Abstract
CCG repeats are highly over-represented in exons of the human genome. Usually they are located in the 5′ UTR but are also abundant in translated sequences. The CCG repeats are associated with three tri-nucleotide repeat disorders: Huntington’s disease, myotonic dystrophy type 1 and chromosome X-linked mental retardation (FRAXE). In this study, we present two crystal structures containing double-stranded CCG repeats: one of an RNA in the native form, and one containing LNA nucleotides. Both duplexes form A-helices but with strands slipped in the 5′ (native structure) or the 3′ direction (LNA-containing structure). As a result, one of two expected C-C pairs is eliminated from the duplex. Each of the three observed C-C pairs interacts differently, forming either one weak H-bond or none. LNA nucleotides have no apparent effect on the helical parameters but the base stacking is increased compared to the native duplex and the distribution of electrostatic potential in the major groove is changed. The CCG crystal structures explain the thermodynamic fragility of CCG runs and throw light on the observation that the MBNL1 protein recognises CCG runs, as well as CUG and CAG, but not the relatively stable CGG repeats.
INTRODUCTION
The repeated CCG motifs are frequent in genomes of many organisms. As shown for the human genome, the CCG repeats are highly over-represented in exons and this positive selection in coding sequences implies a biological function (1). However, the specific functions in genes and transcripts remain poorly understood. Messenger RNAs derived from as many as 139 human genes harbour CCG repeats composed of six or more repeated units (1). About half of them are located in the 5′ untranslated regions (UTR), which can be explained by their involvement in the regulation of transcription and/or in the initiation step of translation. The CCG repeats are abundant also in translated sequences but are rare in the 3′ untranslated region. The longest CCG tracts in mature mRNAs are typically shorter than 15 repeats.
A well-known example of a transcript containing CCG repeats is the mRNA from human huntingtin gene (HTT). In the normal HTT transcript, the polymorphic CCG repeat, composed of 6–12 units, is separated by 12 nt of HTT-specific sequence from a more polymorphic CAG repeat (6–35 CAG) (2). Expansion of the CAG repeat is the underlying cause of Huntington’s disease but the potential role of the CCG repeat as a disease modifying factor has not yet been satisfactorily resolved (3). A recent study showed that both the repeated sequences and their spacer form a single hairpin structure in which the CCG repeats interact with a portion of the CAG repeats (4). CCG repeats occur also within expanded CUG repeats of mutant dystrophia myotonica-protein kinase (DMPK) gene transcript in some patients suffering from myotonic dystrophy type 1 (5). It is likely that the CCG repeats have an impact on the structure of the CUG repeats and on their interactions with cellular proteins, and this contributes to unusual symptoms observed in some myotonic dystrophy patients (5).
The expansion of a CCG repeat in exon 1 of human FMR2 gene plays a direct role in the FRAXE locus associated with one of the forms of chromosome X-linked mental retardation (6). The normal gene contains 3–40 CCG repeats which undergo transcription, genes with up to 200 CCG repeats, classified as pre-mutations, are also transcribed, while expression of genes with the full mutation (200–750 CCGs) is transcriptionally inhibited (7). Although no phenotype has yet been assigned to human FMR2 pre-mutation carriers, 90 CCG repeats were shown to be as toxic as 90 CGG repeats in the Drosophila model of the fragile-X associated tremor ataxia syndrome (FXTAS) which is another triplet repeat disease caused by pre-mutated CGG repeats (7).
The biomedical importance of CCG runs in transcripts stimulated efforts to characterize these sequences structurally. Short repeats (CCG)2–4 were shown by UV melting to form duplex structures (8). Longer repeats (CCG)17,20,25 formed hairpins, according to UV melting, gel mobility analysis as well as chemical and biochemical structure probing (9,10). These hairpins contained either 4 nt or 7 nt terminal loops and their stem portion was composed of the reiterated G-C and C-G pairs and C-C non-canonical base pairs. The methods used did not allow, however, for a more detailed characterization of these structures, especially the exact nature of the C-C pairs, their impact on RNA helix geometry and consequences for interactions of double-stranded CCG regions with proteins.
This work is the final in our crystallographic studies of CNG repeats, following CUG, CAG and CGG structures (11–13). Here, we present two crystal structures containing double-stranded CCG repeats: one of an RNA in the native form, and one containing LNA (‘locked’) nucleotides.
MATERIALS AND METHODS
Synthesis, purification and crystallization of the CCG oligomers
GCCGCCGC and GCCGLCCGC oligomers were synthesized on an Applied Biosystems DNA/RNA synthesiser, using cyanoethyl phosphoramidite chemistry. Commercially available A, C and G phosphoramidites with 2′-O-tertbutyldimethylsilyl were used for the synthesis of RNA (Glen Research, Azco, Proligo). The phosphoramidite GL was synthesized according to Pasternak et al. (14) and Koshkin et al. (15). The details of deprotection and purification of oligoribonucleotides were described previously (16). The RNA oligomers were dissolved in 200 or 400 mM ammonium acetate to the final concentration of 1 mM and annealed for 10 min at 95°C, then cooled slowly to ambient temperature within 1 h. Before the crystallization, 25 mM spermine–HCl was added to the native oligomer solution. Crystals were grown by the hanging drop method at 30°C in 10 mM magnesium acetate, 50 mM MES (pH 5.6) and 2.5 M ammonium sulphate. The GCCGLCCGC oligomer crystals were obtained by the sitting drop method at 19°C. The crystallization medium contained 10 mM MgCl2, MES (pH 5.6) and 1.8 M Li2SO4. RNA was mixed with the screen solution 1:1.
X-ray data collection, structure solution and refinement
X-ray diffraction data were collected at the BESSY synchrotron in Berlin: on the BL 14.2 beam line for native GCCGCCGC crystal to the resolution of 1.54 Å and on the BL 14.1 beam line for the GCCGLCCGC crystal to the resolution of 1.95 Å. Both crystals were cryoprotected by 20% glycerol (v/v) in the mother liquor. The data were integrated and scaled using the program suite DENZO/SCALEPACK (17). The space group was assigned as R3 (native RNA) and P4322 (LNA-containing structure). The structures were solved by molecular replacement, using PHASER (18). In the case of the GCCGLCCGC oligomer, it was crucial to cut the resolution to 3.2 Å during phasing. Early stages of the refinement were done using the program Refmac5 (19) from the CCP4 program suite (20), then refinement was carried out with PHENIX (21). Approximately 500 reflections were set aside for the Rfree statistic. The program Coot (22) was used for visualization of electron density maps calculated with coefficients 2Fo − Fc and Fo − Fc and for manual rebuilding of the atomic model. The last few cycles were performed using all data, including the Rfree set. The models are summarized in Table 1 and in Supplementary Table S1.
Table 1.
Summary of the models and refinement statistics
| (GCCGCCGC)2 | (GCCGLCCGC)2 | |
|---|---|---|
| Overall mean B-factor (Å2) | 27.79 | 49.4 |
| Number of reflections: work/test | 4572/501 | 5155/562 |
| R-value (%) | 25.5 | 25.8 |
| R-free (%) | 30.4 | 30.1 |
| RNA atoms | 300 | 486 |
| Water molecules | 16 | 14 |
| No. sulphate ions | – | 2 |
| RMSD in bonds/target (Å) | 0.01/0.022 | 0.01/0.022 |
| RMSD in angles/target (°) | 1.3/3.0 | 1.2/3.0 |
| PDB code | 4E59 | 4E58 |
The helical parameters were calculated using 3DNA (23). Sequence-independent measures were used, based on vectors connecting the C1′ atoms of the paired residues, to avoid computational artefacts arising from non-canonical base-pairing. Program PBEQ-Solver (24) was used to calculate the electrostatic potential map. All pictures were drawn using UCSF Chimera (25) and PyMOL v0.99rc6 (26). The coordinates of both crystallographic models have been deposited with the Protein Data Bank (PDB). The accession codes are 4E58 and 4E59.
RESULTS
RNA models
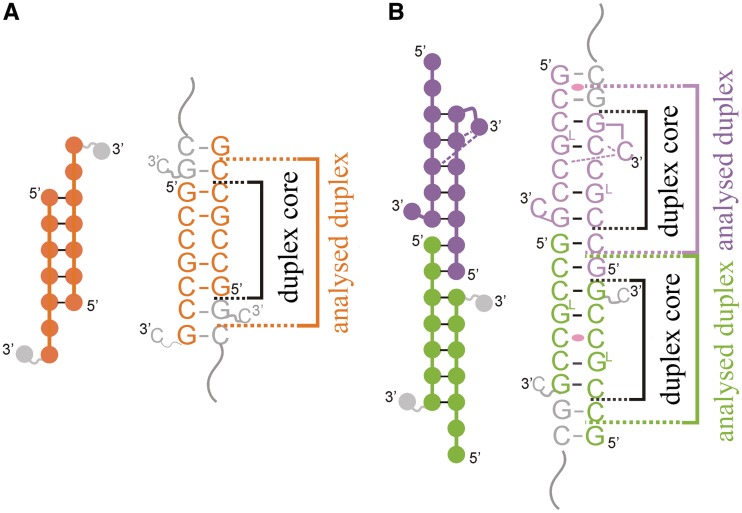
The GCCGCCGC oligomer forms a slippery duplex (strands A + B) with three 3′-overhanging nucleotides (Figure 1A). The terminal residues, 8C, of both strands are disordered and have not been modelled into the electron density map. The remaining two overhanging nucleotides, 6C and 7G, of each strand are paired with two residues of the symmetry-related duplex, forming C-G and G-C pairs. The overlapping oligomers form semi-infinite helices parallel to the c cell edge (Supplementary Figure S1A). The fold of the (GCCGLCCGC)2 structure is different from the native duplex. The asymmetric unit contains three RNA strands. Two distinct double helices are formed: C + D and E with its symmetry-related strand E′. These oligomers also form slippery duplexes but have two dangling nucleotides, 1G and 2C, at the 5′ end of each strand (Figure 1B). Moreover, the last nucleotide, 8C, is folded back. In the C + D duplex, both the cytosine residues are ordered and interact with the minor groove of the duplex (see the Supplementary Data). In the E + E′ helix the terminal residue is disordered, similar to the native structure. The 1G and 2C residues form Watson–Crick pairs with overhanging nucleotides of neighbouring oligomers. In the tetragonal crystal lattice, two distinct columns of semi-infinite helices are present. Both of them are parallel to the a–b plane but perpendicular to each other (Supplementary Figure S1B). The only crystal contacts between the columns are between the flipped bases and the minor grooves of neighbouring helices (see the Supplementary Data). The native and the LNA-containing models also include 14 and 16 water molecules, respectively. The modified structure also contains two sulphate anions (see the Supplementary Data).
Figure 1.
Scheme of base pairing of the native (A) and the LNA-containing duplexes (B). Brackets indicate segments used in calculating the helical parameters and the inner core regions around the C-C pairs.
The RNA duplex conformation
Although in the two crystal forms the strand slippage occurs in different directions, each of the three distinct duplexes consists of seven base pairs, including one non-canonical C-C pair flanked on each side by three C-G and G-C pairs. The core of each duplex is 5 nt long, having unbroken backbone (Figure 1). The A + B and E + E′ duplexes have the A-form with typical helical parameters (Supplementary Tables S2 and 3). All the residues show C3'-endo pucker and Zp values of 2.5 Å (the displacement of the phosphorus atom from the xy-plane of the ‘middle frame’ between neighbouring base-pairs). In the C + D duplex, the sugar conformation of 11 residues is C3′-endo, one is C2'-exo and two are C2'-endo (typical for the B-form). Nevertheless, the Zp values are still above 1.6 Å, thus the overall form of the C + D helix is assigned as A-RNA. Calculated helical parameters of the C + D duplex differ from typical values or show irregularities (Supplementary Tables S2 and 3). For instance, the average value of helical twist is typical, 33.5° (28.4° for the duplex core), but the values are scattered in the range 15.0–49.3° (the standard deviation = 11.5° and 9.8° for the duplex core). The buckle parameter also shows elevated variability (−3.7 ± 14.7°) compared with other duplexes (−0.5 ± 2.9° for A + B and 0.1 ± 4.3° for E + E′).
The non-canonical C-C pair
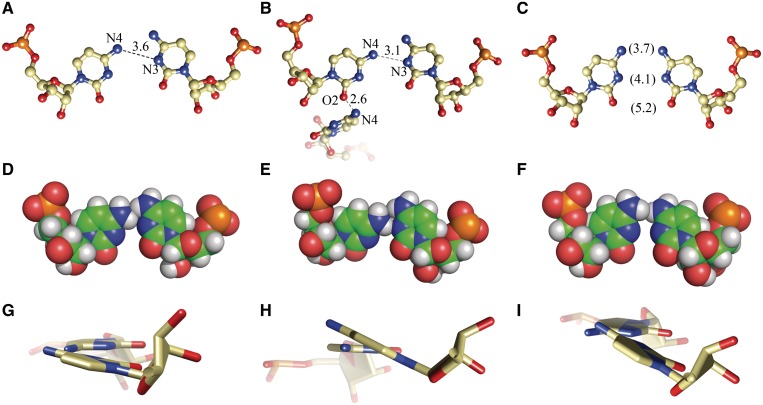
In each helix, there is only one C-C pair and each has a different conformation (Figure 2 and Supplementary Figure S2). In the A + B duplex, one of the cytidine residues is inclined toward the minor groove, indicated by the angle λ = 38.9° (the angle between the glycosidic bond and the line joining the base-paired C1′ atoms) and opening = −11.5° (Supplementary Table S3). The cytidines probably form one weak H-bond (3.6 Å) between the exo-amino group of the inclined residue and the N3 atom of the other C (Figure 2A). The exo-amino groups of the two residues are in close proximity. The H42 atom of the non-inclined C is wedged between the hydrogen atoms of the exo-amino group of the other cytidine (Figure 2D). The C-C pair of the C + D duplex shows a similar interaction between the exo-amino group and the N3 atom (3.1 Å) but one of the cytidines is inclined more toward the minor groove (λ = 30.8° and opening = −22.3°). This conformation is stabilized by a H-bond between the O2 carbonyl atom and N4 of the folded back 8C residue of chain C (Figure 2B). The exo-amino groups are close as in the A + B duplex but the pyrimidine bases are twisted relative to each other (propeller twist = −20.5°; Supplementary Table S3) thus avoiding a clash between the hydrogen atoms (Figure 2E and H). The cytidines of the third C-C pair, in the E + E′ duplex, are related by crystallographic symmetry. Both residues are slightly inclined towards the minor groove (λ = 46.5°, opening = −7.9°). Their Watson–Crick edges are aligned with the same functional groups vis-à-vis (Figure 2C). It is possible that the exo-amino groups form a weak interaction. Their H42 atoms are very close (1.9 Å; Figure 2F) but a clash is avoided due to the relative twist of the bases (Figure 2I). The propeller for the C-C pair is −20.4°. All the C-C pairs fit well within the double-stranded helix. The distance between the C1′ atoms of the paired C is in the range 10.7–10.9 Å, compared to average value of 10.6 Å for the flanking G-C pairs in both structures.
Figure 2.
The three C-C pairs: from the native duplex A + B (A, D, G) and the LNA-containing duplexes C + D (B, E, H) and E + E′ (C, F, I). The first row shows the possible H-bonds. The second row shows the Van der Waals interactions, especially the close proximity of the exo-amine groups. The third row shows the propeller twist of the bases.
GL residues in the modified structure
In the modified structure, 4G is an LNA residue. The sugar ring contains a methylene bridge that enforces the C3'-endo conformation and constrains the range of ribose’s torsion angles. On the other hand, the flexibility of the rest of the backbone, primarily the phosphodiester bond, is retained. In chains C and E, values of the torsion angles of the GL are in the typical range. For the modified guanosine in chain D, the α (rotation about P–O5′ bond), β (O5′–C5′) and γ angles (C5′–C4′) take unusual values. α = 113° (typical is −68°), β is −136.1° (typical value is 178°) and γ has the value of −157.5° (typical is 54°). Distortion of the backbone is due to flipping of the O5′–C5′ bond and shifting of the P and C5′ atoms. Typically, the P, O5′, C5′ and C4′ atoms form a zigzag and are approximately co-planar, whereas in GL the zigzag is inverted and the atoms deviate from the plane to the extent that if three consecutive atoms are placed on a plane, the forth will deviate by approximately 50° (Supplementary Figure S4). Due to this conformation of the backbone, the LNA is shifted toward the major groove, which amounts to a low helical twist of 15°. The local unwinding is compensated in other parts of the duplex. One of the sulphate ions is associated with the 4G LNA residue (see the Supplementary Data). The only parameter clearly distinct in all the LNA residues is roll (Supplementary Table S4). Its value, calculated for the CGL/CG step is greater by 9.9° than the average value (4.1°) of other steps.
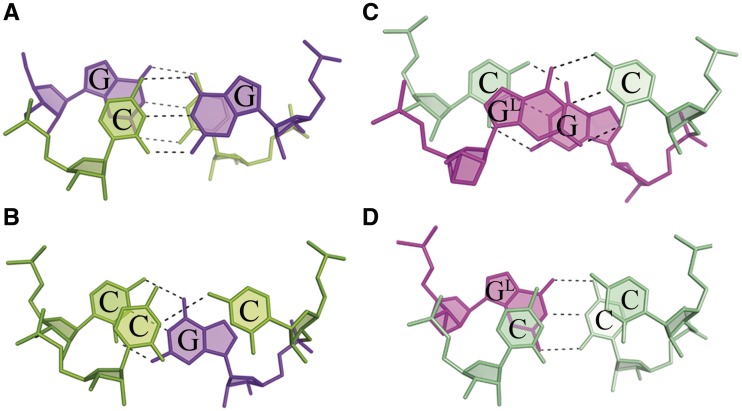
Stacking interactions
Native and modified structures have different distributions of stacking interactions. In the unmodified duplex, they are similar to those observed in previously reported CNG structures (11–13): the steps involving only Watson–Crick pairs show extensive overlaps (overlap area >5.6 Å2), while those involving non-canonical base pairs show limited interactions (the largest overlap area = 0.9 Å2; Figure 3A and B). In the modified structure, the base rings stack more evenly than in the native RNA. In chain C and E, the stacking interactions between GL and the adjacent 5C (from the C-C pair) are the most extensive (7.3 and 5.6 Å2; Figure 3D). At the 5′ site, the LNA interacts cross-strand with 7G (Figure 3C). In chain D, where the conformation around GL is distorted, the residue shows similar overlaps but less extensive. The cytosines of non-canonical pairs interact with GL at the 5′ side and to a lesser extent with C of the same strand.
Figure 3.
Stacking interactions between the Watson–Crick pairs and those involving the C-C pairs, for the native duplex (A, B) and the LNA-containing duplex (C, D).
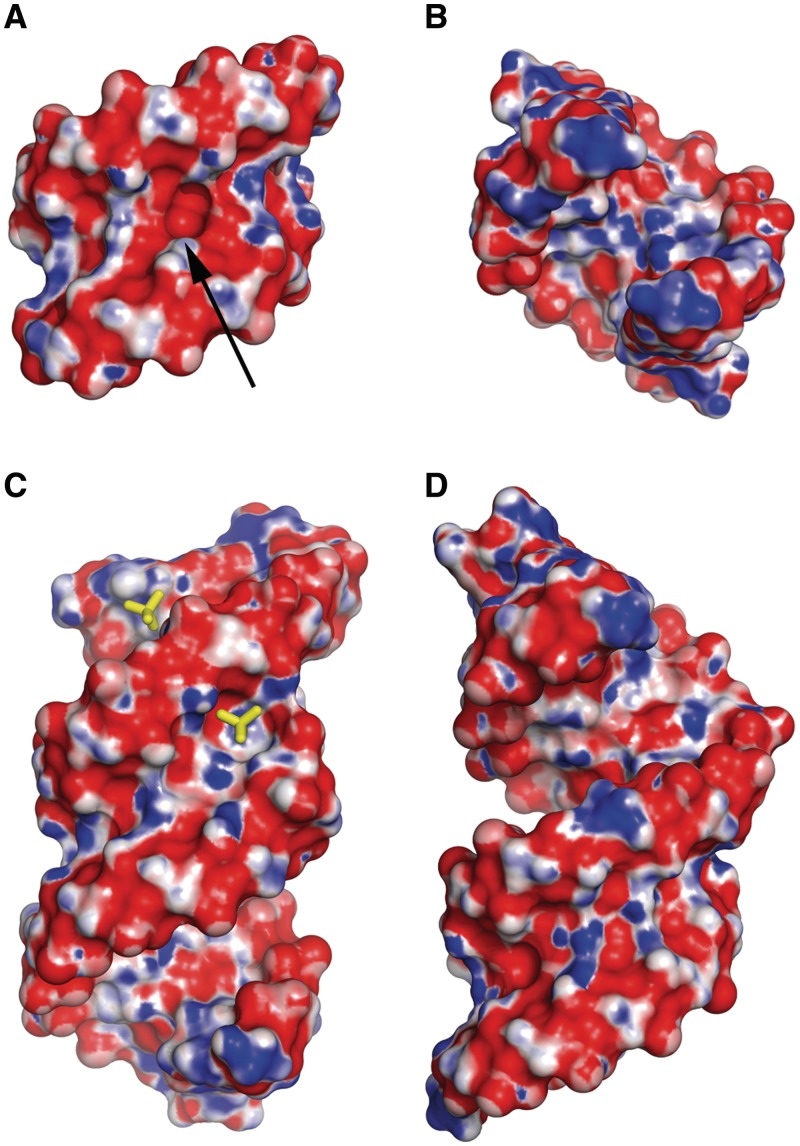
Electrostatic surface potential
The distribution of electrostatic potential is similar in both structures. In the minor groove, the alternating stripes of negative and positive potential are observed—a characteristic feature of all CNG repeats (Figure 4A and C). The stripes are generated by functional groups of the G-C and C-G pairs. In the middle of the helix, one positive band is missing due to the presence of carbonyl groups of the C-C pair. There is an associated indentation between the Watson–Crick edges of the paired cytidine residues (Figure 4A). In the C + D duplex, flipping of the two 8C residues results in a more complex surface and distribution of the electrostatic potential. Nevertheless, the indentation around the C-C pair and the stripes of surface potential are retained. The main difference is the presence of a sulphate-occupied niche between 4GL and 8C of the opposite strand. The shape and the distribution of the electrostatic potential in the cavity are complementary to the bound sulphate ion (Figure 4C). In the major grooves of all the duplexes, the positive and negative bands form a chequered pattern (Figure 4B and D). This is associated with the alternating arrangement of the two types of nucleotide. In the native structure, the positive patches are more pronounced, indicating a higher electropositive potential compared to the modified structure.
Figure 4.
The electrostatic potential surface for the native duplex (A: minor groove; B: major groove) and the two LNA-containing duplexes stacking end-to-end (C, D). Red is negative, blue is positive. The arrow points to an indentation in the minor groove between two paired cytosines. Two sulphate ions (sticks) are shown, one in the minor groove, another in the major groove, interacting with patches of positive potential.
DISCUSSION
What distinguishes CCG repeats from other CNG structures?
Earlier biochemical results suggested that CCG repeats are similar to other CNG runs in that they form duplexes containing blocks of C-G and G-C pairs with C-C pairs in between (9–10). This study shows that short sequences containing CCG repeats do form A-helices but with strands having a tendency to slip in either direction, resulting in 5′ or 3′ overhangs. The sticky ends associate in the crystal lattice so that the RNA duplexes form semi-continuous columns which can be divided into core segments and the overlapping parts. In addition, the 3′ terminal C is expelled from the double helix. The result of the strand slippage, regardless of whether it takes place in the 3′ or the 5′ direction, is the elimination of one of two expected C-C pairs. The resulting helices have a single C-C pair surrounded by C-G and G-C pairs. This is more than has been observed in the NMR structure of (CCGCCG)2 DNA, in which all four cytidines, which could potentially form C-C pairs, have been expelled from the duplex by means of strand slippage and bulging, leaving only C-G pairs (27).
The C-C interaction is the weakest of the N-N pairs (8, 10) and the observed strand slippage could be the result of their destabilizing effect on the double helix. It appears as if the system acts to minimize unfavourable interactions.
One could ask why cytosines are poor partners compared to other non-canonical pairs in the CNG runs? One distinguishing feature of the C-C pairs is their lack of common conformation. Each of the three observed C-C pairs interacts differently. One of them forms no hydrogen bonds, another has one weak H-bond and the third shows a conformation similar to the U-U pairs in the CUG structures (11). In this case, one cytosine is inclined toward the minor groove and its exo-amino group interacts with O4 of the opposite C. A search using FRABASE (28) showed that C-C pairs are relatively rare. The ribosomal subunit of Deinococcus radiodurans contains a 1219C–1253C pair localized between the C-G and G-C pairs. In the 30 models of this structure deposited in the PDB, one of the cytosine residues is relatively constant while the second is shifted to various extents toward the minor groove while the C1′–C1′ distance is more than 11.1 Å and the backbone is distorted relative to a typical A-helix (Supplementary Table S5). In comparison to the ribosomal motif, we see in the helical structures that the C-C pair still shows conformational variability but the inclination of C toward the minor groove is limited. The minimum λ angle (30.8°) in the C + D duplex in the LNA-containing structure could be the result of a stabilizing interaction with the folded back 8C. The C1′–C1′ length for the C-C pairs is 10.8 Å, which is slightly longer than the average value for canonical pairs. This indicates that in the double-helical context the C-C pair conforms to the A-form rather than optimises its pairwise interactions.
An explanation of why the C-C pairing potential is not realized can be obtained from a comparison with U-U pairs in a similar helical environment (11). The most apparent difference is that the exo-amino group in C is larger than the corresponding carbonyl oxygen atom in U. Thus uracil residues easily form stable pairs, whereas cytosines, even when H-bonded, have close contacts between the hydrogen atoms on their Watson–Crick edges.
What effect has LNA on the CCG structure?
Locked nucleic acids have received much attention because of their stabilizing effect on duplexes of RNA or DNA, an increased affinity for complementary sequences, compared with native oligomers and promising properties as therapeutics, such as their relative resistance to enzymatic degradation. We decided to use LNA for the above reasons and also because we had problems with crystallizing the unmodified sequences. One LNA guanosine residue was introduced in position 4 and this resulted in the tetragonal crystal structure.
The distinguishing property of LNA to constrain the ribose ring to the C3′-endo conformation corresponds well with the A form expected for RNA. In the duplex E + E′, the modified nucleotide has no apparent effect on the helical parameters, compared with the unmodified structure (Supplementary Tables S2 and 3). The observed deviations from typical values in propeller and buckle are probably due to the conformation of the C-C pair. The C + D duplex is more distorted and although the average values of the helical parameters (twist, rise, angle and displacement) are typical, the standard deviations are elevated. This is probably related to the folding back of the 8C residues and the associated ribose C2′-endo conformation of the 7C residues. The flipping of the O5′-C5′ bond of 4GL in chain D can be associated with the presence of a sulphate ion in the minor groove.
It has been found that LNA generally increases the thermodynamic stability of RNA duplexes (29), and while it is easiest to assume that this is due to the fixed conformation of the ribose ring, one has to allow that other effects are at play. In the examined structures, the conformation and ordering of the sugar rings in the LNA residues are unambiguously C3′-endo, even when the phosphodiester backbone shows unusual torsions. A general ordering effect is also indicated by a greater proportion of the modified molecule being ordered in the crystal structure (the 8C residues) and also by a greater ease with which the modified oligomer crystallized. The stabilizing effect of LNA has been explained in earlier studies in terms of stacking interactions (30,31). We also observe that in both the modified helices the base stacking is increased compared to the native duplex, but it is not clear how this is connected to the LNA residue. The change in stacking interactions can change the distribution of electrostatic potential in the major groove (Figure 4B and D). The correlation seems to be that when bases associate extensively they exhibit less electrostatic potential.
It is not clear why the native and modified duplexes differ in the direction of the slippage. The dangling of the 3′ ends in the native structure is consistent with thermodynamic findings that an unpaired C at the 3′ end stabilized an RNA duplex by approximately −0.8 kcal/mol (32,33). It is harder to explain the overhang of the 5′ ends in the modified duplexes. Perhaps the GL residues stabilize this form, or it is due to crystal lattice effects, or both. One has to remember that the overhanging nucleotides do not really dangle, but form stable C-G and G-C pairs with adjacent oligomers.
Biological implications
Biochemical probing of secondary structure shows that CCG repeats form extended hairpins, like other CNG tracks (9). Both the presented oligomers were designed as representative fragments of a long stem of a CCG hairpin. This time, however, the obtained structures differ from our previous studies of CNG duplexes (11–13). The mechanism of eliminating C-C pairs from the duplex is likely to be a result of the short length of the oligomer. Nevertheless, it is likely that the core of the crystallized duplexes is relevant to CCG runs in vivo. This implies that the hairpin stems have the form of A-RNA with canonical C-G and G-C pairs flanking unsteady C-C pairs forming one weak H-bond or none. The apparent weakness of the C-C pair explains the thermodynamic fragility of CCG runs and throws light on the observation that the MBNL1 protein recognises CCG runs, as well as CUG and CAG, but not the relatively stable CGG repeats (34–36). MBNL1 is thought to bind single-stranded RNA or duplexes whose strands can be parted easily (37). Perhaps other specific structural features among those observed are also important for RNA/protein interactions, but this remains to be seen.
Expanded CCG repeats have been detected in studies of three neurological diseases, but their role in pathogenesis has been difficult to determine. The proposed models of pathogenesis of TREDs involve interactions of extended RNA hairpins with proteins, and their precipitation as nuclear foci. Is it possible that the relatively unstable CCG runs are relatively poor targets for such interactions, hence their correspondingly mild and ill-defined pathogenic properties?
The CCG structures complete the gallery of structural profiles of the stems of RNA hairpins related to TREDs. The crystal structures are consistent with biochemical and biophysical data and the high-resolution atomic models could stimulate and assist further research. The key structural features of the CCG motifs and the other CNG repeats have been compiled in Supplementary Table S6.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6, Supplementary Figures 1–5, Supplementary Information and Supplementary References [11–13,38,39].
FUNDING
Ministry of Science and Higher Education [Poland, N-N301-0171634], National Science Centre [Poland, UMO-2011/01/B/NZ1/04429] the EU structural funds [POIG.01.03.01-30-098/08], the European Community—Research Infrastructure Action under the FP6 ‘Structuring the European Research Area’ Programme (through the ‘Integrated Infrastructure Initiative’ Integrating Activity on Synchrotron and Free Electron Laser Science—Contract R II 3-CT-2004-506008); Scholarship START of the Foundation for Polish Science (to A.K.); Fellowship of the Foundation for Polish Science (to R.K.). Funding for open access charge: National Science Centre.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kozlowski P, de Mezer M, Krzyzosiak WJ. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson DL. FRAXE mental retardation and other folate-sensitive sites. In: Wells RD, Warren ST, Sarmiento M, editors. Genetic Instabilities and Hereditary Neurological Diseases. San Diego, CA: Academic Press; 1998. pp. 65–74. [Google Scholar]
- 3.Zhang BR, Tian J, Yan YP, Yin XZ, Zhao GH, Wu ZY, Gu WH, Xia K, Tang BS. CCG polymorphisms in the huntingtin gene have no effect on the pathogenesis of patients with Huntington's disease in mainland Chinese families. J. Neurol. Sci. 2012;312:92–96. doi: 10.1016/j.jns.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 4.de Mezer M, Wojciechowska M, Napierala M, Sobczak K, Krzyzosiak WJ. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braida C, Stefanatos RK, Adam B, Mahajan N, Smeets HJ, Niel F, Goizet C, Arveiler B, Koenig M, Lagier-Tourenne C, et al. Variant CCG and GGC repeats within the CTG expansion dramatically modify mutational dynamics and likely contribute toward unusual symptoms in some myotonic dystrophy type 1 patients. Hum. Mol. Genet. 2010;19:1399–1412. doi: 10.1093/hmg/ddq015. [DOI] [PubMed] [Google Scholar]
- 6.Gu Y, Shen Y, Gibbs RA, Nelson DL. Identification of FMR2, a novel gene associated with the FRAXE CCG repeat and CpG island. Nat. Genet. 1996;13:109–113. doi: 10.1038/ng0596-109. [DOI] [PubMed] [Google Scholar]
- 7.Sofola OA, Jin P, Botas J, Nelson DL. Argonaute-2-dependent rescue of a Drosophila model of FXTAS by FRAXE premutation repeat. Hum. Mol. Genet. 2007;16:2326–2332. doi: 10.1093/hmg/ddm186. [DOI] [PubMed] [Google Scholar]
- 8.Broda M, Kierzek E, Gdaniec Z, Kulinski T, Kierzek R. Thermodynamic stability of RNA structures formed by CNG trinucleotide repeats. Implication for prediction of RNA structure. Biochemistry. 2005;44:10873–10882. doi: 10.1021/bi0502339. [DOI] [PubMed] [Google Scholar]
- 9.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the ‘stretched U-U wobble': implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases. Nucleic Acids Res. 2010;38:8370–8376. doi: 10.1093/nar/gkq700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 2011;39:7308–7315. doi: 10.1093/nar/gkr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasternak A, Kierzek E, Pasternak K, Turner DH, Kierzek R. A chemical synthesis of LNA-2,6-diaminopurine riboside, and the influence of 2′-O-methyl-2,6-diaminopurine and LNA-2,6-diaminopurine ribosides on the thermodynamic properties of 2′-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2007;35:4055–4063. doi: 10.1093/nar/gkm421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 16.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 27.Zheng M, Huang X, Smith GK, Yang X, Gao X. Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding. An NMR and UV spectroscopic study. J. Mol. Biol. 1996;264:323–336. doi: 10.1006/jmbi.1996.0643. [DOI] [PubMed] [Google Scholar]
- 28.Popenda M, Szachniuk M, Blazewicz M, Wasik S, Burke EK, Blazewicz J, Adamiak RW. RNA FRABASE 2.0: an advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures. BMC Bioinformatics. 2010;11:231. doi: 10.1186/1471-2105-11-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaur H, Arora A, Wengel J, Maiti S. Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes. Biochemistry. 2006;45:7347–7355. doi: 10.1021/bi060307w. [DOI] [PubMed] [Google Scholar]
- 30.Eichert A, Behling K, Betzel C, Erdmann VA, Furste JP, Forster C. The crystal structure of an ‘All Locked' nucleic acid duplex. Nucleic Acids Res. 2010;38:6729–6736. doi: 10.1093/nar/gkq505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebars I, Richard T, Di Primo C, Toulme JJ. NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer. Nucleic Acids Res. 2007;35:6103–6114. doi: 10.1093/nar/gkm655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole AS, Miller S, Serra MJ. Stability of 3′ double nucleotide overhangs that model the 3′ ends of siRNA. RNA. 2005;11:512–516. doi: 10.1261/rna.7254905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugimoto N, Kierzek R, Turner DH. Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry. 1987;26:4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- 34.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 35.Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 36.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y, Ramisetty SR, Hussain N, Baranger AM. MBNL1-RNA recognition: contributions of MBNL1 sequence and RNA conformation. Chembiochem. 2012;13:112–119. doi: 10.1002/cbic.201100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bloomfield VA, Crothers DM, Tinoco I. Nucleic Acids: Structures, Properties, and Functions. Sausalito: University Science Books; 2000. [Google Scholar]
- 39.Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu XJ, Neidle S, Shakked Z, et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.