Abstract
Stress granules (SGs) are large cytoplasmic ribonucleoprotein complexes that are assembled when cells are exposed to stress. SGs promote the survival of stressed cells by contributing to the reprogramming of protein expression as well as by blocking pro-apoptotic signaling cascades. These cytoprotective effects implicated SGs in the resistance of cancer cells to radiation and chemotherapy. We have found that sodium selenite, a selenium compound with chemotherapeutic potential, is a potent inducer of SG assembly. Selenite-induced SGs differ from canonical mammalian SGs in their morphology, composition and mechanism of assembly. Their assembly is induced primarily by eIF4E-binding protein1 (4EBP1)-mediated inhibition of translation initiation, which is reinforced by concurrent phosphorylation of eIF2α. Selenite-induced SGs lack several classical SG components, including proteins that contribute to pro-survival functions of canonical SGs. Our results reveal a new mechanism of mammalian SG assembly and provide insights into how selenite cytotoxicity may be exploited as an anti-neoplastic therapy.
INTRODUCTION
Stress-induced translational repression is a consequence of reduced translation initiation. This is achieved by inhibiting the assembly of eIF4F (i.e. eIF4E: eIF4G: eIF4A) and 43S pre-initiation complexes (1). Assembly of eIF4F complexes is inhibited by eIF4E-binding proteins (4EBPs) that block interactions between eIF4E and eIF4G to down-regulate 5′-cap-dependent translation initiation. Typically, dephosphorylation of 4EBPs by stress-induced inactivation of the PI3K-mTOR (mammalian target of rapamycin) pathway promotes 4EBP-mediated translational repression. Assembly of 43S complexes is inhibited by stress-induced activation of PKR, PERK, GCN2 and HRI, kinases that phosphorylate eIF2α, a component of the eIF2-GTP-tRNAMet ternary complex that is required for 43S pre-initiation complex assembly. These complementary mechanisms are largely responsible for the general translational repression observed in cells exposed to adverse environmental conditions.
Non-translating messenger RNA (mRNAs) that accumulate as a result of stress-induced translational repression are frequently compartmentalized into cytoplasmic foci known as stress granules (SGs). SGs are large ribonucleoprotein (RNP) complexes composed of abortive translation initiation complexes and an eclectic assortment of RNA-binding proteins and signaling proteins involved in various aspects of cellular metabolism (2). These include tumor necrosis factor receptor-associated factor 2 (TRAF2), receptor for activated C kinase 1 (RACK1) and plakophilin3, which are routed to SGs as a consequence of interactions with core SG components (3–5). Current evidence suggests that SGs, together with a related RNA granule known as the processing (P-) body, play important roles in determining the fate of mRNAs in stressed cells. Some RNAs are stored in the granules, others are degraded and still others are returned to the cytoplasm for translation (6). SGs have also been implicated in signaling cascades that determine whether stressed cells live or die. Thus, the recruitment and sequestration of TRAF2 and RACK1 were reported to inhibit inflammatory signaling and stress-induced apoptotic signaling (3,4), respectively. Furthermore, cells impaired in the ability to assemble SGs invariably show reduced survival upon exposure to environmental stress (7–10). Thus, SGs are thought to promote cell survival under stress conditions through modulation of various aspects of cell metabolism.
Although the cytoprotective functions of SGs should be beneficial to individual cells or unicellular organisms, this may not be the case for multicellular organisms, in which the proper execution of apoptosis is required for differentiation and general survival. The cytoprotective functions of SGs may also benefit tumor cells by allowing the survival of poorly vascularized tumor cells. Indeed, several studies now implicate SGs in cancer biology. Aside from the inhibition of apoptosis by sequestration of signaling molecules as described above, SGs assembled in hypoxic tumor cells have been shown to inhibit the translation of angiogenic factors and promote resistance to radiotherapy (11). In addition, SG assembly is induced by bortezomib (commercially known as Velcade), a proteasome inhibitor whose anti-tumor effects are inversely correlated with SG assembly (12). It is therefore likely that SGs promote resistance to radio- and chemotherapy in cancer cells.
Selenium is an essential micronutrient that has both chemopreventive and chemotherapeutic properties (13). At nutritional levels (∼50 nM), selenium is incorporated into selenoproteins that function as anti-oxidants. At supranutritional levels (>1 µM), selenium acts as a pro-oxidant through the activities of its metabolites, which promote the production of reactive oxygen species (ROS). Importantly, high doses of selenium compounds are selectively toxic to cancer cells. Although the mechanism of this tumor cell-specific cytotoxicity is not fully understood, cancer cells may be particularly susceptible to ROS induced by selenium metabolism (14). A recent study also demonstrated that cancer cells specifically display increased uptake of selenium compounds (15). Interestingly, the efficiency of uptake was positively correlated with the expression level of multi-drug resistance (MDR) proteins, a hallmark of chemoresistance and tumor malignancy; increased expression of MDR proteins leads to increased uptake of selenium compounds and increased toxicity (15). These results suggest that selenium compounds could be used to specifically target tumor cells that cannot be eliminated by conventional chemotherapy.
In addition to the induction of ROS, selenium compounds trigger endoplasmic reticulum (ER) stress, which results in phosphorylation of eIF2α (16,17). Since eIF2α phosphorylation is the major trigger of SGs, we tested the ability of selenium compounds to induce the assembly of SGs. In this study, we focused on sodium selenite, the most common dietary form of water-soluble selenium compound which was also the most effective anti-tumor compound (15). We found that selenite potently inhibits translation and induces the ROS-dependent assembly of SGs that lack the pro-survival proteins found in typical SGs. Our results uncover a novel aspect of selenite-induced cytotoxicity that is relevant to the use of selenite as an anti-cancer drug.
MATERIALS AND METHODS
Reagents, antibodies and small interfering RNA
Sodium arsenite, sodium selenite, emetine, cycloheximide and rapamycin were purchased from Sigma-Aldrich. MnTMPyP (manganese (III) tetrakis (1-methyl-4-pyridyl) porphyrin) was obtained from Enzo Life Sciences. The following antibodies were used for western blotting and immunofluorescence: anti-eIF3b (goat polyclonal, N-20), eIF3e (goat polyclonal, C-20), eIF4E (mouse monoclonal, P-2), eIF4G1 (rabbit polyclonal, H-300), G3BP1 (mouse monoclonal, TT-Y), FMR1 (mouse monoclonal, 148.1), Hedls (mouse monoclonal, originally intended to react with S6K1 (18), H-9), HDAC6 (rabbit polyclonal, H-300), HuR (mouse monoclonal, 3A2), PABP (mouse monoclonal, 10E10), RACK1 (mouse monoclonal, B-3) and TIAR (goat polyclonal, C-18) from Santa Cruz Biotechnology; anti-eIF4E-binding protein1 (4EBP1) (rabbit polyclonal), phospho-4EBP1 (Thr37/46, rabbit monoclonal), eIF4A1 (rabbit monoclonal), ribosomal S6 (mouse monoclonal), phospho-ribosomal S6 (rabbit polyclonal) and Rsk2 (rabbit monoclonal) from Cell Signaling Technology; anti-β-actin (mouse monoclonal) from Chemicon; anti-eIF5A (mouse monoclonal) from BD Biosciences; anti-G3BP1 (rabbit polyclonal) from Bethyl; anti-phospho-eIF2α (rabbit polyclonal) from Assay Designs; anti-Ago2 (mouse monoclonal) from Wako Bioproducts; anti-PCBP2 (mouse monoclonal) from Abnova; anti-O-GlcNAc (mouse IgM, CTD110.6) from Covance; anti-importin β1 (mouse monoclonal) from Pierce; anti-small ribosomal S14 (rabbit polyclonal) from Proteintech group. Anti-Rch1/importin α1 (mouse monoclonal) antibody was kindly provided by Dr. Jun Katahira (Osaka University). Secondary antibodies used for immunofluorescence (Cy2-, Cy3- and Cy5-conjugated) and western blotting (horseradish peroxidase-conjugated) were from Jackson Immunoresearch Laboratories. Biotinylated oligo-dT18 probe was purchased from IDT. Non-targeting, control small interfering RNA (siRNA) (U0) was as described previously (19). siRNA targeting human 4EBP1 was purchased from QIAGEN; the target sequence was 5′-TCGGAACTCACCTGTGACCAA-3′.
Cell culture, drug treatment and transfections
Human osteosarcoma U2OS cells, human prostatic carcinoma DU145 cells, mouse embryonic fibroblasts (MEFs) with or without the S51A mutation of eIF2α and TP53−/− MEFs with or without TSC2 deficiency were maintained at 37°C in a CO2 incubator in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin/streptomycin. For SG induction, cells grown to ∼70% confluency were treated with arsenite (100 µM, 1 h) or selenite (1 mM, 2 h) unless otherwise indicated. Emetine and cycloheximide treatment was done as described previously (20). siRNA transfection was performed using Lipofectamine 2000 (Invitrogen), basically according to the manufacturer’s instructions. Briefly, 20 pmol of siRNA and 1 µl of Lipofectamine 2000, each diluted in 50 µl of OPTI-MEM, were mixed and added to cells plated at the density of 1.5 × 104 cells/well in a 24-well plate. Cells were subjected to assays 72 h after transfection.
Western blotting
Following drug treatment, cells were washed with phosphate buffered saline (PBS) and directly lysed in Laemmli’s sample buffer supplemented with 100 mM dithiothreitol (DTT), boiled and stored at −20°C until use. The samples were resolved using a gradient 4–20% Tris–Glycine gel (Invitrogen), transferred to a nitrocellulose membrane, which was probed with the indicated primary antibodies (used at the dilution of 1:2000, except for anti-β-actin antibody, which was used at the dilution of 1:5000) using 5% normal horse serum diluted in PBS (normal human serum/PBS) as a blocking reagent. Following extensive washes, the membrane was incubated with appropriate secondary antibodies (at the dilution of 1:5000), and antibody detection was performed using SuperSignal West Pico Chemiluminescent Substrate (Themo Scientific).
Metabolic labeling
Cells grown in six-well plates (1.5 × 105 cells/well) were treated either with arsenite or selenite for 1 h, followed by 1 h of labeling with 0.5 µCi of C14-lysine (American Radiolabeled Chemicals, Inc.; ARC0459) and C14-leucine (Moravek Biochemicals, Inc.; MC175W) in the presence or the absence of drugs. Afterwards, cells were washed and harvested in PBS and split evenly into two tubes. Perchloric acid precipitation was performed for one of the pools to measure the amount of incorporated C14-labeled amino acids into proteins; the other pool was suspended in 0.5 M NaOH and the total protein amount was measured by Bio-Rad Protein Assay kit.
m7GTP-sepharose pulldown assay
Cells grown on 10-cm dishes were lysed in 0.5 ml/dish of lysis buffer (Tris–HCl pH7.4, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 0.5% NP-40, supplemented with a protease inhibitor tablet (Roche) just before use), and centrifuged for 15 min at 13 000 rpm at 4°C. The supernatant containing 1 mg of total protein was transferred to a clean tube and incubated with prewashed 15 µl suspension of m7GTP-sepharose (GE Healthcare) for 2 h at 4°C with rotation. The beads were washed extensively with the lysis buffer and cap-bound materials were eluted by boiling in 60 µl of 2 × Laemmli’s sample buffer supplemented with 100 mM DTT.
Immunofluorescence and in situ hybridization
Immunofluorescence was performed essentially as described previously (19). Briefly, cells were fixed in paraformaldehyde, permeabilized with methanol at −20°C and then blocked with normal human serum/PBS for at least 30 min. For staining of eIF5A, importin β1 and HDAC6, permeabilization was performed by treating cells with 0.2% Triton X-100 in PBS for 10 min. Subsequently, primary antibodies were added at the dilution of 1:300, incubated for 1 h at room temperature, followed by incubation with secondary antibodies at the dilution of 1:300. Eventually, cells were washed extensively, stained with Hoechst to reveal nuclei and mounted using Vinol mounting media. Wide-field fluorescence microscopy was performed using an Eclipse E800 microscope (Nikon) equipped with epifluorescence optics and digital camera (CCD-SPOT RT; Diagnostic Instruments). Image acquisition was done with a 40× objective (PlanApo; Nikon). Acquired images were compiled using Adobe Photoshop. Quantification of the percentage of SG-positive cells was done by counting the number of cells with at least two discrete TIAR- and G3BP-positive foci from >100 cells, from at least three independent experiments. For in situ hybridization, cells were processed as described previously (21), and hybridization was performed using a biotinylated oligo-dT18 probe and in situ hybridization buffer (Ambion) at 37°C. After extensive washes with 0.5 × saline-sodium citrate (SSC), probe was revealed using Cy2-conjugated streptavidin (Jackson Immunoresearch Laboratories), followed by immunostaining of TIAR and G3BP. For the measurement of SG size, the ‘Analyze particle’ tool of the ImageJ software (NIH) was used.
Cell viability assay
Cell viability assay was performed essentially as described previously (22). Briefly, U2OS cells transfected either with control or 4EBP1 siRNA were seeded into 48-well plates (1 × 104 cells/well) 48 h after transfection. After overnight culture, cells were treated with arsenite (100 µM for 2 or 4 h) or selenite (either at 1 mM for 1 h, or at 10 µM and 20 µM for 24 h) and cell viability was measured using CellTiter-Glo (Promega) and Veritas Microplate Luminometer (Turner Biosystems) following the manufacturer’s instructions. The experiments were done at least three times.
RESULTS
Composition of selenite-induced SGs
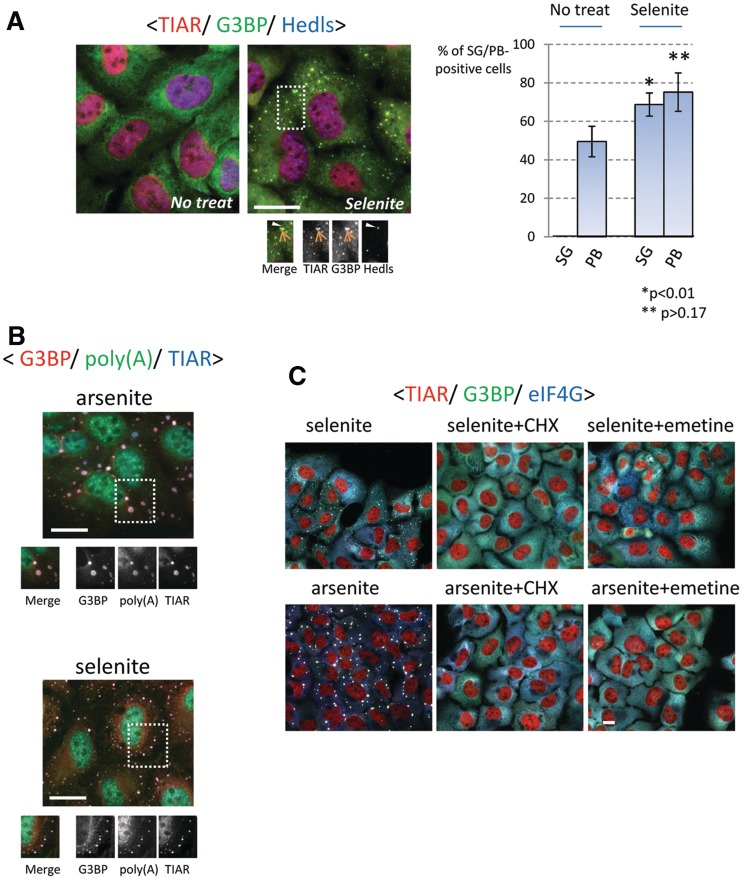
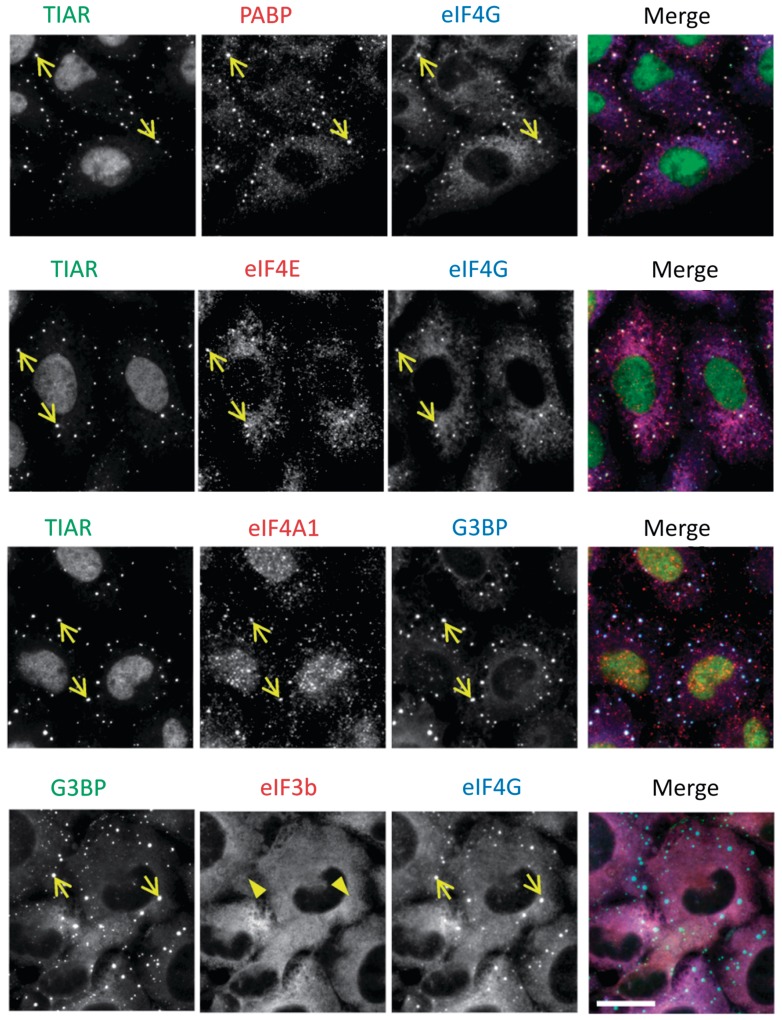
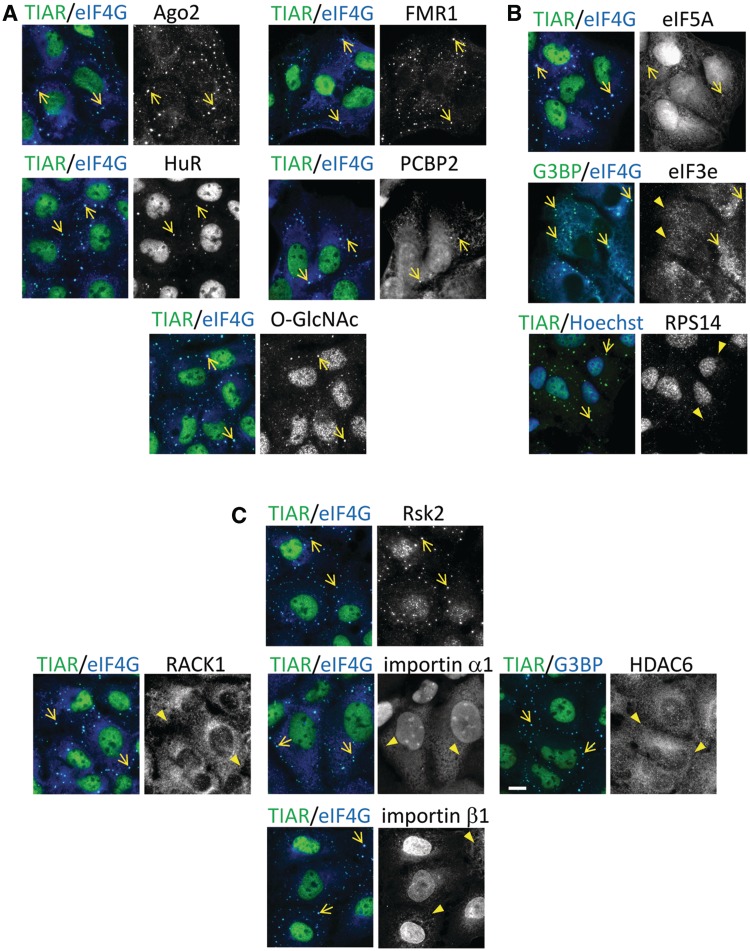
To determine whether selenite has the ability to induce SGs, U2OS cells were treated with various concentrations of sodium selenite, followed by immunostaining for the SG markers, TIAR and G3BP. We found that SGs are assembled in cells exposed to 5 µM selenite for 24 h (Supplementary Figure S1A). In most of our experiments, cells were exposed to 1 mM selenite for 2 h, as this gave the most robust results in the shortest time. As shown in Figure 1A (see Supplementary Figure S1B for unmerged figures), co-staining with the P-body marker Hedls/Ge-1 revealed that selenite-induced SGs are often found in close physical apposition to P-bodies. Although selenite treatment also appeared to increase the percentage of P-body-positive cells, this difference was not statistically significant (Figure 1A, bar graph). In situ hybridization using an oligo-dT18 probe demonstrated strong accumulation of poly-A-containing mRNA in selenite-induced SGs (Figure 1B). Furthermore, emetine and cycloheximide, drugs that arrest translation elongation to stabilize polysomes, dissolved SGs in both arsenite- and selenite-treated cells (20) (Figure 1C). Collectively, these results reveal that selenite induces bona fide SGs in which TIAR, G3BP and poly-(A) mRNAs are selectively concentrated. We noticed that selenite-induced SGs are generally smaller (see Supplementary Figure S1C for size comparison) and found closer to the cell periphery than arsenite-induced SGs (20). To compare the compositions of selenite- and arsenite-induced SGs, we first stained for core SG components, namely, translation initiation factors that constitute the 48 S pre-initiation complex (23). As shown in Figure 2, PABP (poly-A binding protein), eIF4G, eIF4E and eIF4A are recruited to both arsenite- and selenite-induced SGs. In contrast, eIF3b, a protein that is one of the most reliable markers of mammalian SGs and is recruited to SGs under various stresses (18), is selectively excluded from selenite-induced SGs (Figure 2, bottom panel; compare with Supplementary Figure S2A). The lack of eIF3b suggests that the selenite-induced SGs are different from not only arsenite-induced SGs but also SGs triggered by many other stimuli in mammalian cells (18). The lack of eIF3b localization in selenite-induced SGs was also observed with the human prostate cancer cell line DU145 (Supplementary Figure S2B), confirming that this is not a phenomenon specific to U2OS cells. We went on to test the localization of other established components of ‘canonical’ SGs, as shown in Figure 3. Most of the canonical SG components are efficiently recruited to selenite-induced SGs, including Ago2, FMR1, HuR and PCBP2 (Figure 3A) (24–27). O-GlcNAc-modified proteins were also found in selenite-induced SGs (19). In contrast, eIF5A (28), eIF3e/Int6 and the small ribosomal protein Rps14 are only weakly recruited or absent in selenite-induced SGs (compare with arsenite-induced SGs shown in Supplementary Figure S2 and Figure 3B). Among SG components that are not explicitly involved in RNA metabolism, Rsk2 is recruited to selenite-induced SGs, possibly through its interaction with TIA-1 (9). In contrast, RACK1, importin α1/β1 and HDAC6, which are known components of arsenite-induced SGs, are excluded from selenite-induced SGs, at least in U2OS cells (Figure 3C, compare with Supplementary Figure S2) (3,8,22,29). Thus, selenite-induced SGs are morphologically and compositionally distinct from canonical SGs.
Figure 1.
Induction of bona fide SGs by sodium selenite. (A) (Left panel) Selenite triggers TIAR- and G3BP-positive SGs juxtaposed to P-bodies. U2OS cells without or with selenite treatment (1 mM, 2 h) were immunostained for TIAR (red), G3BP (green) and Hedls (blue, cytoplasmic staining). (Right panel) Percentage of SG- or P-body-positive cells in the presence or absence of selenite, as quantified from three independent experiments. Error bars = SD; * and ** denote P-value relative to non-treated cells. (B) Selenite-induced SGs contain poly-A mRNAs. U2OS cells exposed to arsenite (top) or selenite (bottom) were processed for in situ hybridization to detect poly-A containing mRNA (green), followed by staining of TIAR (red) and G3BP (blue). (C) Selenite-induced SGs are disassembled by cycloheximide (CHX) and emetine. U2OS cells were treated with arsenite or selenite for 1 hr, followed by co-treatment with CHX or emetine for an additional 1 hr. Subsequently, cells were immunostained for TIAR (red), G3BP (green) and eIF4G (blue). Bars = 10 µm.
Figure 2.
Accumulation of major translation initiation factors in selenite-induced SGs. The localization of PABP, eIF4G (top row, red and blue, respectively), eIF4E (second row, red), eIF4A1 (third row, red) and eIF3b (bottom row, red) was probed by immunostaining of selenite-treated U2OS cells. Arrows indicate SG localization, whereas arrowheads indicate the lack of enrichment in SGs. Bar = 10 µm.
Figure 3.
Accumulation of various canonical SG components in selenite-induced SGs. The accumulation of canonical SG components, including (A) RNA-binding proteins (Ago2, FMR1, HuR and PCBP2) and O-GlcNAc-modified proteins, (B) translation initiation factors other than the ones tested in Fig. 2 (eIF5A and eIF3e) and the small ribosomal S14 protein (Rps14) and (C) SG constituents that do not possess obvious RNA-binding activity (Rsk2, RACK1, importin α1/β1 and HDAC6) were probed for SG localization by immunostaining. Arrows indicate localization to SGs, whereas arrowheads indicate the lack of enrichment. Bar = 10 µm.
Phophorylation of eIF2α is dispensable for selenite-induced SG assembly
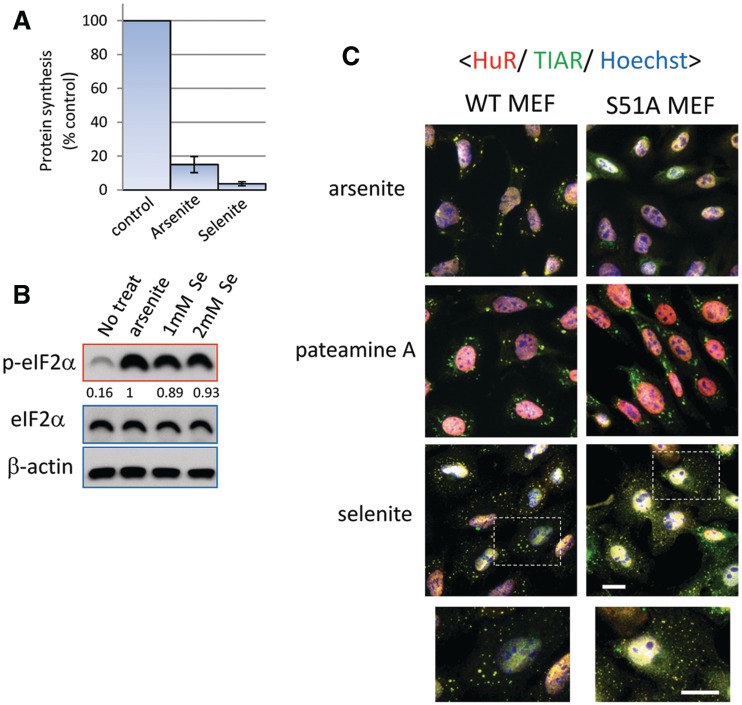
To examine the effect of selenite on translation, we measured the rate of de novo protein synthesis using isotopically labeled amino acids. As shown in Figure 4A, both arsenite and selenite strongly down-regulate global protein synthesis. In accordance with previous reports (16,17), selenite induces the phosphorylation of eIF2α, which probably contributes to translational repression (Figure 4B). In order to determine whether selenite-induced SG assembly requires eIF2α phosphorylation, we used a MEF cell line in which wild-type (WT) eIF2α is replaced with a non-phosphorylatable mutant (eIF2αS51A) (30). Although arsenite treatment does not trigger SG formation in these knock-in (S51A) MEFs, pateamine A, which induces SGs in a phospho-eIF2α-independent manner (31), does (Figure 4C). Although selenite-induced SGs are observed in both WT and S51A MEFs, the SGs assembled in S51A MEFs are smaller than those observed in WT MEFs (Figure 4C). Thus, eIF2α phosphorylation contributes to proper SG assembly by selenite, but it is not indispensable for this process.
Figure 4.
Selenite potently triggers translational repression and increases phospho-eIF2α level, although eIF2α phosphorylation is not essential for selenite-induced SG biogenesis. (A) Selenite inhibits translation even more potently than arsenite. De novo protein synthesis rate was measured by labeling with C14-lysine and C14-leucine in untreated, arsenite-treated and selenite-treated cells. Error bars = SD. (B) Increase of phospho-eIF2α levels by selenite. U2OS cells treated without or with arsenite or selenite (1 mM and 2 mM) were lysed and subjected to western blotting using anti-eIF2α, phospho-eIF2α and β-actin antibodies. Densitometric quantification of relative eIF2α phosphorylation levels is indicated under each lane of the phospho-eIF2α panel. (C) Wild-type MEFs (WT) or MEFs bearing S51A mutant eIF2α (S51A) were treated with selenite and immunostained for HuR (red), TIAR (green), with nuclei revealed with Hoechst (blue). Since S51A MEFs were particularly susceptible to selenite, the drug treatment condition was modified to 0.5 mM, 2 h, to allow both cell lines to remain attached to the coverslip. Bar = 10 µm.
Selenite disrupts eIF4F complex formation through 4EBP1, which is at least partially responsible for SG assembly
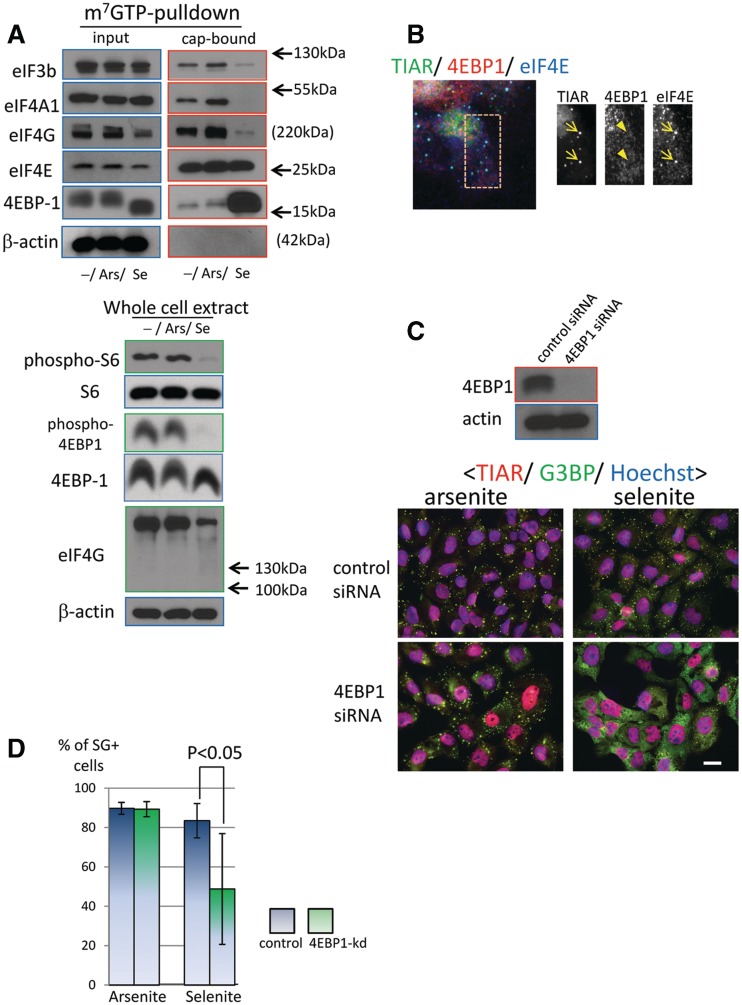
Given that eIF2α phosphorylation is not absolutely required for selenite-induced SG assembly, we compared the integrity of the eIF4F complex in arsenite- and selenite-treated cells. As shown in Figure 5A, eIF4E is pulled down by m7GTP sepharose to a similar extent in selenite- and arsenite-treated cells, suggesting that the ability of eIF4E to recognize the 5′-cap is not impaired. However, other components of the eIF4F complex, including eIF4G, eIF4A and eIF4G/A-interacting protein eIF3b, are significantly reduced in selenite-treated cells, but not arsenite-treated cells. The association of 4EBP1 with the cap is selectively increased by selenite, suggesting that selenite disrupts the eIF4F complex by enhancing binding of 4EBP1 to eIF4E. We also noticed a decrease in eIF4G protein levels upon selenite treatment in the input lysate. Using whole-cell extracts, we confirmed that the total protein level of eIF4G is indeed down-regulated (Figure 5A, lower panel). This is unlikely to be due to apoptosis, since apoptosis-induced proteolysis of eIF4G should produce a 120-kDa cleavage product that is recognized by the antibody that we are using (32). We also observed that 4EBP1 in selenite-treated cells migrates faster than that in control or arsenite-treated cells. This result suggests that 4EBP1 is dephosphorylated upon selenite treatment, which typically indicates inhibition of mTOR activity (1). In fact, levels of phospho-4EBP1 and ribosomal protein S6 are strongly reduced by selenite treatment in whole-cell extracts, further suggesting that selenite inhibits mTOR activity in these cells (Figure 5A, lower panel).
Figure 5.
Involvement of 4EBP1 in selenite-induced eIF4F complex disruption and SG formation. (A) Selenite disrupts eIF4F complex formation and enhances eIF4E-4EBP1 interaction. (Top) U2OS cells without or with drug treatment (arsenite and selenite) were lysed and subjected to m7GTP-sepharose pulldown to isolate cap-associated proteins. Both the input and the precipitate were processed for western blotting and probed for the presence of eIF4E, eIF4G, eIF4A, eIF3b and 4EBP1. (Bottom) Whole-cell extracts from non-treated or drug-treated cells were prepared by directly lysing cells with SDS sample buffer, and protein levels of eIF4G, ribosomal protein S6, phospho-S6 and β-actin were examined. (B) 4EBP1 itself is not recruited to selenite SGs. U2OS cells exposed to selenite were immunostained for 4EBP1 (red), TIAR (green) and eIF4E (blue). Arrows indicate SG localization, whereas the arrowheads indicate the lack of SG accumulation. (C) 4EBP1 depletion by siRNA compromises selenite-induced SG, but not arsenite-induced SG biogenesis. U2OS cells were transfected with control or 4EBP1-directed siRNA for 72 h and processed for western blotting (top, probed for 4EBP1 and β-actin) or immunostaining (bottom, stained for TIAR, G3BP and treated with Hoechst). Bar = 10 µm. (D) The percentage of cells bearing SGs induced by arsenite (left) or selenite (right) without (control) or with 4EBP1 knockdown (4EBP-kd). siRNA-transfected U2OS cells were processed as above, and the percentage of SG-positive cells was calculated from four independent experiments. Error bars = SD; P < 0.05 by unpaired t-test.
We next examined the role of 4EBP1 in selenite-induced SG assembly. Immunofluorescence microscopy revealed that 4EBP1 is not a component of selenite-induced SGs (Figure 5B) (4EBP1 antibody was validated for use in immunofluorescence in Supplementary Figure S2C, where 4EBP1 signal was strongly reduced by siRNA-mediated depletion of this protein). However, siRNA-mediated depletion of 4EBP1 clearly inhibits selenite-induced SG assembly, but not arsenite-induced SG assembly (Figure 5C and D). The percentage of SG-positive cells decreased significantly upon selenite treatment, and even when SGs were assembled, they were far less discrete and the accumulation of individual granule components was strongly compromised. Collectively, these results indicate that, unlike canonical SGs, 4EBP1 protein plays a regulatory role in selenite-induced SG assembly, possibly by feeding the pool of non-translating mRNAs by disrupting the eIF4F complex.
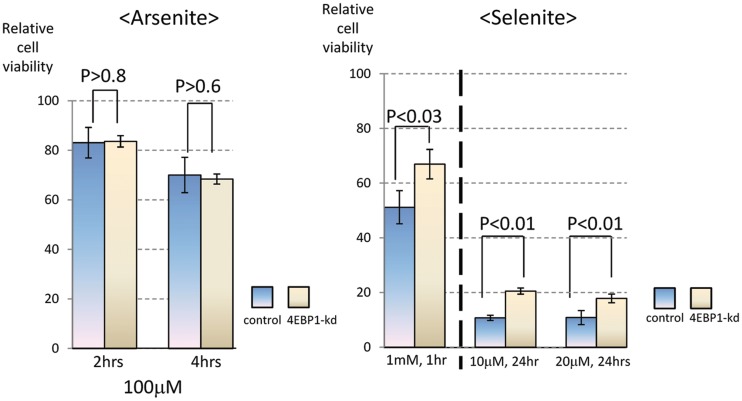
To examine the physiological role of 4EBP1 in the selenite-induced stress response, we compared the viability of U2OS cells treated with 4EBP1-specific cells or control siRNA-transfected cells. Cell viability was assessed by measuring the amount of intracellular ATP. As shown in Figure 6, depletion of 4EBP1 significantly enhanced the viability of selenite-treated cells (right panel). In contrast, 4EBP-1 depletion did not affect the viability of arsenite-treated cells (Figure 6, left panel). These results indicate that 4EBP1 contributes to the cytotoxic effect of selenite, but not arsenite.
Figure 6.
Effect of 4EBP1 knockdown on the viability of stressed cells. U2OS cells transfected with control or 4EBP1 siRNA were treated either with arsenite (at 100 µM for 0, 2 and 4 h; left panel) or selenite (left, at 1 mM for 1 h; right, 10 µM or 20 µM selenite for 24 h; right panel). Subsequently, cell viability was assessed by measuring the amount of intracellular ATP levels. Data represent relative viability expressed as % control relative to non-treated cells.
ROS production by selenite triggers SG formation
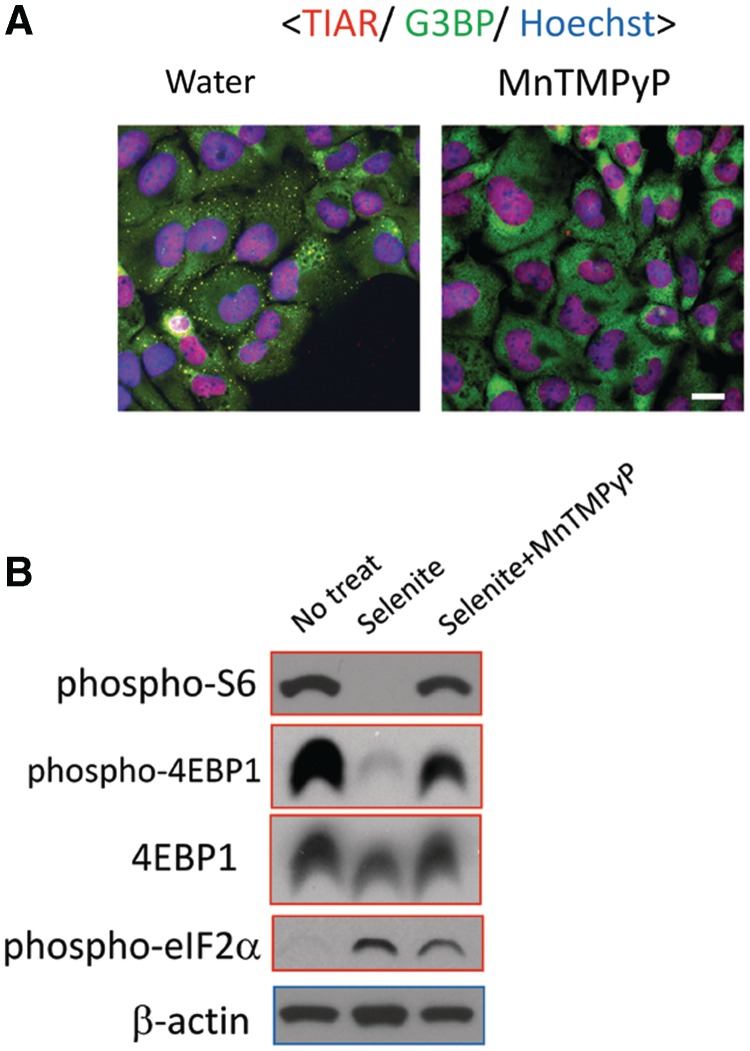
Selenite-induced cytotoxicity is a consequence of ROS that accumulate during selenite metabolism (13,14,33). To test the effect of ROS on SG assembly, cells were pretreated with MnTMPyP, a cell permeable superoxide dismutase mimetic, and then exposed to selenite. As shown in Figure 7A, pretreatment with this ROS scavenger completely suppresses selenite-induced SG assembly, suggesting that ROS production is critical for this phenomenon. Inhibition of ROS generation by MnTMPyP significantly restored the levels of phosphorylated S6 protein and 4EBP1, whereas selenite-induced phosphorylation of eIF2α was only weakly attenuated (Figure 7B). Thus, selenite-induced ROS production is primarily responsible for the suppression of S6- and 4EBP1 phosphorylation, but this is not the case for the induction of eIF2α phosphorylation.
Figure 7.
Selenite-induced SG biogenesis depends on ROS production. (A) ROS scavenger MnTMPyP suppresses selenite-induced SGs. U2OS cells were pre-treated with doubly distilled water or MnTMPyP (10 µM, 1 h), followed by selenite treatment. Subsequently, cells were fixed and stained for TIAR (red) and G3BP (green). The nuclei were revealed by Hoechst staining (blue). Bar = 10 µm. (B) U2OS cells without treatment (No treat), with selenite treatment (selenite) or with selenite and MnTMPyP as above (selenite + MnTMPyP) were lysed and probed for phospho-eIF2α, phospho-S6, 4EBP1, phospho-4EBP1 and β-actin levels.
DISCUSSION
SG assembly is a conserved phenomenon observed in a wide range of eukaryotic species (2). However, species-specific differences in the mechanism of SG assembly, and the composition of SGs, have been noted. In mammalian cells, SGs typically contain eIF4F components (eIF4E, eIF4G and eIF4A), eIF3 and 40 S ribosomal subunits, and their assembly is strongly dependent upon individual eIF3 subunits (19). In the budding yeast Saccharomyces cerevisiae, glucose deprivation induces mRNA-containing cytoplasmic granules that accumulate eIF4E, eIF4G and PABP (EGP-bodies), but not eIF3 or 40 S ribosomal subunits (34,35). Although their lack of eIF3 subunits and 40 S ribosomal subunits and their non-reliance on eIF2α phosphorylation for assembly highlight the differences between yeast EGP-bodies and mammalian SGs, some evidence suggests that EGP-bodies may be functionally analogous to mammalian SGs. Unlike SGs induced by glucose deprivation, yeasts treated with sodium azide assemble SGs that more closely resemble their mammalian counterparts (36), indicating that yeast cells can assemble more than one class of SGs. Here, using sodium selenite as a trigger, we have found that mammalian cells can also assemble more than one class of SGs. The absence or reduced accumulation of ‘later stage’ translation initiation factors, such as eIF3 and eIF5A and the small ribosomal subunit Rps14 in selenite-induced SGs is analogous to EGP-bodies, suggests that selenite stalls messenger RNP remodeling at an earlier step in translation initiation compared with arsenite. Unlike EGP-bodies, however, eIF4A is modestly accumulated in selenite-induced SGs, and O-GlcNAc-modified proteins, which are lacking in S. cerevisiae, are found in selenite-induced SGs (19,37), indicating that selenite-induced SGs are not an exact counterpart of yeast EGP-bodies. Moreover, SGs induced in cells exposed to both selenite and arsenite resemble arsenite-induced SGs with respect to their large size and the recruitment of eIF3b (Supplementary Figure S3). In contrast, yeasts exposed to both glucose deprivation and azide assemble SGs with the glucose deprivation phenotype (i.e. small size, lack of eIF3) (36). In addition, the combined stress resulted in less SG assembly than that observed with selenite or arsenite alone. Although we cannot fully explain these phenomena, there may be a competition between selenite and arsenite regarding a biochemical event upstream of SG formation.
Another interesting aspect of selenite-induced SGs is that their biogenesis appears to involve 4EBP, which has not previously been implicated in SG assembly. Given that the involvement of 4EBPs in EGP-bodies has never been tested, it would be of interest to see if Caf20p and Eap1p, the 4EBP homologues in S. cerevisiae, are involved in EGP-body biogenesis. We also observed selenite-specific loss of phospho-ribosomal S6 protein; considering the increased eIF4E-4EBP1 interaction and apparent dephosphorylation of 4EBP1 (Figure 5A), it seems likely that mTOR activity is down-regulated by selenite, a possible consequence of selenite-induced ROS (Figure 7B). This is in line with a recent report, which monitored mTOR activity through phospho-S6 kinase levels in the context of autophagy (38). Dephosphorylation of neither 4EBP1 nor ribosomal S6 protein was observed by arsenite (Figure 5A), and the superoxide dismutase mimetic MnTMPyP failed to inhibit arsenite-induced SG assembly (Supplementary Figure S4A), in stark contrast to selenite-induced SG assembly (Figure 7A). Thus, although arsenite is a known inducer of oxidative stress, the species of intracellular ROS generated by arsenite and selenite treatment appear to be different; both trigger the phosphorylation of eIF2α, but only the latter diminishes the phosphorylaton of 4EBP1 and S6, possibly by down-regulation of mTOR. The involvement of mTOR may explain the lack of eIF3 accumulation considering a previous study showing the mTOR-stimulated association between eIF4G and eIF3 (39). Meanwhile, in line with several previous studies (40,41), rapamycin did not induce SG assembly in our system (Supplementary Figure S5), indicating that selenite may affect mTOR in a manner distinct from rapamycin. We further attempted to link mTOR to selenite-induced SG assembly, by comparing selenite-induced SG assembly in Tsc2 null MEFs (Tsc2−/−TP53−/−), in which the mTOR activity is constitutively up-regulated (42), and control MEFs (Tsc2+/+TP53−/−) (Supplementary Figure S4). We found that both SG assembly and the loss of phospho-S6 as well as dephosphorylation of 4EBP1 are similar in control and Tsc2-deficient MEFs (Supplementary Figure S4). These results rule out a direct role for Tsc2 in selenite-induced SG assembly. Nevertheless, knock-down experiments clearly implicate 4EBP1, and likely its translational repression activity, in selenite-induced SG assembly. The reason why 4EBP1 knockdown did not completely inhibit SG formation might have been because there are at least two other known 4EBPs (4EBP2 and 4EBP3) that can substitute for 4EBP1. In addition, although eIF2α phosphorylation is not an absolute requirement, it appears to contribute to the proper assembly of selenite-induced SGs (Figure 4C). It may be that phosphorylation of eIF2α provides a pool of translationally repressed mRNPs, from which 4EBP-mediated sequestration of eIF4E from eIF4F complex and subsequent aggregation into discrete foci preferentially occurs. In addition, there was an apparent decrease in total eIF4G (Figure 5A), which may contribute to selenite-triggered translational repression (Figure 4A). Thus, all these elements may collectively contribute to selenite-induced inhibition of protein synthesis and SG assembly (Figure 5A).
An important observation regarding selenite-induced SGs is their lack of accumulation of several canonical SG components. The lack of eIF3 components, HDAC6 and importin β is particularly striking, since these proteins are essential for SG assembly by arsenite (8,19). More intriguingly, among the four canonical SG components whose recruitment to SGs promotes cell survival (Rsk2, RACK1, importin α1 and HDAC6) (4,8,22), three of them (RACK1, importin α1 and HDAC6) failed to accumulate in selenite-induced SGs. These results raise the possibility that selenite-induced SGs may be impaired in their ability to increase cellular resistance to stress. Consistent with this hypothesis, cells that are depleted of 4EBP1 and thus compromised in the ability to assemble selenite-induced SGs exhibited higher cell viability in response to selenite stress (Figure 6). It is also possible, however, that depletion of 4EBP1 promotes cell survival by a mechanism that is unrelated to SG assembly. This limits the interpretation of experiments in which knockdown of multifunctional proteins that are required for SG assembly is performed.
Taken together, our data suggest that mammalian cells assemble two classes of SGs with different components, mechanisms of assembly and physiological roles: Type 1 SGs, induced by arsenite and other types of stress, typically contain eIF3 subunits and small ribosomal proteins, are dependent on eIF2α phosphorylation or eIF4A activity and function to promote cell survival during stress. Type 2 SGs, induced by selenite, lack eIF3 subunits and several other critical Type 1 SG components, rely primarily on 4EBP for assembly and inhibit cellular survival during stress. The possible concurrent involvement of both eIF2α phosphorylation and mTOR down-regulation in selenite-induced SG assembly is somewhat reminiscent of amino acid starvation, where mRNAs bearing 5′-terminal oligopyrimidine tracts (5′TOP) are selectively released from polysomes and sequestered into SGs (43). It will be of great interest to determine whether SGs induced by amino acid starvation are Type 2 SGs, and whether selenite-induced SGs are selectively enriched in 5′TOP mRNAs. Our results also imply that selenite might be a unique anti-cancer drug candidate in that it efficiently represses protein synthesis, which may enable this compound to target cancer cells reliant on aberrantly active translation, without invoking cytoprotective functions of canonical SGs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
National Institutes of Health (NIH) [AI033600 and AI065858 to P.A.]; Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad (to K.F.); JSPS Postdoctoral Fellowship for Research Abroad, Kanae Foundation for Research Abroad and a Genentech Fellowship (to A.T.S.). Funding for open acess charge: NIH.
Conflict of interest statement None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jun Katahira (Osaka University) for kindly providing anti-importin α1 antibody.
REFERENCES
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanism and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson P, Kedersha N. Stress granules: the tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Kim WJ, Back SH, Kim V, Ryu I, Jang SK. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 2005;25:2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK signaling. Nat. Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman I, Casella M, Schnolzer M, Schlechter T, Spring H, Franke WW. Identification of the junctional plaque protein plakophilin 3 in cytoplasmic particles containing RNA binding proteins and the recruitment of plakophilins 1 and 3 to stress granules. Mol. Biol. Cell. 2006;17:1388–1398. doi: 10.1091/mbc.E05-08-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen J-J, Anderson P, Kaufman RJ. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol. Chem. 2005;280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 8.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisinger-Mathason TS, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, et al. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell. 2008;31:722–736. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim B, Cooke HJ, Rhee K. DAZL is essential for stress granule formation implicated in germ cell survival upon heat stress. Development. 2012;139:568–578. doi: 10.1242/dev.075846. [DOI] [PubMed] [Google Scholar]
- 11.Moeller BJ, Cao Y, Liu CY, Dewhirst MW. Radiation activates HIF1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 12.Fournier MJ, Gareau C, Mazroui R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int. 2010;10:12. doi: 10.1186/1475-2867-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selenius M, Rundlof A-K, Olm E, Fernandes AP, Bjornstedt M. Selenium and selenoprotein thioredoxin reductase in the prevention, treatment and diagnostics of cancer. Antioxid. Redox Signaling. 2010;12:867–880. doi: 10.1089/ars.2009.2884. [DOI] [PubMed] [Google Scholar]
- 14.Husbeck B, Nonn L, Peehl DM, Knox SJ. Tumor-selective killing by selenite in patient-matched pairs of normal and malignant prostate cells. Prostate. 2006;66:218–225. doi: 10.1002/pros.20337. [DOI] [PubMed] [Google Scholar]
- 15.Olm E, Fernandes AP, Hebert C, Rundlof A-K, Larsen EH, Danielsson O, Bjornstedt M. Extracellular thiol-assisted selenium uptake dependent on the xc- cystine transporter explains the cancer-specific cytotoxicity of selenite. Proc. Natl. Acad. Sci. USA. 2009;106:11400–11405. doi: 10.1073/pnas.0902204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan L, Han B, Li Z, Hua F, Huang F, Wei W, Yang Y, Xu C. Sodium selenite induces apoptosis by ROS-mediated endoplasmic reticulum stress and mitochondrial dysfunction in human acute promyelocytic leukemia NB4 cells. Apoptosis. 2009;14:218–225. doi: 10.1007/s10495-008-0295-5. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki M, Endo M, Shinohara F, Echigo S, Rikiishi H. Differential apoptotic response of human cancer cells to organoselenium compounds. Cancer Chemother. Pharmacol. 2010;66:475–484. doi: 10.1007/s00280-009-1183-6. [DOI] [PubMed] [Google Scholar]
- 18.Kedersha N, Tisdale S, Hickman T, Anderson P. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 2008;448:521–551. doi: 10.1016/S0076-6879(08)02626-8. [DOI] [PubMed] [Google Scholar]
- 19.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimura K, Katahira J, Kano F, Yoneda Y, Murata M. Microscopic dissection of the process of stress granule assembly. Biochim. Biophys. Acta. 2009;1793:1728–1737. doi: 10.1016/j.bbamcr.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Fujimura K, Suzuki T, Yasuda Y, Murata M, Katahira J, Yoneda Y. Identification of importin α1 as a novel constituent of RNA stress granules. Biochim. Biophys. Acta. 2010;1803:865–871. doi: 10.1016/j.bbamcr.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 23.Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung AKL, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute proteins reveal microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazroui R, Huot M-E, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by fragile mental X retardation into cytoplasmic granules induces translational repression. Hum. Mol. Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 26.Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimura K, Kano F, Murata M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA. 2008;14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C-H, Ohn T, Ivanov P, Tisdale S, Anderson P. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS One. 2010;5:e9942.s. doi: 10.1371/journal.pone.0009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang W-L, Tarn W-Y. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucl. Acids Res. 2009;37:6600–6612. doi: 10.1093/nar/gkp717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 31.Dang Y, Kedersha N, Low W-K, Romo D, Gorospe M, Kaufman RJ, Anderson P, Liu JO. Eukaryotic initiation factor 2 α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 32.Marissen WE, Lloyd RE. Eukaryotic translation initiation factor 4 G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao R, Xiang N, Domann FE, Zhong W. Expression of p53 enhances selenite-induced superoxide production and apoptosis in human prostate cancer cells. Cancer Res. 2006;66:2296–2304. doi: 10.1158/0008-5472.CAN-05-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchan JR, Yoon JH, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 2011;124:228–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci. STKE. 2005;312:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 38.Ren Y, Huang F, Liu Y, Yang Y, Jiang Q, Xu C. Autophagy inhibition through PI3K/Akt increases apoptosis by sodium selenite in NB4 cells. BMB Rep. 2009;42:599–604. doi: 10.5483/bmbrep.2009.42.9.599. [DOI] [PubMed] [Google Scholar]
- 39.Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC., Jr mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J. 2006;25:1659–1668. doi: 10.1038/sj.emboj.7601047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong L, Livingstone M, Sukarieh R, Petroulakis E, Gingras A-C, Crosby K, Smith B, Polakiewicz RD, Pelletier J, Ferraiuolo MA, et al. Control of eIF4E cellular localization by eIF4E-binding proteins, 4 E-BPs. RNA. 2008;14:1318–1327. doi: 10.1261/rna.950608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagner PR, Mazan-Mamczarz K, Dai B, Balzer EM, Corl S, Martin SS, Zhao XF, Gartenhaus RB. Ribosomal protein S6 is highly expressed in non-Hodgkin lymphoma and associates with mRNA containing a 5′ terminal oligopyrimidine tract. Oncogene. 2011;30:1531–1541. doi: 10.1038/onc.2010.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Invest. 2003;112:1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Damgaard CK, Lykke-Andersen J. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25:2057–2068. doi: 10.1101/gad.17355911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.