assessed using Fisher’s PLSD.
Oxygen consumption analysis
N2 nematodes were treated with 10 J/m2 UVC as described earlier in the text (Larval Arrest Protocol) and oxygen consumption was measured at 0, 24 and 48 h post the third UVC exposure using a Mitocell (MT200) respiration chamber with magnetic stirrer and 1302 Clark-type microcathode oxygen electrode attached to a 782 oxygen meter (Strathkelvin Instruments, Glasgow, UK) as described in (65). At 0 and 24 h, 1000 nematodes were sorted into four 1.5 ml microcentrifuge tubes using a COPAS Biosort (Union Biometrica, Holliston, MA, USA) with the same gate used for untreated and treated samples. At 48 h, 500 nematodes were sorted. The UVC-exposed group was sorted into two populations: large (non-arrested) and small (arrested). The large or non-arrested gate was defined using the control population that is at L4 or older. The small or arrested gate was defined as any nematode smaller than the control population. Immediately before measurement worms were centrifuged, 100 µl of pelleted worms were added to the Mitocell chamber and oxygen consumption was measured for 4 min once linear. The rate of oxygen consumption was calculated within the linear range for each sample and two-way ANOVA followed by Fisher’s PLSD (if applicable) were used to identify significant effects of treatment and recovery (Statview 5.0.1).
RESULTS
UVC-induced mtDNA damage is removed over time
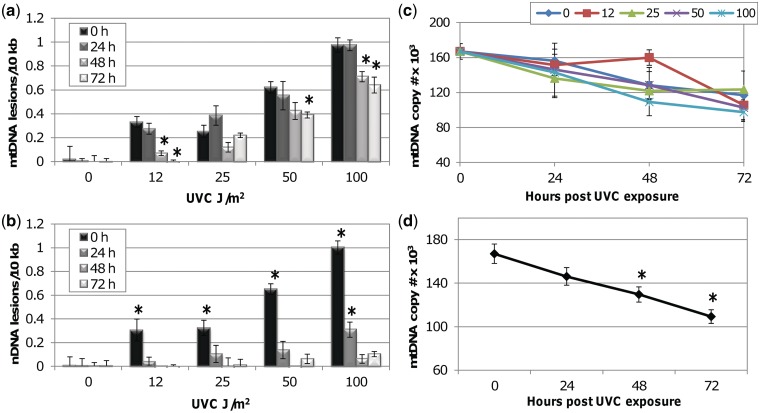
Studies in cell culture and C. elegans have established the persistence of mtDNA damage up to 48 h post exposure to UVC (17,66,67) but further time points have not been investigated, partially because of the confounding effect of dilution of mtDNA damage encountered with proliferating cells in culture. We utilized the germ-line proliferation defective mutant strain glp-1(q224) to investigate the persistence of UVC-induced mtDNA damage. Adult glp-1 raised at 25°C have post-mitotic somatic cells and few (6–8) germ cells (68), such that changes in DNA damage can be attributed to repair or removal rather than dilution following cell replication (67). Radiation (UVC) of 254 nm serves as an excellent model for bulky adducts because it similarly induces helix-distorting lesions and it is primarily absorbed by nucleic acids, with little damage to other macromolecules; thus we utilized UVC to induce helix-distorting mtDNA lesions throughout our studies. Post-mitotic young adult glp-1 nematodes raised at 25°C were exposed to 0–100 J/m2 UVC and both nuclear and mtDNA damage was analyzed at 0, 24, 48 and 72 h post exposure via a highly sensitive QPCR assay that does not require differential extraction of nuclear and mtDNA (55,56,69). Over 72 h, 30–40% removal of mtDNA damage was observed at 50 and 100 J/m2 (Figure 1a). nDNA damage was repaired to control level by 72 h (Figure 1b). mtDNA replication occurs independently of nDNA replication, and while UV-induced lesions are likely to block replication (26), it is unlikely that all of the mitochondrial genomes per cell were damaged even at our highest doses. Assuming a Poisson distribution of lesions and a similar mtDNA copy number per cell, even at 50 and 100 J/m2 ∼35 and 20%, respectively, of mtDNA genomes would be predicted to remain undamaged. Thus, in principle, the reduction in mtDNA damage observed here could result from dilution of damage rather than removal; however, mtDNA copy number did not increase during the recovery period at any UV dose as measured by real-time PCR (Figure 1c). In fact, there was a significant decrease in copy number across treatments by 48 h post exposure (including controls; Figure 1d); this decrease was not altered by the level of damage (P = 0.3246 for treatment × time interaction). Therefore, the decrease in mtDNA damage levels cannot be explained by dilution alone, and must, at least in part, be a result of removal of damaged genomes.
Figure 1.
mtDNA lesion frequency slowly decreases after a single dose of UVC in post-mitotic adult C. elegans (glp-1). Nematodes were exposed to UVC and analyzed for DNA damage immediately (0 h), or after 24, 48 or 72 h via QPCR. (a) mtDNA damage is removed by ∼40% over 72 h at 50 and 100 J/m2. Two-way ANOVA indicated a significant effect of recovery (P < 0.0001) and a recovery × treatment interaction (P < 0.05). Asterisks denote a significant difference compared with 0 h lesion frequency within each UVC treatment (Fisher’s PLSD, P < 0.05). (b) nDNA damage is repaired following a single dose of UVC. Two-way ANOVA indicated a significant effect of treatment (P < 0.0001) and a recovery × treatment interaction (P < 0.0001). Asterisks denote a significant difference compared with undosed control lesion frequency at each recovery timepoint (Fisher’s PLSD, P < 0.05). (c) No significant increase in mtDNA copy number was observed during the recovery period. (d) mtDNA copy number significantly decreased during the recovery period irrespective of UVC exposure. Two-way ANOVA indicated a significant effect of time (P < 0.0001) but no significant treatment effect (P = 0.4714) or treatment × time interaction (P = 0.3246). Asterisks denote a significant difference compared with 0 h mtDNA copy number (Fisher’s PLSD, P < 0.05). Bars ± SEM.
Mitochondrial dynamics and autophagy are involved in removal of persistent mtDNA damage
We next tested whether mitochondrial fusion, fission and autophagy assist in removal of mtDNA damage. Fusion, fission and autophagy genes were knocked down using RNAi in adult glp-1 C. elegans. Adult C. elegans were then dosed with 50 J/m2 and nuclear and mtDNA damage was measured immediately and 120 h post exposure. These RNAi experiments were carried out at 15°C except for a 15-h sterilization period at 25°C because we observed higher RNAi efficiency in our RNAi-positive control at the lower incubation temperature. We extended the DNA damage removal period from 72 to 120 h because the lower incubation temperature slowed removal. We considered examining removal at later time points, but chose not to based on our previous observation that mtDNA copy number declines with age (52,67), as does mitochondrial function in general (70), effects that could confound our measurements. Although the damage removal observed during this timecourse was not complete, it was sufficient for analysis and replicable in multiple experiments.
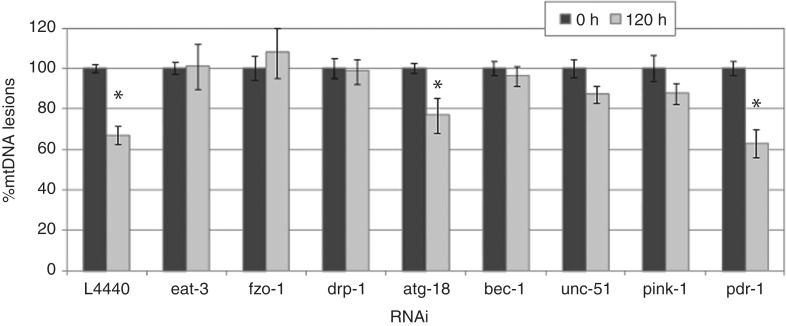
In our empty vector control (L4440), there was ∼30% removal of mtDNA damage 120 h after a single UVC exposure (Figure 2). RNAi knockdown of fusion genes fzo-1 and eat-3, fission gene drp-1, and autophagy/mitophagy genes bec-1, unc-51 and pink-1 abolished detectable removal of UVC-induced mtDNA damage. No increase in mtDNA copy number was observed in any sample as measured by real-time QPCR (Supplementary Figure S1), and decreases were observed in some cases. Repair of UVC-induced nDNA damage was not affected by RNAi (Supplementary Figure S2). These data demonstrate that fusion, fission and autophagy are required for removal of UVC-induced mtDNA damage.
Figure 2.
Fusion, fission and autophagy gene knockdown inhibits mtDNA damage removal. mtDNA lesions are removed in empty vector control (L4440) by 30–40% 120 h post a single UVC exposure in post-mitotic adult C. elegans (glp-1). RNAi knockdown of eat-3, fzo-1, drp-1, bec-1, unc-51 and pink-1 inhibited mtDNA damage removal (inhibition of removal was determined by a lack of a significant difference between 0 and 120 h lesion frequency within each RNAi treatment). Two-way ANOVA indicated a significant interaction between RNAi and recovery (P < 0.05). Asterisks denote a significant difference between 0 h and 120 h mtDNA lesions within RNAi treatment (Fisher’s PLSD, P < 0.05). Percent mtDNA lesions remaining after 120 h was calculated based on 0 h lesion frequency within each RNAi treatment. Bars ± SEM.
Persistent mtDNA damage induces autophagy without detectable changes in mitochondrial morphology
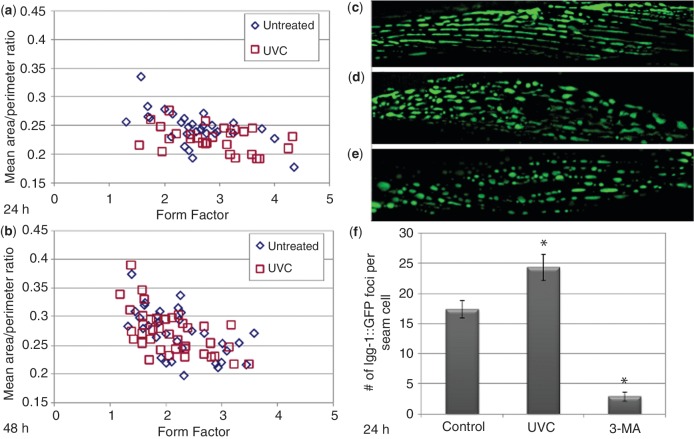
Given the requirement for mitochondrial dynamics and autophagy genes in removal of mtDNA damage, we evaluated the effect of persistent mtDNA damage on mitochondrial morphology and autophagy following UVC exposure in adult C. elegans. To assess mitochondrial morphology, young adult wild-type nematodes containing an extrachromosomal Pmyo-3::matrixGFP, which express GFP in the mitochondrial matrix of muscle cells (62), were exposed to a single dose of 50 J/m2 and mitochondrial morphology was assessed via confocal fluorescence microscopy at 24 and 48 h post exposure. Mitochondrial morphology was quantified using form factor as a measure of mitochondrial elongation and mean area/perimeter ratio as a measure of mitochondrial interconnectivity (63,64). As shown in Figure 3a–b, no clear differences were observed in mitochondrial elongation or interconnectivity between control and UVC exposed nematodes at any time point. We observed that within individual nematodes and across treatments there was significant variability in mitochondrial morphology (Figure 3c–e).
Figure 3.
UVC exposure induces autophagy but no detectable changes in mitochondrial morphology in adult C. elegans. Mitochondrial morphology was assessed using form factor as a measure of elongation (perfect circles = 1) and mean area/perimeter ratio as a measure of interconnectivity. No detectable differences were observed at (a) 24 h or (b) 48 h following UVC exposure. Each data point represents the mean area/perimeter ratio and form factor of mitochondria within a single muscle cell. Mitochondrial morphology was highly variable within each treatment group. Representative images show a (c) tubular, (d) intermediate and (e) fragmented mitochondrial morphology in muscle cells of untreated controls 48 h after UVC exposure. (f) The number of LGG-1::GFP foci per seam cell increased 24 h following UVC exposure compared with untreated controls (Fisher’s PLSD, P = 0.004). Inhibition of autophagy with 3-MA exposure reduced the formation of LGG-1::GFP foci compared with untreated controls (Fisher’s PLSD, P < 0.0001). An overall effect of treatment was determined by ANOVA (P < 0.0001). Bars ± SEM.
To evaluate induction of autophagy following UVC exposure, young adult nematodes that express a GFP-tagged LGG-1 protein (71) were exposed to 50 J/m2, and LGG-1::GFP foci were quantified in hypodermal seam cells 24 h after the exposure. Lgg-1 is the C. elegans homolog to mammalian MAP-LC3 and is incorporated into autophagosomal membranes (71). Appearance of LGG-1::GFP foci in hypodermal seam cells has been extensively used as an indicator of autophagy in C. elegans (71–75). UVC treatment induced a mild but statistically significant increase in LGG-1::GFP foci 24 h after exposure (Figure 3f). Interestingly, we observed a much greater number of LGG-1::GFP foci in untreated N2 nematodes than has been previously reported (71–75). To confirm that foci were in fact autophagosomal structures, we inhibited autophagosome formation with 10 mM 3-MA, a widely used class III phosphatidylinsositol-3 kinase inhibitor (76), previously shown to inhibit autophagy in C. elegans at this concentration (77). The number of LGG-1::GFP foci decreased significantly with 3-MA treatment, indicating that the foci observed in our untreated and UVC-treated nematodes did represent induction of autophagy. Therefore we conclude that UVC exposure induced a mild increase in autophagy without persistent changes in mitochondrial morphology. Whether this increase in autophagy signifies a greater mitochondrial clearance requires further research.
Serial UVC exposure results in accumulated mtDNA damage and dose-dependent larval arrest
We next sought to investigate the in vivo importance of persistent mtDNA lesions and removal of those lesions. Caenorhabditis elegans develop through four larval stages before reaching adulthood, and mitochondrial function and mtDNA copy number throughout development have been well-characterized (52). Development from L3 to L4 entails a dramatic 3- to 5-fold increase in mtDNA copy number and a switch from anaerobic respiration to OXPHOS, both of which are associated with somatic and germ cell development (52,57). Several researchers demonstrated that either knockdown of ETC components or chemical inhibition of mtDNA replication or protein translation early in larval development (L1 and L2) results in developmental arrest at or before L3 (51–53). The authors concluded that larval development serves as an indicator of overall mitochondrial function. This conclusion was further supported by Addo et al. who utilized L3 larval arrest as a screenable phenotype for genes involved in mtDNA maintenance (50).
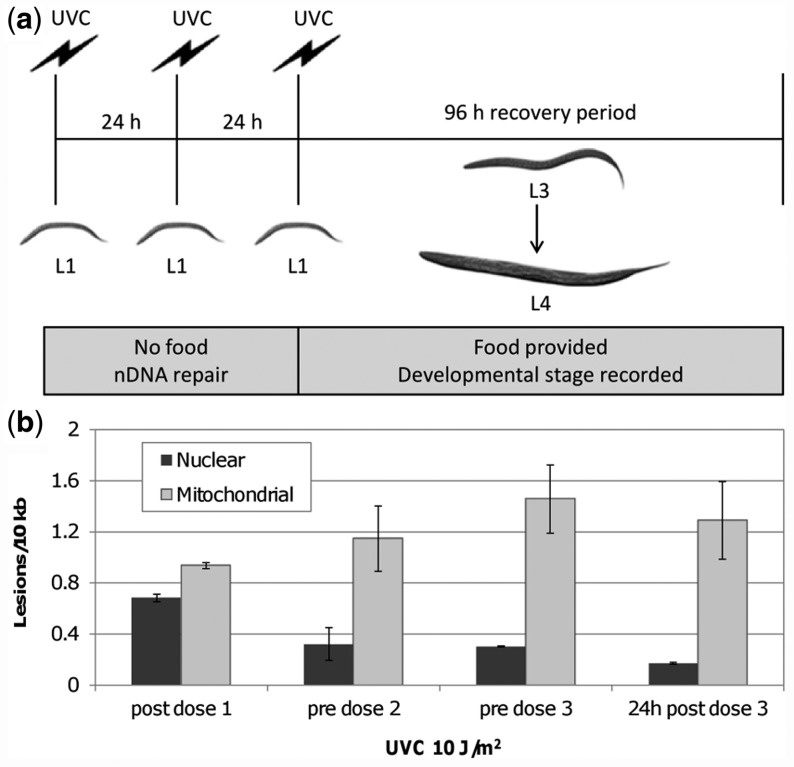
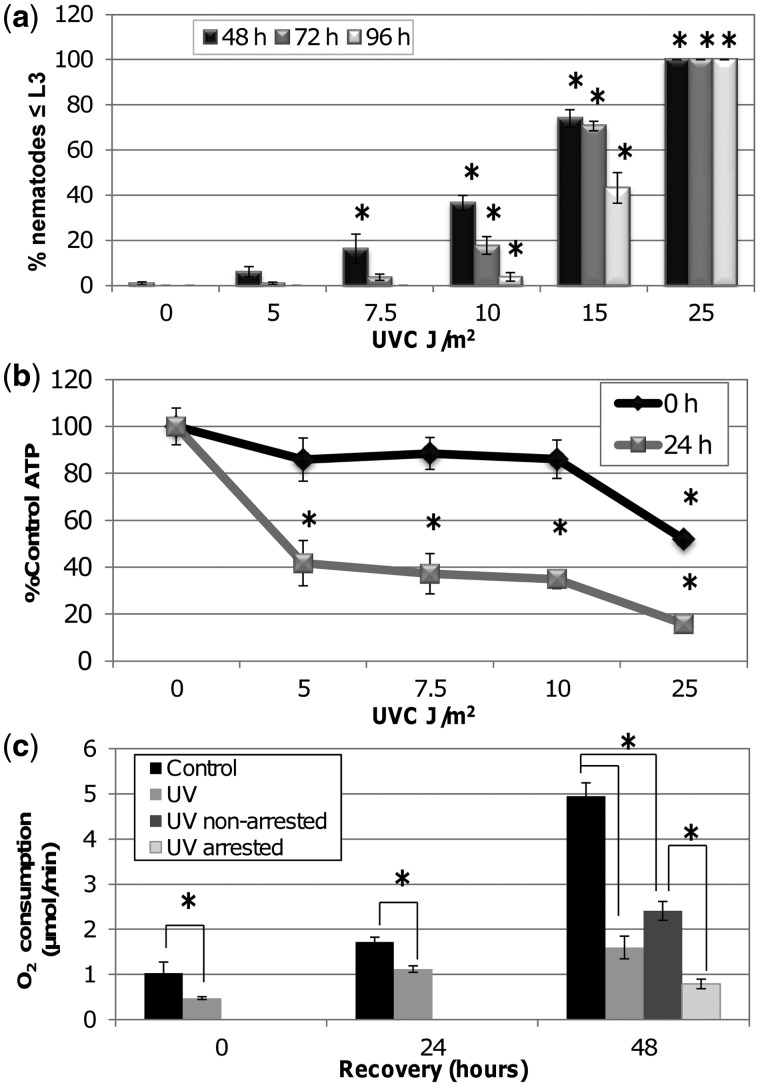
We utilized this L3 arrest phenotype to address the role of removal of persistent mtDNA damage in recovery of mitochondrial function following UVC exposure. To address specifically the effects of mtDNA damage and not nDNA damage, we developed an experimental protocol (Figure 4a) that results in accumulation of mtDNA damage in early larval development but permits repair of nDNA damage. In this protocol, age-synchronized L1 nematodes are dosed three times with UVC over the course of 48 h, with 24 h recovery periods to allow nDNA damage repair. This larval arrest protocol results in time-dependent accumulation of mtDNA damage to a level similar to that induced in adult experiments given earlier in the text, whereas nDNA damage levels were reduced to close to the limit of detection (0.1 lesions/10 kb; (56)) (Figure 4b). Following the third exposure to UVC, L1 nematodes were provided with food and screened at 48, 72 and 96 h to assess the frequency and duration of L3 arrest. A UVC dose-dependent increase in L3 arrest was observed with significant inhibition starting at 7.5 J/m2 (Figure 5a). Steady state ATP levels, quantified 24 h after the third dose and prior to detectable larval arrest, were significantly lower at all UVC exposures (Figure 5b).
Figure 4.
Serial UVC exposure results in mtDNA damage accumulation. (a) Schematic of serial UVC protocol. L1 nematodes on unseeded plates are dosed with UVC at 254 nm every 24 h for a total of three doses. Following the third dose, nematodes are provided with food and developmental stage recorded at 48, 72 and 96 h post UVC exposure. (b) mtDNA damage accumulates over serial UVC exposure and persists 24 h after the last UVC exposure and addition of food. Following 24 h recovery periods, nDNA damage was repaired. Bars ± SEM.
Figure 5.
Serial UVC exposure results in dose-dependent L3 arrest, lower steady state ATP level and reduced O2 consumption. (a) Larval arrest increased in a dose-dependent manner following serial UVC exposure. (b) Steady state ATP levels were significantly lower by 24 h after UVC exposure and addition of food. Asterisks denote a significant treatment effect compared with untreated control (Fisher’s PLSD, P < 0.05). (c) Nematodes exposed to serial UVC 10 J/m2 had significantly lower O2 consumption compared with untreated nematodes at 0, 24 and 48 h post exposure and addition of food. At 48 h, O2 consumption was further decreased in arrested nematodes compared with non-arrested nematodes and both were decreased compared with the untreated group. Two-way ANOVA indicated a significant treatment × recovery interaction (P < 0.0001) and asterisks denote significant differences at each time point (Fisher’s PLSD, P < 0.05). Bars ± SEM.
As a more specific measure of mitochondrial respiration, oxygen consumption was assessed at 0, 24 and 48 h after exposure to 10 J/m2 UVC and addition of food. UVC-exposed nematodes displayed significantly lower oxygen consumption by 24 h (Figure 5c) with a more significant effect in treated nematodes at 48 h. At 48 h when larval arrest is first quantified, control nematodes are at young adult stage while ∼40% of nematodes in the UVC treated group are L2 and L3. We separated these two populations based on size in the UVC exposed group using a COPAS Biosort, defining large or non-arrested nematodes as those which were the same size as untreated nematodes and smaller or ‘arrested’ nematodes as those smaller than untreated nematodes. Both the arrested and non-arrested nematodes had significantly lower oxygen consumption compared with controls, but oxygen consumption in arrested nematodes was significantly lower than non-arrested UVC-treated nematodes.
Thus, accumulated mtDNA damage resulted in an overall decrease in mitochondrial function as indicated by increased L3 arrest, lower steady state ATP level and reduced oxygen consumption. Although nDNA damage is repaired throughout larval exposure and the recovery period, we cannot entirely rule out the possibility that L3 arrest is influenced by lingering low (below our limit of detection) levels of nDNA damage. We note, however, that in a previous study that employed a single 10 J/m2 dose of UVC, which should result in a similar maximal level of nDNA damage to that which we measured throughout the larval arrest protocol (∼0.8 lesions/10 kb) but a much lower level of mtDNA damage (also ∼0.8 lesions/10 kb), no effect on larval development was observed (78).
mtDNA replication is critical for recovery from persistent mtDNA damage
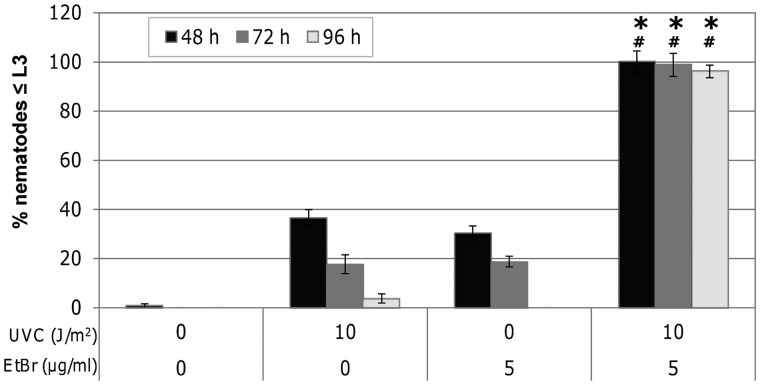
At serial UVC doses <25 J/m2, nematodes recover from larval arrest over time (Figure 5a). This recovery may be facilitated by replacement of UVC-damaged mitochondrial genomes. Ethidium bromide is a DNA intercalating agent that preferentially targets mtDNA over nDNA and blocks replication (79). Previous studies indicate that inhibition of mtDNA replication by ethidium bromide results in L3 arrest and depletion of mtDNA (52). If recovery from UVC-induced L3 arrest is aided by dilution of damaged mtDNA via replication of undamaged genomes, then co-exposure to UVC and ethidium bromide should exacerbate larval arrest. We selected an ethidium bromide concentration (5 µg/ml) that results in a similar frequency and duration of larval arrest as UVC 10 J/m2. Co-exposure with UVC (10 J/m2) and ethidium bromide (5 µg/ml) dramatically exacerbated L3 arrest across all time points compared with UVC or chemical alone (Figure 6). This demonstrates the importance of mtDNA replication in recovery of mitochondrial function resulting from persistent mtDNA damage.
Figure 6.
Co-exposure to UVC and ethidium bromide exacerbates L3 arrest. Co-exposure to UVC (10 J/m2) and ethidium bromide (5 µg/ml) further exacerbates L3 arrest compared with UVC (asterisks) or chemical (hash) exposure alone at every time point (Fisher’s PLSD, P < 0.05). Bars ± SEM.
Recovery from UVC-induced mtDNA damage involves mitochondrial fusion and autophagy
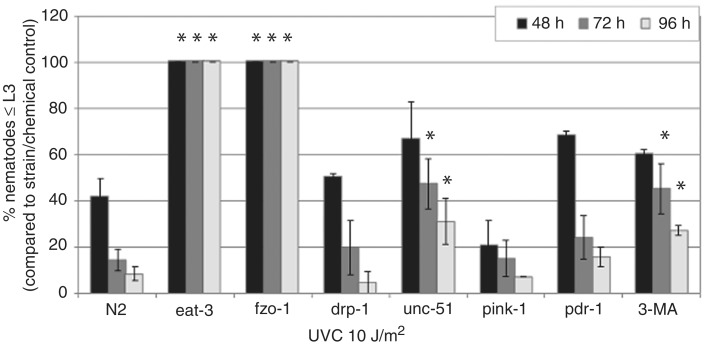
To better understand the role of removal of mtDNA damage in recovery from UVC-induced L3 arrest, we compared L3 arrest in wild-type (N2) C. elegans with strains carrying mutations in fusion, fission and autophagy genes. If removal of persistent mtDNA damage plays a significant role in vivo in recovery from UVC-induced mitochondrial dysfunction then knockout of these genes involved in removal will increase the frequency and duration of larval arrest compared with wild-type. As shown in Figure 7, significant exacerbation of L3 arrest was observed in the fusion mutants fzo-1 and eat-3 and the autophagy mutant unc-51. Mutations in fission gene drp-1 or mitophagy genes pink-1 and pdr-1 did not significantly exacerbate L3 arrest at any time point.
Figure 7.
Mutations in fusion and autophagy genes exacerbate L3 arrest following serial UVC exposure. Mutations in fusion genes fzo-1 and eat-3 and autophagy gene unc-51, as well as inhibition of autophagy with 3-MA, exacerbated L3 arrest compared with wild-type at 72 and 96 h. Asterisks denote a significantly different effect of treatment on mutant compared with wild-type strain (two-way ANOVA, P < 0.05). Bars ± SEM.
We attempted to test other autophagy mutants but were unsuccessful because of embryonic lethality in homozygotes [as in the case of bec-1 (ok961)] or an inability of the mutant strain to recover from the 48-h L1 starvation period [as in the case of atg-18 (gk378)]. Since we were limited in our ability to test genetically for a role of autophagy in recovery from persistent mtDNA damage because of mutant inviability, we chemically inhibited autophagy with 3-MA. N2 nematodes were exposed to 10 mM 3-MA following the third UVC exposure and placement on food. At this dose, 3-MA exposure alone did not result in significant larval arrest (data not shown); however, 3-MA exposure did exacerbate UVC-induced L3 arrest (Figure 7).
It is interesting that knockout of fusion resulted in complete larval arrest with no recovery, whereas knockout of autophagy led to a more moderate increase in larval arrest with delayed recovery. If removal of UVC-induced mtDNA damage were the sole driver of recovery then knockout of these two pathways would lead to similar results. Mitochondrial fusion has been shown to be critical for mtDNA integrity and recovery from mitochondrial dysfunction in part because it aids in mtDNA replication, which is critical for recovery from UVC-induced larval arrest as shown earlier in the text and in functional complementation (80) that results from mixing damaged mtDNA among undamaged mtDNA. These results suggest that removal of damaged mtDNA does aid in recovery from mitochondrial dysfunction; however, mtDNA replication and mitochondrial fusion are necessary for recovery. We suggest that enhanced mitochondrial fusion as a result of drp-1 or pink-1 knockout may explain why knockout of these genes did not affect larval arrest or recovery although both of these are involved in mtDNA damage removal.
As previously mentioned, measuring removal of mtDNA damage in proliferating cells is confounded by dilution of damage associated with cell replication. C. elegans larval development is marked by a 2-fold increase in somatic cells, a 5-fold increase in mtDNA copy number and development of approximately 250 germ cell nuclei (52). Therefore, we were not able to directly test if mutations in these genes prevented removal of damage or if arrested nematodes had more persistent mtDNA damage compared with non-arrested nematodes because the developmental changes would have confounded those measurements. Nonetheless, our genetic and pharmacologic data allow us to conclude that mitochondrial fusion and autophagy play an important role in recovery from UVC-induced mitochondrial dysfunction.
DISCUSSION
Mitochondrial dynamics and autophagy play critical roles in response to persistent mitochondrial DNA damage
MtDNA integrity is critical to mitochondrial function and is more vulnerable than nDNA to common types of environmentally induced damage (6–11). This vulnerability is exacerbated by the fact that mitochondria lack NER, the pathway required to repair helix distorting and bulky DNA damage induced by important environmental genotoxins including UVC, mycotoxins and PAHs (12–15). Thus, at least in most species (fission yeast is an exception (81)), mtDNA damage induced by such agents is irreparable. Our experiments indicate that mitochondrial dynamics and autophagy protect against persistent mtDNA damage; we suggest that this results both from a role in removal of damaged mtDNAs and from a role in protecting mitochondrial function in the face of mtDNA damage.
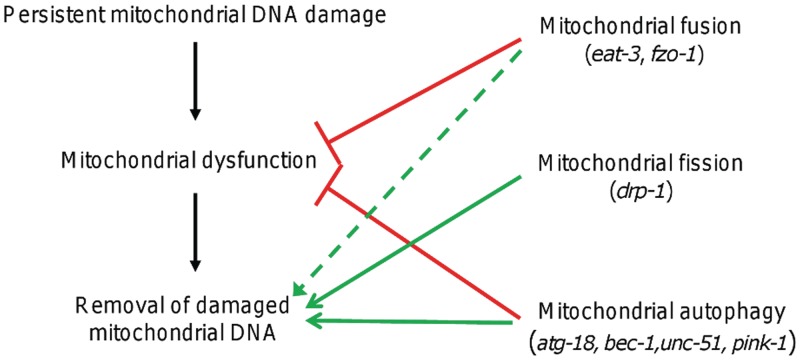
We propose a model (Figure 8) whereby persistent mtDNA damage leads to mitochondrial dysfunction. Mitochondrial fusion protects against dysfunction, as does autophagy, presumably by promoting functional complementation, mtDNA replication and removal of dysfunctional mitochondria (these processes are further discussed below). Mitochondrial genomes carrying persistent damage can be removed by mitochondrial fission and subsequent autophagy of dysfunctional daughter mitochondria, which is observed in our experiments as a mitochondrial fission- and autophagy-dependent removal of mtDNA damage. It is not intuitive that mitochondrial fusion would be required for the removal of damaged mtDNAs, but we propose that fusion is so critical for mitochondrial function (as supported by our larval arrest results) and mitochondrial biogenesis that the lack of fusion results in severe mitochondrial dysfunction that incapacitate the processes required for removal. Although additional experiments will be required to fully test this model, it is consistent with our data and supported by a significant body of literature, as discussed later in the text.
Figure 8.
Proposed model of the effect of mitochondrial fusion, fission and autophagy processes on persistent mtDNA damage-induced mitochondrial dysfunction and removal of mtDNA damage. Persistent mtDNA damage leads to mitochondrial dysfunction (e.g. reduced ATP levels and larval arrest). Mitochondrial fusion protects against dysfunction, as does autophagy, presumably by promoting functional complementation, mtDNA replication and removal of dysfunctional mitochondria (red solid lines). Mitochondrial fission and autophagy promote mtDNA damage removal (green dashed lines), whereas mitochondrial fusion is indirectly required for mtDNA damage removal because it is required to preserve basic mitochondrial function and mtDNA replication (dashed green line denotes the indirect effect of fusion on mtDNA damage removal).
Mitochondrial fusion, fission and autophagy are required for removal of persistent mtDNA damage
Irreparable, helix-distorting mtDNA damage is persistent but our results indicate that this damage is removed slowly in vivo with significant removal observed by 72 h post exposure. These data are consistent with other literature indicating the persistence of UV-induced photodimers up to 48 h (15,17,18,66,67), although one study reported a 40% reduction in UVC-induced photodimers in mtDNA of mouse cells by 24 h (82). We considered the role of mitochondrial dynamics and degradation via autophagy in this removal. Autophagy is the primary mechanism of removal for mitochondria and can be both non-selective, such as during starvation (31), or selective meaning that isolated, dysfunctional mitochondria with low MP are degraded preferentially (30,39,43). Selective autophagy of mitochondria or mitophagy under certain conditions is mediated by accumulation of PINK1, a serine/threonine–protein kinase on dysfunctional mitochondria and subsequent recruitment of Parkin, an E3 ubiquitin ligase to those mitochondria (44–46). However, the necessity and sufficiency of these events to induce mitophagy in vivo is unclear (47,83).
UNC51 is a serine/threonine kinase with a role in autophagy induction; BEC1, a class III phosphatidylinositol 3-kinase (PI3K), is involved in recruiting other autophagy proteins to pre-autophagosomal structures; and ATG18, which binds phosphoinositol-3-phosphate, is required for autophagosomes formation; mutation or knockdown of these genes results in abnormal autophagy in C. elegans (71). We found that RNAi knockdown of autophagy/mitophagy genes bec-1, unc-51 and pink-1 inhibited removal of UVC-induced mtDNA damage, suggesting that autophagy is an important mechanism of degradation of damaged mtDNA. The lack of an effect of knockdown of atg-18 may result from insufficient knockdown or a degradation mechanism independent of atg-18. We noted that knockdown of autophagy proteins bec-1 and unc-51 prevented the trend of an age-dependent decrease in mtDNA copy number; however, atg-18 knockdown did not prevent this decrease. This is consistent with incomplete knockdown of atg-18, which might permit residual mitochondrial turnover and explain why knockdown did not inhibit damage removal. Interestingly, knockdown of pink-1 but not pdr-1, the C. elegans Parkin homolog, inhibited mtDNA damage removal. Parkin is thought to act downstream of PINK1 and in other model systems compromised function of either reduces mitophagy (44,45). In C. elegans, few studies have investigated the role of these genes in relation to mitochondrial function. pdr-1 mutants have reduced oxygen consumption and are sensitive to mitochondrial complex I inhibitors (84), pink-1 mutants have reduced cristae length and are sensitive to oxidative stress (85) and expression of wild-type pink-1 and pdr-1 rescued α-synuclein induced mitochondrial fragmentation (86). These reports are consistent with those in other models suggesting conserved function of these proteins between C. elegans and mammals (85). Further research is needed to better understand mitophagy in C. elegans and the role of pink-1 and pdr-1 in that process.
Knockdown of fusion genes fzo-1 and eat-3 and fission gene drp-1 also inhibited removal of UVC-induced mtDNA damage. The role of mitochondrial morphology and dynamics in mediating mitophagy is unclear. Inhibition of fission via manipulation of fission genes Fis1 and Drp1 results in reduced mitophagy, whereas overexpression of fission proteins induces mitophagy and decreases mitochondrial mass (87). Consistent with these studies, our results indicate the Drp-1 is required for removal of damaged mtDNAs. It may be tempting then to assume that knockdown of fusion genes would enhance mitophagy and removal of mtDNAs. Certainly OPA1 deficiency results in a general increase in autophagy, and mitochondria targeted for degradation exhibit OPA1 depletion (39,88). However, loss of mitochondrial mass resulting from nicotinamide treatment is partially inhibited by fusion knockdown rather than enhanced as might be predicted (89), and studies that inhibit fusion without compromising MP or mitochondrial function show that fission alone without accompanying mitochondrial dysfunction does not result in increased mitophagy (43,90). Consistent with our data, human fibroblasts harboring a missense mutation in the fusion gene MFN2 exhibit significantly reduced repair efficiency of oxidative mtDNA damage and DNA instability (91). Our data suggest that fusion is required for removal of mtDNA damage; whether this is by facilitating autophagy or another mechanism requires further research.
Persistent mtDNA damage induces autophagy without detectable changes in mitochondrial morphology
We considered that slow removal of UVC-induced mtDNA damage may reflect the natural turnover rate of mitochondria and not an induction in autophagy. We therefore evaluated the level of autophagy following UVC exposure in LGG-1::GFP expressing C. elegans. We observed a mild increase in the number of autophagosomes in the UVC-exposed nematodes 24 h after treatment, suggesting that UVC exposure does induce autophagy. Whether this increase in autophagy signifies a greater mitochondrial clearance requires further research.
Mitochondrial dynamics are responsive to mitochondrial function. Loss of MP inhibits fusion and results in an overall fragmented morphology and treatment with pro-oxidants or mitochondrial respiratory chain uncouplers triggers fragmentation (92–94). On the other hand, mitochondrial hyperfusion has been observed following treatment with DNA intercalators or UVC (94,95) highlighting the complex responses of mitochondrial dynamics to mitochondrial stressors. We evaluated the effect of UVC exposure on mitochondrial morphology. We did not observe a difference in mitochondrial interconnectivity or elongation with UVC treatment at 24 or 48 h after treatment. There remains the possibility that changes in morphology occurred between the time points analyzed; however, given the persistence of this damage we would expect any morphological changes induced by this damage would also be persistent. Thus, while our genetic data demonstrate the importance of mitochondrial dynamics in responding to persistent mtDNA damage, such damage did not lead to changes in mitochondrial morphology that were easily observable by microscopy.
Recovery from UVC-induced larval arrest requires mtDNA replication and mitochondrial fusion and is aided by removal of damaged mtDNA via autophagy
Mitochondria contain multiple copies of DNA; thus recovery from helix-distorting mtDNA damage could be achieved by both removal of the damaged copies and dilution of damaged copies with undamaged genomes. In adult glp-1 C. elegans raised at 25°C [(52,67) and Figure 1d] and 15°C (Supplementary Figure S1), mtDNA copy number decreases with age. This indicates that degradation of mtDNA is occurring and the resultant decline in copy number is not compensated for by increased biogenesis. This decrease in copy number likely relates to the decreased metabolic rate and ATP production shown in aging nematodes (70). We therefore used developing rather than adult C. elegans to determine the significance of removal of damaged mtDNAs in recovery from UVC exposure. During the L3 and L4 transition, nematodes switch to OXPHOS to produce energy; this is accompanied by a significant increase in energy demand and mtDNA copy number (52). Inhibition of mitochondrial replication or translation or mutations in mitochondrial electron transport chain components all result in arrest at or before the L3 stage (51–53). Using a novel protocol designed to permit accumulation of a high level of mtDNA damage early in L1 (Figure 4a), we showed that UVC-induced mtDNA damage caused dose-dependent increases in mitochondrial dysfunction as indicated by L3 arrest, reduced steady state ATP level and reduced oxygen consumption. Treatment with low levels of ethidium bromide nearly abolished recovery from UVC-induced L3 arrest suggesting that recovery of mitochondrial function is facilitated by dilution of damaged mtDNA via mtDNA replication.
unc-51 mutants displayed a significant increase in L3 arrest as did nematodes in which autophagy was inhibited with 3-MA. These data indicate that autophagy (and presumably removal of damaged mtDNA via autophagy) is involved in functional recovery from persistent mtDNA damage. Autophagy provides protection against mitochondrial dysfunction, apoptosis and cell death following toxicant exposure or cell stress (40,41,96,97), and abnormal mitochondrial autophagy coupled with mitochondrial dysfunction occurs in a variety of pathologies including mitochondrial diseases (40), lysosomal storage disorders (98) and neurodegenerative disorders including Parkinson’s, Alzheimer’s and Huntington’s disease (99–101). It is not unlikely that autophagy protects against mitochondrial stressors, in part, by removing mitochondria that would otherwise generate reactive oxygen species and trigger apoptosis or necrosis (48,49). Thus, the reduction in autophagy with age particularly in post-mitotic cells may result in persistence of unstable mitochondria potentially leading to development of age-related disorders (102). Surprisingly, pink-1 mutants did not exhibit increased larval arrest despite the role of this protein in removal of damaged mtDNA in adult C. elegans. This may indicate that removal of damage in replicating cells is not PINK-1 dependent. On the other hand, PINK1 knockdown in C. elegans increases mitochondrial fusion (Unpublished observations made by Maxwell C.K. Leung, Duke University) and in other model organisms is associated with increased mitochondrial fusion (103,104) which is critical for recovery from L3 arrest as discussed below.
Mutations in the fusion genes fzo-1 and eat-3 resulted in complete arrest of larval development following serial UVC, suggesting that mitochondrial fusion is critical in recovery of mitochondrial function following UVC-induced mtDNA damage. Although it is likely that the complete larval arrest observed in these mutants is partially because of inhibition of damage removal, the degree of exacerbation suggests failure of other recovery mechanisms. Fusion is critical for recovery of mitochondrial function following toxicant exposure and cell stress. Loss of fusion capacity increases susceptibility to loss of MP (105), respiratory deficiency (105,106), oxidative stress (61,107), apoptosis and cell death (61,108,109). Additionally, enhanced mitochondrial fusion has been shown to be protective against UV irradiation-induced depletion of ATP (94,95) and blockage of mtDNA replication by mtDNA intercalators (110).
Fusion provides protection against the effects of mtDNA mutations by facilitating ‘functional complementation’ (111,112), the mixing of lipids, proteins and mtDNA to compensate for mutated DNA (80) and this mechanism may explain the lack of phenotypic expression of mtDNA mutations until the occurrence is high (>40–90%) (113). This function of fusion also equally distributes proteins involved in mtDNA replication and repair (80). Mice harboring mutations in fusion proteins exhibit severe depletion of mtDNA (114) because of defects in mtDNA replication (80), and polg-1–mutant C. elegans exhibit increased mitochondrial fusion in response to mtDNA depletion (57). As described earlier in the text, mtDNA replication is a particularly important mechanism of recovery from L3 arrest. Therefore, mitochondrial fusion may aid in recovery from UVC-induced larval arrest by promoting functional complementation, mtDNA replication and removal of damaged mtDNAs.
Consistent with this idea, mutation in the fission gene drp-1 did not increase L3 arrest. Although this could suggest that mtDNA damage removal is DRP-1 independent in developing C. elegans, we hypothesize that enhanced fusion resulting from the drp-1 mutation may compensate for lack of mtDNA damage removal. Decreased mitochondrial fragmentation has been shown to be protective against toxicant-induced mitochondrial dysfunction, ATP depletion and apoptosis (89,90,94). Conversely, Yang et al. found that drp-1 nematodes are more sensitive to heat stress and paraquat (115) and, in mammalian cells, knockdown of drp1 results in mitochondrial dysfunction, loss and disorganization of mtDNA and decreased membrane fluidity (92,116).
We were unable to directly test in developing C. elegans if mutations in fusion, fission and autophagy/mitophagy genes inhibit removal of UVC-induced mtDNA damage or if lack of damage removal correlates with the frequency and duration of L3 arrest because of developmental changes (i.e. somatic and germ cell proliferation and substantial increases in mtDNA copy number) that would have confounded those measurements. Assuming that larval C. elegans employ the same mechanism as adults to remove UVC-induced mtDNA damage, our genetic and pharmacologic data suggest that removal of mtDNA damage aids in recovery from UVC-induced mitochondrial dysfunction facilitated by autophagy and mitochondrial fusion.
Potential role of NER proteins and mitochondrial nucleases
Our model assumes that mitochondria in C. elegans, like other metazoans studied to date, lack NER such that UVC-induced mtDNA damage is irreparable and persistent (12–15). Recently, however, NER proteins CSA and CSB have been shown to localize to mitochondria and to have a direct role in sensing and responding to oxidative mtDNA damage (117,118), as well as inducing mitochondrial turnover via autophagy in response to cell stress (119). Based on these studies, an interesting future research direction will be to determine if CSA/CSB aid in the response to helix-distorting mtDNA damage and facilitate the removal of these lesions via autophagy.
There is evidence mtDNA harboring double strand breaks resulting from high levels of oxidative lesions can be degraded in vitro by nucleases residing in mitochondria (120–123) of which endonuclease G, involved in nDNA degradation during apoptosis and EXOG, involved in mitochondrial long patch BER (123), remain the primary candidates. Additionally, fission yeast possesses a UV-damaged DNA endonuclease-dependent excision repair that drives repair of UV photodimers in mtDNA by nicking the phosphodiester bond 5′ to the UV-induced photodimers to initiate BER (81). This repair mechanism has not been observed in higher organisms. Still, it is possible that degradation of mtDNA harboring irreparable UVC-induced photodimers may in part involve nuclease-dependent degradation. We considered this less likely than our current model for two reasons. Firstly, the degradation rate reported for mtDNA with strand breaks is significantly faster than that reported here, with significant degradation reported within an hour of oxidant exposure (122). Secondly, when we screened viable C. elegans mutants of known endonucleases including cps-6, the C. elegans homolog of endonuclease G using the larval arrest protocol, we found that mutations in cps-6, crn-6 and nuc-1 did not exacerbate larval arrest (Supplementary Figure S3). This indicates that these nucleases are not necessary for recovery from UVC-induced mtDNA damage in larval C. elegans and suggests that they are not involved in removal of UVC-induced mtDNA damage. However, because of the potential redundancy in mitochondrial nucleases, we cannot exclude a role of nucleases in recovery from and removal of UV-induced mtDNA damage.
Broader implications of persistent mtDNA damage removal
In summary, our results suggest a model whereby mitochondrial dynamics and autophagy play a role both specifically in removal of mtDNA damage, and more broadly in the response to mtDNA damage and mitochondrial dysfunction (Figure 8). This research shows that UVC-induced mtDNA damage can be removed, and furthermore demonstrates that mitochondrial dynamics and autophagy are part of a novel pathway for the removal of otherwise irreparable mtDNA damage. Based on this and published literature, we hypothesize that initial recovery of mitochondrial function from such mtDNA damage requires mitochondrial fusion and mtDNA replication, and is assisted by removal of this damage via autophagy. Upon recovery from mtDNA damage, mitochondrial fission is restored creating a heterogeneous population of mitochondria, some with a high proportion of damaged mtDNA, which are removed by autophagy, potentially in a selective fashion involving PINK1. This pathway is of potentially great significance because of the fact that many common environmental contaminants cause high levels of mtDNA damage that, like UVC-induced damage, is not repaired in the mitochondrial genome. This pathway may also be important for removal of a subset of oxidative mtDNA damage (124) and damage induced by certain biological aldehydes (125) that are repaired in the nucleus by NER, extending the importance of our findings to damage caused by normal metabolism. Finally, mutations in genes in these pathways are associated with human disease states, demonstrating the potential for important gene–environment interactions affecting mitochondrial health after genotoxin exposure.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
National Institute of Environmental Health Sciences [1 P30 ES-011961-01A1, 1R01-ES017540-01A2]; National Institute of Neurological Disorders and Stroke [1 R21-NS065468-01]. Funding for open access charge: National Institute of Environmental Health Sciences [1 P30 ES-011961-01A1, 1R01-ES017540-01A2].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Alexander van der Bliek, Dr Ding Xue, Adam Knight, Dr Guy Caldwell and Dr Christina Lagido for providing mutant and transgenic C. elegans strains, Dr Windy Boyd for assistance with RNAi, Alex Ji for assistance with oxygen consumption experiments and Dr Alicia Melendez for advice on LGG-1::GFP analysis.
REFERENCES
- 1.Greaves LC, Reeve AK, Taylor RW, Turnbull DM. Mitochondrial DNA and disease. J. Pathol. 2012;226:274–286. doi: 10.1002/path.3028. [DOI] [PubMed] [Google Scholar]
- 2.Bossy-Wetzel E, Barsoum MJ, Godzik A, Schwarzenbacher R, Lipton SA. Mitochondrial fission in apoptosis, neurodegeneration and aging. Curr. Opin. Cell Biol. 2003;15:706–716. doi: 10.1016/j.ceb.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 4.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 5.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 6.Backer JM, Weinstein IB. Interaction of Benzo(a)pyrene and its dihydrodiol-epoxide derivative with nuclear and mitochondrial DNA in C3H10T½ cell cultures. Cancer Res. 1982;42:2764–2769. [PubMed] [Google Scholar]
- 7.Mita S, Monnat RJ, Jr, Loeb LA. Resistance of HeLa cell mitochondrial DNA to mutagenesis by chemical carcinogens. Cancer Res. 1988;48:4578–4583. [PubMed] [Google Scholar]
- 8.Stairs PW, Guzelian PS, Van Tuyle GC. Benzo[a]pyrene differentially alters mitochondrial and nuclear DNA synthesis in primary hepatocyte cultures. Res. Commun. Chem. Pathol. Pharmacol. 1983;42:95–106. [PubMed] [Google Scholar]
- 9.Jung D, Cho Y, Collins LB, Swenberg JA, Di Giulio RT. Effects of benzo[a]pyrene on mitochondrial and nuclear DNA damage in Atlantic killifish (Fundulus heteroclitus) from a creosote-contaminated and reference site. Aquat. Toxicol. 2009;95:44–51. doi: 10.1016/j.aquatox.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Backer JM, Weinstein IB. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980;209:297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- 11.Allen JA, Coombs MM. Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA. Nature. 1980;287:244–245. doi: 10.1038/287244a0. [DOI] [PubMed] [Google Scholar]
- 12.Chen RH, Maher VM, Brouwer J, Vandeputte P, McCormick JJ. Preferential repair and strand-specific repair of benzo[a]pyrene diol epoxide adducts in the HPRT gene of diploid human fibroblasts. Proc. Natl Acad. Sci. USA. 1992;89:5413–5417. doi: 10.1073/pnas.89.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedard LL, Massey TE. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006;241:174–183. doi: 10.1016/j.canlet.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 14.Sinha RP, Hader DP. UV-induced DNA damage and repair: a review. Photochem. Photobiol. Sci. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 15.LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 16.Salazar I, Tarrago-Litvak L, Gil L, Litvak S. The effect of benzo[a]pyrene on DNA synthesis and DNA polymerase activity of rat liver mitochondria. FEBS Lett. 1982;138:45–49. doi: 10.1016/0014-5793(82)80391-8. [DOI] [PubMed] [Google Scholar]
- 17.Clayton D, Doda J, Friedberg E. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pascucci B, Versteegh A, van Hoffen A, van Zeeland AA, Mullenders LH, Dogliotti E. DNA repair of UV photoproducts and mutagenesis in human mitochondrial DNA. J. Mol. Biol. 1997;273:417–427. doi: 10.1006/jmbi.1997.1268. [DOI] [PubMed] [Google Scholar]
- 19.Niranjan BG, Bhat NK, Avadhani NG. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982;215:73–75. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- 20.Sajan MP, Satav JG, Bhattacharya RK. Activity of some respiratory enzymes and cytochrome contents in rat hepatic mitochondria following aflatoxin B1 administration. Toxicol Lett. 1995;80:55–60. doi: 10.1016/0378-4274(95)03256-k. [DOI] [PubMed] [Google Scholar]
- 21.Ko C-B, Kim S-J, Park C, Kim B-R, Shin C-H, Choi S, Chung S-Y, Noh J-H, Jeun J-H, Kim N-S, et al. Benzo(a)pyrene-induced apoptotic death of mouse hepatoma Hepa1c1c7 cells via activation of intrinsic caspase cascade and mitochondrial dysfunction. Toxicology. 2004;199:35–46. doi: 10.1016/j.tox.2004.01.039. [DOI] [PubMed] [Google Scholar]
- 22.Shen HM, Shi CY, Lee HP, Ong CN. Aflatoxin B1-induced lipid peroxidation in rat liver. Toxicol. Appl. Pharmacol. 1994;127:145–150. doi: 10.1006/taap.1994.1148. [DOI] [PubMed] [Google Scholar]
- 23.Baird WM, Hooven LA, Mahadevan B. Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ. Mol. Mutagen. 2005;45:106–114. doi: 10.1002/em.20095. [DOI] [PubMed] [Google Scholar]
- 24.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat. Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 25.Graziewicz MA, Sayer JM, Jerina DM, Copeland WC. Nucleotide incorporation by human DNA polymerase gamma opposite benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyguanosine and deoxyadenosine. Nucleic Acids Res. 2004;32:397–405. doi: 10.1093/nar/gkh213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasiviswanathan R, Gustafson MA, Copeland WC, Meyer JN. Human mitochondrial DNA polymerase gamma exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J. Biol. Chem. 2012;287:9222–9229. doi: 10.1074/jbc.M111.306852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Partridge MA, Huang SX, Kibriya MG, Ahsan H, Davidson MM, Hei TK. Environmental mutagens induced transversions but not transitions in regulatory region of mitochondrial DNA. J. Toxicol. Environ. Health A. 2009;72:301–304. doi: 10.1080/15287390802526381. [DOI] [PubMed] [Google Scholar]
- 28.Bogenhagen DF. Repair of mtDNA in vertebrates. Am. J. Hum. Genet. 1999;64:1276–1281. doi: 10.1086/302392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croteau DL, Stierum RH, Bohr VA. Mitochondrial DNA repair pathways. Mutat. Res. 1999;434:137–148. doi: 10.1016/s0921-8777(99)00025-7. [DOI] [PubMed] [Google Scholar]
- 30.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J. Cell. Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 33.Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 34.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Ney PA. Autophagy-dependent and -independent mechanisms of mitochondrial clearance during reticulocyte maturation. Autophagy. 2009;5:1064–1065. doi: 10.4161/auto.5.7.9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elmore S, Qian T, Grissom S, Lemasters J. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 37.Mijaljica D, Prescott M, Devenish RJ. Different fates of mitochondria: alternative ways for degradation? Autophagy. 2007;3:4–9. doi: 10.4161/auto.3011. [DOI] [PubMed] [Google Scholar]
- 38.Tolkovsky AM, Xue L, Fletcher GC, Borutaite V. Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease? Biochimie. 2002;84:233–240. doi: 10.1016/s0300-9084(02)01371-8. [DOI] [PubMed] [Google Scholar]
- 39.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotan D, Cordero MD, Garrido-Maraver J, Oropesa-Avila M, Rodriguez-Hernandez A, Gomez Izquierdo L, De la Mata M, De Miguel M, Lorite JB, Infante ER, et al. Secondary coenzyme Q10 deficiency triggers mitochondria degradation by mitophagy in MELAS fibroblasts. FASEB J. 2011;25:2669–2687. doi: 10.1096/fj.10-165340. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Hernandez A, Cordero MD, Salviati L, Artuch R, Pineda M, Briones P, Gomez Izquierdo L, Cotan D, Navas P, Sanchez-Alcazar JA. Coenzyme Q deficiency triggers mitochondria degradation by mitophagy. Autophagy. 2009;5:19–32. doi: 10.4161/auto.5.1.7174. [DOI] [PubMed] [Google Scholar]
- 42.Kim I, Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am. J. Physiol. Cell. Physiol. 2011;300:C308–C317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell. Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 46.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilkerson RW, De Vries RL, Lebot P, Wikstrom JD, Torgyekes E, Shirihai OS, Przedborski S, Schon EA. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum. Mol. Genet. 2012;21:978–990. doi: 10.1093/hmg/ddr529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boya P, Gonzalez-Polo R-A, Casares N, Perfettini J-L, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malhi H, Gores GJ, Lemasters JJ. Apoptosis and necrosis in the liver: a tale of two deaths? Hepatology. 2006;43:S31–S44. doi: 10.1002/hep.21062. [DOI] [PubMed] [Google Scholar]
- 50.Addo MG, Cossard R, Pichard D, Obiri-Danso K, Rotig A, Delahodde A. Caenorhabditis elegans, a pluricellular model organism to screen new genes involved in mitochondrial genome maintenance. Biochim. Biophys. Acta. 2010;1802:765–773. doi: 10.1016/j.bbadis.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsang WY, Lemire BD. Mitochondrial genome content is regulated during nematode development. Biochem. Biophys. Res. Commun. 2002;291:8–16. doi: 10.1006/bbrc.2002.6394. [DOI] [PubMed] [Google Scholar]
- 53.Tsang WY, Sayles LC, Grad LI, Pilgrim DB, Lemire BD. Mitochondrial respiratory chain deficiency in Caenorhabditis elegans results in developmental arrest and increased life span. J. Biol. Chem. 2001;276:32240–32246. doi: 10.1074/jbc.M103999200. [DOI] [PubMed] [Google Scholar]
- 54.Boyd WA, Crocker TL, Rodriguez AM, Leung MCK, Wade Lehmann D, Freedman JH, Van Houten B, Meyer JN. Nucleotide excision repair genes are expressed at low levels and are not detectably inducible in Caenorhabditis elegans somatic tissues, but their function is required for normal adult life after UVC exposure. Mutat. Res. 2009;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter SE, Jung D, Di Giulio RT, Meyer JN. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods. 2010;51:444–451. doi: 10.1016/j.ymeth.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 57.Bratic I, Hench J, Henriksson J, Antebi A, Burglin TR, Trifunovic A. Mitochondrial DNA level, but not active replicase, is essential for Caenorhabditis elegans development. Nucleic Acids Res. 2009;37:1817–1828. doi: 10.1093/nar/gkp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 60.Kodoyianni V, Maine EM, Kimble J. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell. 1992;3:1199–1213. doi: 10.1091/mbc.3.11.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanazawa T, Zappaterra MD, Hasegawa A, Wright AP, Newman-Smith ED, Buttle KF, McDonald K, Mannella CA, van der Bliek AM. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 2008;4:e1000022. doi: 10.1371/journal.pgen.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 63.Dagda RK, Cherra SJ, III, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merrill RA, Dagda RK, Dickey AS, Cribbs JT, Green SH, Usachev YM, Strack S. Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol. 2011;9:e1000612. doi: 10.1371/journal.pbio.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grad LI, Sayles LC, Lemire BD. Isolation and functional analysis of mitochondria from the nematode Caenorhabditis elegans. Methods Mol. Biol. 2007;372:51–66. doi: 10.1007/978-1-59745-365-3_4. [DOI] [PubMed] [Google Scholar]
- 66.Rochette PJ, Brash DE. Human telomeres are hypersensitive to UV-induced DNA damage and refractory to repair. PLoS Genet. 2010;6:e1000926. doi: 10.1371/journal.pgen.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 69.Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis A-K, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braeckman BP, Houthoofd K, De Vreese A, Vanfleteren JR. Assaying metabolic activity in ageing Caenorhabditis elegans. Mech. Ageing Dev. 2002;123:105–119. doi: 10.1016/s0047-6374(01)00331-1. [DOI] [PubMed] [Google Scholar]
- 71.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 72.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morck C, Pilon M. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev. Biol. 2006;6:39. doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy. 2007;3:569–580. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 75.Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blommaart EFC, Krause U, Schellens JPM, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 77.Samokhvalov V, Scott BA, Crowder CM. Autophagy protects against hypoxic injury in C. elegans. Autophagy. 2008;4:1034–1041. doi: 10.4161/auto.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Astin JW, O'Neil NJ, Kuwabara PE. Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues, RAD-3 and WWP-1. DNA Repair. 2008;7:267–280. doi: 10.1016/j.dnarep.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 79.Gaines G, Attardi G. Intercalating drugs and low temperatures inhibit synthesis and processing of ribosomal RNA in isolated human mitochondria. J. Mol. Biol. 1984;172:451–466. doi: 10.1016/s0022-2836(84)80017-0. [DOI] [PubMed] [Google Scholar]
- 80.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yasuhira S, Yasui A. Alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe operates both in nucleus and in mitochondria. J. Biol. Chem. 2000;275:11824–11828. doi: 10.1074/jbc.275.16.11824. [DOI] [PubMed] [Google Scholar]
- 82.Kalinowski DP, Illenye S, Van Houten B. Analysis of DNA damage and repair in murine leukemia L1210 cells using a quantitative polymerase chain reaction assay. Nucleic Acids Res. 1992;20:3485–3494. doi: 10.1093/nar/20.13.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum. Mol. Genet. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ved R, Saha S, Westlund B, Perier C, Burnam L, Sluder A, Hoener M, Rodrigues CMP, Alfonso A, Steer C, et al. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of α-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J. Biol. Chem. 2005;280:42655–42668. doi: 10.1074/jbc.M505910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J. Biol. Chem. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kamp F, Exner N, Lutz AK, Wender N, Hegermann J, Brunner B, Nuscher B, Bartels T, Giese A, Beyer K, et al. Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, parkin and DJ-1. EMBO. J. 2010;29:3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.White KE, Davies VJ, Hogan VE, Piechota MJ, Nichols PP, Turnbull DM, Votruba M. OPA1 deficiency associated with increased autophagy in retinal ganglion cells in a murine model of dominant optic atrophy. Invest. Ophthalmol. Vis. Sci. 2009;50:2567–2571. doi: 10.1167/iovs.08-2913. [DOI] [PubMed] [Google Scholar]
- 89.Kang HT, Hwang ES. Nicotinamide enhances mitochondria quality through autophagy activation in human cells. Aging Cell. 2009;8:426–438. doi: 10.1111/j.1474-9726.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- 90.Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim. Biophys. Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- 91.Rouzier C, Bannwarth S, Chaussenot A, Chevrollier A, Verschueren A, Bonello-Palot N, Fragaki K, Cano A, Pouget J, Pellissier JF, et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain. 2012;135:23–34. doi: 10.1093/brain/awr323. [DOI] [PubMed] [Google Scholar]
- 92.Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, Rossignol R. Mitochondrial bioenergetics and structural network organization. J. Cell Sci. 2007;120:838. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 93.Malka F, Guillery O, Cifuentes-Diaz C, Guillou E, Belenguer P, Lombès A, Rojo M. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep. 2005;6:853. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zanchetta LM, Garcia A, Lyng F, Walsh J, Murphy JE. Mitophagy and mitochondrial morphology in human melanoma-derived cells post exposure to simulated sunlight. Int. J. Radiat. Biol. 2011;87:506–517. doi: 10.3109/09553002.2011.556175. [DOI] [PubMed] [Google Scholar]
- 96.Marino ML, Fais S, Djavaheri-Mergny M, Villa A, Meschini S, Lozupone F, Venturi G, Della Mina P, Pattingre S, Rivoltini L, et al. Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis. 2010;1:e87. doi: 10.1038/cddis.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hamacher-Brady A, Brady N, Gottlieb R. The interplay between pro-death and pro-survival signaling pathways in myocardial ischemia/reperfusion injury: apoptosis meets autophagy. Cardiovasc. Drugs Ther. 2006;20:445–462. doi: 10.1007/s10557-006-0583-7. [DOI] [PubMed] [Google Scholar]
- 98.Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, de Pablo R, Tacchetti C, Rubinsztein DC, Ballabio A. A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 99.Vila M, Ramonet D, Perier C. Mitochondrial alterations in Parkinson's disease: new clues. J. Neurochem. 2008;107:317–328. doi: 10.1111/j.1471-4159.2008.05604.x. [DOI] [PubMed] [Google Scholar]
- 100.Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington's disease. Eur. J. Neurosci. 2008;27:2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 101.Santos RX, Correia SoC, Xinglong W, Perry G, Smith MA, Moreira PI, Xiongwei Z. A synergistic dysfunction of mitochondrial fission/fusion dynamics and mitophagy in Alzheimer's disease. J. Alzheimers Dis. 2010;20:401–412. doi: 10.3233/JAD-2010-100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Terman A, Kurz T, Navratil M, Arriaga EA, Brunk UT. Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid. Redox Signal. 2010;12:503–535. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gispert S, Ricciardi F, Kurz A, Azizov M, Hoepken HH, Becker D, Voos W, Leuner K, Muller WE, Kudin AP, et al. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS One. 2009;4:e5777. doi: 10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu W, Sun Y, Guo S, Lu B. The PINK1/Parkin pathway regulates mitochondrial dynamics and function in mammalian hippocampal and dopaminergic neurons. Hum. Mol. Genet. 2011;20:3227–3240. doi: 10.1093/hmg/ddr235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 107.Zanna C, Ghelli A, Porcelli A, Karbowski M, Youle R, Schimpf S, Wissinger B, Pinti M, Cossarizza A, Vidoni S. OPA 1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131:352. doi: 10.1093/brain/awm335. [DOI] [PubMed] [Google Scholar]
- 108.Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 109.Arnoult D, Grodet A, Lee YJ, Estaquier J, Blackstone C. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J. Biol. Chem. 2005;280:35742–35750. doi: 10.1074/jbc.M505970200. [DOI] [PubMed] [Google Scholar]
- 110.Ashley N, Poulton J. Anticancer DNA intercalators cause p53-dependent mitochondrial DNA nucleoid re-modelling. Oncogene. 2009;28:3880–3891. doi: 10.1038/onc.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoneda M, Miyatake T, Attardi G. Complementation of mutant and wild-type human mitochondrial DNAs coexisting since the mutation event and lack of complementation of DNAs introduced separately into a cell within distinct organelles. Mol. Cell. Biol. 1994;14:2699–2712. doi: 10.1128/mcb.14.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ono T, Isobe K, Nakada K, Hayashi JI. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet. 2001;28:272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- 113.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem. J. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 115.Yang CC, Chen D, Lee SS, Walter L. The dynamin-related protein DRP-1 and the insulin signaling pathway cooperate to modulate C. elegans longevity. Aging Cell. 2011;10:724–728. doi: 10.1111/j.1474-9726.2011.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM, III, Stevnsner T, et al. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dolle ME, Kuiper R, Majora M, et al. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson Iii DM, Cooper M, Spencer R, de Cabo R, et al. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J. Exp. Med. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Davies AM, Hershman S, Stabley GJ, Hoek JB, Peterson J, Cahill A. A Ca2+-induced mitochondrial permeability transition causes complete release of rat liver endonuclease G activity from its exclusive location within the mitochondrial intermembrane space. Identification of a novel endo-exonuclease activity residing within the mitochondrial matrix. Nucleic Acids Res. 2003;31:1364–1373. doi: 10.1093/nar/gkg205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ikeda S, Ozaki K. Action of mitochondrial endonuclease G on DNA damaged by l-ascorbic acid, peplomycin, and cis-diamminedichloroplatinum (II) Biochem. Biophys. Res. Commun. 1997;235:291–294. doi: 10.1006/bbrc.1997.6786. [DOI] [PubMed] [Google Scholar]
- 122.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5'-EXO/endonuclease) in their repair. J. Biol. Chem. 2011;286:31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brooks PJ. The case for 8,5'-cyclopurine-2'-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience. 2007;145:1407–1417. doi: 10.1016/j.neuroscience.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jeong YC, Nakamura J, Upton PB, Swenberg JA. Pyrimido[1,2-a]-purin-10(3H)-one, M1G, is less prone to artifact than base oxidation. Nucleic Acids Res. 2005;33:6426–6434. doi: 10.1093/nar/gki944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.