Abstract
Trypanosomatid parasites provide an extreme model for the posttranscriptional control of eukaryotic gene expression. However, most analysis of their differential gene regulation has focussed on comparisons between life-cycle stages that exist in the blood of mammalian hosts and tsetse flies, the parasite’s vector. These environments differ acutely in their temperature, and nutritional, metabolic and molecular composition. In the bloodstream, however, a more exquisitely regulated developmental step occurs: the production of transmissible stumpy forms from proliferative slender forms. This transition occurs in the relatively homogenous bloodstream environment, with stumpy-specific gene expression being repressed until accumulation of a proposed parasite-derived signal, stumpy induction factor. Here, we have dissected the regulatory signals that repress the expression of the stumpy-specific surface transporter PAD1 in slender forms. Using transgenic parasites capable of stumpy formation we show that PAD1-repression is mediated by its 3′-untranslated region. Dissection of this region in monomorphic slender forms and pleomorphic slender and stumpy forms has revealed that two regulatory regions co-operate to repress PAD1 expression, this being alleviated on exposure to SIF in pleomorphs or cAMP analogues that act as stumpy induction factor mimics in monomorphs. These studies identify elements that regulate trypanosome gene expression during development in their mammalian host.
INTRODUCTION
Cell-type differentiation is usually driven by an external cue. In the developmental events within multicellular organisms, soluble signals such as growth factors, cytokines and hormones can generate paracrine and autocrine signalling systems that trigger cell-type specialisation (1). Developmental events can similarly be stimulated in unicellular organisms by cell-derived signals, such as the yeast mating pheromone (2) and the DIF1 stalk cell differentiation signal in Dictyostelium discoideum (3). However, unicellular organisms also respond to environmental cues such as pH, temperature and osmolarity. This is particularly the case for those organisms that encounter extreme environmental instability such as Chlamydomonas spp. that undergo sexual development in response to nitrogen starvation (4), and dimorphic fungi that alternate between mould and yeast forms dependent on temperature (5). Transduction of the resulting signals generates specific changes in gene expression that elicit the cellular events associated with developmental adaptation.
Kinetoplastid parasites, infectious agents responsible for a variety of important tropical and subtropical diseases, provide important models for development and developmental gene expression for three reasons. First, these parasites were among the earliest branching eukaryotic organisms (6), such that their developmental events can provide insight into the processes underlying the differentiation of all eukaryotic organisms. Second, the genome organisation of these parasites is highly unusual (7). Specifically, genes are arranged in large groups of co-transcribed cistrons (polycistronic arrays) whereby pre-mRNAs are transcribed from often-distant upstream promoters, individual mRNAs being generated after a concerted trans splicing and polyadenylation reaction (8,9). This organisation dictates that differential gene expression is controlled predominantly at the posttranscriptional level, through regulated mRNA stability and translational mechanisms (10). Third, kinetoplastid parasites undergo complex developmental pathways, being transmitted between mammalian hosts by blood feeding arthropods (11). These developmental events require elaborate changes in the parasite’s morphology, metabolism, and surface protein expression, each being governed by differential gene expression (12). The important cue for these changes in different kinetoplastid parasites is the change in temperature associated with passage from a homoeothermic to poikilothermic carrier (13,14). Progression from the bloodstream of mammalian hosts to the alimentary canal of arthropod vectors is also associated with major changes in available glucose, pH and osmolarity, as well as exposure to the proteolytic and immunological environment of the insect gut (15,16). These environmental changes stimulate altered gene expression, the best characterized being the regulation of procyclin surface antigens on African trypanosomes as they establish in their tsetse fly vector (17). Here, surface protein expression is controlled by exposure to glycerol or low oxygen content (GPEET procyclin) (18) or a temperature reduction of 15°C, or more (EP procyclin) (14).
Most studies of developmental gene expression in trypanosomes have focused on the differentiation from bloodstream to procyclic forms in culture, using ‘monomorphic’ bloodstream parasite lines selected for their uncontrolled growth in vitro and in vivo (19). However, in natural infections, the transition to procyclic forms from bloodstream forms requires the production of specialized transmission stages, called stumpy forms, which arise in the bloodstream from proliferative slender forms. Slender forms cannot differentiate in the tsetse midgut because they are rapidly killed by its digestive environment (20) and because they cannot detect the differentiation signal, which comprises citrate/cis aconitate (14,21). This signal is detected in stumpy forms because they express a carboxylate surface transporter family, called PAD proteins, of which PAD1 is only expressed at significant levels in the transmission stage (22). The transition from slender forms to stumpy forms is believed to be triggered by a parasite-derived factor, stumpy induction factor (SIF) (23,24), which has thus far eluded identification. Nonetheless, in response to accumulating SIF, slender cells stop proliferating and differentiate to stumpy forms, which are characterized by their morphology, limited mitochondrial elaboration, resistance to proteases and pH and their expression of PAD proteins (22,25–28). Since this transformation occurs in the mammalian blood and is triggered by a parasite-derived factor, it must be stringently regulated without the extreme environmental cues that characterize the transition to procyclic forms. As such, it represents an exquisitely specific form of developmental regulation, whereby stumpy expressed genes must be held silent until repression is released by the accumulation, and detection, of SIF.
In this article, we have exploited our identification of PAD1 as the first molecular marker for the parasite transmission stage to examine the gene regulatory signals that are responsible for stumpy-specific gene expression. We find that PAD1 is regulated through elements in its 3′-untranslated region, which repress its expression in slender forms, this being alleviated in stumpy forms and when monomorphic slender forms are exposed to cell permeable cAMP analogues believed to mimic activation of the SIF pathway. This provides a first dissection of gene expression in the parasite’s transmission stage, and its regulation in response to highly specific, and parasite-derived, developmental cues.
MATERIALS AND METHODS
Trypanosome culturing and chemicals
Monomorphic bloodstream form trypanosomes were Trypanosoma brucei Lister 427. Pleomorphic bloodstream form trypanosomes used to harvest stumpy RNA were AnTat1.1, whereas those used in transfections were T. brucei AnTat1.1 90:13 (14). Parasites were grown routinely in HMI-9 at 37°C in 5% CO2 (29). Slender forms were harvested from MF1 mice at 3 days post infection and stumpy forms were harvested from cyclophosphamide-treated MF1 mice at 5 and 6 days post infection. Trypanosomes were purified from blood by DEAE cellulose purification (30). For parasite transfection, 10–15 µg of linearized DNA was electroporated using an Amaxa Nucleofector II. Monomorphic parasites were selected using 0.5 µg/ml puromycin (pHD617 CAT-PAD1 3′-UTR), 3 µg/ml hygromycin (pHD617 GUS Actin 3′-UTR) or 0.5 µg/ml phelomycin (all CAT449 reporter constructs). Pleomorphic parasites were selected with 1.5 µg/ml phleomycin.
Differentiation of bloodstream forms to procyclic forms was induced by addition of 6 mM cis-acconitate and a temperature change from 37°C to 27°C. Differentiation capacity was assessed by EP procyclin expression, measured by antibody staining as described in the study by Dean et al. (22).
8pCPT-cAMP was purchased from Sigma Aldrich (UK), 8pCPT-2′-O-Me-cAMP was purchased from BioLog Life Science Institute (Germany). Troglitazone was purchased from Biomol (Germany).
Analysis of mRNA polyadenylation site
To map the site of PAD1 mRNA polyadenylation, reverse transcriptase–polymerase chain reaction on stumpy-form RNA was carried out. The 3′-end of the PAD1 transcript was amplified using a gene-specific oligonucleotide hybridizing close to the 3′-end of the coding region (5′-GAC CAA AGG AAC CTT CTT CCT-3′) and a 3′ oligo-dT ADAPT oligonucleotide hybridizing to the poly (A) tail (5′-GGC CAC GCG TCG ACT AGT ACT TTT TTT TTT TTT TT-3′) (31). Amplified products were then subjected to a second round of amplification using an oligonucleotide hybridizing to the 5′-end of the PAD1 3′-UTR (5′-TTA GGA TCC GCT TAG GGG AGC CAG TGG AGG GC-3′) and the primer AUAP (5′-GGC CAC GCG TCG ACT AGT AC-3′), which binds to the specific oligonucleotide sequence incorporated into the 5′-end of the ADAPT primer (31). The resulting products were gel purified and sequenced to determine the site of polyadenylation.
DNA cloning and trypanosome transfection
Plasmid constructs in this study were based on the trypanosome expression construct pHD617 (32) and a CAT 449 reporter construct previously described (31). Precise details of each construct and the associated primers used for their construction and analysis are provided in the Supplementary Materials.
CAT and GUS reporter assays
CAT protein levels were determined by CAT ELISA assay (Roche) according to manufacturer’s instructions, with absorbance being measured using a BioTek ELx808 Absorbance Microplate Reader. Duplicate wells for each assay were included to ensure consistency. In all cases, a CAT standard curve was constructed to ensure all sample readings were within the linear range of the assay. CAT standard curves had a linear regression value of 0.99 or greater.
GUS enzyme activity was measured using 4-methylumbelliferyl-β-d-glucopyranosiduronic acid (MUG) as a fluorescent substrate. Trypanosome cultures were incubated 1:1 with the MUG substrate (1 mM MUG, 0.82M Tris–HCl pH8, 0.6% SDS, 0.3 mg/ml BSA) at 37°C for 2 hours. Fluorescence was measured with an excitation of 355 nm and emission of 460 nm on a BioTek Flx800 Fluorescent Microplate Reader.
Northern blotting and quantification
RNA was extracted from trypanosomes using an RNeasy Mini Kit (Quiagen). Between 0.15 and 1.5 µg purified RNA was resolved on formaldehyde agarose gels in MOPS buffer and transferred to nylon membranes by capillary blotting. A digoxigenin (DIG)-labelled riboprobe was hybridized to the membrane and the transcript was detected using an anti-DIG antibody (Roche) with a chemiluminescent CDP-star substrate (Roche). The transcript levels were quantified using a Syngene Gbox with GeneTools software, using rRNA levels to normalize for loading.
Data analysis
For all analyses, error bars represent the SEM calculated using two biological replicates (n = 2), whereas the solid bars represent the mean of those biological replicates, this providing an intuitive visual representation of the data. These biological replicates represent two independently isolated transfectant cell lines derived from transfection with the same construct. For both CAT and GUS assay, values were normalized to cell number in all cases.
RESULTS
Identification of the endogenous polyadenylation site for PAD1
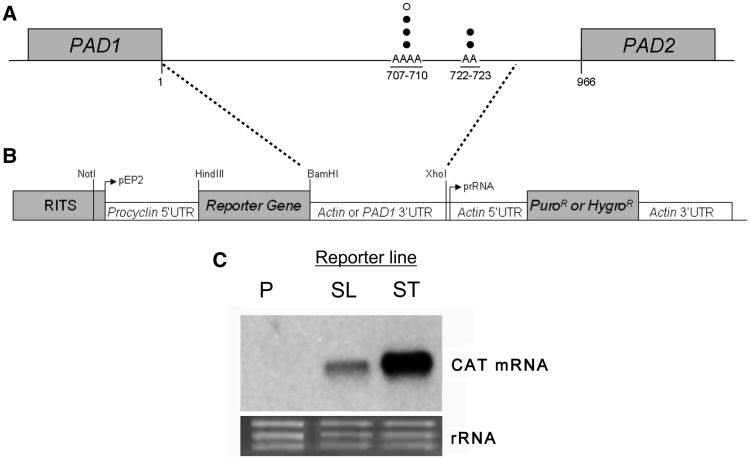
To identify any regulatory regions in the PAD1 3′-UTR, it was first necessary to identify its polyadenylation site and thereby define the boundaries of regulatory sequences within the PAD1 mRNA. Initially, we analysed existing data describing the polyadenylation sites of almost 6000 genes in slender and procyclic form trypanosomes (33). Three potential polyadenylation sites were predicted downstream of the PAD1 gene in procyclic forms, these being positioned at 428, 572 and 835 nt after the PAD1 termination codon (33). Contrasting with this data, a 273 nt EST sequence was available (accession number W00184) from a T. brucei rhodesiense cDNA library which aligned from positions 437 to 709 in the intergenic region downstream of PAD1, suggesting that the PAD1 3′-UTR polyadenylation site extended at least 709 nt after the termination codon. In order to resolve this apparent discrepancy, and to identify the polyadenylation site used in the relevant life-cycle stage, the PAD1 polyadenylation site was mapped experimentally using bloodstream stumpy form cDNA as a template, as previously described (31). This analysis confirmed positions 707–710 as a PAD1 polyadenylation site, with a further site at positions 722–723 being identified (Figure 1A). This sequence is contained within a region also found downstream of PAD5 and PAD7. These intergenic regions contain several potential alternative processing sites that may generate non-coding intergenic region-derived RNAs responsible for the sites mapped in whole transcriptome studies. The entire region downstream of the PAD1 coding region to its polyadenylation site did not contain any motifs previously observed as being present in some stumpy-enriched transcripts identified by microarray analysis (34).
Figure 1.
Polyadenylation and devlopmental regulation of PAD1. (A) Identification of the PAD1 polyadenylation sites in stumpy form T. brucei. Sites mapped experimentally by reverse transcriptase–polymerase chain reaction are shown by black dots, whereas the site indicated by EST analysis of T. brucei rhodesiense is shown by a white dot. The diagram is not drawn to scale. (B) Developmental regulation of gene expression by the PAD1 3′-UTR. Representation of the expression construct used to analyse PAD1 3′-UTR effects on gene expression in pleomorphic cells (Figure 2B and C) or during evaluation of reported chemical inducers of stumpy formation (Figure 6). (C) Slender and stumpy populations of the AnTat1.1 90:13 CAT-PAD1 3′-UTR cell line were isolated from mouse infections either on Day 3 (‘Slender’) or Day 6 (‘Stumpy’) post infection. CAT mRNA levels were elevated in the stumpy population. rRNA labelled with ethidium bromide is shown in the bottom panel to demonstrate relative loading. ‘P’ untransfected trypanosomes, ‘SL’ slender forms, ‘ST’ stumpy forms.
The PAD1 3′-UTR controls stumpy-specific gene expression
To identify whether the PAD1 3′-UTR was responsible for stage-specific expression in stumpy forms, it ability to appropriately regulate a heterologous marker gene encoding chloramphenicol acetyl transferase was investigated. To achieve this, a variant of the expression vector, pHD617 (32), lacking the tetracycline operator sequence and conferring resistance to puromycin, was created, to allow transgene expression independently of the presence of tetracycline induction. Thereafter, the CAT coding region linked to the PAD1 3′-UTR construct was integrated, and the resulting construct, CAT-PAD1 3′-UTR (Figure 1B), transfected into the T. brucei AnTat1.1 90:13 cell line (a kind gift of Michael Boshart and Markus Engstler) (14). This is a pleomorphic cell line, capable of generating uniform populations of stumpy forms when grown in rodents.
To determine whether the AnTat1.1 90:13 CAT-PAD1 3′-UTR reporter cell line exhibited developmentally regulated CAT expression, two mice were infected with the cell line. Trypanosomes were harvested from one mouse on Day 3 post-infection when the population was predominantly slender, whereas trypanosomes were harvested from the second mouse on Day 6 post-infection when the population was predominantly stumpy in morphology. Figure 1C demonstrates that slender form AnTat1.1 90:13 CAT-PAD1 3′-UTR cells generated very little CAT mRNA, consistent with PAD1 3′-UTR-mediated repression of gene expression at this life-cycle stage. In contrast, the transgenic stumpy forms expressed a large amount of CAT mRNA at equivalent cell numbers. This demonstrated that in pleomorphic trypanosomes, the PAD1 3′-UTR is capable of directing the appropriate life-cycle stage regulation of a heterologous reporter gene.
Mapping of the PAD1 3′-UTR silencing elements in monomorphic slender forms
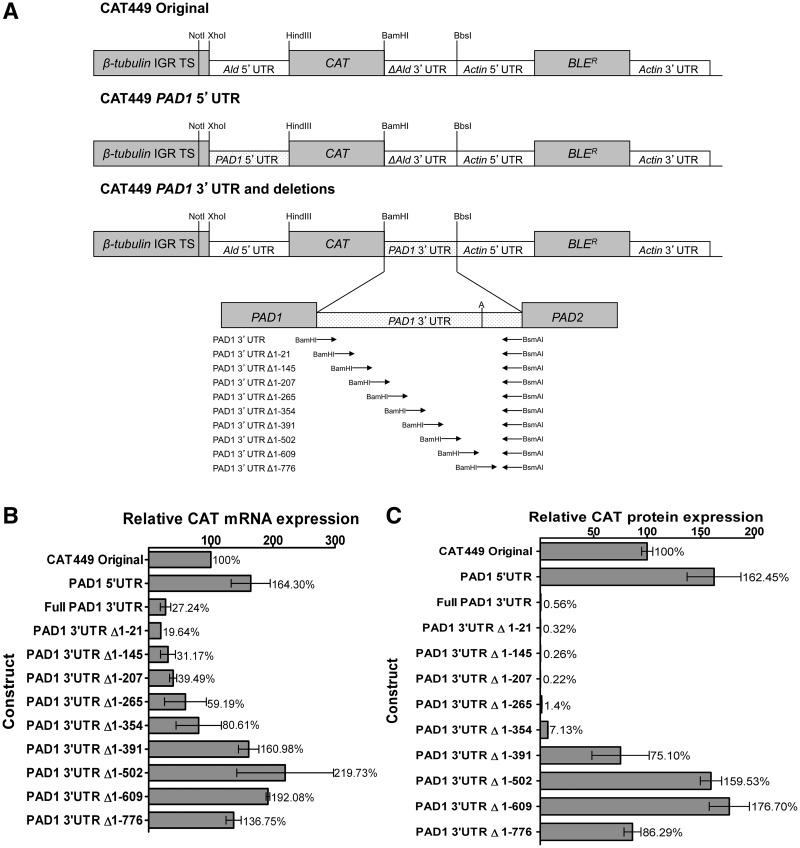
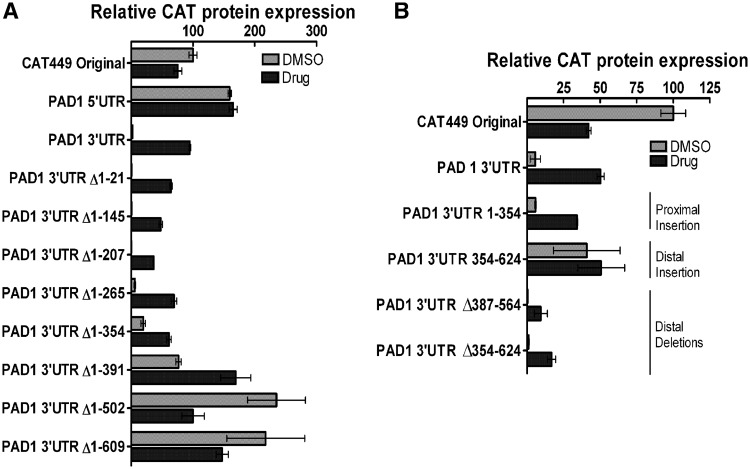
Given the difficulty in generating transgenic cell lines from trypanosomes capable of developing to stumpy forms, we initially investigated the regulatory elements in the PAD1 3′-UTR in more detail using transgenic monomorphic slender lines. These cell lines are readily grown in culture and transfected and were expected to repress gene expression of the CAT reporter when linked to the PAD1 3′-UTR, matching the scenario in pleomorphic slender cells. Thus, the full-length PAD1 5′-UTR, PAD1 3′-UTR, as well as nine sequential deletions of the PAD1 3′-UTR were coupled to the chloramphenicol acetyl transferase (CAT) reporter gene and incorporated into an expression vector [CAT449 (31), derived from pHD449 (32)] designed to integrate into the trypanosome tubulin locus, ensuring constitutive read-through transcription within a polycistronic polII transcription unit (Figure 2A). The sequence of the 3′-UTR is shown in Supplementary Figure 1, whereas the predicted Sfold structures of the intact and deletions of the PAD1 3′-UTR are shown in Supplementary Figures S2 and S3. These constructs were then transfected into monomorphic trypanosomes, and two stable cell lines derived for each. Our expectation was that as sequences were deleted in the PAD1 3′-UTR, an alleviation of reporter gene repression would be observed that would identify elements that repress gene expression in slender forms.
Figure 2.
Deletion analysis of regulatory elements in the PAD1 3′-UTR. (A) Schematic diagram of the CAT449 PAD1 3′-UTR deletion constructs used to map control elements in the PAD1 3′-UTR. The ‘A’ in the PAD1 3′-UTR depicts the 707-710 nt and 722-723 nt polyadenylation sites. This diagram is not to scale. (B) Quantification of the mRNA abundance in two independent clones for each cell line represented in panel A. Values are normalized to the CAT mRNA abundance derived from the CAT449 control construct. Deletion 1–776 nt deletes beyond the mapped polyadenylation sites. (C) Quantification of the CAT protein in two independent clones for each cell line represented in panel A. Values are normalized to the CAT protein abundance derived from the CAT449 control construct. Deletion 1–776 nt deletes beyond the mapped polyadenylation sites.
Figure 2B shows the CAT mRNA levels derived from each construct, values representing the mean and S.E.M. of two independent cells lines for each of 11 distinct PAD1 UTR-linked constructs and normalized to the CAT449 control, in which the CAT gene is linked to a truncated Aldolase 3′-UTR. First, matching the earlier observations using a pHD617-based construct in pleomorphic cells, inclusion of the full length PAD1 3′-UTR caused an approximately 3.7-fold decrease in CAT mRNA abundance compared with the control construct (i.e. 27.2% CAT with respect to CAT449, normalized to 100%). In contrast, replacement of the Aldolase 5′-UTR in the CAT449 control construct with the PAD1 5′-UTR did not repress mRNA expression, with mRNA levels being 164% of control levels. These results confirmed that repression was mediated in the 3′-UTR of the PAD1 transcript.
Progressive deletions into the PAD1 3′-UTR from the 5′-end initially caused very little effect on CAT mRNA abundance, such that deletion of the first 207 nt of the PAD1 3′-UTR generated CAT mRNA levels equivalent to the intact PAD1 3′-UTR. Further deletions beyond 207 nt, however, resulted in a progressive increase in CAT mRNA levels with deletion of 1-502 nt of the PAD1 3′-UTR causing an approximately 8.1-fold increase in reporter mRNA abundance compared to the full-length PAD1 3′-UTR, consistent with sequences between 207 and 502nt containing a slender-form repression element. The final PAD1 3′-UTR deletion construct (Δ1-776 nt) removed the sequences containing the experimentally mapped polyadenylation site for the PAD1 gene in stumpy forms. Consistent with this, by northern blot analysis (Supplementary Figure S4), it was observed that the progressive deletions of the full length PAD1 3′-UTR up to 609 nt caused equivalent decreases of the CAT transcript size, indicating use of the same polyadenylation sites. In contrast, transcripts from the PAD1 3′-UTR Δ1–776 construct were longer than those from the previous deletion. This likely represents the use of an alternative polyadenylation site and indicates that the dominant polyadenylation sites used in monomorphic slender forms coincides with those mapped in stumpy forms, rather than those reported in earlier RNAseq analyses (33).
To assess regulation at the protein level, at least two CAT ELISA assays were carried out for each of the two replicate cell lines described earlier, and these normalized to the CAT449 control (Figure 2C). Matching the analysis at the mRNA level, replacement of the Aldolase 5′-UTR with the PAD1 5′-UTR caused an approximately 1.6-fold increase in CAT protein expression, eliminating a function for this element in silencing gene expression in slender forms. In contrast, CAT protein was almost undetectable when the aldolase 3′-UTR of the original CAT449 construct was replaced by the full-length PAD1 3′-UTR. Indeed, while there was a 3.7-fold decrease in mRNA expression, there was an almost 180-fold decrease in protein expression using the PAD1 3′-UTR, indicating that repression operates at both the level of transcript abundance and protein expression in slender forms.
Protein level control was also more stringent as deletion constructs of the PAD1 3′-UTR were analysed. As with the mRNA analysis, deletion of the first 207 nt of the PAD1 3′-UTR caused no discernable effect on reporter protein expression compared with the full-length PAD1 3′-UTR. Hence, it appears that the first 207 nt is not necessary for repression of mRNA abundance or protein expression in slender forms at least in the presence of distal regions of the 3′-UTR. Discrepancy between mRNA and protein levels was observed for deletions between 207 and 354 nt, however, whereby mRNA levels reached 59% (Δ1–265) and 80% (Δ1–354) of CAT449 levels while CAT protein levels were 1.4% and 7.1% of CAT449, respectively (Figure 2C). This indicates the presence of a dominant repressive element operating at the protein level between 207 and 354 nt in the PAD1 3′-UTR. Deletion from 354 to 502 nt generated further increases in CAT protein levels, similar in overall trend to the mRNA profile. This indicated the presence of a further repressive element at the distal end of the 3′-UTR.
Operation of identified regulatory regions in isolation
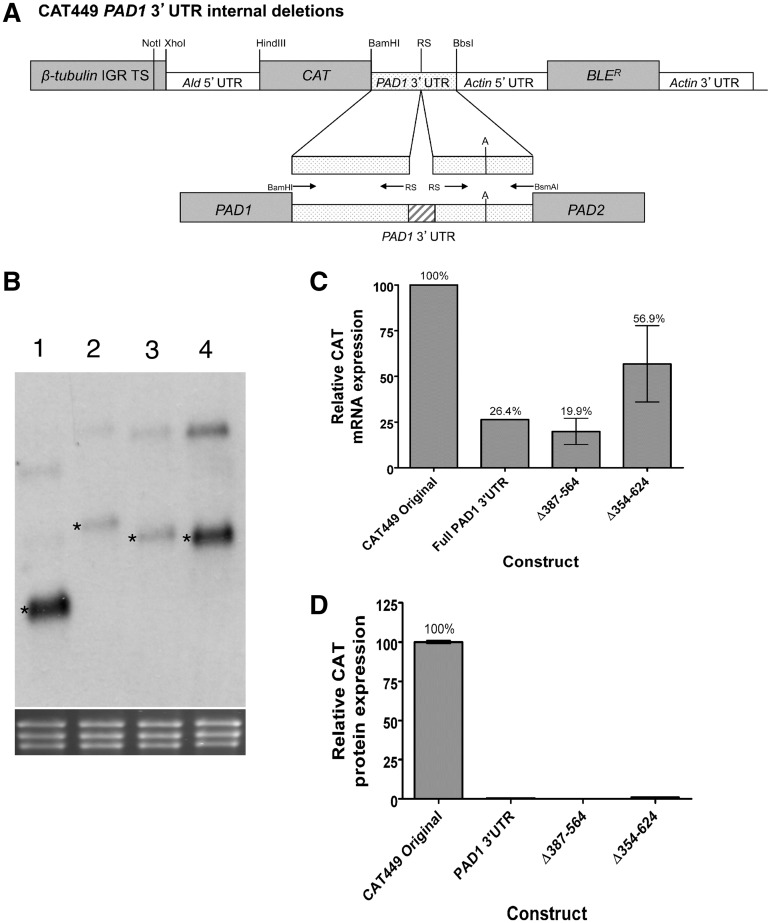
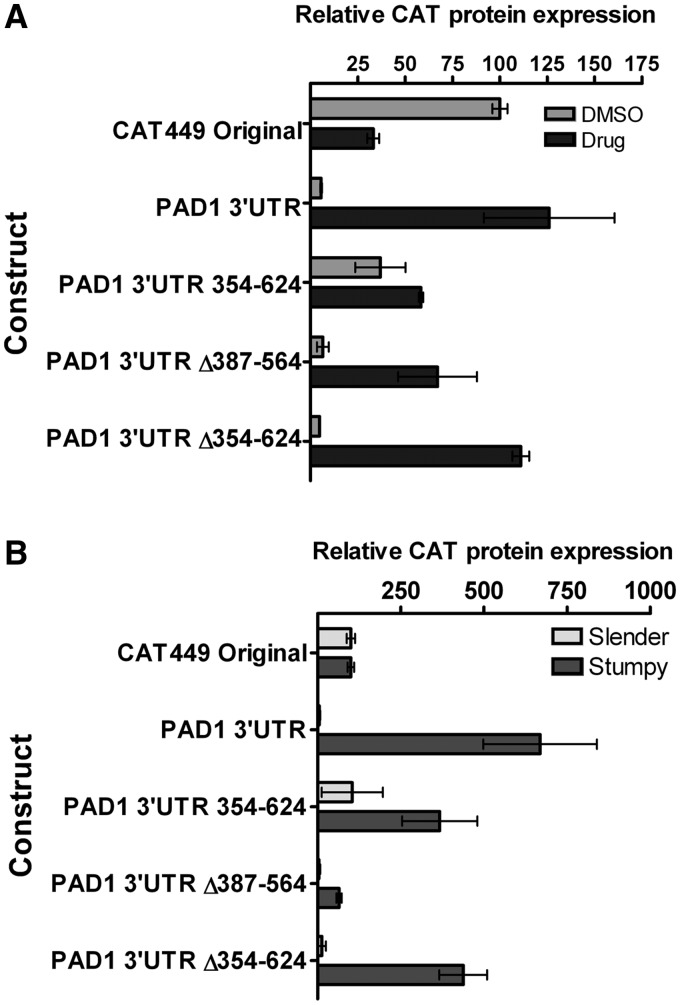
To further investigate the contributions of subregions within the PAD1 3′-UTR, two constructs containing internal deletions were created to specifically remove 387–564 nt and 354–624 nt from the intact PAD1 3′-UTR (Figure 3A). Both deletions were also designed to preserve the overall secondary structure of the entire PAD1 3′-UTR predicted using S-fold (Supplementary Figure S5). From the analysis of two independent cell lines, the PAD1 3′-UTR Δ387–564 deletion generated no detectable loss of repression of the mRNA with respect to the full length PAD1 3′-UTR (Figure 3B and C). In contrast, the PAD1 3′-UTR Δ354–624 construct generated a 2-fold increase (56% of CAT499) in CAT mRNA abundance over the full-length PAD1 3′-UTR (26% of CAT 449) (Figure 3B and C), although this was considerably less than observed when individual sequential deletions were analysed in this region (e.g. PAD1 3′-UTR Δ1–502 was 219.73% of CAT449, Figure 2B). However, when the CAT protein levels were assayed, negligible expression was detected with either the PAD1 3′-UTR Δ386–564 or PAD1 3′-UTR Δ354–624 deletion constructs, demonstrating that in isolation neither could relieve the PAD1 3′-UTR-mediated repression (Figure 3D). This indicated that a proximal element must also operate in the PAD1 3′-UTR to silence expression even in the absence of the distal silencing element positioned beyond nt 354.
Figure 3.
Analysis of specific deletions in the PAD 3′-UTR for their effects on gene silencing in slender forms. (A) Representation of the expression construct used to analyse specific PAD1 3′-UTR elements for their contribution to gene silencing in slender forms. (B) A northern blot of the cell lines used to analyse the contribution of specific PAD1 3′-UTR elements to reporter gene expression. rRNA stained with ethidium bromide is shown in the bottom panel to demonstrate relative loading. Lane 1: CAT449 Original; Lane 2: CAT449 PAD1 3′-UTR; Lane 3: CAT449 Δ387–564 PAD1 3′-UTR; Lane 4: CAT449 Δ354–624 PAD1 3′-UTR. The size of the relevant bands (marked with an asterisk) are Lane 1, 862 nt; Lane 2, 1449 nt; Lane 3, 1278 nt and Lane 4, 1185 nt. (C) Quantitation of the CAT mRNA generated from the PAD1 3′-UTR deletion constructs detailed in Panels A and B. The data represent a quantitation based on independent northern blots of each of two cell lines derived for each expression construct. (D) Quantitation of the CAT protein generated from the PAD1 3′-UTR deletion constructs detailed in Panels A and B. The data represents a quantitation based on independent assays of each of two cell lines derived for each expression construct.
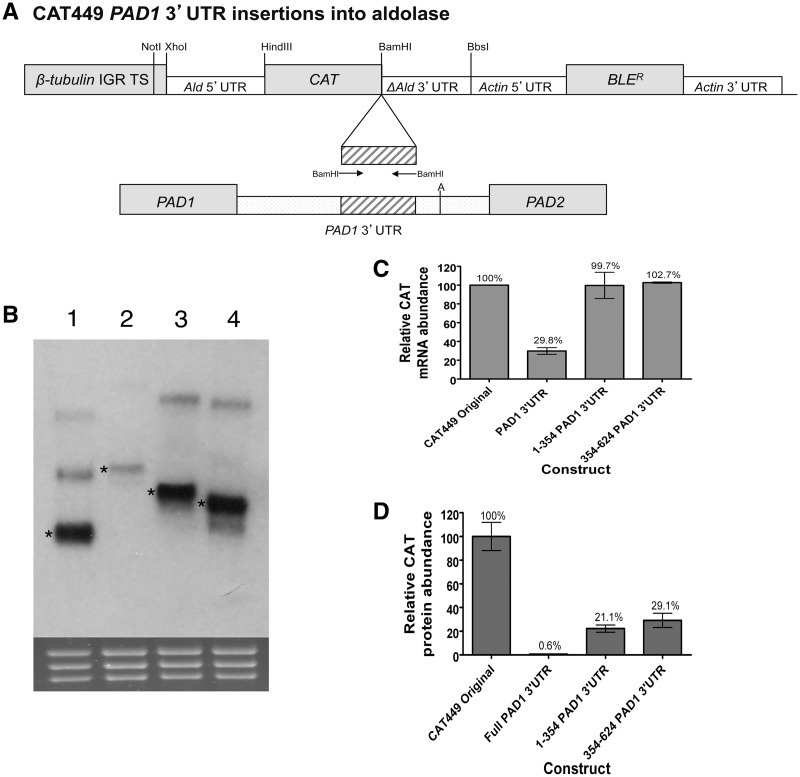
Having generated evidence for proximal and distal silencing elements in the PAD1 3′-UTR, we investigated whether either region could function when inserted independently into the 3′-UTR of a constitutively expressed gene. Therefore, the proximal region (1–354 nt) and distal region (354–624 nt) of the PAD1 3′-UTR were integrated into the truncated aldolase 3′-UTR of the CAT449 control construct, directly after the CAT gene termination codon (Figure 4A). Two transgenic cell lines were then isolated for each construct and these cell lines analysed for their CAT mRNA and protein expression. Figure 4B shows that the integration of each element did not alter the preferred polyadenylation site in the aldolase 3′-UTR because the size increase of the generated CAT mRNA was consistent with the size of each integrated element (Figure 4B, compare lanes 1, 3 and 4). Nonetheless, whilst neither the proximal or distal insertion elements caused repression of CAT mRNA abundance from the reporter construct (Figure 4B and C), the expression of CAT protein was reduced to 21% and 29% of the CAT449 using the proximal and distal element, respectively (Figure 4D). Apparently in the context of the aldolase 3′-UTR both proximal and distal elements can act to repress upstream gene expression at the protein level, but neither can act independently to reduce mRNA abundance.
Figure 4.
Out of context regulation of gene silencing by PAD1 3′-UTR control elements. (A) Representation of the expression construct used to analyse specific PAD1 3′-UTR elements inserted into the aldolase 3′-UTR for their contribution to gene silencing in slender forms. (B) A northern blot of the cell lines used to analyse the contribution of specific PAD1 3′-UTR elements to reporter gene expression. The sizes of the CAT transcripts demonstrate that the endogenous aldolase polyadenylation site is preserved after insertion of the PAD1 3′-UTR elements. rRNA stained with ethidium bromide is shown in the bottom panel to demonstrate relative loading. Lane 1: CAT449 Original; Lane 2: CAT449 PAD1 3′-UTR; Lane 3: CAT449 1-354 PAD1 3′-UTR; Lane 4: CAT449 354-624 PAD1 3′-UTR. The relevant band in each lane is marked with an asterisk. The predicted size of each band is Lane 1, 862 nt; Lane 2, 1449 nt; Lane 3, 1216 nt and Lane 4, 1132 nt. (C) Quantitation of the CAT mRNA generated from the PAD1 3′-UTR deletion constructs details in Panels A and B. The data represent a quantitation based on independent northern blots of each of two cell lines derived for each expression construct. (D) Quantitation of the CAT protein generated from the PAD1 3′-UTR deletion constructs detailed in Panels A and B. The data represent a quantitation based on independent assays of each of two cell lines derived for each expression construct. In some samples, the presence of DMSO apparently altered the basal level of CAT protein expression, although the effect did not alter the overall interpretation of the analysis.
Mapping elements that respond to inducers of stumpy formation
Upon differentiation to stumpy forms, the silencing of stumpy-specific proteins is alleviated in response to the parasite-derived signal, stumpy induction factor. However, this factor is unidentified and monomorphic cell lines are not responsive to it. Therefore, to investigate the gene regulation on activation of the stumpy induction pathway we examined the response of the PAD1 reporter lines to treatment with compounds proposed to mimic the SIF response.
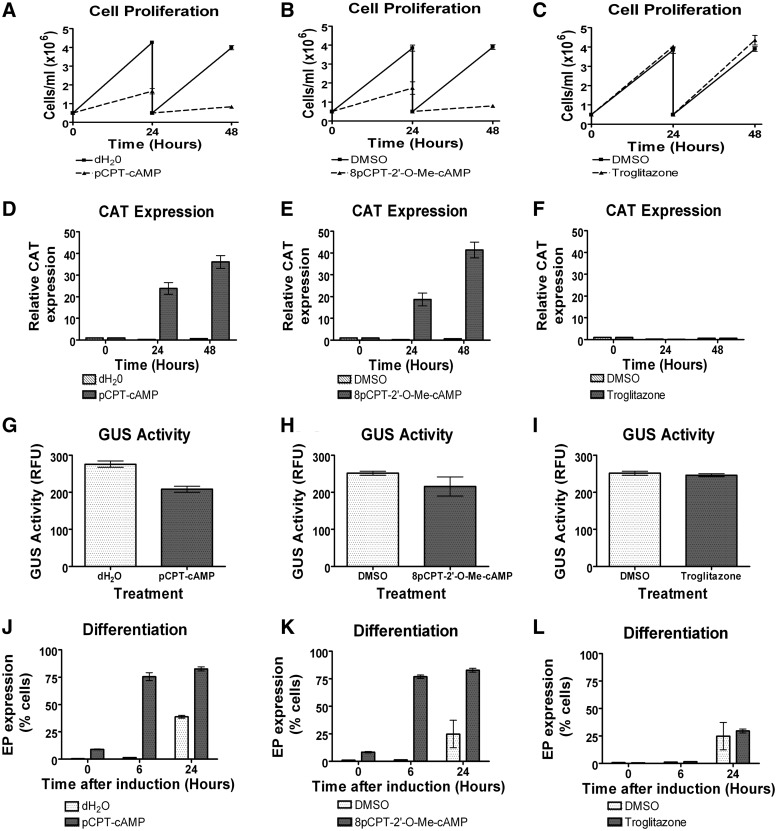
Initially, we assayed the ability of the cell permeable cAMP analogues, pCPTcAMP (24), 8pCPT-2′-O-Me-cAMP (35) or troglitazone (36), to induce hallmarks of stumpy formation in monomorphic lines, namely the inhibition of cell growth and improved efficiency of differentiation to procyclic forms. These validation experiments were carried out using a monomorphic reporter line transfected with both the pHD617-CAT-PAD1 3′-UTR expression vector and pHD617-GUS-actin 3′-UTR (Figure 1A), enabling gene repression and alleviation via the PAD1 3′-UTR to be monitored in the context of a constitutively expressed Actin 3′-UTR control. Matching the observation in pleomorphic slender cells (Figure 1C) and the monomorphic lines harbouring the CAT PAD1 3′-UTR deletion series (Figure 2), the levels of CAT protein expression were very low in the resulting transgenic monomorphic line (Figure 5D–F, 0 h time point), confirming repression by the PAD1 3′-UTR.
Figure 5.
Effect of chemical inducers of stumpy formation on monomorphic PAD1 reporter lines. Analysis of the response of the CAT-PAD1 3′-UTR, GUS-actin 3′-UTR reporter line to 100 µM pCPTcAMP (panels A, D, G and J), 10 µM 8pCPT-2′-O-Me-cAMP (panels B, E, H and K) and 5 µM troglitazole (panels C, F, I and L). Panels A–C show the growth of cells under each drug regimen, Panels D–F represent the CAT protein expression over 48 h of drug treatment relative to zero hours, Panels G–I show the GUS protein expression after 48 h of treatment whereas Panels J and K represent the percentage of cells expressing EP procyclin protein determined by flow cytometry after exposure to each drug for 48 h followed by incubation with 6 mM cis-aconitate for 6 or 24 h.
The reporter line was then exposed to 100 µM pCPTcAMP, 10 µM 8pCPT-2′-O-Me-cAMP or 5 µM troglitazone (36) doses being determined via titration experiments or literature analysis (data not shown). After treatment with each compound, the cells were investigated for different parameters of stumpy formation, including cell growth arrest (Figure 5A–C), their capacity for differentiation to procyclic forms (Figure 5J–L; Supplementary Figure S6) and their expression of the CAT (Figure 5D–F) and GUS (Figure 5G–I) reporter proteins. Figure 5 shows that both cAMP analogues induced a potent growth arrest within 24 h (Figure 5A and B) and more efficient differentiation to procyclic forms after treatment with cis-aconitate, such that 75% (pCPTcAMP) and 83% (8pCPT-2′-O-Me-cAMP) of cells expressed procyclin within 6 h compared with <2% in the untreated populations (Figure 5J and K; Supplementary Figure S6). 8pCPT-2′-O-Me-cAMP induced some morphological transformation in the treated cells, although there also appeared to be ‘unhealthy’ cells in the population. Nonetheless, when the response of the reporter genes was analysed after treatment with either cAMP analogue, the level of the constitutive GUS reporter was essentially unchanged or somewhat decreased (Figure 5G and H), whereas the expression of the PAD1 3′-UTR linked CAT gene was strongly elevated (pCPTcAMP, 24-fold, Figure 5D; 8pCPT-2′-O-Me-cAMP, 18-fold, Figure 5E) after 24h. This indicated that both compounds promoted some stumpy form characteristics in the treated monomorphic cells, supportive of an (incomplete) developmental response that also relieves repression of the CAT-PAD1 3′-UTR reporter. In contrast, troglitazone did not activate CAT (Figure 5F) or induce growth arrest (Figure 5C) under the assay conditions used. This indicates that this compound may not act on the stumpy formation pathway, despite its reported anti-proliferative and morphological effects (36).
Having validated the ability of cAMP analogues to derepress CAT PAD1 3′-UTR in monomorphic forms, we used the cell lines transfected with the panel of PAD1 3′-UTR deletion constructs (Figure 2) to investigate whether the regions responsible for repression in slender forms coincided with those responsible for the alleviation of repression in response to these compounds. Initially, treatment of cells containing the control CAT449 construct with 10 µM 8pCPT-2′-O-Me-cAMP was found to cause a small decrease in CAT protein expression (as was also seen with this compound in Figure 5H), perhaps caused by the cells arresting growth after treatment, or exhibiting some translational repression (37) (Figure 6A). Interestingly, however, replacement of the aldolase 5′-UTR with the PAD1 5′-UTR eliminated this effect, perhaps because the PAD1 5′-UTR contributes to stabilizing the transcript, or assisting translation, as the cells progress to ‘stumpy-like’ forms. As expected, the PAD1 3′-UTR responded effectively to 8pCPT-2′-O-Me-cAMP treatment such that CAT protein became elevated at least 40-fold over untreated cells. Hence both repression and the alleviation of that repression are each dependent on the 3′-UTR of PAD1.
Figure 6.
Effect of PAD1 3′-UTR control elements on inducibility in response to 8pCPT-2′-O-Me-cAMP. Response of different deletion constructs of the PAD1 3′-UTR (A), or specific 3′-UTR deletions or 3′-UTR element inserts into the aldolase 3′-UTR (B) to incubation with 8pCPT-2′-O-Me-cAMP or with an equivalent amount of DMSO as control. Individual graphs show the CAT protein expression normalized to the control CAT449 construct, each bar representing the mean and standard error of the mean of the analysis of two independent cell lines for each construct.
When cells containing constructs with deletions of the PAD1 3′-UTR were analysed, the overall CAT expression caused by 8pCPT-2′-O-Me-cAMP treatment diminished progressively with removal of the first 207 nt of the PAD1 3′-UTR, with even a deletion of the first 21 nt generating a reproducible reduction (Figure 6A). Nonetheless, the CAT expression was in all cases clearly inducible in response to the treatment. Deletions beyond position 265 nt, however, reduced the effect of 8pCPT-2′-O-Me-cAMP, with an 11-fold increase in CAT protein expression in the Δ1-265 cell lines and a 3- and 2-fold increase in the Δ1-354 and Δ1-391 cell lines, respectively. Deletion beyond position 502 nt actually caused decreased protein expression on 8pCPT-2′-O-Me-cAMP treatment, indicating that at this point, responsiveness had been completely lost. Combined, these data indicate that the element (s) responsible for up-regulation of protein expression on stumpy induction is located 5′ to position ∼391 nt, essentially coincident with the proximal region responsible for repression in slender forms. Supporting this, constructs where there was specific deletion of nucleotides 387–564 and 354–624 from the PAD1 3′-UTR remained responsive to 8pCPT-2′-O-Me-cAMP (Figure 6B, PAD1 3′-UTR Δ387–564 and PAD1 3′-UTR Δ354–624), although the overall CAT protein expressed was less than with the intact PAD1 3′-UTR.
To dissect in more detail the regions responsible for the responsiveness to 8pCPT-2′-O-Me-cAMP within the PAD1 3′-UTR and their ability to operate out of context, cell lines in which the proximal (nucleotides 1–354) and distal silencing elements (nucleotides 354–564) were inserted individually into the aldolase 3′-UTR (described in Figure 4) were exposed to this compound. Figure 6B (PAD1 3′-UTR 1–354, PAD1 3′-UTR 354–624) demonstrates that the distal element alone was not responsive to 8pCPT-2′-O-Me-cAMP, whereas the proximal element was strongly inducible. We conclude that in monomorphic forms the proximal region of the PAD1 3′-UTR contributes to mRNA and protein repression in slender forms and the activation of PAD1 expression on treatment with a cAMP analogue, whereas the distal region contributes only to gene repression.
To validate these observation in the physiologically relevant pleomorphic cells, the proximal and distal regions were analysed after treatment with 8pCPT-2′-O-Me-cAMP and in true slender and stumpy forms. Thus, transgenic pleomorphic trypanosome cell lines were generated transfected with the CAT-449 construct, CAT-PAD1 3′-UTR, CAT-PAD1 3′-UTRΔ387–564, CAT-PAD1 3′-UTR Δ354–624 and the CAT-PAD1 3′-UTR 354–624 element inserted into the truncated aldolase 3′-UTR. Each construct was inserted into the pleomorphic AnTat1.1 90:13 line and two independent cell lines were isolated for each construct. Once these transfectants were derived, each was inoculated into mice and infections allowed to progress for either 3 or 4 days, to derive predominantly slender cell populations. For 8pCPT-2′-O-Me-cAMP treatment, the slender cells were then maintained in culture by daily passage to 1 × 105 cells/ml and then exposed to drug (or DMSO) for 48 h. Alternatively, infections in mice were maintained for 6 and 7 days, to isolate true stumpy-enriched cell populations. Thereafter, protein samples were derived and assayed for their CAT protein expression in either slender forms, 8pCPT-2′-O-Me-cAMP treated slender forms (‘stumpy-like’ forms) or in vivo generated stumpy forms (‘true stumpy forms’).
Figure 7 shows CAT protein levels from 8pCPT-2′-O-Me-cAMP treated slender forms (Figure 7A) or in vivo generated slender and stumpy populations (Figure 7B) of pleomorphic cells transfected with each construct. In each case, CAT-PAD1 3′-UTR demonstrated a strongly elevated CAT expression in 8pCPT-2′-O-Me-cAMP treated slender forms or stumpy forms. The response of true stumpy forms was considerably greater than the response of either pleomorphic slender or monomorphs to 8pCPT-2′-O-Me-cAMP, indicating a more robust response in the pleomorphic lines generated in vivo than in response to drug. When the distal deletion constructs were analysed (PAD1 3′-UTRΔ387–564; Δ354–624), the repression of CAT expression was preserved in true slender forms and the induction of expression in true stumpy forms was also retained (most strongly in the Δ354–624 construct), confirming that the proximal element alone can provide stumpy-specific expression. Insertion of the 354–624 element of the PAD1 3′-UTR into the aldolase 3′-UTR also revealed elevated expression in true stumpy forms compared to slender forms, contrasting with the weak inducibility of the same construct in pleomorphic or monomorphic cells exposed to 8pCPT-2′-O-Me-cAMP (Figures 6B and 7A). This, however was less than the change seen with the intact 3′-UTR or deletions removing the distal element.
Figure 7.
Effect of PAD1 3′-UTR control elements on reporter expression in pleomorphic slender and stumpy forms. (A) AnTat1.1 90:13 pleomorphic transfectants were harvested from mice infections when a slender parasitaemia was apparent (Days 3 or 4 post infection). Thereafter, cells were maintained in culture and passaged daily to 1 × 105/ml. Cells were treated with 10 µM 8pCPT-2′-O-Me-cAMP, or with an equivalent volume of DMSO, for 48 h before the expression of CAT protein was determined. Error bars represent the SEM calculated from two biological replicates (n = 2). (B). AnTat1.1 90:13 pleomorphic lines were transfected with each reporter construct and then inoculated in to mice. Thereafter, the expression of CAT protein was determined in slender (Day 3 post-infection) and stumpy (Days 6–7 post-infection) cells. Values represent the mean and SEM of two cell lines analysed in independent infections. Values are normalized to the expression of the CAT-449 construct in slender cells.
Combined, this indicated that the response of monomorphic and pleomorphic slender cells to cAMP analogues broadly mimicked that during the in vivo transition from pleomorphic slender to stumpy cells, confirming the physiological relevance of assays using this compound. However, the overall amplitude of differential response was greatly enhanced in true stumpy forms and the distal element remained able to confer stage-specific expression, supporting the concept that the response to cAMP analogues is incomplete with respect to in vivo developmental responses that generate true stumpy forms. Overall, the proximal element provided the most stringent regulation, but both proximal and distal elements cooperate to achieve developmental gene expression.
DISCUSSION
When trypanosomes prepare for transmission they undergo a precise developmental programme involving cell cycle arrest, morphological transformation and the expression of proteins specific to the stumpy form. Of these, PAD1 is linked to transmission competence, being the transporter through which the citrate/cis-aconitate signal is transduced (22), with this stimulating the transformation to procyclic forms. Unlike differentiation in the tsetse midgut, however, the generation of stumpy forms occurs in the mammalian bloodstream, a relatively stable environment in which a parasite-derived signal provides the essential cue for the development of transmission stages. Hence, the gene expression changes that accompany stumpy formation are driven by a soluble signal in many ways analogous to the precisely controlled and programmed differentiation responses characteristic of metazoan cell specialisation. This contrasts with the differentiation of parasites when they enter an arthropod vector, where extreme changes in the external environment, such as in temperature, might be predicted to have wide-ranging consequences for gene expression at the post transcriptional level, for example by altering the respective conformation of different RNA secondary structures (14).
PAD1 expression represents the first molecular hallmark of stumpy cells, tightly regulated at both the mRNA and protein level (22,26). It therefore provided an excellent tool to dissect, for the first time, gene expression control signals regulating transmission competence, the gene being tightly repressed in slender forms but highly expressed in stumpy forms. Our analysis in both pleomorphic and monomorphic parasite lines established that PAD1 expression is controlled through sequences in its 3′-untranslated region, matching the paradigm for most regulated gene expression identified thus far in these parasites (38). Moreover, dissection of the sequences within the 3′-UTR has identified a distal repression element, which operates in concert with a proximal element to repress PAD1 expression in slender forms.
In addition to identifying repression elements that prevent PAD1 expression in slender forms, our analyses also exploited chemical inducers to drive the production of ‘stumpy-like’ forms in monomorphic parasites. Several studies in the literature have used a number of chemical agents to provoke the generation of ‘stumpy-like’ forms. However the interpretation of the cellular outcomes has been restricted by the limitation of morphology as a marker for stumpy formation, even when coupled with mitochondrial activation, or cell-cycle perturbation, both of which are common in stressed cells. In the study used here we showed that a cell permeable cAMP analogue, pCPTcAMP, and a hydrolysable cAMP analogue, 8pCPT-2′-O-Me-cAMP, were both capable of inducing cell-cycle arrest and promoting synchronized differentiation to procyclic forms in response to cis aconitate as measured by EP procyclin expression. More importantly, however, both compounds were also shown to activate the expression of the PAD1 reporter gene, revealing that they are likely operating through, or intersecting with, a physiologically relevant pathway to generate stumpy-like responses. Despite this, neither compound generated populations of cells that had a fully stumpy morphology and both produced high levels of misshapen and likely ‘unhealthy’ cells. Moreover, the amplitude of PAD1 regulation in response to cAMP regulation was less than during the transition from slender to stumpy forms in vivo. Hence, although useful for mapping inducible control regions in the PAD1 3′-UTR, neither compound could be used to reproducibly generate populations of stumpy cells representative of those generated in vivo using pleomorphic populations.
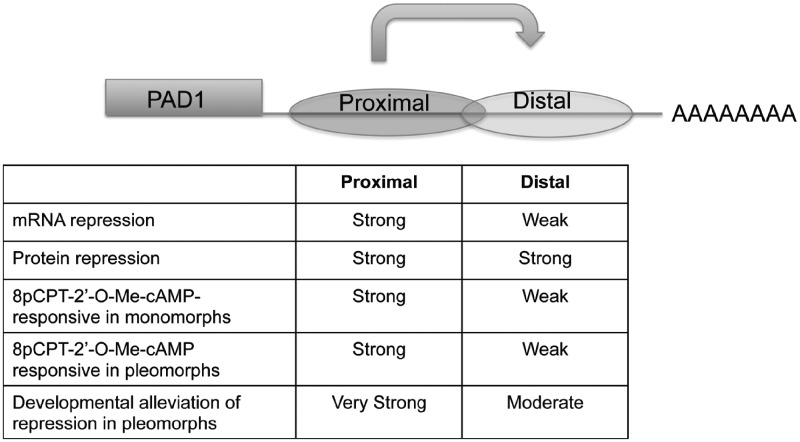
Figure 8 shows a model for the regulation of gene expression mediated via the PAD1 gene 3′-UTR based on our analysis of its component sequences in monomorphic and pleomorphic trypanosomes. At the distal end of the 3′-UTR, within 300 nt of the mapped polyadenylation site, is an mRNA silencing element that limits both mRNA and protein expression. However, specific deletion of this section of the 3′-UTR does not alleviate protein expression when the remainder of the 3′-UTR is intact, suggesting a contribution from gene proximal regions of the 3′-UTR. Consistent with this, when the proximal or distal region of the PAD1 3′-UTR were integrated independently into the 3′-UTR of a constitutively expressed gene, neither reduced mRNA levels although both reduced the levels of expressed reporter protein. Therefore, repression is exerted throughout distinct regions of the PAD1 3′-UTR and protein control is more stringent than mRNA repression. This matches analysis of PAD1 mRNA and protein in pleomorphic cells, whereby PAD mRNA is detected early in intermediate forms (26) whereas protein expression is restricted to fully differentiated stumpy forms (22).
Figure 8.
A model of the regulation by the PAD1 3′-UTR based on the deletion constructs generated in this study. The PAD1 3′-UTR contains proximal and distal silencing elements that repress gene expression at both the RNA and protein level in slender forms. In the absence of the proximal region, silencing is achieved through mRNA regulation, but more prominently through protein regulation. In the absence of the distal region, control is predominantly exerted through the proximal region at the protein level. The proximal region is responsible for regulation in response to 8pCPT-2′-O-Me-cAMP, this generating an alleviation of repression that must also operate to alleviate repression mediated through the distal region. In pleomorphic cells, both proximal and distal elements exhibit an alleviation of repression in stumpy forms.
In contrast to the repression of the gene expression mediated by proximal and distal regions within the PAD1 3′-UTR, regulatable expression induced by 8pCPT-2′-O-Me-cAMP was only associated with the proximal 354 nt, although the distal region was regulatable during development to stumpy forms in vivo. This highlights the proximal region as being more responsive to cAMP, whereas both proximal and distal regions can each contribute to the developmental control of stumpy-specific gene expression in pleomorphs. This further emphasises that the exposure to cAMP analogues does not fully reproduce the complete development to stumpy forms. In terms of the mechanisms of stumpy-specific gene regulation, in the simplest scenario, the proximal and distal regions would be the target of a single negative factor (or mRNP) responsible for repression through binding to the PAD1 3′-UTR, with this binding being reversed by the response to 8pCPT-2′-O-Me-cAMP or SIF (for example, via phosphorylation as the product of a signal transduction pathway). Alternatively, there may be negative and positive factors, such that an 8pCPT-2′-O-Me-cAMP-induced or SIF-induced activator displaces any silencing factors through competing at the same binding site (s). Changes in the affinity of components of an mRNP spanning several regions of the 3′-UTR could also relieve mRNA silencing, this potentially explaining the different sensitivities of the proximal and distal regions after 8pCPT-2′-O-Me-cAMP.
Although the resolution of the precise mechanisms awaits the isolation of the responsible protein factors, the identification of the regulatable defined elements in the PADI 3′-UTR allows the pursuit of experimental approaches using such ligands to dissect the gene expression processes necessary for parasite transmission. Through manipulating these processes by targeted drug approaches, novel routes to prevent disease spread or to limit trypanosome virulence through accelerated parasite development (25) are possible.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6, Supplementary Methods and Supplementary References [31,39–41].
FUNDING
Wellcome Trust [080460 to P.M.], [088293 to K.R.M.], [095831MA to K.R.M.]; Centre for Immunity, Infection and Evolution, this being supported by a strategic award from the Wellcome Trust [095831MA]. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Deborah Hall and Julie Wilson for assistance with mouse infections for the generation of pleomorphic slender and stumpy forms, Dr Sam Dean for the modification of the pHD617 vector and Prof Isabel Roditi for advice on the GUS assay.
REFERENCES
- 1.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marsh L, Neiman AM, Herskowitz I. Signal transduction during pheromone response in yeast. Annu. Rev. Cell. Biol. 1991;7:699–728. doi: 10.1146/annurev.cb.07.110191.003411. [DOI] [PubMed] [Google Scholar]
- 3.Kay RR, Jermyn KA. A possible morphogen controlling differentiation in Dictyostelium. Nature. 1983;303:242–244. doi: 10.1038/303242a0. [DOI] [PubMed] [Google Scholar]
- 4.Pan J, Snell WJ. Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr. Opin. Microbiol. 2000;3:596–602. doi: 10.1016/s1369-5274(00)00146-6. [DOI] [PubMed] [Google Scholar]
- 5.Madhani HD, Fink GR. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 6.Sogin ML, Elwood HJ, Gunderson JH. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc. Natl. Acad. Sci. USA. 1986;83:1383–1387. doi: 10.1073/pnas.83.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 8.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly (A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 9.Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 10.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Matthews KR. Controlling and coordinating development in vector-transmitted parasites. Science. 331:1149–1153. doi: 10.1126/science.1198077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr. Opin. Microbiol. 2007;10:539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zilberstein D, Shapira M. The Role Of Ph and Temperature In the Development Of Leishmania Parasites. Annu. Rev. Microbiol. 1994;48:449–470. doi: 10.1146/annurev.mi.48.100194.002313. [DOI] [PubMed] [Google Scholar]
- 14.Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18:2798–2811. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia ES, Genta FA, de Azambuja P, Schaub GA. Interactions between intestinal compounds of triatomines and Trypanosoma cruzi. Trends Parasitol. 26:499–505. doi: 10.1016/j.pt.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Roditi I, Lehane MJ. Interactions between trypanosomes and tsetse flies. Curr. Opin. Microbiol. 2008;11:345–351. doi: 10.1016/j.mib.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Roditi I, Liniger M. Dressed for success: the surface coats of insect-borne protozoan parasites. Trends Microbiol. 2002;10:128–134. doi: 10.1016/s0966-842x(02)02309-0. [DOI] [PubMed] [Google Scholar]
- 18.Vassella E, VandenAbbeele J, Butikofer P, Renggli CK, Furger A, Brun R, Roditi I. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post- transcriptionally by glycerol or hypoxia. Genes Dev. 2000;14:615–626. [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel TN, Gunasekera K, Cross GA, Ochsenreiter T. Gene expression in Trypanosoma brucei: lessons from high-throughput RNA sequencing. Trends Parasitol. 2011;27:434–441. doi: 10.1016/j.pt.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolan DP, Rolin S, Rodriguez JR, Van Den Abbeele J, Pays E. Slender and stumpy bloodstream forms of Trypanosoma brucei display a differential response to extracellular acidic and proteolytic stress. Eur. J. Biochem. 2000;267:18–27. doi: 10.1046/j.1432-1327.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 21.Czichos J, Nonnengaesser C, Overath P. Trypanosoma brucei: cis-aconitate and temperature reduction as triggers of synchronous transformation of bloodstream to procyclic trypomastigotes in vitro. Exp. Parasitol. 1986;62:283–291. doi: 10.1016/0014-4894(86)90033-0. [DOI] [PubMed] [Google Scholar]
- 22.Dean SD, Marchetti R, Kirk K, Matthews K. A surface transporter family conveys the trypanosome differentiation signal. Nature. 2009;459:213–217. doi: 10.1038/nature07997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuner B, Vassella E, Yutzy B, Boshart M. Cell density triggers slender to stumpy differentiation of Trypanosoma brucei bloodstream forms in culture. Mol Biochem. Parasitol. 1997;90:269–280. doi: 10.1016/s0166-6851(97)00160-6. [DOI] [PubMed] [Google Scholar]
- 24.Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J. Cell Sci. 1997;110 (Pt 21):2661–2671. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- 25.MacGregor P, Matthews KR. New discoveries in the transmission biology of sleeping sickness parasites: applying the basics. J. Mol. Med. 2010;88:865–871. doi: 10.1007/s00109-010-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macgregor P, Savill NJ, Hall D, Matthews KR. Transmission Stages dominate trypanosome within-host dynamics during chronic infections. Cell Host Microbe. 2011;9:310–318. doi: 10.1016/j.chom.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vickerman K. Polymorphism and mitochondrial activity in sleeping sickness trypanosomes. Nature. 1965;208:762–766. doi: 10.1038/208762a0. [DOI] [PubMed] [Google Scholar]
- 28.Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Brit. Med. Bull. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- 29.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 30.Lanham SM. Separation of trypanosomes from the blood of infected rats and mice by anion-exchangers. Nature. 1968;218:1273–1274. doi: 10.1038/2181273a0. [DOI] [PubMed] [Google Scholar]
- 31.Mayho M, Fenn K, Craddy P, Crosthwaite S, Matthews K. Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 2006;34:5312–5324. doi: 10.1093/nar/gkl598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biebinger S, Wirtz LE, Lorenz P, Clayton C. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- 33.Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabani S, Fenn K, Ross A, Ivens A, Smith TK, Ghazal P, Matthews K. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics. 2009;10:427. doi: 10.1186/1471-2164-10-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laxman S, Riechers A, Sadilek M, Schwede F, Beavo JA. Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. Proc. Natl. Acad. Sci. USA. 2006;103:19194–19199. doi: 10.1073/pnas.0608971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denninger V, Figarella K, Schonfeld C, Brems S, Busold C, Lang F, Hoheisel J, Duszenko M. Troglitazone induces differentiation in Trypanosoma brucei. Exp Cell Res. 2007;313:1805–1819. doi: 10.1016/j.yexcr.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Brecht M, Parsons M. Changes in polysome profiles accompany trypanosome development. Mol. Biochem. Parasitol. 1998;97:189–198. doi: 10.1016/s0166-6851(98)00151-0. [DOI] [PubMed] [Google Scholar]
- 38.Clayton CE. NEW EMBO MEMBER'S REVIEW Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.