Abstract
The transcription factor Sp2 is essential for early mouse development and for proliferation of mouse embryonic fibroblasts in culture. Yet its mechanisms of action and its target genes are largely unknown. In this study, we have combined RNA interference, in vitro DNA binding, chromatin immunoprecipitation sequencing and global gene-expression profiling to investigate the role of Sp2 for cellular functions, to define target sites and to identify genes regulated by Sp2. We show that Sp2 is important for cellular proliferation that it binds to GC-boxes and occupies proximal promoters of genes essential for vital cellular processes including gene expression, replication, metabolism and signalling. Moreover, we identified important key target genes and cellular pathways that are directly regulated by Sp2. Most significantly, Sp2 binds and activates numerous sequence-specific transcription factor and co-activator genes, and represses the whole battery of cholesterol synthesis genes. Our results establish Sp2 as a sequence-specific regulator of vitally important genes.
INTRODUCTION
Specificity proteins (Sps) are sequence-specific transcription factors that share a conserved DNA-binding domain of three consecutive C2H2 zinc fingers that bind to GC-boxes (GGGGCGGGG) and closely related motifs that are present in a variety of housekeeping, tissue-specific, developmental-specific and cell-cycle-regulated genes (1). It is therefore generally believed that Sp proteins are involved in regulating these genes.
In mammals, nine closely related Sp proteins, designated Sp1 to Sp9, have been identified (2). On the basis of their sequence homology and structural features they can be divided into two subfamilies (3). Sp2 along with Sp1, Sp3 and Sp4 constitute the glutamine-rich subfamily. Sp1, Sp2 and Sp3 are ubiquitously expressed whereas expression of Sp4 is largely restricted to cells of neuronal origin. Despite their structural similarities and largely overlapping expression patterns there appears to be little functional redundancy between individual Sp family members as gene targeting of all four Sp family members in mice revealed unambiguously strong and distinct phenotypes. In brief, Sp1 null embryos die around Embryonic Day 10 (4), whereas Sp3 null embryos develop until the end of pregnancy but die immediately after birth due to various developmental defects including impaired lung and cardiac development, skeletal bone ossification, tooth development (5–7) and placenta organization (8). Consistent with the more restricted expression pattern of Sp4, Sp4 null mice are born alive (9,10) but two-thirds die within the first month after birth. Sp4 appears to be required for specification of the cardiac conduction system (11,12) and normal brain development (13,14).
Constitutive Sp2 knockout embryos are severely retarded in growth, show a broad range of phenotypic abnormalities and die before E9.5 of gestation (15). Moreover, mouse embryonic fibroblasts (MEFs) derived from E9.5 Sp2 null embryos fail to grow, and Cre-mediated ablation of Sp2 in conditional Sp2cko MEFs carrying floxed Sp2 alleles results in a strong decrease of proliferation (15). The latter findings strongly suggest a role of Sp2 in the control of cellular processes affecting proliferation. Sp2 also has an impact on cellular differentiation programs. Ectopic expression of Sp2 in basal keratinocytes prevents keratinocyte differentiation and promotes carcinogen-induced tumorigenesis in transgenic mice (16).
Despite the well established physiological significance of Sp2, its biochemical and molecular properties remain enigmatic. A preferred binding site of Sp2 identified by cyclic amplification and selection of target oligonucleotides is very similar to the binding sites of other Sp family members (17). However, no DNA-binding activity of Sp2 is detected in nuclear extracts of Sp2-expressing cells using electrophoretic mobility shift assays (EMSA) (17). Furthermore, in transient reporter gene assays Sp2 has little or no capacity to stimulate transcription from promoters that are activated by other Sp family members (17). Since Sp2 was found to be associated with the nuclear matrix and to be localized predominantly within subnuclear foci (18), it was even proposed that Sp2 might have functions that are not directly associated with regulation of gene expression (18).
In the present study, we have investigated the role of Sp2 in MEFs and human HEK293 cells. By combining in vitro protein–DNA analysis, genome-wide identification of Sp2-binding sites by chromatin immunoprecipitation (ChIP)-Seq, and global expression profiling we identified transcriptional networks that are regulated by Sp2. Our results establish Sp2 as a sequence-specific master regulator of multiple genes essential for fundamental cellular processes.
MATERIALS AND METHODS
Antibodies
The following antibodies were used for EMSA supershift, western blotting and ChIP experiments: Rabbit anti-Sp1, home-made (19) and Millipore 07-645; rabbit anti-Sp2, home-made (15) and Santa Cruz sc-643; rabbit anti-Sp3, home-made (19) and Santa Cruz sc-644; rabbit anti-Mi2α/β, Santa Cruz sc-11378; mouse anti-tubulin, Millipore MAB3408; rat anti-HA, Roche 11867423; rabbit IgG control, Diagenode kch-504-250.
Electrophoretic mobility shift assay
EMSAs were essentially performed as described (20) with 0.2 ng of 32P-labelled double-stranded GC-box oligonucleotides (21). For supershift experiments, 1 µl of crude anti-Sp1, anti-Sp2 or anti-Sp3 sera (19) were included in the binding reaction.
DNA affinity precipitation assay
DNA affinity precipitation assays (DAPAs) were performed essentially as described (22) in the presence of 250 mM NaCl with 1 µg of biotinylated double-stranded oligonucleotides and 100 µg of nuclear extract. Bound proteins were recovered with Streptavidin MagneSphere® paramagnetic particles (Promega) and subsequently analysed by western blotting. The upper strand sequences of the biotinylated GC and GCm oligonucleotides are:
5′-GATCTCCGAAGTGGGCGGGACAAGCCTCTAG-3′ (GC) and 5′-GATCTCCGAAGTGGtttGGACAAGCCTCTAG-3′ (GCm).
Knockdown of Sp2 and determination of growth rates
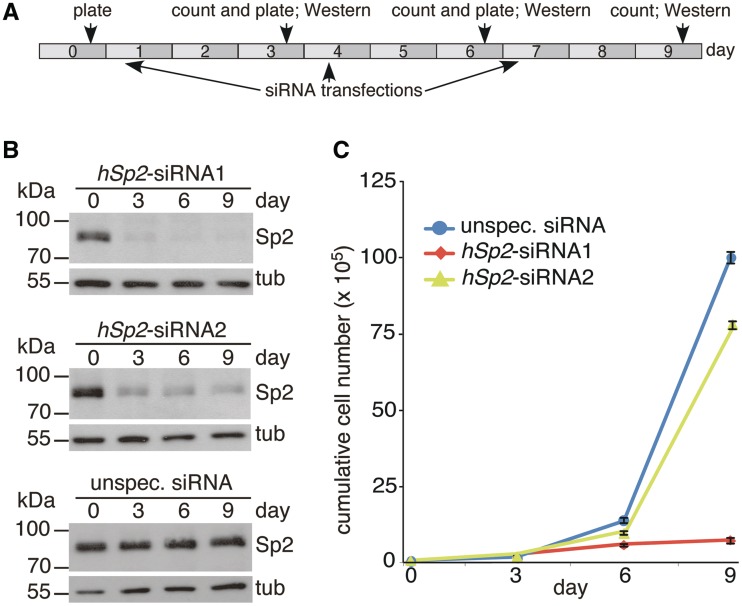
The siRNAs J-015955-07 (siRNA1) and J-015955-08 (siRNA2) of the ON-TARGETplus set from Thermo Scientific Dharmacon were used for RNAi-mediated knockdown of Sp2 in HEK293 cells, and the ON-TARGETplus Non-targeting Pool D-001810-10-05 was used as an unspecific siRNA control. Triplicates of 0.5 × 105 HEK293 cells on six-well plates were transfected with 10 nM hSp2-siRNA1, 20 nM hSp2-siRNA2 or corresponding unspecific siRNAs using OligofectamineTM (Invitrogen). Three days post-transfection cells were counted, 0.5 × 105 cells were re-plated and 16 h later transfected again with siRNAs as indicated in Figure 1A. Knockdown efficiency was monitored by western blot analysis.
Figure 1.
Knockdown of Sp2 impairs proliferation of HEK293 cells. (A) Experimental design of growth rate determination. A total of 0.5 × 105 HEK293 cells were plated at Day 0, 3 and 6. Transfection of siRNA was performed 16 h post-plating on Day 1, 4 and 7. (B) Knockdown efficiency of Sp2. Western blotting was performed at Day 0, 3, 6 and 9. To control for loading, the blots were re-probed for tubulin. (C) Growth rates of HEK293 cells transfected with the indicated siRNAs. Cumulative cell numbers were calculated by multiplying the number of plated cells with the fold increase in cell number at the end of each 3-day interval resulting in theoretical total numbers of proliferated cells arising from cells plated on Day 0 without contact inhibition due to restricted culture vessel size. Presentation is as mean ± SD (n = 3).
Western blot analysis
Whole-cell SDS-extracts, nuclear extracts of MEFs and HEK293 cells or DNA affinity precipitated proteins were separated through SDS–PAGE blotted on PVDF membranes and probed with antibodies as indicated in the figures. Antibodies were visualized using the ImmobilonTM western chemiluminiscent HRP substrate (Millipore).
ChIP–qPCR and ChIP-Seq
ChIP–qPCR experiments were performed essentially as described previously (23) using the OneDay ChIP kit (Diagenode) in accordance to the manufacturer’s instructions. Briefly, MEFs and HEK293 cells were fixed with 1% formaldehyde for 10 min at room temperature followed by quenching with 125 mM glycine for 5 min. Immunoprecipitation was performed with 100 µg of chromatin using 2 µg of affinity-purified home-made Sp2 antibody or unspecific rabbit IgG as a control. The gene-specific primers used for ChIP–qPCRs are listed in Supplementary Table S1. For ChIP-Seq, 10 immunoprecipitates were pooled and purified by the QIAquick® PCR purification kit (Qiagen). Sequencing libraries were prepared from 10 ng of immunoprecipitated DNA using the Illumina ChIP-Seq DNA Sample Prep Kit according to the manufacturer’s instructions. Libraries were sequenced on the Illumina Genome Analyzer IIx, single-read 36 bp.
Expression profiling
Total RNA of wild-type (wt) (Sp2cko) and Sp2ko MEFs was extracted using the RNeasy Mini kit (Qiagen). RNA was labelled with the two-colour Quick-Amp Labelling kit (Agilent) and hybridized against Agilent G4122F-014868 (short-term Sp2 depletion experiment) and G2519F-026655 (long-term Sp2 depletion experiment) microarrays according to the manufacturer’s instructions (Agilent). Raw microarray data were normalized using the ‘loess’ method implemented within the marray package of R/Bioconductor (24). Agilent probes were reassigned to the Ensembl genome annotations by aligning sequences with a short read aligner (Bowtie against both, the transcriptome and the genome). Microarray analysis after short- and long-term depletion of Sp2 in MEFs was performed in duplicate, and genes were considered regulated between Sp2wt and Sp2ko MEFs when their expression level showed a ≥1.5-fold change, had an average log fluorescence intensity of 5 and the fluctuation between replicates was ≤50%.
Quantitative real-time PCR
Two micrograms of total RNA was used for cDNA synthesis with the SuperScript II Reverse Transcriptase kit (Invitrogen) using 0.5 µg oligo(dT) as primer according to the manufacturer’s instruction. Quantitative RT–PCR was performed in triplicate with 1 µl of 1:20 diluted cDNA and gene-specific primers (Supplementary Table S2) on the Mx3000P real-time PCR system (Stratagene) using ImmoMixTM (Bioline) and SYBR Green (Invitrogen). Values were normalized to Gapdh mRNA content.
Retroviral transductions
The generation of retroviral expression plasmids for murine Sp2, the production of virus stocks and the selection of transduced Sp2ko MEFs were performed as described (15).
ChIP-Seq data analysis
Raw ChIP-Seq data were deduplicated, and aligned to the appropriate genome with Bowtie version 0.12.7 (25) allowing for two mismatches in the seed and a total mismatch quality sum score of 70 (−n 2 −e 70). Reads mapping to multiple locations on the genome were discarded (−m 1 −k 1), and Bowtie output was converted to BAM format. The following amounts of deduplicated and uniquely matching reads were obtained: wt MEFs Ab1, 23 843 938; wt MEFs Ab2, 21 835 314; Sp2ko MEFs Ab1, 24 352 769; Sp2ko MEFs Ab2, 19 196 445; HEK293 Ab1, 7 033 072; HEK293 IgG, 10 942 303.
Peak calling was performed with MACS version 1.4.0rc2 20110214 (26) modified for reading BAM files via pysam (http://code.google.com/p/pysam/). This yielded 5999 (MEF, Ab1), 6330 (MEF, Ab2) and 3344 (HEK293, Ab1) putative Sp2-binding sites, of which 3290, 2687 and 2380, respectively, had a MACS defined ‘False Discovery Rate’ (FDR) of ≤ 0.001. MEF FDR ≤ 0.001 peaks obtained from either antibody were pooled (interval union). Peaks that were not present in both unfiltered datasets were removed resulting in 3261 high confidence Sp2-binding sites. Genes and Sp2-binding sites were associated by testing for overlap between peak regions and transcription start sites ±500 bp. Assignment of homology between human and mouse genes was on the basis of bi-directionally associated genes in Ensembl Compara.
RNA polymerase II GRO-Seq (accession no. GSE27037) and H3K4me3 (accession no. GSE11074) data sets were retrieved from the Gene Expression Omnibus. Both datasets were aligned to the mouse genome and sites of paused polymerase II and enriched H3K4me3 signals were identified with MACS.
De novo motif search was performed with MEME version 4.3.0 (27) using all sequences surrounding Sp2 peak summits (±50 bp). Summits were defined by pooling Sp2wt-Ab1 and Sp2wt-Ab2 reads, elongating them to 200 bp and determining the position of highest overlap. Motifs retrieved by MEME were compared to known motifs using TOMTOM (28).
Pathway analysis
Functional enrichment analysis was performed with ChIP-Seq- and microarray-derived gene lists versus predefined gene sets using Fisher’s exact test. The Benjamini-Hochberg procedure (29) was used to correct for multiple hypothesis testing.
Databases and data deposition
Genome sequences, annotation and homology data were retrieved from Ensembl revision 64 (http://sep2011.archive.ensembl.org/) (30). Known transcription factor motifs were retrieved from Jaspar (31), UniProbe (32) and Matbase (Genomatix). For pathway analysis gene sets from DAVID 6.7 (33), the pathway interaction database (34) and MsigDB (35) were used.
The microarray data from this publication have been submitted to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) and assigned the identifier E-MTAB-1015. The ChIP-Seq data from this publication have been submitted to the ArrayExpress and the European Nucleotide Archive database (http://www.ebi.ac.uk/arrayexpress/, http://www.ebi.ac.uk/ena/home) and assigned the identifier E-MTAB-994 and ERP001252, respectively.
RESULTS
Depletion of Sp2 in HEK293 cells impairs proliferation
MEFs derived from E9.5 Sp2 null embryos do not divide, whereas wt and heterozygous MEFs derived from siblings grow normally. Moreover, Cre-mediated ablation of Sp2 in MEFs carrying floxed Sp2 alleles impairs proliferation relative to control retrovirus-infected cells (15). To analyse whether Sp2 is also essential for normal proliferation of other cell lines, we performed siRNA-mediated knockdown studies with HEK293 cells (Figure 1). Two different siRNAs interfering with Sp2 expression were used. Transfection of siRNA1 in HEK293 cells resulted in an almost complete loss of Sp2 protein 3 days post-transfection, whereas siRNA2 was less efficient but nevertheless caused a significant reduction of Sp2 (Figure 1B). Ablation of Sp2, particularly after repeated transfection of siRNA1, resulted in strongly reduced proliferation relative to control siRNA transfected cells (Figure 1C). This finding reinforces the conclusion that Sp2 plays an important role in the control of cellular processes affecting proliferation.
Detecting a specific Sp2 DNA-binding activity in nuclear extracts of mammalian cell lines
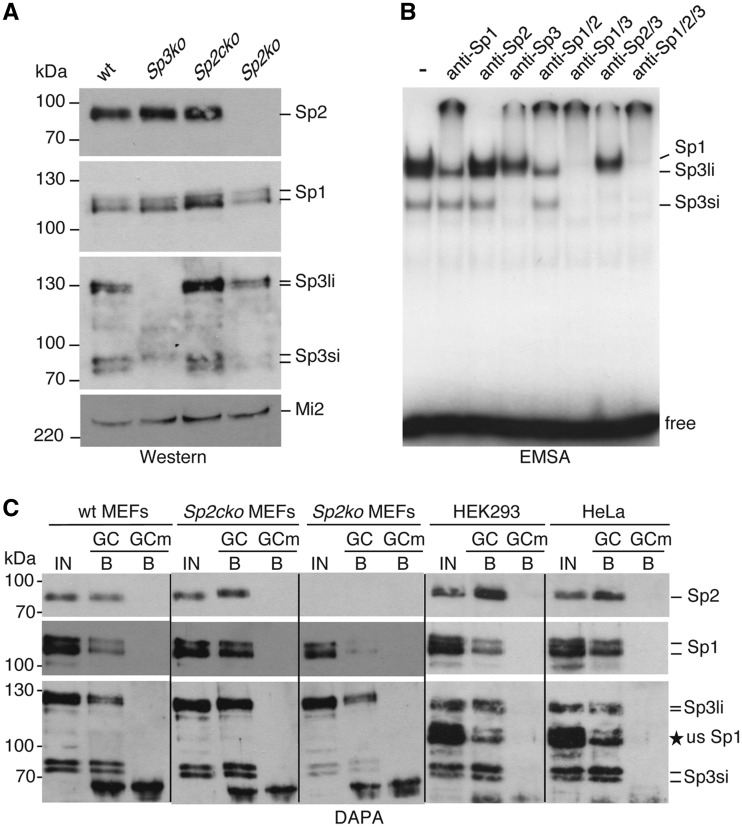
Towards understanding the molecular properties of Sp2 we sought to analyse the DNA-binding properties of Sp2 using EMSAs. Sp2 is present in extracts of wt MEFs, in Sp3 knockout MEFs (Sp3ko) as well as in extracts of MEFs with floxed Sp2 alleles (Sp2cko) but not in MEFs in which Sp2 was depleted upon Cre transduction (Sp2ko) (15) (Figure 2A). For reasons of clarity we will refer to Sp2cko MEFs from hereon also as wt MEFs. EMSAs failed to detect any Sp2 binding to a classical GC-box oligonucleotide that was readily bound by Sp1 and Sp3 (Figure 2B). The lack of DNA binding of Sp2 in nuclear extracts is consistent with previously published results that showed expression of Sp2 in several mammalian cell lines but failed to detect Sp2 DNA binding by EMSAs (17). We also performed EMSAs with recombinant Sp2 expressed in insect Schneider cells (SL2 cells) lacking endogenous Sp proteins. Again, full-length Sp2 did not bind to DNA. Strikingly, a number of N-terminal- and internal Sp2 deletion mutants bound to the Sp1-binding site oligonucleotide with high affinity (Supplementary Figure S1), However, by analysing additional Sp2 mutants we could not delineate a particular region or motif in the N-terminal region of Sp2 that is responsible for interfering with DNA binding.
Figure 2.
DNA-binding activity of endogenous Sp1, Sp2 and Sp3 in nuclear extracts. (A) Expression of Sp1, Sp2 and Sp3 in MEFs. Nuclear extracts of wt MEFs (wt), Sp3-deficient MEFs (Sp3ko), MEFs with floxed Sp2 alleles (Sp2cko) and Sp2-deficient MEFs (Sp2ko) were resolved by SDS–PAGE and successively subjected to western blot analysis for Sp1, Sp2 and Sp3 (Sp3li, long Sp3 isoforms; Sp3si, short Sp3 isoforms). Mi2 was used as a loading control. (B) EMSA with nuclear extracts of wt MEFs. Extracts were incubated with 0.2 ng of 32P-labelled GC oligonucleotide. Antibodies against Sp1, Sp2 and Sp3 were included in the binding reactions as indicated. Sp1, Sp3li and Sp3si containing DNA protein complexes are indicated on the right side. (C) Nuclear extracts of MEFs with the indicated genotype, and of HEK293 and HeLa cells were analysed by DAPA using a biotinylated GC box oligonucleotide (GC) and an oligonucleotide with a mutation in the Sp recognition sequence (GCm). Bound proteins were successively analysed for the presence of Sp2, Sp1 and Sp3 proteins by western blotting. IN, input 15%; B, bound protein 80%. The asterisk marked usSp1 next to the Sp3 blot of HEK293 and HeLa cells denotes unstripped Sp1 signals that were removed incompletely prior to the re-probing for Sp3.
We noticed that the N-terminal region of Sp2 preceding the zinc-finger region, in contrast to Sp1, Sp3 and Sp4, has a very high isoelectric point (IEP for hSp1 amino acids 1–596: 4.4; hSp2 amino acids 1–498: 10.5; hSp3 amino acids 1–592: 3.9 and hSp4 amino acids 1–617: 4.4) raising the possibility that the positive charge of Sp2 affects the stability of the Sp2–DNA complex during electrophoresis. To test this hypothesis we performed DAPAs. The DAPA method does not include a native electrophoresis step and is therefore suited to this task. A biotinylated GC-box oligonucleotide was incubated with nuclear extracts of SL2 cells containing either full-length Sp2 that was not detectable by EMSA, or Sp2 deletion mutants that bound the GC-box oligonucleotide in EMSAs with high affinity (Supplementary Figure S1). Bound Sp2 proteins were subsequently analysed by western blotting. Full-length Sp2 and the Sp2 deletion mutants bound to the GC-box oligonucleotide with similar affinity (Supplementary Figure S2) strongly supporting the notion that electrophoresis impedes detection of a full-length Sp2–DNA complex by EMSA. Next, we analysed nuclear extracts of wt MEFs, MEFs with floxed Sp2 alleles, Sp2ko MEFs, HEK293 and HeLa cells by DAPA. Endogenous Sp2 as well as endogenous Sp1 and Sp3 of all three cell types bound to the GC-box oligonucleotide but not to an oligonucleotide with a mutation in the Sp1 recognition sequence (Figure 2C). The absence of any Sp2 signal in the nuclear extract of Sp2ko cells (Figure 2C) further confirmed the specificity of the Sp2 signal. We conclude that Sp2 is a bona fide DNA-binding protein that recognizes GC-box motifs similar to Sp1 and Sp3. The failure to detect DNA binding of endogenous Sp2 by EMSA is likely due to the high isoelectric point of Sp2 leading to de-stabilization of the Sp2–DNA complex during electrophoresis.
Genome-wide identification of Sp2 target genes
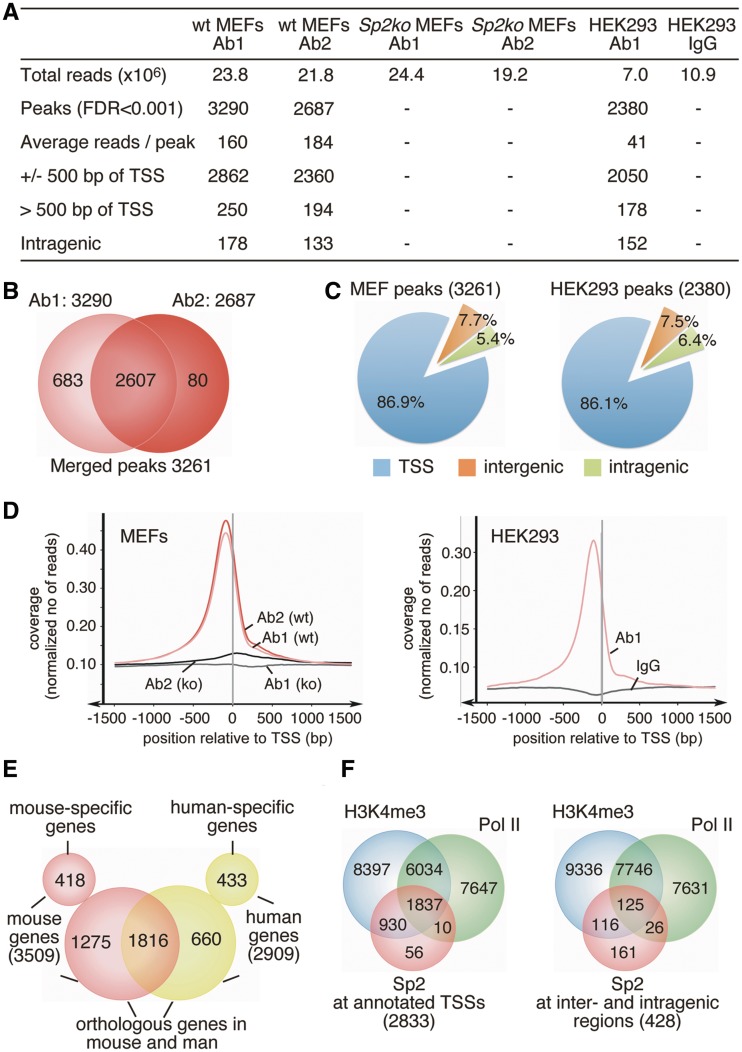
Given that Sp2 is a DNA-binding transcription factor essential for cell proliferation we sought to identify genes regulated by Sp2, and performed ChIP followed by deep sequencing. Chromatin was precipitated from MEFs that contain floxed Sp2 alleles (wt MEFs), and from MEFs in which Sp2 was deleted upon Cre transduction (Sp2ko MEFs). Two different antibodies that are specific for Sp2 and do not cross-react with other Sp family members were used for ChIP. An overview of the ChIP-Seq results is presented in Figure 3A.
Figure 3.
Overview of ChIP-Seq results. (A) Summary of Sp2 ChIP-Seq analysis. (B) Venn diagram showing the overlap of Sp2 peaks obtained with two different Sp2 antibodies (Ab1 and Ab2). The total of 3261 high-confident sites were obtained by merging Sp2-binding sites that had at least an FDR < 0.001 with one antibody and an FDR < 1 with the other antibody. (C) Distribution of Sp2 occupancy in MEFs and HEK293 cells relative to annotated genes. (D) Averaged Sp2 coverage around TSSs in MEFs and HEK293 cells normalized to 1 million reads. (E) Venn diagram showing the overlap of genes with Sp2-binding sites in MEFs and HEK293 cells. (F) Venn diagrams showing the overlap of Sp2-binding sites with the presence of H3K4me3 signatures (37,38) and promoter-proximal paused RNA polymerase II (36) in MEFs. Left diagram, overlap at annotated TSSs; right diagram, overlap at inter- and intragenic sites.
ChIP-Seq with antibody 1 (Ab1) yielded 23.8 million (Ab2: 21.8 million) unique genomic mouse reads with chromatin of wt MEFs and 24.4 million (Ab2: 19.2 million) with chromatin of Sp2ko MEFs. Using a FDR of 0.001, MACS peak calling (26) identified a total of 3290 peaks with Ab1 and 2687 peaks with Ab2 with an overlap of 2607 peaks. Merging binding sites that had at least an FDR < 0.001 with one antibody and an FDR < 1 with the other antibody yielded 3261 high-confident Sp2-binding sites (Figure 3B and Supplementary Dataset S1). Remarkably, the large majority of the Sp2-binding sites (86.9%) were located close to the 5′-end (±500 bp) of annotated transcripts encompassing 3509 genes (Figure 3C and D). Note that the number of genes assigned to Sp2-binding sites was higher than the number of peaks at transcriptional start sites (TSSs) due to nearby and overlapping promoters. 5.4% of the peaks were located downstream of TSSs within the body of genes, and did not overlap with annotated alternative promoters. Only 7.7% of Sp2-binding sites were located at intergenic regions (Figure 3C).
To further substantiate the conclusion that Sp2 is a transcription factor that binds predominantly to proximal promoters, we also performed ChIP-Seq with chromatin of HEK293 cells (Figure 3A). Analysis of this data set identified 2380 high-confident Sp2 peaks (Supplementary Dataset S2) of which 2050 (86.1%) were located within ±500 bp of annotated 5′-ends of transcripts (Figure 3C and D) encompassing 2909 genes. 152 binding sites (6.4%) were located within transcribed regions and 178 peaks (7.5%) at a distance >500 bp upstream of annotated transcripts. In conclusion, the overall genomic distribution of Sp2-binding sites relative to annotated genes is very similar in MEFs and in HEK293 cells (Figure 3C and D).
Furthermore, 2476 of the 2909 human genes occupied by Sp2 at their promoters do have an orthologous gene in the mouse genome. Sp2 is present at 1816 (73%) of these conserved genes (Figure 3E). The large overlap of promoters that are occupied by Sp2 in murine fibroblasts as well as in human HEK293 cells indicates that Sp2 may regulate general cellular functions.
We compared the Sp2-binding sites with sites occupied by transcriptionally engaged, paused RNA polymerase II mapped by genome-wide nuclear run-on (GRO-Seq) in MEFs (36), and with sites that contain H3K4me3 patterns in MEFs (37,38). H3K4me3 marks are signatures for TSSs and are associated with gene activation. 98% of the Sp2-binding sites at annotated TSSs contained either H3K4me3 patterns or were occupied by transcriptionally engaged RNA polymerase II (Figure 3F) showing that Sp2 is bound at promoters of active genes. Moreover, H3K4me3 patterns or stalled RNA polymerase II were also present at 62% of the Sp2-binding sites that do not overlap with an annotated TSS (Figure 3F) strongly suggesting that these Sp2 sites are also in close proximity to transcribed regions. Likely, these inter- and intragenic Sp2 sites mirror unknown alternative promoters and/or promoters of thus far unknown non-coding RNA genes. The close proximity of Sp2-binding sites to H3K4me3 patterns and paused RNA polymerase II further suggests that Sp2 acts predominantly from proximal promoters and not from remote positions.
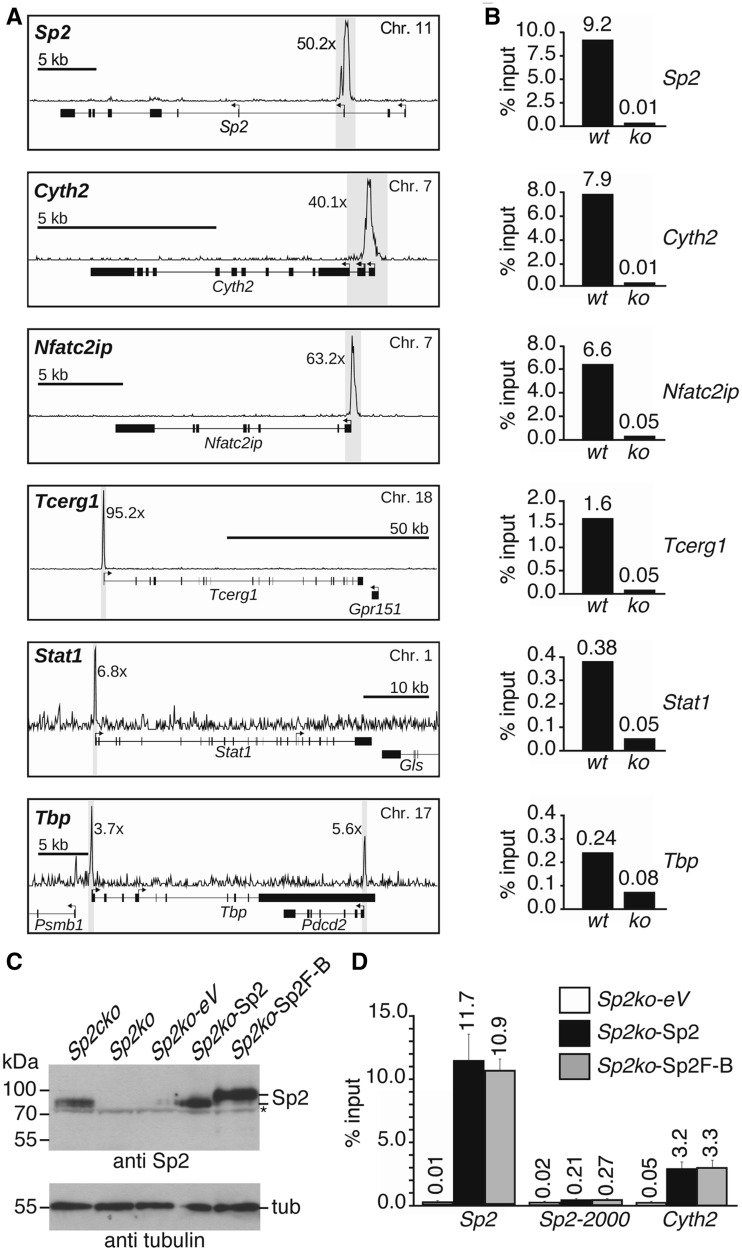
ChIP-Seq read counts at different Sp2 peaks exhibited a striking variation across two orders of magnitude. Snapshots of representative Sp2 target promoters in MEFs are shown in Figure 4A. These strong differences in Sp2 occupancy at different promoters were also seen in corresponding ChIP–qPCR control experiments (Figure 4B). For instance, ChIP–qPCR at the Sp2, Cyth2 and Nfat2cip promoters resulted in enrichment of >5% of input whereas qPCR at the Stat1 and the Tbp promoters yielded <0.5% of input (Figure 4B). Specific localization of Sp2 at the TSS of its own gene was further validated by a ChIP–qPCR promoter walk with eight primer pairs encompassing a chromosomal region from −2500 bp to +1000 bp relative to the TSS (Supplementary Figure S3). Finally, we confirmed specificity of Sp2 binding by rescue experiments. Sp2ko cells were transduced with retroviral vectors expressing untagged Sp2 or a C-terminally tagged Sp2 version. Both proteins were expressed at levels similar to endogenous Sp2 in wt MEFs (Figure 4C), and specifically re-occupied Sp2 target promoters as exemplified by binding to the Sp2 and Cyth2 promoters (Figure 4D).
Figure 4.
Validation of representative Sp2 targets identified by ChIP-Seq analysis of MEFs. (A) Snapshot signal maps of representative Sp2 target genes (Sp2, Cyth2, Nfat2cip, Tcerg1, Stat1 and Tbp) along with their exon–intron structure and TSSs indicated by arrows. The numbers next to the peaks indicate the enrichment in wt MEFs relative to Sp2ko MEFs. (B) ChIP–qPCR analysis of Sp2 occupancy in wt and Sp2ko MEFs (ko) at high (Sp2, Cyth2, Nfat2cip and Tcerg1) and low (Stat1 and Tbp) ChIP-Seq hits. (C) Western blot analysis of Sp2 in Sp2cko and Sp2ko MEFs transduced with an empty control (Sp2ko-eV) or Sp2-expressing retroviruses (Sp2ko-Sp2, untagged Sp2 and Sp2ko-Sp2F-B, C-terminal Flag-Biotin-tagged Sp2). The asterisk marks a cross-reacting protein. To control for loading, the same blot was re-probed for tubulin. (D) ChIP–qPCR analysis of Sp2 occupancy at the Sp2 gene (Sp2, promoter region; Sp2-2000, 2000-bp upstream of the TSS) and at the Cyth2 gene in rescued MEFs. The percent of input values are mean ± SD (n = 3). A list of gene abbreviations is found in Supplementary Table S3.
Computational mining of Sp2-binding sites
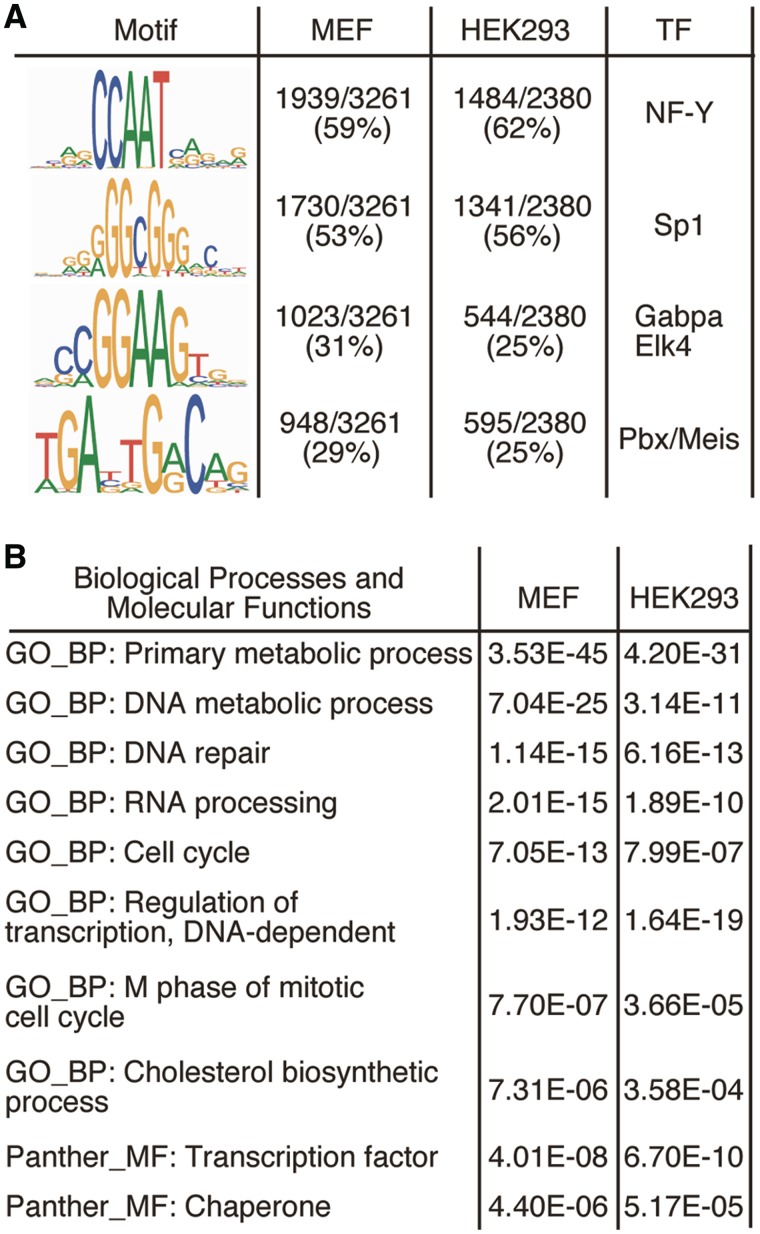
We employed the motif discovery algorithm MEME (27,39) for de novo motif search within the Sp2-occupied regions in MEFs and in HEK293 cells. MEME identified known Sp factor-binding motifs with the GGCGG core sequence (Figure 5A). The second motif identified by this analysis, which was even more prevalent, is the CCAAT motif, a binding site for the transcription factor NF-Y. MEME reported also two additional motifs, which according to the TOMTOM Motif Comparison Tool (28) represent binding sites for the Ets family members Gabpa and Elk4, and the heterodimeric homeobox Pbx/Meis complex (Figure 5A). The presence of NF-Y, Ets family and Pbx/Meis-binding motifs at Sp2 target promoters may indicate that Sp2 cooperates with these transcription factors in gene regulation.
Figure 5.
Computational mining of Sp2-binding sites and target genes. (A) Sequence logos of motifs present in Sp2 ChIP-Seq peaks. Logos were obtained by running MEME (39) with 100 bp summits of all FDR < 0.001 sites. TOMTOM (28) identified these motifs as binding sites for the transcription factors Sp1, NF-Y, Gabpa and Elk4 and Pbx/Meis. (B) Cellular roles of Sp2 target genes. Shown are 10 representative top scoring hits of the pathway analyses along with their Benjamini values.
To explore the cellular processes that are linked to Sp2, we examined the functional categories associated with Sp2 target genes using DAVID 6.7 (33), the pathway interaction database (34) and MsigDB (35) (Supplementary Dataset S3). GO terms encompassing broad biological categories such as ‘regulation of biological process’ and ‘primary metabolic process’ were highly over-represented. Examples for more specific GO_BP terms associated with Sp2 target genes were ‘DNA repair’, ‘RNA processing’, ‘cell cycle’, ‘M phase of mitotic cell cycle’ and ‘DNA-dependent regulation of transcription’ (Figure 5B). Of note, genes of the latter category included the Sp2 paralogous genes Sp1 and Sp3. Taken together, Sp2 is associated with a wide range of primary cellular biological processes and molecular functions including RNA and DNA metabolic processes, DNA repair, transcriptional- and cell-cycle regulation as well as intracellular signalling. Essentially the same categories of biological and molecular functions were obtained with genes occupied by Sp2 in HEK293 cells (Figure 5B and Supplementary Dataset S4). The assignment of Sp2 target genes in MEFs and HEK293 cells to largely identical categories of biological and molecular functions suggests that Sp2 is engaged in the regulation of fundamental cellular processes. However, despite the large overlap of evolutionary conserved promoters occupied by Sp2 in MEFs and HEK293 cells, there were also a few striking differences. For instance, Sp2 was present at the murine Stat1 promoter in MEFs but not at the Stat2 promoter. Vice versa, Sp2 occupied the human STAT2 promoter in HEK293 cells but was absent at the STAT1 promoter.
Expression analysis of Sp2 target genes
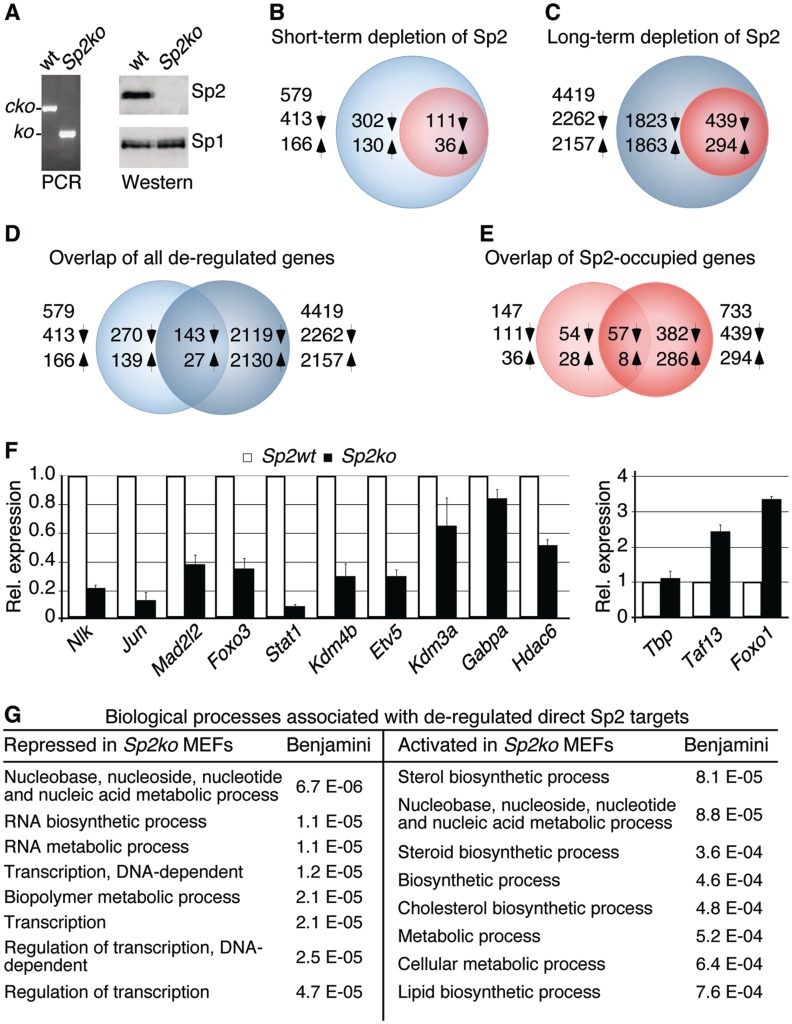
To assess to which extent ChIP-Seq peaks pinpoint Sp2-regulated genes in MEFs, we profiled gene expression in wt MEFs and in Sp2ko MEFs. Depletion of the Sp2 protein by Cre-mediated deletion of the floxed Sp2 alleles resulted in reduced proliferation relative to control retrovirus-transduced cells (15). However, proliferation of Sp2-deficient MEFs did not cease completely, and Sp2ko cells could be kept in culture for prolonged time. Periodical PCR and western blot analyses revealed complete absence of Sp2 (Figure 6A) precluding over-growth of Sp2ko cells by cells that had escaped recombination of the floxed Sp2 alleles.
Figure 6.
Gene-expression profiling of Sp2 target genes in MEFs. (A) PCR and western blot analyses of Sp2cko MEFs transduced with an empty retrovirus (wt) or a Cre-expressing vector (Sp2ko) after several weeks in culture. The western blot was re-probed for Sp1. (B, C) Venn diagrams showing de-regulated genes after short-term (B) and long-term depletion (C) of Sp2. Blue circles denote indirect and red circles direct Sp2 target genes. (D, E) Overlaps of all genes (D) and direct Sp2 target genes (E) that were de-regulated after short-term and long-term depletion of Sp2. Up arrows indicate activated and down arrows repressed genes. (F) Quantitative RT–PCR analysis of ten randomly chosen repressed and three activated direct Sp2 target genes. Normalized mRNA levels are presented relative to Sp2cko MEFs infected with an empty vector (Sp2wt) arbitrarily set to 1. Data are expressed as mean ± SD (n = 3). (G) Classification of direct Sp2 target that was repressed and activated in Sp2ko MEFs. Shown are the eight top-scoring hits of the GO analysis along with their Benjamini values.
We performed microarray-based gene-expression profiling of MEFs at 7 days post-Cre transduction (15) and of Sp2-deficient MEFs after several weeks in culture (Figure 6). Short-term depletion of Sp2 revealed 413 repressed and 166 activated genes (>1.5-fold change, Figure 6B and Supplementary Dataset S5). As expected, the number of de-regulated genes increased markedly after long-term depletion of Sp2 yielding 2262 repressed and 2157 activated genes (Figure 6C, D and Supplementary Dataset S6).
Merging ChIP-Seq and profiling data sets revealed 111 repressed (26.9%) and 36 activated (21.7%) direct Sp2 target genes in short-term Sp2-depleted, and 439 repressed (19.4%) and 294 activated (13.6%) genes in long-term Sp2-depleted MEFs (Figures 6B and C). Approximately 44% of the direct target genes that were de-regulated after short-term depletion of Sp2 were also found to be de-regulated after several weeks in culture (Figure 6E). De-regulation of 12 out of 13 randomly chosen direct Sp2 target genes was confirmed by qRT–PCR with independent RNA preparations (Figure 6F) validating the microarray data sets. Notably, ranking the list of de-regulated direct Sp2 target genes according to the enrichment of Sp2 at their promoters revealed no significant correlation: similar numbers of Sp2 target genes with low tag and high tag counts were de-regulated in Sp2ko MEFs.
We performed biological pathway analysis with the bona fide direct Sp2 target genes that were repressed and activated in Sp2ko MEFs. Most strikingly, 20% of the Sp2 target genes that were repressed after short-term depletion of Sp2 including 15 zinc-finger genes are linked to regulation of transcription. Moreover, several genes such as Msh2, Mad2l2 and Ung are involved in DNA damage response pathways (40). Pathway analysis of repressed Sp2 target genes after long-term depletion of Sp2 reported also several highly significant gene ontology terms related to regulation of gene expression (Figure 6G and Supplementary Dataset S7). For instance, 73 out of the 416 repressed genes are linked to the GO term ‘transcription, DNA-dependent’. This list of genes includes direct DNA-binding factors such as Crem, Etv5, Foxo3, Gabpa, Irx2, Jun, Klf9, Mxd1, Nfia, Pax9, Sp3, Stat1 and Tbx3, the transcriptional cofactors Mllt1, Ncoa2, Tle1 and Trp53bp1, the chromatin-modifying enzymes Hltf, Baz1b, L3mbtl2, Hdac6, Rsf1, Kdm3a and Kdm4b and the protein kinase Nlk.
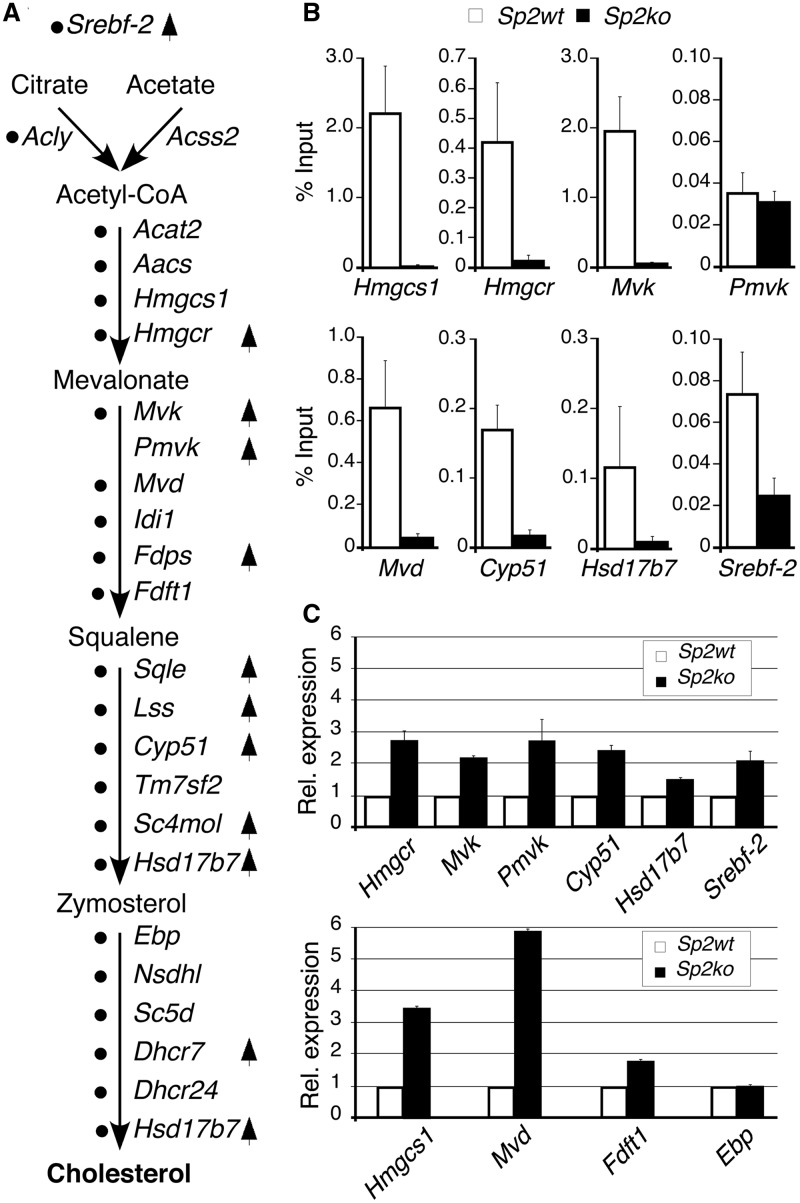
Direct Sp2 target genes that were activated in Sp2ko MEFs are largely linked to anabolic processes including nucleotide, sterol, lipid and amino acid biosynthetic pathways (Figure 6G and Supplementary Dataset S8). Most strikingly, 10 out of 22 genes in this data set are directly involved in cholesterol synthesis (Figure 7A). Inspection of the ChIP-Seq data set revealed that Sp2 is present at the promoters of additional 12 genes of this pathway (Acly, Acat2, Aacs, Hmgcs1, Mvd, Idi1, Fdft1, Tm7sf2, Ebp, Nsdhl, Sc5d and Dhcr24) (Figure 7A). The only gene involved in cholesterol synthesis that was not bound by Sp2 is Pmvk (Figure 7A and B). Expression analysis by qRT–PCR of nine randomly selected genes confirmed activation of genes found by microarray analysis (Hmgcr, Cyp51, Mvk, Pmvk and Hsd17b7), and identified additional genes of this pathway (Hmgcs1, Mvd and Fdft1) that were also activated but had escaped microarray detection (Figure 7C). We also analysed expression of the Srebf-2 gene, which encodes Srebp-2, a master regulator of cholesterol metabolism (41). Srebf-2 mRNA was increased in Sp2ko MEFs as well (Figure 7C). Furthermore, inspection of our ChIP-Seq data set revealed binding of Sp2 to the Srebf-2 promoter in wt MEFs, which we also confirmed by ChIP–qPCR (Figure 7B). Of note is that the Srebf-2 promoter is not listed in our original data set due to an FDR = 0.0066 that is above the stringent threshold that we had chosen for the primary analysis. Taken together, enhanced expression of cholesterol synthesis genes and its master regulator Srebf-2 concomitant with the presence of Sp2 at their promoters suggests that Sp2 contributes to cholesterol homeostasis by directly repressing these genes in MEFs.
Figure 7.
Sp2 negatively regulates cholesterol biosynthesis. (A) The cholesterol biosynthesis pathway with key intermediates and enzymes indicated by their gene symbols. Black dots mark genes that were occupied by Sp2 at their promoters in MEFs. Arrows indicate genes that were activated in Sp2ko MEFs revealed by microarray-based expression profiling. (B) ChIP–qPCR analysis of Sp2 occupancy at selected target promoters in MEFs. (C) qRT–PCR analysis of ten randomly chosen cholesterol biosynthesis pathway genes including four genes (Hmgcs1, Mvd, Fdft1 and Ebp) that were not found by the microarray-based expression profiling. Normalized mRNA levels in Sp2ko MEFs are presented relative to Sp2wt MEFs arbitrarily set to 1. Data are expressed as mean ± SD (n = 3).
Deletion of Sp2 does not lead to increased occupancy of Sp1 at Sp2 target promoters
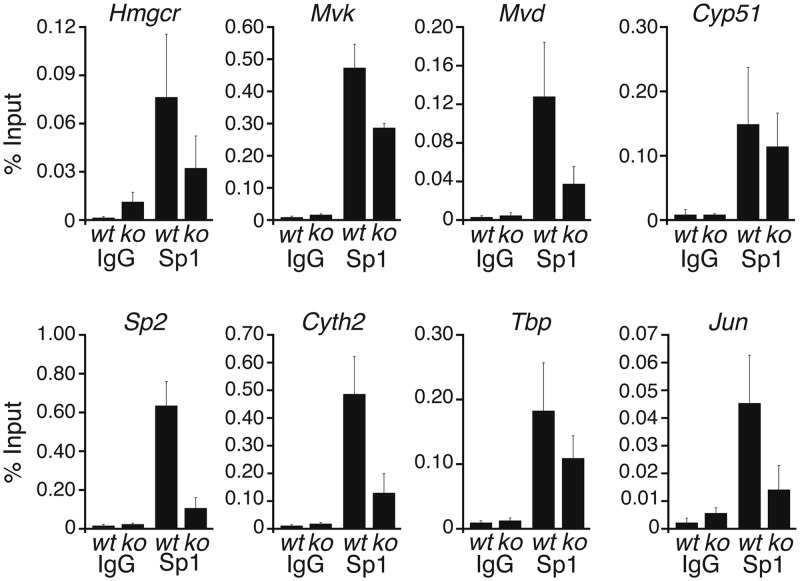
Sp2 recognized the same DNA motif as Sp1 in vitro (Figure 2). Arguing that Sp1 and Sp2 may compete for the same sites in vivo, increased recruitment of Sp1 to Sp2 target promoters could contribute to activation of cholesterol pathway genes in Sp2ko MEFs as well. We performed ChIP–qPCR analysis of Sp1 on selected bona fide Sp2 target promoters with chromatin of wt MEFs and of Sp2ko MEFs (Figure 8). Sp1 was bound to the Hmgcr, Mvk, Mvd and Cyp51 promoters in wt as well as in Sp2ko MEFs. However, Sp1 occupancy at these promoters was not increased but rather reduced in Sp2ko cells. Thus, increased binding of Sp1 does not contribute to the activation of cholesterol pathway genes in Sp2ko cells.
Figure 8.
Binding of Sp1 to Sp2 target promoters in wt and Sp2ko MEFs. Sp1 occupancy at the indicated promoters in wt and Sp2ko (ko) MEFs was analysed by ChIP–qPCR in duplicate with three independent chromatin preparations and three independent ChIPs. ChIP–qPCR values are represented as % of input ± SD.
Following the startling observation that the absence of Sp2 led to reduced occupancy of Sp1 at cholesterol synthesis genes in Sp2ko cells, we analysed additional Sp2 target promoters. We chose promoters at which Sp2 ChIP-Seq yielded high tag counts exemplified by the Sp2 and Cyth2 promoters, and promoters with relatively low tag counts exemplified by Tbp and Jun. Sp1 was present at these promoters in wt MEFs and in Sp2ko MEFs. As in the case of the cholesterol pathway genes the abundance of Sp1 at these promoters was reduced in Sp2ko cells (Figure 8) suggesting that Sp2 and Sp1 do not compete for steady-state binding to their common site in vivo. Decreased occupancy of Sp1 at target promoters in Sp2ko MEFs is consistent with the observation that expression of Sp1, which by itself is a direct Sp2 target gene, is slightly reduced in Sp2ko MEFs (Figure 2).
DISCUSSION
In this study we establish Sp2 as a sequence-specific master transcription factor regulating the expression of multiple genes essential for diverse fundamental cellular processes including protein synthesis, replication, metabolism and signalling. This conclusion is supported by the following findings. (i) DAPAs revealed specific DNA binding of endogenous Sp2 present in nuclear extracts of various cell lines. (ii) RNAi-mediated depletion of Sp2 in HEK293 cells as well as previously reported Cre-mediated ablation of Sp2 in MEFs with floxed Sp2 alleles impaired cell proliferation. (iii) Genome-wide analysis of Sp2 chromatin occupancy in MEFs and HEK293 cells revealed binding of Sp2 primarily to active proximal promoters of genes essential for fundamental regulatory and metabolic cellular processes. (iv) Global expression analysis of wt and Sp2 knockout MEFs identified important Sp2 key target genes and cellular pathways exemplified by the large set of regulatory genes that were repressed, and the cholesterol synthesis genes that were concertedly activated upon Sp2 depletion.
Specific DNA binding of Sp2 present in high-salt nuclear extract preparations was established by DAPA. In this experimental setting, Sp2 bound to the same sequence that was also bound by Sp1 and Sp3. Strikingly, and in accordance with previously published observations (17) DNA-binding of full-length Sp2 was not detected by EMSA, which is the most commonly used experimental approach to analyse protein–DNA interactions in vitro. The failure to detect DNA-binding of Sp2 in nuclear extracts by EMSAs explains why this biologically important protein largely escaped attention after its initial discovery 20 years ago (42).
Our genome-wide analysis of Sp2 chromatin occupancy identified the sequence DDRGGHGGRNC (Figure 5A), where Gs dominate at the D and R positions, and a C dominates at the H position yielding the consensus sequence (G/A)GGCGGGNC. This sequence matches the Sp2 consensus DNA-binding site (GGGCGGGAC) previously identified in vitro by cyclic amplification and selection of targets from an unbiased set of degenerate oligonucleotides (17). As Sp1 and Sp3 also bind to this sequence motif, it remains to be determined whether there exist binding sites that are bound preferentially by individual Sp factors in vivo. Future analysis of Sp1 and Sp3 occupancy by ChIP-Seq in the same cell lines should clarify this point. Our motif search with Sp2 peak regions revealed also a particular high recurrence of the NF-Y recognition sequence CCAAT suggesting a cis regulatory network involving Sp2 and NF-Y. Of note is that CCAAT motifs are present at Sp2 peaks of promoters that were activated as well as of promoters that were repressed upon Sp2 depletion in MEFs. Interestingly, a high degree of overlap was previously also reported for NF-Y and Sp1 occupancy at promoters in HepG2 cells using ChIP–chip (43) suggesting that NF-Y co-operates with different Sp factors.
ChIP-Seq using two different antibodies identified 3261 high-confidence Sp2-binding sites in MEFs and 2380 binding sites in HEK293 cells. Concurrent ChIP-Seq with chromatin of Sp2ko MEFs as well as re-expression of Sp2 in Sp2ko MEFs followed by ChIP–qPCR confirmed the high fidelity of the ChIP-Seq data sets. The great majority of the Sp2-binding sites are localized within 500 bp of annotated TSSs strongly suggesting that Sp2 acts primarily at proximal promoters and not from remote positions. Moreover, a comparison of our Sp2 ChIP-Seq data set from HEK293 cells with recently released ChIP-Seq data sets of the ENCODE consortium (44) showed that 1927 (81%) and 1646 (70%) of the Sp2 peaks in HEK293 cells also occur in K562 cells and in HepG2 cells, respectively. The large overlap of Sp2-binding sites at orthologous promoters in MEFs and different human cell lines further substantiates the notion that Sp2 is regulating primarily general cellular processes.
We previously reported impaired proliferation of MEFs after depletion of Sp2 by retroviral transduction of Cre recombinase in MEFs carrying floxed Sp2 alleles (15). In this report we show that Sp2 is also necessary for normal proliferation of HEK293 cells. Impaired proliferation of HEK293 cells upon siRNA-mediated depletion of Sp2 is consistent with the results of a genome-wide RNAi screen for genes essential for mitosis (45). RNAi-mediated depletion of Sp2 in HeLa cells resulted in phenotypes such as ‘strange nuclear shape’, ‘segregation problems’, ‘metaphase delay’ and ‘mitotic delay’ (see http://www.mitocheck.org/cgi-bin/mtc?query=MCG_0009279) (45). Remarkably, knockdown of none of the other Sp transcription factors including Sp1 and Sp3 led to an overt phenotype in this screen. The fact that solely depletion of Sp2 impaired proliferation reinforces the exceptional importance of Sp2. The phenotypic specificity of Sp2 in cultured cells further suggests that there is little redundancy between Sp2 and other Sp factors. We consider it unlikely that impaired proliferation in general and the occurrence of mitotic defects in particular is attributed to the misregulation of a single or a few genes. In this report we identified multiple Sp2-regulated genes involved in diverse fundamental cellular processes suggesting that, rather than deregulation of a few individual genes, a broad impact on pivotal biological pathways leads to the observed cellular phenotypes.
The conclusion that Sp2 regulates primarily basic cellular functions is also consistent with the Sp2 knockout phenotype. Before their death at E9.5 of gestation, Sp2 null mouse embryos are already severely growth-retarded but no particular cell lineage or developmental process is affected (15). Nevertheless, Sp2 could also negatively regulate differentiation programs as transgenic mice over-expressing Sp2 in the epidermal basal layer of the skin are devoid of terminally differentiated keratinocytes (16).
Mining ChIP-Seq and transcriptional profiling datasets revealed a rich set of repressed and induced direct Sp2 target genes in MEFs, which were validated with high reliability. Most strikingly, genes that were repressed in Sp2ko MEFs encode numerous sequence-specific transcription factors including Foxo3, Gabpa, Jun, Klf9, Sp3, Stat1 and many uncharacterized zinc-finger proteins as well as co-activators such as Baz1b, Brd8, Hdac6 or the histone demethylases Kdm3a and Kdm4b. Reduced expression of the broad range of transcriptional regulators in Sp2-deficient MEFs justifies the assignment of Sp2 as a master regulator of basic cellular processes.
An intriguing finding is the concerted activation of cholesterol synthesis genes in Sp2-deficient MEFs. Sp2 occupies promoters of 22 genes that catalyse various steps of cholesterol synthesis suggesting that Sp2 plays a repressive role at these promoters. Since we also found enhanced expression of the Srebf-2 gene, activation of cholesterol pathway genes in Sp2ko MEFs could be the consequence of increased activation by Srebp-2, the master regulator of the cholesterol pathway (41). In human hepatocyte cells, many genes are co-occupied by Srebp-1 and Sp1 (43) and it is generally accepted that Sp1 co-operates with Srebps in the activation of cholesterol pathway genes. Sp1 expression is not increased in Sp2ko MEFs; in contrast there is less Sp1 protein in Sp2ko MEFs and reduced binding of Sp1 to Sp2 target promoters. Thus, increased occupancy of Sp1 at cholesterol pathway genes does not contribute to the activation of these genes. Reduced binding of Sp1 to Sp2 target promoters in Sp2ko MEFs implies that Sp2 and Sp1 do not compete for steady-state binding to DNA target sites in vivo, which is in agreement with the prediction that transcription factor-binding sites are unsaturated during physiological transcriptional activation (46). Whether the reduced abundance of Sp1 solely accounts for its reduced binding to target promoters in Sp2ko MEFs is unclear. An alternative, admittedly very speculative explanation would be that the presence of Sp2 facilitates binding of Sp1 to its target sites in vivo.
Taken together, the data provided in this report establish Sp2 as a key transcription factor regulating vital cellular functions. The detection of Sp2 DNA-binding activity in nuclear extracts and the global identification of its conserved targets in murine and human cells unveiled to a large degree the enigmatic nature of this physiologically important transcription factor. Further mining of the resources provided in this report is expected to reveal additional insights into cellular processes regulated by Sp2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–3 and Supplementary Datasets 1–8.
FUNDING
Deutsche Forschungsgemeinschaft (DFG Su 102/8-1 to G.S.); and the priority research program LOEWE ‘Tumor and Inflammation’ to FF. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Iris Rohner is gratefully acknowledged for excellent technical assistance, and Sjaak Philipsen, Alexander Brehm and Martha Kalff-Suske for many helpful discussions and for critical reading of the manuscript. The authors also thank Wolfgang Meissner for RNA quality control and the technical staff members of the Genomics Core Facility at the IMT for help with the gene-expression profiling.
REFERENCES
- 1.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 2.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Schaeper ND, Prpic NM, Wimmer EA. A clustered set of three Sp-family genes is ancestral in the Metazoa: evidence from sequence analysis, protein domain structure, developmental expression patterns and chromosomal location. BMC Evolut. Biol. 2010;10:88. doi: 10.1186/1471-2148-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 5.Bouwman P, Göllner H, Elsässer HP, Eckhoff G, Karis A, Grosveld F, Philipsen S, Suske G. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 2000;19:655–661. doi: 10.1093/emboj/19.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Göllner H, Dani C, Phillips B, Philipsen S, Suske G. Impaired ossification in mice lacking the transcription factor Sp3. Mech. Dev. 2001;106:77–83. doi: 10.1016/s0925-4773(01)00420-8. [DOI] [PubMed] [Google Scholar]
- 7.Van Loo PF, Mahtab EA, Wisse LJ, Hou J, Grosveld F, Suske G, Philipsen S, Gittenberger-de Groot AC. Transcription factor Sp3 knockout mice display serious cardiac malformations. Mol. Cell Biol. 2007;27:8571–8582. doi: 10.1128/MCB.01350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krüger I, Vollmer M, Simmons DG, Elsässer HP, Philipsen S, Suske G. Sp1/Sp3 compound heterozygous mice are not viable: impaired erythropoiesis and severe placental defects. Dev. Dyn. 2007;236:2235–2244. doi: 10.1002/dvdy.21222. [DOI] [PubMed] [Google Scholar]
- 9.Supp DM, Witte DP, Branford WW, Smith EP, Potter SS. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev. Biol. 1996;176:284–299. doi: 10.1006/dbio.1996.0134. [DOI] [PubMed] [Google Scholar]
- 10.Göllner H, Bouwman P, Mangold M, Karis A, Braun H, Rohner I, Del Rey A, Besedovsky HO, Meinhardt A, van den Broek M, et al. Complex phenotype of mice homozygous for a null mutation in the Sp4 transcription factor gene. Genes Cells. 2001;6:689–697. doi: 10.1046/j.1365-2443.2001.00455.x. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen-Tran VT, Kubalak SW, Minamisawa S, Fiset C, Wollert KC, Brown AB, Ruiz-Lozano P, Barrere-Lemaire S, Kondo R, Norman LW, et al. A novel genetic pathway for sudden cardiac death via defects in the transition between ventricular and conduction system cell lineages. Cell. 2000;102:671–682. doi: 10.1016/s0092-8674(00)00089-1. [DOI] [PubMed] [Google Scholar]
- 12.St Amand TR, Lu JT, Zamora M, Gu Y, Stricker J, Hoshijima M, Epstein JA, Ross JJ, Jr, Ruiz-Lozano P, Chien KR. Distinct roles of HF-1b/Sp4 in ventricular and neural crest cells lineages affect cardiac conduction system development. Dev. Biol. 2006;291:208–217. doi: 10.1016/j.ydbio.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Long JM, Geyer MA, Masliah E, Kelsoe JR, Wynshaw-Boris A, Chien KR. Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Mol. Psychiatry. 2005;10:393–406. doi: 10.1038/sj.mp.4001621. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Qyang Y, Kelsoe JR, Masliah E, Geyer MA. Impaired postnatal development of hippocampal dentate gyrus in Sp4 null mutant mice. Genes Brain Behav. 2007;6:269–276. doi: 10.1111/j.1601-183X.2006.00256.x. [DOI] [PubMed] [Google Scholar]
- 15.Baur F, Nau K, Sadic D, Allweiss L, Elsasser HP, Gillemans N, de Wit T, Kruger I, Vollmer M, Philipsen S, et al. Specificity protein 2 (Sp2) is essential for mouse development and autonomous proliferation of mouse embryonic fibroblasts. PloS One. 2010;5:e9587. doi: 10.1371/journal.pone.0009587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TH, Chiera SL, Linder KE, Trempus CS, Smart RC, Horowitz JM. Overexpression of Transcription Factor Sp2 Inhibits Epidermal Differentiation and Increases Susceptibility to Wound- and Carcinogen-Induced Tumorigenesis. Cancer Res. 2010;70:8507–8516. doi: 10.1158/0008-5472.CAN-10-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moorefield KS, Fry SJ, Horowitz JM. Sp2 DNA binding activity and trans-activation are negatively regulated in mammalian cells. J. Biol. Chem. 2004;279:13911–13924. doi: 10.1074/jbc.M313589200. [DOI] [PubMed] [Google Scholar]
- 18.Moorefield KS, Yin H, Nichols TD, Cathcart C, Simmons SO, Horowitz JM. Sp2 localizes to subnuclear foci associated with the nuclear matrix. Mol. Biol. Cell. 2006;17:1711–1722. doi: 10.1091/mbc.E05-11-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagen G, Müller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garner MM, Revzin A. The use of gel electrophoresis to detect and study nucleic acid-protein interactions. TIBS. 1986;11:395–396. [Google Scholar]
- 21.Hagen G, Müller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decesse JT, Medjkane S, Datto MB, Cremisi CE. RB regulates transcription of the p21/WAF1/CIP1 gene. Oncogene. 2001;20:962–971. doi: 10.1038/sj.onc.1204169. [DOI] [PubMed] [Google Scholar]
- 23.Stielow B, Sapetschnig A, Wink C, Krüger I, Suske G. SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep. 2008;9:899–906. doi: 10.1038/embor.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. ISMB. 1994;2:28–36. [PubMed] [Google Scholar]
- 28.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controllng the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser B. 1995;57:289–300. [Google Scholar]
- 30.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman HB. DNA damage repair and response proteins as targets for cancer therapy. Curr. Medicin. Chem. 2008;15:360–367. doi: 10.2174/092986708783497328. [DOI] [PubMed] [Google Scholar]
- 41.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kingsley C, Winoto A. Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol. Cell. Biol. 1992;12:4251–4261. doi: 10.1128/mcb.12.10.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 45.Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.