Abstract
Dna2 nuclease/helicase is a multitasking protein involved in DNA replication and recombinational repair, and it is important for preservation of genomic stability. Yeast Dna2 protein contains a conserved putative Fe–S (iron–sulfur) cluster signature motif spanning the nuclease active site. We show that this motif is indeed an Fe–S cluster domain. Mutation of cysteines involved in metal coordination greatly reduces not just the nuclease activity but also the ATPase activity of Dna2, suggesting that the nuclease and helicase activities are coupled. The affinity for DNA is not significantly reduced, but binding mode in the C to A mutants is altered. Remarkably, a point mutation (P504S), proximal to the Fe–S cluster domain, which renders cells temperature sensitive, closely mimics the global defects of the Fe–S cluster mutation itself. This points to an important role of this conserved proline residue in stabilizing the Fe–S cluster. The C to A mutants are deficient in DNA replication and repair in vivo, and, strikingly, the degree to which they are defective correlates directly with degree of loss of enzymatic activity. Taken together with previous results showing that mutations in the ATP domain affect nuclease function, our results provide a new mechanistic paradigm for coupling between nuclease and helicase modules fused in the same polypeptide.
INTRODUCTION
The identification of a network of proteins interacting with Dna2 in the maintenance of genomic stability pointed to its central role in DNA replication and recombination (1). This genetic road map determined most of the partners of Dna2 involved in carrying out its essential cellular functions. Not surprisingly, follow-up mechanistic studies have confirmed the importance of this network in many aspects of DNA metabolism (2). Major roles of Dna2 in Okazaki fragment processing, double-strand break repair and telomere maintenance are well documented, whereas an additional role in mitochondrial DNA maintenance is less well understood (3; for review).
In recent years, different classes of Fe–S (iron–sulfur) clusters have been identified as essential components of a wide variety of DNA and RNA processing enzymes (4). DNA glycosylases, nucleases, helicases, eukaryotic primase and eukaryotic B family DNA polymerases have all been shown to have Fe–S clusters. They fall into more than one structural class and play diverse roles in DNA binding, structural stability, coupling of ATPase and translocation and interaction between subunits. Several are redox active when bound to DNA, and this feature has been suggested to constitute a new type of signaling pathway in transcription and repair [see (5) for review, (6)]. One class of enzyme that has not yet been extensively studied has nuclease and helicase domains fused in the same polypeptide. This class includes type 1 restriction endonucleases, RecBCD, AddAB and Werner protein. Such proteins abound in evolution, but the functional relationship between the helicase, or translocase, and nuclease domains has remained elusive. It has been proposed that nucleases and helicases linked in this way function in a coupled manner, making the whole work better than the sum of the parts. Although not all of these helicase/nuclease proteins contain Fe–S clusters, to explore this coupling model, we have examined the role of a putative Fe–S domain in coupling between helicase and nuclease in the yeast Dna2 helicase/nuclease (Figure 1).
Figure 1.
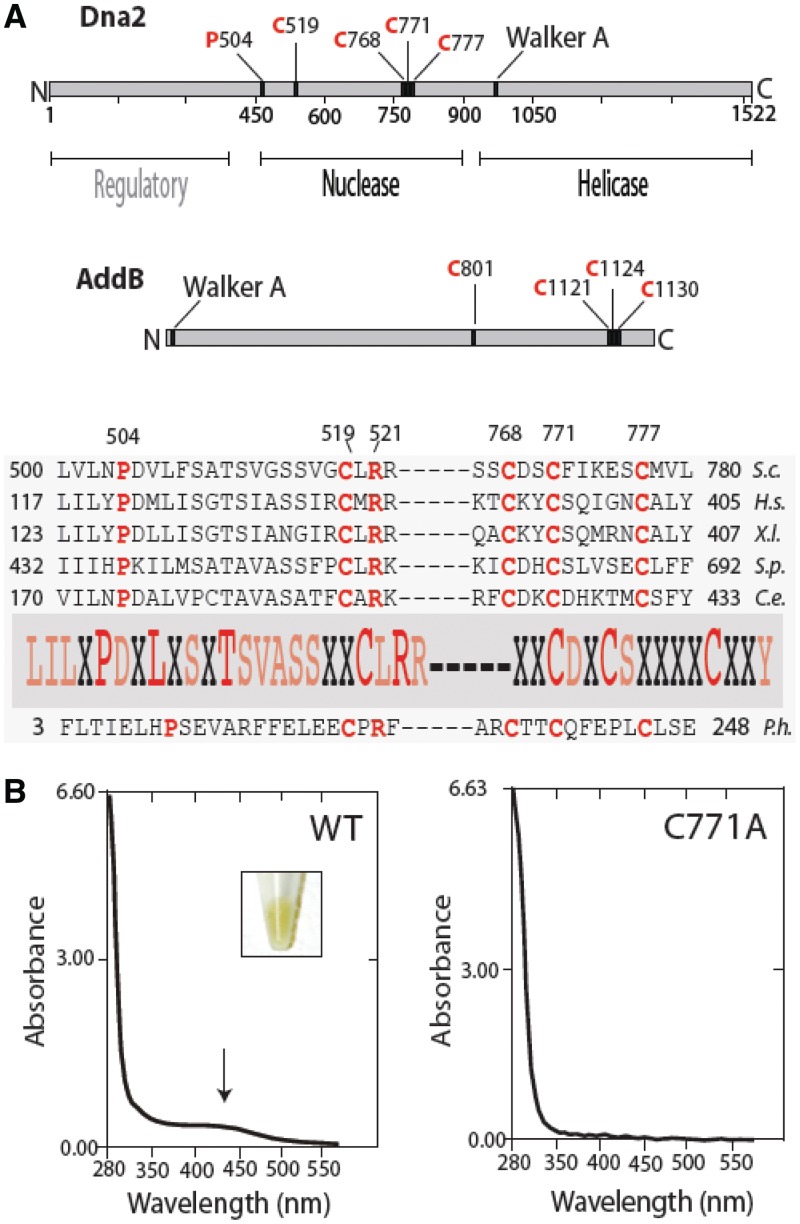
Dna2 protein contains an Fe–S cluster. (A) Primary structure and conservation of the Dna2 Fe–S motifs. In eukaryotes, Dna2 is well conserved within the N-terminal nuclease and C-terminal helicase domains. Yeast Dna2 has an extra approximately 400 amino acids at the N-terminal, which is absent in the other metazoan orthologs such as human and Xenopus. Although not conserved, the deletion of the N-terminal 405 amino acids of Dna2 results in temperature-sensitive growth of mutant yeast cells, indicating its regulatory roles (26). Yeast dna2-1, with a point mutation at P504S, is temperature sensitive and cannot grow at 37°C (13). As the P504 residue is conserved and close to the first cysteine residue (C519) of the conserved Fe–S motif, the mutation may disrupt the conserved motif, leading to temperature sensitivity in yeast. (B) Ultraviolet–visible spectra of WT Dna2 and Dna2 C771A. Insert: Dna2 preparations (15 mg/ml) show yellow color, whereas C771A does not (data not shown). S.c., Saccharomyces cerevisiae; H.s., human; X.l., Xenopus; S.p., Schizosaccharomyces pombe; C.e., worm.
Dna2 helicase/nuclease is now recognized as a RecB-like protein with a C-terminal SF1B 5′- to 3′ helicase and an N-terminal dual specificity nuclease (both 5′- to 3′ and 3′- to 5′) joined by a nonconserved linker domain (7). Dna2 and RecB contain four highly conserved motifs in the nuclease active site, including the Q/Y motif that defines the family and is thought to stabilize the sizile bond. Early sequence alignments revealed a three-cysteine putative metal-binding cluster, CX2CX5C, in Dna2 downstream of the nuclease active site motifs, with homology to clusters present in a subfamily of small RecB nucleases (8,9). The AddAB protein of Gram-positive bacteria is also a helicase/nuclease. In this protein, the helicase is encoded in the AddA subunit, along with a 3′- to 5′ nuclease, whereas the AddB subunit contains a RecB-like 5′ nuclease and a RecA-like domain, albeit lacking all helicase motif consensus sequences, except for a close match to the Walker A box. The AddB nuclease domain is spanned by a 4F–4S cluster. Disruption of this cluster abolishes the ability of AddB to bind dsDNA ends and reduces nuclease activity and dsDNA-dependent ATPase in the AddAB complex but has no effect on protein/protein interaction or single-stranded DNA-dependent ATPase activity (10). The motif has been called an iron staple domain because the cysteine ligands span the nuclease site, and their mutation leads to structural alterations resulting in increased sensitivity to trypsin. Recently, this structural role for the Fe–S cluster has been supported by the X-ray crystal analysis of Bacillus subtilis AddAB (11). Sequence alignment of AddB and Dna2 revealed an additional conserved cysteine residue in Dna2, located 248-aa upstream of the three-cysteine cluster (10). Taken together, the four conserved cysteines in Dna2 are likely part of an Fe–S motif, CX248CX2CX5C, spanning the nuclease motifs, as in AddB. An interesting feature of the RecB-like enzymes is that they share the rare property for helicases of loading from the end of the substrate (12). Here, we show that the yeast Dna2 cysteine cluster is indeed an Fe–S domain and present biochemical and genetic evidence that this domain plays a role not only in nuclease but also in helicase activity, and that it is critical for the physiological function of Dna2. We also discuss our previous results indicating that mutations in the helicase domain affect nuclease activity.
MATERIALS AND METHODS
Expression constructs
To produce the recombinant yeast Dna2 protein, an N-terminal histidine tag and a tobacco etch virus (TEV) protease recognition site were introduced by PCR. A two-step tandem PCR, which was more efficient for cloning, was used to generate the recombinant yDna2. The first round of PCR was performed by using 5′-(CATCAC)3 GAG AAC CTC TAT TTC CAG GGG TCC AAT TTG AGT AGG CAT and 5′-CCG CGC CTC GAG TCA ACT TTC ATA CTC TTG TAG primers and pGAL18-Dna2HA plasmid (13). After purification, the resulting PCR product was used as a template for the second round of PCR where 5′-CCG CGC CGT CTC GGA TCC GTA ACC ATG TCA (CATCAC)5 and 5′-CCG CGC CTC GAG TCA ACT TTC ATA CTC TTG TAG were used as primers. To construct the Dna2 expression vector, YEpDNA2PGAL1 (Ura+), the resulting PCR product, was digested with the BamHI and XhoI and ligated into a BamHI- and XhoI-digested expression plasmid YEpTOP2PGAL1 (14). The expression vector has a pBR322 backbone and contains a 2 -µm origin of replication, a URA3 gene, an ampicillin resistance marker and a GAL1 promoter immediately upstream of the BamHI cloning site. The fidelity of the insert was confirmed by sequencing. After protein expression, the His-tag was removed using TEV enzyme to yield DNA2 containing an extra N-terminal glycine, a remnant of the TEV recognition site. Additionally, the Dna2 protein is missing 105 amino acids at the N-terminal. The full-length yDNA2 gene, along with N-terminal histidine tag and a TEV recognition site, was also cloned in the same vector backbone using an additional primer 5′-(CATCAC)3 GAG AAC CTC TAT TTC CAG GGG ATG CCC GGA ACG CCA CAG AAG and purified to ensure that the 105 amino acid deletion did not affect function. pRS314-DNA2 (Trp+, CEN) has the full-length Dna2 gene under the control of its endogenous promoter (15).
Site-directed mutagenesis
Each of the four conserved cysteines at positions 519, 768, 771 and 777 was individually substituted with alanine by using site-directed mutagenesis in both YEpDNA2PGAL1 and pRS314-DNA2 plasmids. The 5′-GGA AGT TCA GTA GGT GCT TTA AGA CGT TCA ATT C and 5′-GAA TTG AAC GTC TTA AAG CAC CTA CTG AAC TTC C, 5′-CTG CGC GAT TCA TCT GCT GAT TCA TGT TTC ATC and 5′-GAT GAA ACA TGA ATC AGC AGA TGA ATC GCG CAG, 5′-CAT CTT GTG ATT CAG CTT TCA TCA AAG AAT C and 5′-GAT TCT TTG ATG AAA GCT GAA TCA CAA GAT G and 5′-GTT TCA TCA AAG AAT CAG CCA TGG TGT TGA ATA AGC TAC and 5′-GTA GCT TAT TCA ACA CCA TG GCT GAT TCT TTG ATG AAA C DNA oligomers were, respectively, used to mutate C519A, C768A, C771A and C777A residues. All constructs were confirmed by DNA sequencing.
Purification of Dna2 enzymes
Wild type (WT) yeast Dna2 protein and its C to A mutants were purified from yeast as described previously (16). Mutant proteins were expressed at similar levels to WT protein and purification yields were also similar.
Preparation of radiolabeled substrates for nuclease and gel shift assays
Oligonucleotides used have been described previously: 5′-tail [74 nt (17)], flap [99 nt (17)], ssDNA (D4, 55 nt) (18)], fork [T2, 51 nt and D4, 55 nt (18)] and double-flap [T2, 51 nt; U1, 26 nt and D4, 55 nt (18)]. The corresponding oligodeoxynucleotide was labeled at the 5′-end using polynucleotide kinase and [γ-32P]-ATP. The 5′-tail and flap oligonucleotides fold to form the 5′-flap DNA substrate as depicted in the figures (17).
Nuclease assay
Indicated amounts of wild-type or mutant proteins were incubated at 37°C for 30 min with 20 fmol of radiolabeled oligonucleotide in 20 µl of reaction volume containing 50 mM Tris–HCl (pH 8.0), 2 mM dithiothreitol (DTT), 0.1 mg/ml BSA, 30 mM NaCl, 2 mM MgCl2, 50 µM ATP and 5% glycerol. In this assay, the reactions contain 2, 7, 30 and 100 nM WT or mutant enzyme. For replication protein A (RPA)-stimulated nuclease reactions 5 fmol of WT or 500 fmol of mutant enzymes were used. To stop the reaction, 20 µl of sequencing gel loading buffer was added. Samples were boiled for 5 min at 95°C and loaded onto a 12% sequencing gel. The gel was run for 1 h at 60 W, exposed overnight to a phosphor screen and visualized using a Typhoon 9400 with ImageJ software.
DNA-binding assay
The 5′-flap DNA binding activity of WT and mutant Dna2 proteins was measured using a gel mobility shift assay. Protein was incubated with 20 fmol of radiolabeled DNA substrate in a reaction volume of 20 µl containing 50 mM Tris–HCl (pH 8.0), 2 mM DTT, 0.1 mg/ml BSA, 30 mM NaCl, 50 µM ATP and 5% glycerol. In this assay, each reaction contains 2, 7, 30 and 120 nM protein. The reactions were mixed at room temperature, then incubated on ice for 25 min. The reactions were loaded onto 6% native polyacrylamide gel (0.5X TBE) and separated at 4°C for 90 min at 100 V. The gels were exposed overnight to phosphor screens and visualized using a Typhoon 9400 with ImageQuant software.
Plasmid shuffling
BY4741 dna2Δ::G418 pGAL-DNA2 (URA) strain was transformed with pRS314-DNA2 WT or Fe–S mutant plasmids following the lithium acetate procedure (19). After transformation, yeast were plated directly onto agar containing complete minimal medium lacking tryptophan and uracil and containing 2% glucose. The plates were incubated at 30°C for 3 days. A yeast colony was harvested and placed into 5 ml of the same medium at 30°C for 2 days with shaking. Cells were counted and spotted onto an agar plate containing 5-fluoroorotic acid (5-FOA) (1 µg/µl). Since the 5-FOA kills cells expressing URA3, the surviving yeast cells lose the pGAL-DNA2 plasmid. Only cells carrying the pRS314-Dna2 plasmids and able to complement dna2Δ::G418 will survive on 5-FOA.
Strains and plasmids used
The strain BCY123 (14), a protease-deficient ura3 yeast strain, was used for the expression of Dna2 proteins. The temperature-sensitive dna2-1 strain for complementation assay was as described (13).
Limited subtilisin proteolysis
Lyophilized subtilisin was resuspended in 50 mM Tris–HCl (pH 8.0) and 150 mM NaCl. For a 10 -µl reaction, 10 µg of purified Dna2 and 0.005 µg of subtilisin were mixed in 50 mM NaHCO3 solution. Reactions were incubated at room temperature for the indicated times and were quenched by adding 1 µl of 25 mM phenylmethylsulfonylfluoride and 10 µl of SDS–PAGE loading buffer. Samples were boiled for 5 min at 95°C and resolved in a 4–20% gradient gel (Thermo Scientific), and peptide fragments were visualized by Coomassie Blue.
RESULTS
Dna2 contains an Fe–S cluster
Alignment of amino acid sequences with the AddB iron staple domain suggests that in yeast Dna2, four C (cysteine) residues (C519, C768, C771 and C777) contribute the ligands required for Fe coordination (10). Figure 1 shows a schematic of the Dna2 protein and conservation of the proposed Fe–S cluster ligands. To determine the role of the Fe–S motif in Dna2, we mutated all of the four conserved C ligands individually to A (alanine). We purified WT yeast Dna2 from yeast, along with each of the four mutant proteins. Interestingly, the WT protein displays a yellow to yellow-brown color when isolated in high concentration (Figure 1B). The mutant proteins were colorless. We further analyzed the spectral characteristics of the proteins. The purified WT Dna2 has a broad absorption spectrum of ∼450 nm (Figure 1B), which is characteristic of proteins containing 4Fe–4S or 3Fe–4S clusters (20). The characteristic absorbance spectra was not found in the C771A mutant protein, although due to limited amounts of protein, the sensitivity of this measurement is lower than for wild-type (Figure 1B). For the preparation shown in Figure 1B, assuming an extinction coefficient 17 000 M−1 for the cluster (21), ∼70% of the WT Dna2 contains Fe.
C to A mutations reduce Dna2 nuclease activity
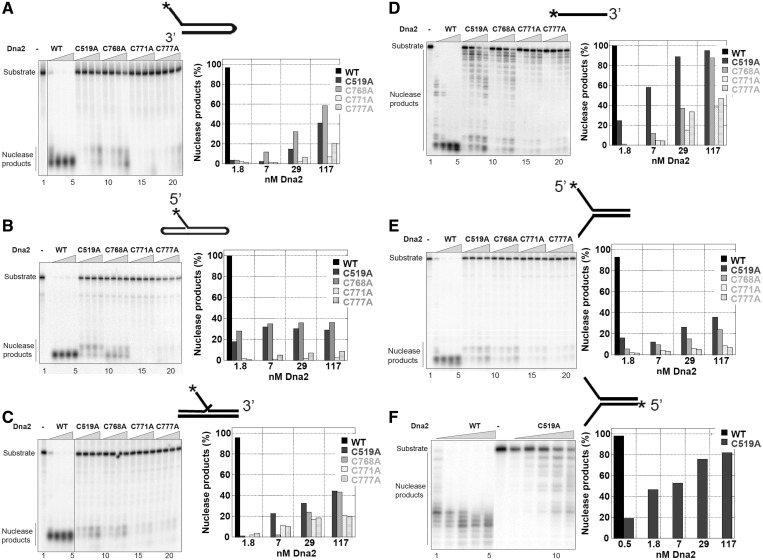
The nuclease activity of each of the C to A mutants was measured using radiolabeled DNA oligonucleotides simulating intermediates in Okazaki fragment processing (double-flap and flap) and DSB repair (fork and ssDNA). For WT Dna2, maximum cleavage (>98% cleavage) was observed in a 30-min reaction at 2–8 nM Dna2 protein (Figure 2A–F). Dna2 protein with C to A mutations in any of the four putative iron coordination sites exhibited significantly reduced nuclease activity for all of the substrates (Figure 2A–F). At 2 nM, the mutant proteins showed no activity. Only 40–50% cleavage of double-flap substrates occurred even at 100 nM protein concentration for C519A and C768A, and <10% was observed for C771A and C777A on double-flap substrates (Figure 2C). The mutant proteins had greater residual activity on ssDNA than on the flap or fork substrates (Figure 2D), indicating an altered substrate specificity. C519A and C768A at 100 nM cleaved 80 and 70% of the ssDNA, and C771A and C777A removed 30–40%, respectively. Both the 5′ to 3′ and 3′ to 5′ cleavage activities were inhibited on a fork substrate (Figure 2E and F). The 3′ to 5′ activity was reduced less than the 5′ to 3′ (Figure 2E and F). Greater residual activity on ssDNA suggests that mutant Dna2 binds differently to ssDNA and ssDNA adjacent to a duplex region. We have shown previously that WT Dna2 prefers a flap substrate to ssDNA (22); therefore, the mutations may affect the structure-specific activities of Dna2 more than the ssDNA activity, though both are reduced.
Figure 2.
The Dna2 Fe–S cluster is important for nuclease activity. Cleavage extent assays were performed in 20 -µl reaction volume with 20 fmol of DNA substrate as described in the ‘Materials and Methods’ section. Enzyme concentration was varied between 2, 7, 30 and 100 nM protein, as indicated by the triangles. Lane 1 or 6 is the substrate-only control. Schematic representations of the substrate structures are depicted above the graph, and the positions of the substrate and nuclease products are indicated on the left. The percentage of nuclease products was calculated by quantifying the gel using the ImageQuant program. The results are presented as histograms. For clarity, the percentage of nuclease products of WT enzyme with excess enzyme concentrations is removed from the graph. Panels A-F, respectively, represent the specific substrates as shown.
C to A mutants are stimulated by RPA
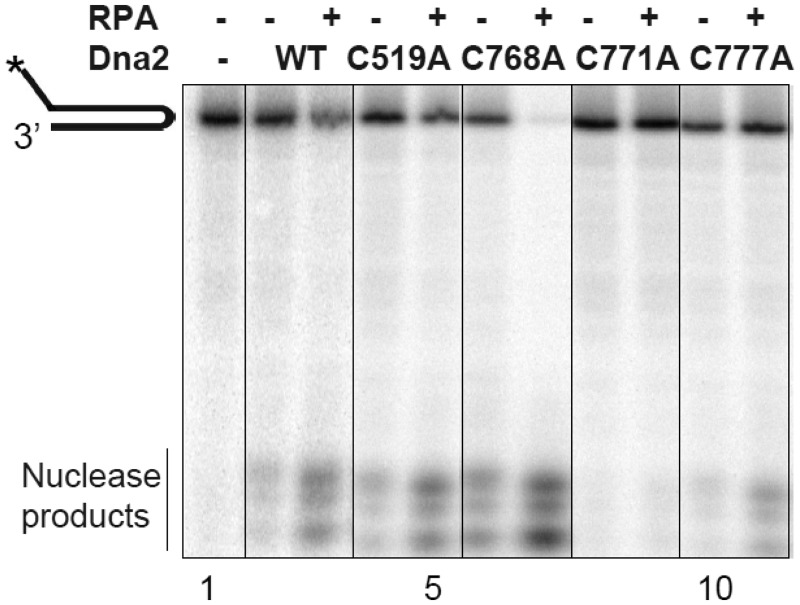
Dna2 activities are stimulated by RPA (23,24), and Dna2 interacts specifically with RPA (23). The precise amino acids involved in the RPA/Dna2 interaction have been not been mapped, although the N-terminus of Dna2 stimulates interaction (23). Since RPA is a natural partner of Dna2, we determined whether the C to A mutations altered RPA interaction. We observed that RPA stimulates the nuclease activity of all mutant proteins to approximately the same extent as WT Dna2 (Figure 3). Quantitation in this experiment shows a 4-fold stimulation of wild-type. Using 100 times higher concentration of each mutant, which was required to see activity, we found a 3-fold stimulation of C519A, 2-fold stimulation of C668A and 2-fold stimulation of C777A. In this experiment, C771A activity was undetectable, in the presence of RPA. Thus, an intact Fe–S domain does not seem to be critical for interaction with RPA, but rather the Fe–S cluster may play a structural role with respect to the nuclease active site. We also note that RPA stimulation of the mutants suggests that the residual nuclease activity observed derives from Dna2, rather than a contaminating nuclease, which would be unlikely to respond to RPA.
Figure 3.
The nuclease activity of the Dna2 Fe–S mutants is stimulated by RPA. Assays measuring extent of degradation were conducted as described in the ‘Materials and Methods’ section and legend to Figure 2. Fold stimulation was quantified using the phosphorimager and is reported in the text.
C to A mutations do not alter the affinity of Dna2 for DNA
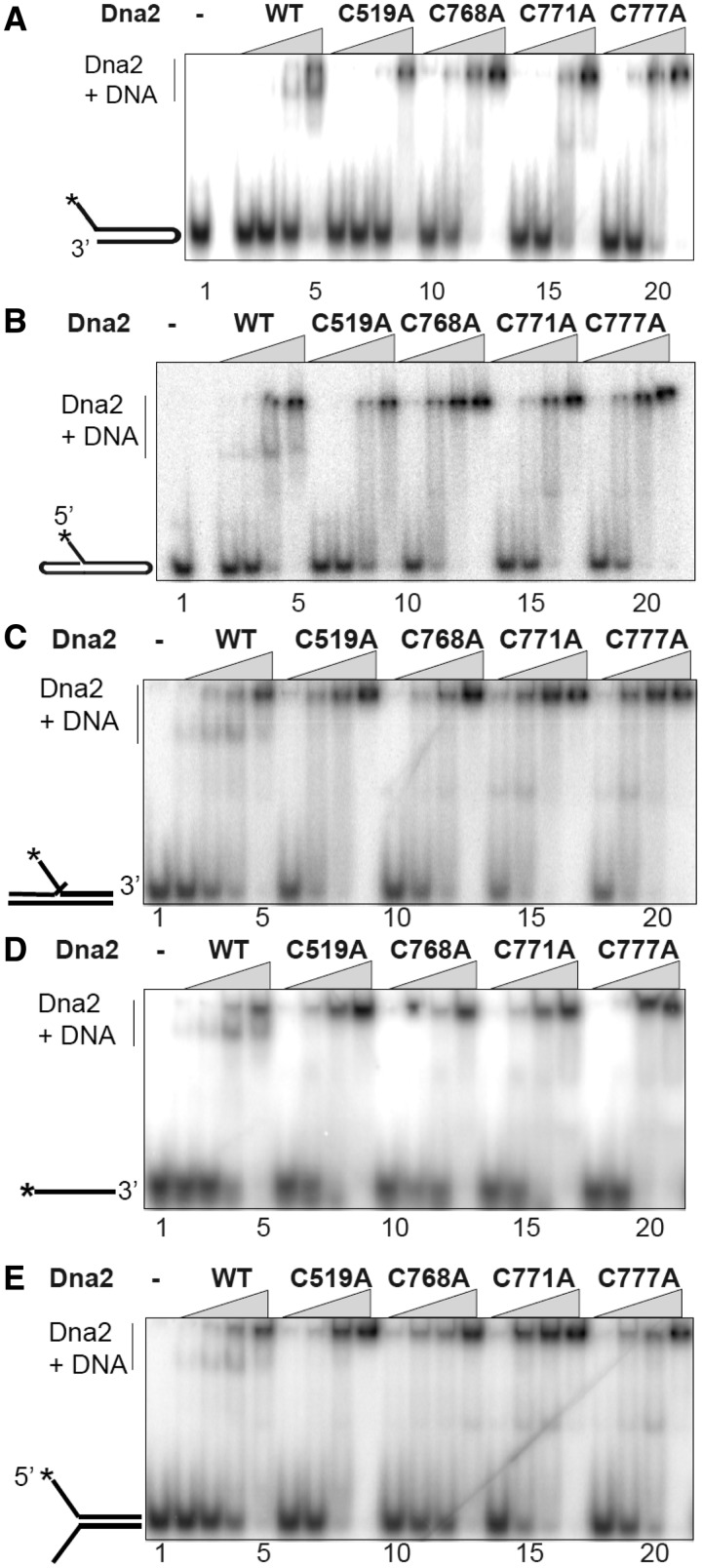
We used an electrophoretic mobility shift assay to determine whether the mutant enzymes had impaired DNA binding, as observed for AddAB. DNA binding is determined by observing the reduction in mobility of a radioactive DNA band in a polyacrylamide gel after addition of protein. The same spectrum of oligonucleotides used for nuclease assays was used for DNA binding. The discrete complexes that were observed to form with the WT protein suggest that this is a valid way of measuring Dna2–substrate affinity (Supplementary Figure S1). Surprisingly, unlike with nuclease activity, the affinity of the mutant enzymes for the various DNA substrates was similar to that of the wild-type. However, the gel pattern of Dna2/DNA complexes was altered by the mutations. The WT enzyme shows two shifted bands when tested with any of the fork, flap or ssDNA substrates, suggesting two different complexes with different molar ratios of Dna2 and DNA. This interpretation is supported by a competition assay, which demonstrated that the slowly migrating species, presumably containing higher order multimers of Dna2, is preferentially converted to the more rapidly migrating species as increasing cold substrate is added (Supplementary Figure S2). In contrast to the WT Dna2, all four mutants exhibit only one predominant shifted band, the more slowly migrating (multimeric) band (Figure 4). We conclude that although the mode of binding may be different between the WT and mutant enzymes, the dissociation constants for the wild-type and the mutants are comparable for all the substrates tested.
Figure 4.
The Dna2 Fe–S cluster is not essential for DNA binding. The electrophoretic mobility shift assays were performed in 20 -µl reaction volume with 20 fmol of DNA substrates. An enzyme titration (2, 7, 30 and 100 nM protein, as indicated by the triangles) was performed as described in the ‘Materials and Methods’ section. Lane 1 is the substrate-only control. Schematic representations of the substrate structures are depicted on the left of the gel.
C to A mutations inhibit ssDNA-dependent ATPase activity
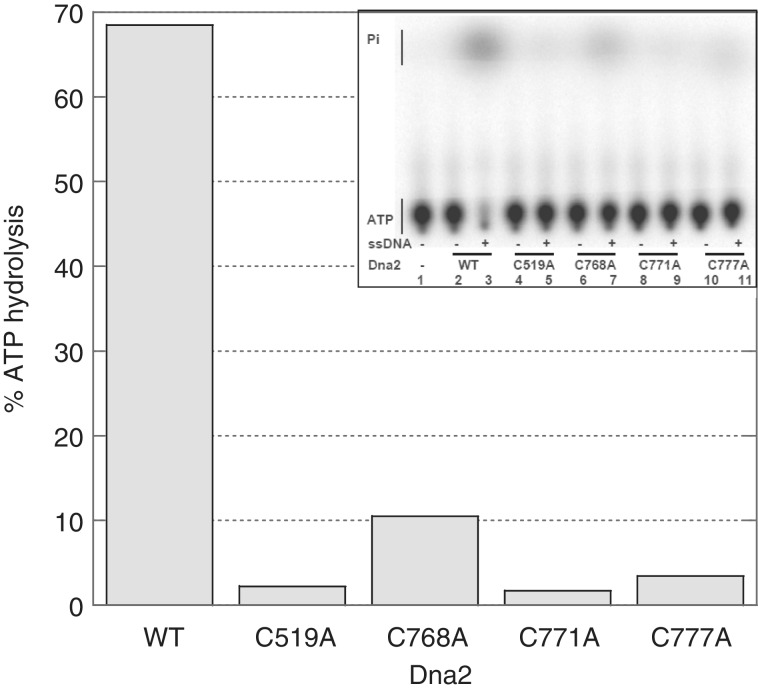
The nature of domain interactions in the helicase/nuclease family of proteins remains enigmatic, and therefore, we were interested to test the effect of the Fe–S mutations on the helicase activity. Although all four C residues responsible for the formation of the metal cluster are located at the N-terminal nuclease domain, we had previously observed that a P504S mutation in the nuclease domain, the originally isolated temperature-sensitive allele of Dna2, caused loss of both nuclease and helicase activities (15). Dna2 uses ssDNA-dependent ATP hydrolysis to unwind adjacent dsDNA. Fully dsDNA does not support the Dna2 ATPase activity. In a ssDNA-dependent ATPase assay, all four Fe–S mutants exhibited greatly reduced ATP hydrolysis compared with the WT enzyme, even at 120 nM concentration in the reaction (Figure 5). Thus, the C to A mutations, like P504S, have an effect on Dna2 ATPase activity at a distance. Since the ssDNA presumably binds to the helicase domain, as in other SF1 helicases, and since nuclease active site mutations, in contrast to the Fe–S mutants, have been reported to have negligible effects on the helicase activity (15,25), we conclude that the Fe–S mutations have a long-range effect on the ATPase domain in addition to causing local disruption of the nuclease domain. Our results provide good evidence that there is cross talk between the structures responsible for nuclease and ATPase/helicase activities, and that the Fe–S cluster is important for this interaction.
Figure 5.
The Dna2 Fe–S cluster is essential for the ssDNA dependent ATPase activity. The ATPase reactions were performed with 117 nM enzyme in the presence or absence of ssDNA. The percentage of ATPase products was calculated by quantifying the gel using the ImageQuant program and plotted in the graph. This assay was done twice. The histogram is the average of two experiments such as the example shown in the insert. The two assays differed by <10%.
C to A mutants do not increase sensitivity to subtilisin
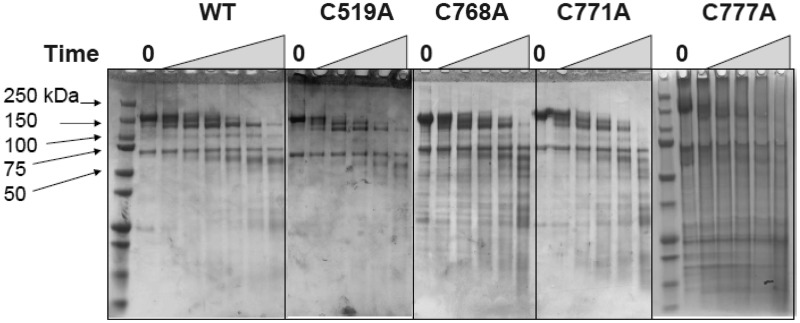
Because of the low nuclease and ATPase activities of the mutants compared with the WT enzyme, we thought that the mutation might have substantially changed the global protein folding pattern. To assess protein folding after mutation, we performed limited proteolysis using subtilisin (Figure 6). Wild-type and mutant proteins digested with subtilisin produced similar fragment patterns indicating no substantial protein structure change resulting from the mutation in the iron binding sites, consistent with the normal expression levels and protein recovery during purification mentioned earlier.
Figure 6.
Subtilisin treatment reveals no major global changes in structure in the Dna2 Fe–S mutants. Partial digestion with subtilisin was carried out as described in ‘Materials and Methods’ section. Dna2 preparations were treated with subtilisin (1, 3, 5, 10 and 40 min, an extra 20 min for the WT, as indicated by the triangles). The reactions were quenched and analyzed by SDS–PAGE gel. The molecular masses of the standards are indicated. The gel for C777A was electrophoresed for a longer time than the other gels, as indicated by the additional lane of markers.
C to A mutants are defective in vivo
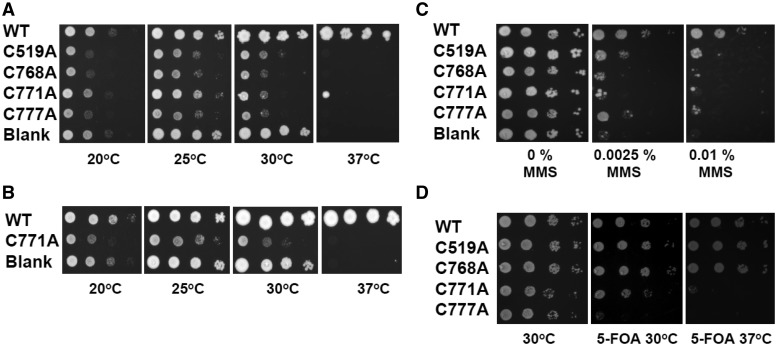
The P504S mutant, dna2-1, is temperature-sensitive and unable to grow at 37°C (13). To test whether the point mutations in the metal-coordinating cysteines are essential in vivo, we assessed their ability to rescue the temperature-sensitive growth defect of dna2-1. The dna2-1 strain of yeast was transformed with plasmids expressing WT Dna2, the C to A mutants or an empty vector at 25°C. Growth of the transformants was then tested at 20, 25, 30 and 37°C. Cells were grown on glucose to keep the concentration of proteins as close to natural levels as possible (15). The strain expressing the WT enzyme suppressed the dna2-1 temperature sensitivity at 37°C, whereas none of the mutant-expressing cells survived at 37°C, just as with the strain transformed with the empty vector (Figure 7A). This result is the same as we obtained earlier with the nuclease active site motif Dna2 mutants, E640A, D657A, E675A and Y693A (15). We conclude that the Fe–S cluster is essential for Dna2 function. We also point out that, as with the nuclease active site mutants (15), dna2-1 transformed with the empty vector exhibited better growth than the C to A mutants at 25 and 30°C, permissive temperatures for dna2-1. Expression of WT Dna2 does not have this deleterious effect. This indicates that the Fe–S mutants are transdominant at 20, 25 and 30°C. Plating of either the WT or mutant Dna2-expressing transformants on galactose containing plates demonstrated, as also previously reported (15), that overproduction even of WT Dna2 is toxic (data not shown).
Figure 7.
The Dna2 Fe–S cluster is important for growth and resistance to DNA damage. (A and B) Growth at various temperatures of the dna2-1 strain expressing the indicated WT DNA2 or mutants in YEpDNA2PGAL1 (Ura+), as labeled. Cells were grown to saturation and aliquots of 10-fold dilutions were placed on plates and incubated at the temperatures indicated. The first spot contains 40 000 cells. Panel B uses full-length forms of WT DNA2 and C771A. (C) Strain dna2-1 strain expressing WT, mutant proteins or no protein were plated on medium containing the indicated amount of MMS at 25°C. (D) Plasmid shuffling assay to test the ability of the C to A mutants to support growth in the absence of any other Dna2 protein in the cell. See ‘Materials and Methods’ section for details.
For Figure 7A, we used Dna2 proteins initiated at the second methionine, 105 amino acids internal to the first methionine, because this gene had been shown to fully complement a dna2Δ strain and behave identically biochemically to wild-type and was available at the time we did the initial C to A mutagenesis. Since the C to A mutants are defective compared with the wild-type in Figure 7A, this experimental design was effective in revealing specific defects in the mutants. To further ensure that defects were not due to the 105 amino acid deletion, we analyzed the full-length Dna2. As a representative, we made the C771A substitution in the full-length DNA2 gene cloned into the same expression vector. This C771A mutant, the full-length WT DNA2 and blank vector were transformed into the dna2-1 yeast strain. The strain expressing the full-length WT enzyme suppressed the temperature sensitivity at 37°C, whereas cells expressing either the C771A mutant protein or transformed with the empty vector failed to survive at 37°C (Figure 7B). Taken together, the expression of neither N-terminal truncated nor full-length Fe–S mutant Dna2 protein suppresses the temperature sensitivity of dna2-1 strain of yeast. Therefore, we conclude that the Fe–S cluster, and not the N-terminal 105 amino acids, is essential for the proper functioning of the Dna2 protein in vivo at 37°C.
We also tested the DNA damage sensitivity of the dna2-1 strain transformed with the various mutants. Mutant dna2-1 is sensitive to methyl methanesulfonate (MMS) at room temperature. As shown in Figure 7C, none of the four C to A mutant enzymes complemented the MMS sensitivity of the dna2-1 mutant strain.
Finally, we carried out plasmid shuffling to determine whether the mutants can support cells with no endogenous expression of Dna2. As shown in Figure 7D, at 30°C, C519 and C768A supported growth of a dna2Δ strain, whereas C771A and C777A do not. At 37°C, C519 and C768 are slightly defective, but after sufficient incubation grew to high density. C771A and C777A were completely defective in supporting growth at 37°C. Since C519 and C768 failed to support growth of the dna2-1 strain but supported the dna2Δ strain at the permissive temperature of 30°C, we propose that they are dominant-negative alleles in the presence of the dna2-1 protein. In the complete absence of Dna2 in the dna2Δ strain, however, another nuclease, such as FEN1 or Exo1, may be able to compensate for absence of Dna2 but may be blocked from doing so in the presence of the defective dna2-1 protein. We conclude that the Fe–S cluster is required for the essential function of Dna2, and that mutations in C519 or C768 are less deleterious than the C771A and C777A mutations. Thus, the degree of physiological defect correlates with the severity of the defect in enzymatic activity.
DISCUSSION
We show here that yDna2 nuclease/helicase contains a four cysteine Fe–S cluster. As the first cysteine residue and the remaining three cysteines are separated by more than 250 amino acids spanning the nuclease active site, the Fe–S cluster may ‘staple’ the nuclease domain to give a unique tertiary structure to Dna2, by analogy to AddB (10,11). Interestingly, although the metal domain is found in the N-terminal nuclease domain, mutation of any of the four cysteines that bind iron not only led to reduction of nuclease activity but also reduced the ATPase of the C-terminal helicase domain. Although the mutations diminished both the nuclease and helicase activities, surprisingly, the DNA binding affinity of the mutants was similar to WT. Additionally, the conformation of the apoprotein was unchanged as determined by protease sensitivity, expression and recovery of purified protein and ability to act as a negative factor in vivo. In view of these observations, we propose that the defect in the Fe–S cluster disrupts obligatory dynamic conformational alterations during coupled cleavage/translocation reactions. An example of coupling is that binding contacts made by the nuclease could increase the translocation processivity of the helicase. The likely interaction and interdependence of nuclease and helicase functions provide an explanation for why we were not able to detect nuclease activity in a fragment of Dna2 containing amino acids 1–963 nor ATPase in a partial protein from amino acids 964–1522 in the past, i.e. because their activities stimulate each other (15). For instance, tightly bound nuclease might increase the processivity of the helicase. Others have also tried and failed to express nuclease and ATPase activities independently (26). Biochemical evidence for coupling has been presented previously by showing that helicase stimulates nuclease activity on flap substrates containing secondary structure (18,27), and the experiments suggested strongly that the active helicase and nuclease have to be in the same polypeptide, supporting conformational coupling (27). However, the structural basis for these observations remains elusive.
Our results define a novel paradigm of Fe–S domain function that not only resembles but also differs from other proteins containing fused nuclease and helicase/translocase domains. Based on the AddB protein (10,11), we had expected that the C to A mutations would seriously impact DNA binding. This was not the case, possibly because the Fe–S cluster of AddB is important for binding dsDNA ends, and Dna2 only binds flaps or unwound ends. The defect in binding dsDNA in AddB mutants thus indirectly inactivates the dsDNA-dependent AddAB ATPase/translocase, associated with AddA. However, the Fe–S mutations do not alter the ssDNA-dependent ATPase of AddAB (10,11). Characteristics of Dna2 more closely resemble those of the HsdR subunit of the type 1 restriction enzymes, which contain N-terminal nuclease and C-terminal translocase domains. In the HsdR subunit of EcoR124I, mutations in the catalytic motifs II and III of the nuclease have dramatic effects on the C-terminal translocase activity. The similarity lies in the implied long-range interaction between the nuclease and translocase activities. Remarkably, this cross-talk is not mediated by and Fe–S cluster in HsdR. A WT Dna2 protein lacking Fe may mimic EcoR124I, in that there may be some residual interaction. The Dna2 Fe–S cluster also differs from that in the xeroderma pigmentosum group D (XPD) and FancJ families, where the cluster contains four closely associated cysteines inserted in helicase domain 1, contributing to an arch that is important for coupling ATPase and unwinding (5,28). Yet, another 4C motif with four closely linked cysteines is located in DNA polymerases; and, in DNA polymerase delta, the cluster stabilizes subunit interactions and replisome stability but does not have a long-range effect on polymerase catalysis (29).
The helicase domain also reciprocally regulates the nuclease domain. A mutation in helicase motif 1, K1080E, which affects but does not abolish the ATP β- and γ-phosphate binding, did not inactivate but significantly changed the nuclease. Normally, the nuclease is inhibited by ATP. However, in the K1080E protein, the nuclease is highly stimulated by ATP (15). This resembles the ATP-dependent stimulation of Dna2 exonuclease in the presence of Mn+2 instead of Mg+2 (16). ATP also inhibited strand annealing in Dna2-K1080E (30). Remarkably, the N-terminal nuclease domain of a bacterial DinG protein has also recently been shown to be regulated by a C-terminal ATPase (31). Finally, in vivo, dna2-D675A, a nuclease active site mutant, is toxic, but dna2-D675A,1080E is viable and X-ray resistant, strongly supporting a model in which helicase must be coordinated with nuclease for viability. Perhaps, helicase makes a product that requires nuclease for resolution (32).
The temperature-sensitive phenotype of the dna2-1 mutation carrying the P504S substitution had always been puzzling, in that the location of the mutation, roughly 250 bp from the nuclease active site and more than 1 kb away from the helicase domain, had a dramatic effect on both nuclease and helicase activities. Other mutations introduced in the nuclease or helicase motifs affected only one, but not the other enzymatic activities. Based on the following current observations, we now propose that the temperature-sensitive phenotype of the dna2-1 mutation is based on destabilization of the Fe–S cluster domain. First, sequence analysis identifies the P504, proximal to the first C of the Fe–S motif, as a conserved residue (Figure 1). Second, mutational disruption of the Fe–S cluster results in major inhibition of both nuclease and helicase activities of Dna2, similar to the dna2-1 mutation. Third, another dna2 mutation that changes R521 (33), which is conserved in both Dna2 and AddB, falls in the highly conserved region of the Fe–S motif (Figure 1) and also results in a temperature-sensitive phenotype. We caution, however, that the R521K substitution also carries a G446A change, and therefore, the temperature-sensitive phenotype may be an effect of two changes. Last, although mutations in the helicase domain render cells sensitive to MMS, most mutations in the nuclease domain do not confer MMS sensitivity. Nevertheless, the dna2-1 mutant is MMS-sensitive, even at the permissive temperature for growth, suggesting effects on the helicase, as well as the nuclease domain.
On all of the substrates tested, WT Dna2 forms two major distinct protein/DNA complexes, whereas the Fe–S mutant proteins show a single complex, corresponding to the most highly retarded and predominant WT Dna2/DNA complex (Figure 5 and Supplementary Figure S2). We propose that the faster migrating complex contains a lower molar ratio of Dna2/DNA, probably 1:1, than the slower Dna2/DNA complex, probably 2:1. We do not know whether more than one molecule of Dna2 binds autonomously to the substrate, or whether Dna2 dimerizes through protein/protein interaction, with possibly only one component actually bound to the substrate. The latter is suggested by the appearance of protein/DNA complexes in similar ratios, regardless of ssDNA length (34). for publication). We also have considered that Dna2 exists in two different forms in cells, each with a specialized function, one would be chelated with a metal ion and one not. In support of this concept, the nuclease activity on the ssDNA substrates was compromised less on flap substrates than in the C to A mutants (Figure 3). This observation clearly indicates that the metal cluster defective enzyme can still have substantial nuclease activity for ssDNA substrates while lacking the ability to act on other configurations, suggestive that both metal-containing and metal-free enzyme have distinct roles in vivo. Indeed, we observed two species of Dna2 protein in our original analysis of the purified protein by gel filtration (15), which might correspond to such species (or to different multimeric species). Possibly, the metal-containing Dna2 is primarily assigned for the 5′- to 3′ nuclease activities in vivo, whereas Dna2 without metal prefers to show 3′- to 5′ nuclease activity.
An important outcome of this work is that we were able to show that the Dna2 Fe–S cluster has a role in vivo. The expression of WT enzyme, but not any of the cluster mutant enzymes, suppresses the temperature sensitivity of the dna2-1 strain of yeast at 37°C (Figure 7). Expression of C519A and C768A was able to support growth in the complete absence of Dna2, but C771A and C777A were defective at both 30 and 37°C. This pattern of defective in vivo function mirrors the degree of defect in the nuclease activities in vitro, lending significance to the results. Another interesting observation is that, at permissive temperatures, the mutants are dominant negative and either outcompete or synergistically inactivate the dna2-1 protein.
The presence of an essential Fe–S cluster in Dna2 also has implications related to the observation that Dna2 is localized to the mitochondrion in organisms from yeast to man (35–38). Formation of Fe–S clusters is not a spontaneous process; a complex biosynthetic assembly machinery is required (29,39). Since mitochondria are critical for the synthesis of Fe–S clusters, we have to consider that Dna2 may also localize to the mitochondrion primarily to load Fe in its metal cluster. Furthermore, the efficiency of metal loading is not perfect for any protein. This additionally supports the idea that there are two pools of Dna2 in cells, one loaded with metal and one without.
Recently, it has been reported that the nucleotide excision repair and transcription helicase, XPD; the MutY glycosylase; EndoIII and the SoxR transcription factor, all Fe–S proteins, exhibit ATP-stimulated and DNA-mediated charge transport (40–42). It has been proposed that this redox activity is important for enzymatic and physiological activity and constitutes a new signaling mechanism in DNA-binding proteins. Investigating the role of the Dna2 Fe–S cluster in potential DNA-mediated charge transport will therefore be interesting.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
National Institutes of Health [GM100196]; Congressionally Directed Medical Research Programs [W81XWH-09-1-0041]; Ellison Medical Foundation [AG-SS-2143-08]. Funding for open access charge: California Institute of Technology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Robert Bambara, University of Rochester, and members of the Campbell laboratory for their discussions and comments on the manuscript.
REFERENCES
- 1.Budd ME, Tong AH, Polaczek P, Peng X, Boone C, Campbell JL. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 2005;1:634–650. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakrishnan L, Bambara RA. The changing view of Dna2. Cell Cycle. 2011;10:2620–2621. doi: 10.4161/cc.10.16.16545. [DOI] [PubMed] [Google Scholar]
- 3.Kang YH, Lee CH, Seo YS. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 4.White MF, Dillingham MS. Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012;22:94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y, Brosh RM., Jr DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res. 2012;40:4247–4260. doi: 10.1093/nar/gks039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sontz PA, Mui TP, Fuss JO, Tainer JA, Barton JK. DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. Proc. Natl Acad. Sci. USA. 2012;109:1856–1861. doi: 10.1073/pnas.1120063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balakrishnan L, Bambara R. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J. Biol. Chem. 2011;286:6865–6870. doi: 10.1074/jbc.R110.209502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind L, Makarova KS, Koonin EV. Survey and summary: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeeles JT, Cammack R, Dillingham MS. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J. Biol. Chem. 2009;284:7746–7755. doi: 10.1074/jbc.M808526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saikrishnan K, Yeeles JT, Gilhooly NS, Krajewski WW, Dillingham MS, Wigley DB. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 2012;31:1568–1578. doi: 10.1038/emboj.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balakrishnan L, Polaczek P, Pokharel S, Campbell JL, Bambara RA. Dna2 exhibits a unique strand end-dependent helicase function. J. Biol. Chem. 2010;285:38861–38868. doi: 10.1074/jbc.M110.165191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budd ME, Campbell JL. A new yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl Acad. Sci. USA. 1995;92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokharel S, Beal PA. High-throughput screening for functional adenosine to inosine RNA editing systems. ACS Chem. Biol. 2006;1:761–765. doi: 10.1021/cb6003838. [DOI] [PubMed] [Google Scholar]
- 15.Budd ME, Choe W-C, Campbell JL. The nuclease activity of the yeast Dna2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 2000;275:16518–16529. doi: 10.1074/jbc.M909511199. [DOI] [PubMed] [Google Scholar]
- 16.Fortini BK, Pokharel S, Polaczek P, Balakrishnan L, Bambara RA, Campbell JL. Characterization of the endonuclease and ATP-dependent flap endo/exonuclease of Dna2. J. Biol. Chem. 2011;286:23763–23770. doi: 10.1074/jbc.M111.243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart JA, Campbell JL, Bambara RA. Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:8283–8291. doi: 10.1074/jbc.M809189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao H-I, Campbell JL, Bambara RA. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 2004;279:50840–50849. doi: 10.1074/jbc.M409231200. [DOI] [PubMed] [Google Scholar]
- 19.Budd M, Campbell JL. Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc. Natl Acad. Sci. USA. 1987;84:2838–2842. doi: 10.1073/pnas.84.9.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruschi M, Hatchikian C, Le Gall J, Moura JJ, Xavier AV. Purification, characterization and biological activity of three forms of ferredoxin from the sulfate-reducing bacterium Desulfovibrio gigas. Biochim. Biophys. Acta. 1976;449:275–284. doi: 10.1016/0005-2728(76)90139-0. [DOI] [PubMed] [Google Scholar]
- 21.Boal AK, Yavin E, Lukianova OA, O'Shea VL, David SS, Barton JK. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry. 2005;44:8397–8407. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- 22.Stewart JA, Campbell JL, Bambara RA. Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Res. 2010;38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae K-H, Kim H-S, Bae S-H, Kang H-Y, Brill S, Seo Y-S. Bimodal interaction between replication-protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Res. 2003;31:3006–3015. doi: 10.1093/nar/gkg422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae SH, Bae K-H, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 25.Lee K-H, Kim DW, Bae S-H, Kim J-A, Ryu G-H, Kwon Y-N, Kim K-A, Koo H-S, Seo Y-S. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 2000;28:2873–2881. doi: 10.1093/nar/28.15.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bae S-H, Kim J-A, Choi E, Lee K-H, Kang H-Y, Kim H-D, Kim J-H, Bae K-H, Cho Y, Park C, et al. Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res. 2001;29:3069–3079. doi: 10.1093/nar/29.14.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bae S-H, Kim DW, Kim J, Kim J-H, Kim D-H, Kim H-D, Kang H-Y, Seo Y-S. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 2002;277:26632–26641. doi: 10.1074/jbc.M111026200. [DOI] [PubMed] [Google Scholar]
- 28.Pugh RA, Honda M, Leesley H, Thomas A, Lin Y, Nilges MJ, Cann IK, Spies M. The iron-containing domain is essential in Rad3 helicases for coupling of ATP hydrolysis to DNA translocation and for targeting the helicase to the single-stranded DNA-double-stranded DNA junction. J. Biol. Chem. 2008;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- 29.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 2012;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda-Sasa T, Polaczek P, Campbell JL. Single strand annealing and ATP-independent strand exchange activities of yeast and human DNA2: possible role in Okazaki fragment maturation. J. Biol. Chem. 2006;281:38555–38564. doi: 10.1074/jbc.M604925200. [DOI] [PubMed] [Google Scholar]
- 31.McRobbie AM, Meyer B, Rouillon C, Petrovic-Stojanovska B, Liu H, White MF. Staphylococcus aureus DinG, a helicase that has evolved into a nuclease. Biochem. J. 2012;442:77–84. doi: 10.1042/BJ20111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Formosa T, Nitiss T. Dna2 mutants reveal interactions with DNA polymerase alpha and Ctf4, a Pol α accessory factor, and show that full DNA2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gloor JW, Balakrishnan L, Campbell JL, Bambara RA. Biochemical analyses indicate that binding and cleavage specificities define the ordered processing of human Okazaki fragments by Dna2 and FEN1. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks388. May 7. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010;11:962–968. doi: 10.1038/embor.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol. Cell. Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 40.Mui TP, Fuss JO, Ishida JP, Tainer JA, Barton JK. ATP-stimulated, DNA-mediated redox signaling by XPD, a DNA repair and transcription helicase. J. Am. Chem. Soc. 2011;133:16378–16381. doi: 10.1021/ja207222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, Barton JK. Redox signaling between DNA repair proteins for efficient lesion detection. Proc. Natl Acad. Sci. USA. 2009;106:15237–15242. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee PE, Demple B, Barton JK. DNA-mediated redox signaling for transcriptional activation of SoxR. Proc. Natl Acad. Sci. USA. 2009;106:13164–13168. doi: 10.1073/pnas.0906429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.