Abstract
Human monocytic ehrlichiosis is an emerging tick-borne disease caused by the rickettsia Ehrlichia chaffeensis. To examine the role of helper T cells in host resistance to this macrophage-tropic bacterium, we assessed E. chaffeensis infections in three mouse strains with differing functional levels of helper T cells. Wild-type, C57BL/6J mice resolved infections in approximately 2 weeks. Major histocompatibility complex class II (MHCII) knockout, B6.129-Abbtm1 mice lacking helper T cells developed persistent infections that were not resolved even after several months. CD4+ T-cell-deficient, B6.129S6-Cd4tm1Knw mice cleared the infection, but the clearance took 2 weeks longer than it did for wild-type mice. C57BL/6J mice resolved infection more rapidly following a second experimental challenge, but B6.129S6-Cd4tm1Knw mice did not. The B6.129S6-Cd4tm1Knw mice also developed active E. chaffeensis-specific immunoglobulin G responses that were slightly lower in concentration and slower to develop than that observed in C57BL/6J mice. E. chaffeensis-specific cytotoxic T cells were not detected following a single bacterial challenge in any mouse strain, including wild-type C57BL/6J mice. However, the cytotoxic T-cell activity developed in all three mouse strains, including the MHCII and CD4+ T-cell knockouts, when challenged with a second E. chaffeensis infection. The data reported here suggest that the cell-mediated immunity, orchestrated by CD4+ T cells is critical for conferring rapid clearance of E. chaffeensis.
Human monocytic ehrlichiosis, an emerging tick-borne disease with influenza-like symptoms, is caused by the macrophage/monocyte-tropic intracellular rickettsial agent Ehrlichia chaffeensis (11, 19, 30). E. chaffeensis also infects a wide range of vertebrate hosts which include white-tailed deer, dogs, goats, and coyotes (4, 12, 13, 15, 25, 26). Human monocytic ehrlichiosis can cause a severe, potentially fatal illness in immunocompromised and elderly people with symptoms such as prolonged fever, renal failure, respiratory distress, seizures, and coma (33, 34).
Tick-transmitted Rickettsiales members, including E. chaffeensis, persist in their vertebrate hosts for long periods despite the active host immune response (1, 16, 36, 49). The relationship between E. chaffeensis and its targeted host cells, macrophages and monocytes, is critical because contrary to their natural function these cells fail to clear E. chaffeensis, allowing it to establish persistence by undefined evasion mechanisms. Studies of the murine host have been valuable in mapping the contributions of the host response to E. chaffeensis infections (20, 41, 46, 47). Immunocompetent mice clear E. chaffeensis infection within 16 days (20, 46), while the absence of macrophage activation results in a prolonged infection that can last over a month (20, 41). Mice deficient in major histocompatibility complex class II (MHCII) antigens do not clear E. chaffeensis (20). Because the CD4+ T cells do not develop in the absence of MHCII, we tested the hypothesis that the deficiency in CD4+ T cells would impact the course of E. chaffeensis infection. To test the hypothesis, we examined infections in three mouse strains with differing functional levels of helper T cells. We report that CD4+ helper T cells, but not cytotoxic T cells, orchestrate the rapid E. chaffeensis clearance in mice.
MATERIALS AND METHODS
Mice. (i) C57BL/6J (B6) mice.
B6 mice were obtained from the Jackson Laboratory (Bar Harbor, Maine) or from the breeding colony at Kansas State University (KSU). The B6 mouse breeding colony at KSU has been maintained by brother-sister matings for approximately 10 years.
(ii) B6.129-Abbtm1 N5F20 (C2D) mice.
The C2D mouse strain lacks MHCII. It has been backcrossed to the B6 mouse background and has been brother-sister mated for over 20 generations over the last 9 years at KSU. These mice do not express MHCII and lack CD4+ T cells (Fig. 1) due to natural and manipulated mutagenesis (21). We established that these mice are unable to clear E. chaffeensis after infection (20).
FIG. 1.
Distribution of CD4+ and CD8+ T cells in B6, CD4D, and C2D mouse thymuses and spleens. Spleen cells and thymocytes were stained with PE-Cy5-conjugated anti-CD3ɛ, PE-conjugated anti-CD8, and fluorescein isothiocyanate-conjugated anti-CD4 monoclonal antibodies. Cells were gated on CD3-positive cells, and profiles of CD4+ and CD8+ cells are shown for the following tissue samples: B6 thymus and spleen, C2D thymus and spleen, and CD4D thymus and spleen.
(iii) B6.129S6-Cd4tm1Knw (CD4D) mice.
CD4D mice were obtained from the Jackson Laboratory. These mice have a targeted disruption in the CD4 gene. We have confirmed the lack of CD4+ T cells in these mice by flow cytometry (Fig. 1).
All mice used in this study at the time of experimental infection were between 8 and 12 weeks old and were of mixed sexes. They were maintained ad libitum on standard lab chow with autoclaved water. Gene-deficient mice were routinely genotyped as described previously to confirm their genetic backgrounds (22, 48).
E. chaffeensis mouse infections.
The E. chaffeensis Arkansas isolate was cultivated in the canine macrophage cell line DH82 as described previously (9). The mouse infections were performed as we recently reported (20). Mice were sacrificed and evaluated on specific postinfection dates as indicated in Results.
Blood collection.
Mice were anesthetized with halothane by the procedure of Huerkamp (23). Blood was collected from the retro-orbital sinus for plasma as described earlier (20). Plasma samples were assayed for the presence of E. chaffeensis-specific immunoglobulin G (IgG) antibodies.
Collection of peritoneal macrophages for monitoring the E. chaffeensis infection by culture isolation or RT-PCR.
Peritoneal cells containing macrophages were collected aseptically from infected and control mice by peritoneal lavage with 12 ml of ice-cold, sterile phosphate-buffered saline (PBS). Peritoneal exudate cells were used to determine the presence of viable rickettsiae by in vitro culture assay as described in reference 20 and by reverse transcription-PCR (RT-PCR) targeted to amplify an E. chaffeensis small-subunit rRNA segment (described below). Detection of viable rickettsiae by culture was monitored for 6 weeks but usually required only a 2-week incubation period. Culture-positive samples were verified by RT-PCR with the total RNA isolated from the cultured organisms.
RNA isolation and E. chaffeensis rRNA gene-specific RT-PCR.
Total RNA from peritoneal wash cells and spleen tissue samples was extracted with RNAwiz (Ambion Inc., Austin, Tex.). RT-PCR was performed using ∼1 μg of RNA and an E. chaffeensis rRNA gene-specific primer pair (E. chaffeensis species-specific forward primer RRG3, 5′CAATTGCTTATAACCTTTTGGTTATAAAT, and Ehrlichia genus-specific reverse primer RRG27, 5′GTATTACCGCGGCTGCTGGCAC). The primers were designed based on the published sequence available in GenBank (GenBank accession no. U60476). Amplicons of 0.43 kb were identified by hybridization with an rRNA gene-specific probe. The hybridization step was included to rule out false positives resulting from nonspecifically amplified, predicted-size products. RNA isolation and RT-PCR setup were performed in an RNA isolation laboratory, while RT-PCRs and the product analyses were done in a separate PCR analysis laboratory. RT-PCR mixtures were prepared in a Clean Spot PCR UV workstation (Coy Laboratory Products, Grass Lake, Mich.). A master mix containing all RT-PCR ingredients (Promega, Madison, Wis.) except the polymerases and template was prepared, divided into multiple aliquots, and stored at −20°C for use in the assays. The assays were performed after adding template and polymerases to freshly thawed aliquots. All RT-PCR assays included a negative control containing RNA template without the reverse transcriptase added and a positive control containing culture-derived E. chaffeensis genomic DNA. RT-PCR cycles were performed in a GenAmp9700 instrument (Applied Biosystems, Foster City, Calif.) with the following temperature cycles: 42°C for 5 min, 48°C for 1 h, and 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min and 1 cycle of 72°C for 3 min. The amplified products were used to detect rRNA gene-specific products as described earlier (20).
C2D macrophage cell line.
The C2D macrophage cell line was created by our group (3). The differentiated cells have macrophage properties (3, 22) and the MHCII−/− and TLR4Lps−n genotype. C2D cells were passaged three times weekly in Dulbecco's modified Eagle's medium (Atlanta Biologicals, Lawrenceville, Ga.) supplemented with 5% fetal bovine serum, 2% Nu Serum 1 (Becton Dickinson Collaborative Biochemicals, Bedford, Mass.), and 0.3% l-glutamine (DMEM5).
C2D cells are permissive for the growth of E. chaffeensis (unpublished data) and could be used to assess cytotoxic T-lymphocyte (CTL) activity. To infect cells, E. chaffeensis-infected DH82 cells were dispersed with glass beads by being rocked for several minutes at 37°C. The suspension was centrifuged at 10,000 × g, and the pellet was resuspended in 3 ml of DMEM5. C2D cells were grown to 60 to 70% confluence in 100-mm-diameter tissue culture dishes. Medium was aspirated off, and the 1-ml bacterial suspension was added to the cells. The plates were rocked for 1 h at 37°C. Afterward, 5 ml of DMEM5 was added, and cells were incubated at 37°C.
Histopathology.
Liver samples were collected from infected mice and were placed in formalin. Formalin-fixed samples were processed, embedded in paraffin, cut at 5-μm thickness, and stained with hematoxylin and eosin. Two cross-sectional areas of liver obtained from each mouse were evaluated for inflammatory foci, and the average of the two slides was used to assign a score for the inflammatory lesion: 0, no inflammatory foci; 1, ≤5 neutrophil foci associated with apoptotic hepatocytes; 2, 5 to 15 neutrophil foci associated with apoptotic cells; 3, 15 to 30 neutrophil foci associated with apoptotic hepatocytes; 4, >30 neutrophil foci associated with apoptotic cells; 5, <15 granulomas (defined as epithelioid macrophages surrounded by mononuclear cells and a few neutrophils); 6, >15 granulomas; and 7, >30 granulomas.
Western blot analysis.
Plasma from E. chaffeensis-infected mice at a 1:64 dilution was assayed for the presence of E. chaffeensis-specific antibody as described earlier (20), except that the antigen for the assay was a whole-cell protein extract (3 μg/well) of purified E. chaffeensis Arkansas isolate and the antigens were resolved on a sodium dodecyl sulfate-12% polyacrylamide gel.
Quantitative ELISA to measure IgG subclasses.
Quantitative enzyme-linked immunosorbent assays (ELISAs) were performed to measure the concentrations of E. chaffeensis-specific IgG subclass antibodies by following the protocol we described previously (20) but using whole-cell antigen as described above (20 ng/well).
Cytotoxicity assays. (i) Chromium release assay.
Infected and uninfected C2D cells were refed with 5 ml of fresh DMEM5 containing 100 to 150 μCi of Na251CrO4 and incubated for 12 to 16 h at 37°C (6). Target cells were dispersed with 0.25% trypsin and 0.02% EDTA, resuspended in DMEM2 (2% serum), and pelleted at 300 × g. Cells were resuspended in DMEM2 and counted using a hemacytometer. Target cells (104) were added to round-bottomed, 96-well plates along with various numbers of spleen cells to give effector/target cell (E/T) ratios ranging from 100:1 to 3:1.
Spleens were removed from euthanized mice, and erythrocytes were removed from spleen cell suspensions by resuspending the cells in 5 ml of lysing solution (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA) for 5 min. Thereafter, 10 ml of DMEM2 was added and cells were pelleted at 300 × g. Spleen cells were resuspended in DMEM2, counted, and seeded into wells of 96-well round-bottomed plates along with targets in a final volume of 250 μl. The assay mixture was incubated for 18 h, and 110 μl of supernatant was counted in a gamma counter. The percent specific release was calculated as follows: % specific release = ([experimental release − spontaneous release]/[maximum release − spontaneous release]) × 100. The maximum and spontaneous release values were determined by incubating 104 target cells in 1 N HCl or DMEM2 medium, respectively. Spontaneous release for C2D cells was less than 40%. Spontaneous release for E. chaffeensis-infected C2D cells ranged from 35 to 55%.
(ii) MTT assay.
Target cells were prepared as described above without prelabeling with Na251CrO4. Target cells (105) were added to round-bottomed, 96-well plates with or without 106 spleen cells to give an E/T ratio of 10:1. Spleen cells were incubated with or without targets for 48 h in a final volume of 200 μl. Cytotoxicity was determined spectrophotometrically (A570) by determining viable cell conversion of 1-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) to formazin. At the end of the 48-h incubation, 50 μl (2 mg/ml) of MTT was added to each assay well for 3 h at 37°C. The medium was removed, and 150 μl of iso-PBS (67% isopropyl alcohol, 33% PBS, and 0.03% 5 N HCl) was added to dissolve crystals. Plates were scanned at 570 nm on a Packard Spectracount microtiter plate spectrophotometer (Packard Instruments, Meriden, Conn.). Cytotoxicity was determined using the following formula: % cytotoxicity = 100 − (MTT conversion by spleen cells + targets/MTT conversion by targets alone) × 100. The MTT assay offers advantages over the 51Cr-release assay in that it eliminates the worry about high spontaneous release values and does not require as many effector cells. However, we realize that the MTT assay does not distinguish between killing and stasis, and so it is used in this study to confirm killing for the 51Cr-release assay. We also accounted for target cell growth in the assay so that the E/T ratios were not skewed over the course of the experiment.
Flow cytometry.
Mouse thymuses and spleens were removed from B6, CD4D, and C2D mice; homogenized; and washed in PBS. Erythrocytes were removed from spleen cell suspensions as described above. Thymocytes and spleen cells were resuspended in DMEM2 and counted, and 106 cells were incubated for 30 min in wells of 96-well plates in 200 μl of goat serum diluted 1:2 in Hanks buffered salt solution (HBSS). Cells were washed in HBSS and stained with 20 μl of mouse-specific, anti-CD3-phycoerythrin (PE)-Cy5.5 or CD4-fluorescein isothiocyanate or CD8-PE antibodies (sc-3970; Santa Cruz Biotechnology, Inc.). Cells were incubated for 1 h, washed two times in HBSS, and fixed with 250 μl of PBS containing 1% formalin. Fluorescence intensity was measured on a FACSCalibur analytical flow cytometer (Becton Dickinson, San Jose, Calif.).
Statistical analysis.
All data presented are representative of multiple, reproducible experiments. Nonparametric Mann-Whitney tests were done using the StatMost statistical software package (Dataxiom, Los Angeles, Calif.). Histopathology lesion scores were compared among three groups of mice using Kruskal-Wallis one-way analysis of variance on ranks.
RESULTS
Evaluation of E. chaffeensis infections by in vitro culture and RT-PCR methods.
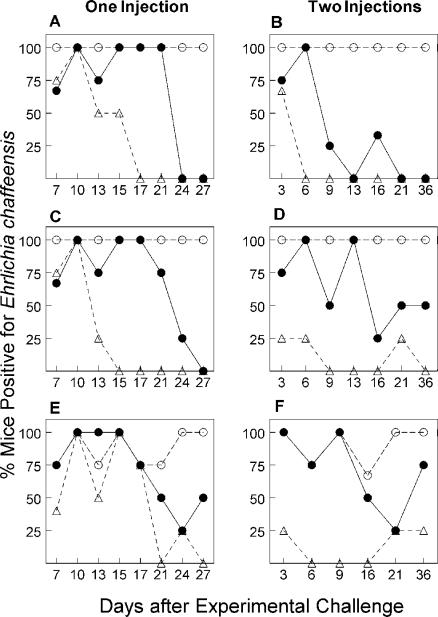
E. chaffeensis persists in mice lacking MHCII genes but is cleared by wild-type, B6 mice in approximately in 2 weeks (20). The kinetics of bacterial clearance suggested that the cell-mediated immune response might be important to resolving the infection. To test the hypothesis that helper T cells are important for host resistance, we evaluated the clearance of E. chaffeensis in three different mouse strains with different levels of T-helper cell function. Single and dual experimental challenges were carried out in B6, C2D, and CD4D mice (Fig. 2). B6 mice have functional CD4+ helper T cells. CD4D mice carry an expanded CD4− CD8− helper T-cell population compared to B6 mice but lack CD4+ helper T cells (Fig. 1) (35). C2D mice lack MHCII and CD4+ helper T cells (Fig. 1). In a preliminary experiment, after a single experimental E. chaffeensis challenge, C2D mice remained infected for the entire study period (71 days) while 100% of the B6 mice cleared E. chaffeensis by 16 days. The CD4D mice also resolved the infection, but clearance took between 24 and 50 days (data not shown). A more detailed examination of the clearance kinetics in the three mouse strains revealed a 100% clearance in the peritoneum of CD4D mice between 24 and 27 days, as judged by culture and RT-PCR methods, respectively (Fig. 2).
FIG. 2.
Comparison of clearance of E. chaffeensis by B6, CD4D, and C2D mice after single and dual E. chaffeensis challenges. The figure shows detection of E. chaffeensis in peritoneal exudate cells (A to D) or spleen (E and F) by RT-PCR (C to F) or culture techniques (A and B) after a single (A, C, and E) or secondary (B, D, and F) experimental challenge. Numbers represent percent mice positive for B6 (▵), C2D (○), or CD4D (•) mice, four to six mice per time point.
Because bacterial growth is associated with active transcription and translation, we compared the E. chaffeensis detection by rRNA-based RT-PCR and in vitro culture methods. We discovered that the RT-PCR assay is as sensitive as and perhaps more sensitive than the culture method because it detected the presence of E. chaffeensis more often (Fig. 2). Interestingly, we found that both B6 and CD4D mice took longer to clear the organisms from the spleen than from the peritoneum after a single challenge (Fig. 2). About 8% of the spleens from B6 mice were bacteria positive when examined from days 21 to 27 compared to 42% of the CD4D mouse spleens. No organisms were detected in the peritoneum of B6 mice at comparable times whereas 25% of the CD4D mouse peritoneal exudates were positive. Regardless of the technique or the site examined, B6 mice cleared bacteria faster than CD4D mice did and C2D mice had a persistent rickettsemia.
In preliminary experiments attempting to generate CTLs, we found that splenic CTLs could be detected only in wild-type B6 mice after a second injection of bacteria. Since one of the hallmarks of cell-mediated immunity is enhanced host resistance after multiple pathogen challenges, we monitored bacterial clearance after mice were infected with E. chaffeensis and rechallenged 10 days later. The C2D mice remained E. chaffeensis positive after the second infection, whereas 50% of the B6 mice cleared the infection by day 3 and greater than 90% were cured by day 9 (Fig. 2). In contrast, the CD4D mice did not have a significant change in the rate of ehrlichial clearance after the second injection. As judged by RT-PCR, E. chaffeensis was detected in 42% of the CD4D mice in the peritoneum (5 of 12 mice) after 16 days and beyond the second injection. These data are similar to the clearance of bacteria after a single injection (Fig. 2).
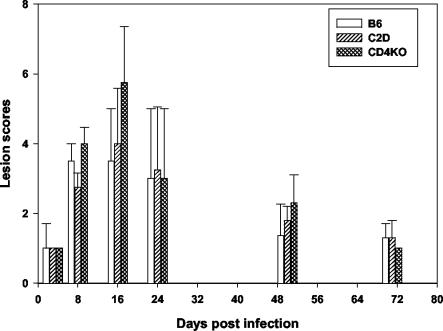
Evaluation of the inflammatory response determined by histopathological analysis of sections of the livers of infected B6, CD4D, and C2D mice following a single injection revealed lesions with scores ranging from 3 to 4 for samples analyzed on day 8 after infection (Fig. 3). The lesions were consistent with our previous observations of the B6, C2D, and other mouse strains (20). Lesion scores by day 8 and day 16 for all mouse strains were significantly greater than that at day 3 (P = 0.04). Although the lesion scores were not statistically different among the mouse groups, granulomatous inflammation was more extensive in CD4D mice at 16 days postinfection than in C2D and B6 mice in this study and in our earlier study (20). Three out of four CD4D mice had lesion scores of 6 to 7, whereas only two of four C2D mice had scores of 5 to 6. Granulomas were noted in the spleen sections of two CD4D mice. Lesions peaked on day 16 postinfection in all three mouse strains and resolved thereafter (Fig. 3).
FIG. 3.
Liver lesion severity in mice infected with E. chaffeensis. Lesions were evaluated at various time intervals and assigned lesion scores (described in Materials and Methods). Data are presented as the mean scores. Error bars represent standard deviations. Analysis included four mice per each postinfection date.
Generation of cytotoxic T cells after E. chaffeensis infection.
To evaluate the contributions of CTLs to the resolution of E. chaffeensis infection, we used the C2D macrophage cell line (3, 22). C2D macrophages are excellent targets because they are MHCI positive and histocompatible (H-2b) with B6 mice and are of macrophage origin. Moreover, we were able to infect them with E. chaffeensis. The data in Table 1 show that uninfected B6 mouse spleen cells do not lyse E. chaffeensis-infected C2D cultures any more effectively than do uninfected targets. B6 mice assayed after a single E. chaffeensis challenge also did not have significant E. chaffeensis-specific cytolytic activity (Table 1). Assessments done as early as 3 days or as late as 19 days after a single ehrlichia injection also did not show cytotoxicity (data not shown). Spleen cells from B6 mice injected with E. chaffeensis twice, however, had significant cytolytic activity (Table 1). Cytotoxicity of infected targets occurred in a dose-dependent manner and was significantly higher than lysis mediated by spleen cells from uninfected or once-infected B6 mice.
TABLE 1.
CTL activity by B6 spleen cells against E. chaffeensis-infected target cells
| E/T ratio | % Specific 51Cr release by spleen cellsa
|
|||||
|---|---|---|---|---|---|---|
| Uninfected spleen cells
|
One immunization
|
Two immunizations
|
||||
| C2D | C2DE | C2D | C2DE | C2D | C2DE | |
| 1.5:1 | 14 ± 1 | 10 ± 2 | 11 ± 3 | 12 ± 4 | 15 ± 1 | 10 ± 4 |
| 3:1 | 17 ± 1 | 17 ± 2 | 14 ± 2 | 13 ± 6 | 14 ± 3 | 6 ± 1 |
| 6:1 | 16 ± 2 | 18 ± 2 | 16 ± 1 | 4 ± 4 | 17 ± 2 | 23 ± 4 |
| 12.5:1 | 11 ± 2 | 15 ± 4 | 16 ± 1 | 12 ± 1 | 13 ± 4 | 38 ± 3 |
| 25:1 | 11 ± 1 | 0 ± 3 | 15 ± 3 | 9 ± 3 | 11 ± 6 | 39 ± 5 |
| 50:1 | 16 ± 1 | 26 ± 8 | 25 ± 4 | 9 ± 3 | 20 ± 3 | 57 ± 5 |
| 100:1 | 22 ± 3 | 21 ± 7 | 31 ± 2 | 9 ± 2 | 27 ± 2 | 76 ± 4 |
Percent specific 51Cr release of target cells was measured in an 18-h assay. C2D macrophages (infected or uninfected) used as targets were prelabeled with 150 μCi of Na251CrO4. Spleen cells were from unimmunized B6 mice (healthy) or mice immunized with E. chaffeensis intraperitoneally (two mice per group). Mice were assayed 7 days following the final E. chaffeensis injection. C2D cells were infected with E. chaffeensis 72 h (C2DE) before the assay or were left uninfected. C2D spontaneous release was 39% ± 3%; C2DE spontaneous release was 54% ± 4%. C2DE cells were >25% infected as determined by inclusion of morulae by light microscopy. Values represent means ± standard errors of the means of triplicate samples. The experiment is representative of more than three experiments performed independently.
CTLs can be generated against macrophage-tropic organisms, such as listerias, in the absence of CD4+ T cells (40). Therefore, we examined the CTL response against E. chaffeensis by C2D and CD4D spleen cells after one and two injections. CTL activity in C2D and CD4D mice was minimal after a single injection, which is similar to B6 mice (Fig. 4). When mice were given a second E. chaffeensis challenge, the B6 mice developed an antigen-specific CTL response 3 days later. Cytotoxicity stayed high at 6 days and declined to baseline levels by 13 days. In contrast, C2D and CD4D mice also developed CTL activity, but it peaked by day 6 to levels comparable to those for B6 mouse CTLs (Fig. 4).
FIG. 4.
Kinetics of a secondary CTL response. The figure shows the CTL response of B6 (top), CD4D (middle), and C2D (bottom) mice against uninfected (left) and E. chaffeensis-infected (right) targets at 3, 6, 8 (B6 mice only), and 13 days after a secondary experimental E. chaffeensis challenge. Numbers represent the mean CTL responses of cells from four mice per time point assayed independently, except for the 8-day B6 data, which were from two mice.
To confirm that cytotoxicity was not an artifact of the higher-than-optimal spontaneous 51Cr-release values in E. chaffeensis-infected C2D macrophage targets, we also assessed CTL activity by using a nonisotopic measurement (MTT) to determine target cell viability. Preliminary experiments established maximal sensitivity of the assay by using E/T ratios of 10:1 with spleen cells being incubated with targets for 48 h. Spleen cells from uninfected B6 and C2D mice were not cytotoxic to either infected or uninfected C2D cells. In contrast, spleen cells from the secondary E. chaffeensis-injected mice killed infected C2D cells (Table 2). Cytotoxicity was comparable to cytotoxicity assessed by the 51Cr-release assay (Table 2).
TABLE 2.
CTL activity against E. chaffeensis-infected cellsc
| Effector cell sourced | % Cytotoxicitya (MTT assay; E/T ratio = 10:1)
|
% Specific 51Cr release at E/T ratioe:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 25:1
|
50:1
|
100:1
|
||||||
| C2D | C2DEb | C2D | C2DE | C2D | C2DE | C2D | C2DE | |
| B6 mouse | ||||||||
| Uninfected | 9 ± 8 | 15 ± 5 | 7 ± 3 | 2 ± 1 | 9 ± 2 | 6 ± 2 | 13 ± 4 | 15 ± 3 |
| Immune | 2 ± 5 | 50 ± 5 | 9 ± 3 | 13 ± 6 | 12 ± 3 | 22 ± 4 | 14 ± 5 | 46 ± 8 |
| C2D mouse | ||||||||
| Uninfected | 4 ± 7 | 4 ± 3 | 6 ± 5 | 11 ± 3 | 10 ± 6 | 10 ± 4 | 9 ± 5 | 13 ± 3 |
| Immune | −9 ± 12 | 42 ± 3 | 9 ± 5 | 31 ± 12 | 9 ± 4 | 33 ± 6 | 5 ± 3 | 53 ± 6 |
Spleen cells (106) were incubated with 105 target cells for 48 h. Cytotoxicity was determined spectrophotometrically (A570) by MTT conversion to formazin.
C2D cells were infected with E. chaffeensis 72 h before the assay.
Values represent means ± standard errors of the means of CTL activity of spleen cells from more than five mice assayed independently.
Spleen cells were from unimmunized B6 or C2D mice (healthy) or mice immunized with E. chaffeensis intraperitoneally two times (immune). Spleen cells were assayed 6 days after the second E. chaffeensis injection.
Specific 51Cr release of C2D target cells was measured in an 18-h assay. C2D macrophages used as targets were prelabeled with 150 μCi of Na251CrO4.
IgG response against E. chaffeensis.
Because CD4D mice resolved E. chaffeensis infection more slowly than did B6 mice, we assessed E. chaffeensis-specific IgG responses by quantitative ELISA following one and two injections of bacteria (Fig. 5). The analysis revealed the predominant expression (>95%) of complement-fixing IgG molecules IgG2a, -2b, and -3 in both B6 and CD4D mice. Moreover, IgG2b was the predominant isotype made among those IgG molecules. We found mouse-to-mouse variations in the concentrations of antibodies made by B6 and CD4D mice. Western blot analysis for total IgG response, described below, showed similar mouse-to-mouse variations (Fig. 6). Although the CD4D mouse antibody response appeared to be lower in quantity and slightly delayed compared to the response observed in the B6 mice, statistical analysis of the data showed marginally significant differences in total IgG and IgG2b at days 13 and 21 (P = 0.08, Mann-Whitney test) following a second bacterial challenge.
FIG. 5.
IgG subclass distribution in B6 and CD4D mice for E. chaffeensis infection. Plasma samples from all postinfection day mice were analyzed by quantitative ELISA using purified whole-cell antigen. The data are presented by day postinfection and are pooled from four mice each from all postinfection dates. Each bar represents the median IgG concentration determined from samples analyzed.
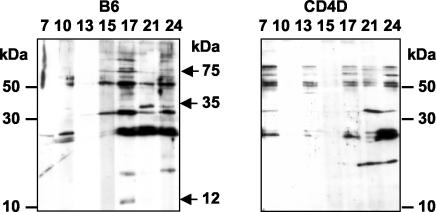
FIG. 6.
Western blot profile showing response to E. chaffeensis whole-cell antigen. Representative data for each postinfection date following a single bacterial injection are presented. Plasma samples were analyzed 7, 10, 13, 15, 17, 21, and 24 days postinfection.
Interestingly, when E. chaffeensis-specific antibodies were examined for specificity by Western blotting of whole-cell antigen (Fig. 6), B6 mice, but not CD4D mice, generated antibodies against an antigen of approximately 35 kDa on days 21 and 24 postinfection. Similarly, B6 mice were unique in their response to 12- and 75-kDa antigens in the antisera on day 17 after infection. Therefore, although the quantities of IgG antibodies may not have appeared to differ between B6 and CD4D mice, there may have been unique specificities generated in B6 mice that also differed over time after infection.
DISCUSSION
We evaluated the clearance of E. chaffeensis in three mouse strains with different levels of helper T-cell function to define the role of helper T cells in host resistance. We confirmed previous work showing that immunocompetent mice resolve a single experimental E. chaffeensis infection in about 2 weeks (20, 46). This study presents a more comprehensive set of data on the kinetics of host clearance in the period of 10 to 21 days and compares bacterial clearance in the spleen to that in the peritoneum. Resolution of the infection often occurred faster in the peritoneum than in the spleen, but the trends were similar and measurement of infection could be done with reasonable accuracy in both sites. We also compared two different methods of detecting E. chaffeensis and concluded that measuring the presence of ehrlichial small-subunit rRNA equals or betters the sensitivity of the tissue culture method, which requires long-term maintenance of cell cultures. Together, the methods facilitate tracking of infections. We previously reported that E. chaffeensis detection by in vitro culture was more sensitive than was the rRNA gene-based PCR assay performed on genomic DNA (20). The higher sensitivity of the RT-PCR assay than of the PCR assay observed here is consistent with a recent report by Felek et al. (17).
The data suggest a role for CD4+ T cells in the clearance of E. chaffeensis and are consistent with studies of other Rickettsiales members. For example, Mwangi et al. (32), working with bovines, found that Ehrlichia ruminantium (formerly known as Cowdria ruminantium) major outer membrane antigen protein MAP1-specific CD4+ T cells produced gamma interferon in response to infection. The presence of CD4+ T cells in β2M knockout mice that lacked MHCI molecules resulted in increased survival after an experimental E. ruminantium challenge (7). Similarly, Brown et al. (5) demonstrated an outer membrane antigen MSP2-specific CD4+ T-lymphocyte recognition of immunodominant epitopes of Anaplasma marginale. We correlated the rapid resolution of infection with the presence of CD4+ T cells. B6 mice have CD4+ T cells, and they clear infections in a manner temporally consistent with the activation of cell-mediated immunity. In contrast, C2D mice completely lack MHCII and helper T cells and do not resolve infections. MHCII knockout mice generally remain bacteria positive regardless of how long after infection we screen them, which has included determinations at more than 120 days postinfection (our unpublished results). The CD4D mice provided the most insightful data in this study. The CD4D mice do not have CD4+ T cells, and they take approximately 2 additional weeks to clear the organisms compared to B6 mice. Their late resistance may be due to the help provided by the CD4− CD8− helper T-cell population that develops in these mice (35). These helper cells have been previously associated with viral resistance in CD4D mice (35). Similarly, the involvement of the CD4− CD8− T cells in the control of Francisella tularensis growth has been reported recently (10). We have confirmed the previous observation that the CD4D mice used in this study have approximately twice the numbers of CD3+ CD4− CD8− thymocytes that B6 and C2D mice do (Fig. 1). Therefore, activation of helper cells, regardless of their phenotype, allows for the development of host immunity to E. chaffeensis.
Faster clearance following a second experimental injection 10 days after the initial exposure also correlated with the presence of CD4+ T cells. Most B6 mice were cured by 6 days after a second E. chaffeensis injection, whereas as many CD4D mice were infected 4 weeks after the second challenge as were infected at similar times following just one injection. Although the short period between the two injections in our study does not allow us to say conclusively that the B6 mice developed memory immunity to E. chaffeensis, the presence of CD4+ T cells clearly allowed for faster clearance, which could be attributable to the expansion of antigen-specific precursors, some of which may have been memory cells. This premise is consistent with the commitment of antigen-specific T cells within the first 24 h of infection (31), the observation that primary effectors undergo apoptosis by 14 days after infection by other macrophage-tropic bacteria (14), and observations that CD4+ T cells are necessary for the generation of memory (8, 24, 38, 40). The quicker production of the E. chaffeensis-specific IgG response after the second E. chaffeensis injection also supports the hypothesis that some memory was generated by day 10. We propose that the E. chaffeensis infection model will be a valuable tool to dissect the generation of memory in B6 and CD4D mice in future experiments.
The activation of CTLs often occurs in response to infection by intracellular bacteria (2, 18, 39, 45). Our data support the previous observations that CTLs can be generated against Rickettsia-infected cells (44). However, we made three observations that support the hypothesis that the CTLs do not play a major role in host resistance to E. chaffeensis. (i) B6 mice clear a single exposure to the bacteria in approximately 2 weeks, during which time very little, if any, splenic CTL activity can be detected. (ii) C2D mice do not resolve E. chaffeensis infections; however, when challenged with the organisms a second time, they do generate CTL activity, indicating that the clearance is not impacted by the presence of the CTL. (iii) There is not a substantial difference in the time it takes to clear an infection in CD4D mice after one or two injections of bacteria, even in the presence of a strong CTL response at 6 days after a second experimental challenge. Our observations are also consistent with the data presented by Byrom et al. (7). They found that β2M knockout mice, deficient for CD8+ T cells, were less susceptible to E. ruminantium infection than were healthy or MHCII knockout mice and suggested that the presence of CTLs may be detrimental in the face of infection.
It is possible that CTLs play a role in resistance that is not reflected in cytotoxic activity (42). Totte et al. (43) found that CD8+ T cells were activated in response to E. ruminantium antigens to produce macrophage inhibitory factor. Alternatively, the antigen specificity of the CTLs generated in C2D mice may differ from CTL specificity in B6 mice, which impacts the effectiveness of the CTLs, or we may have not assessed CTLs in the right host lymphoid organ or compartment (27, 37). We have measured only splenic CTL activity, and it may be that CTLs at other sites may be the critical components. It may be, however, that CTLs augment resistance but are not essential to host resistance.
The E. chaffeensis-specific IgG (total and isotype) response in CD4D mice tended to be lower and slower than that in B6 mice. However, the differences were only marginally statistically significant at days 13 and 21 after the secondary infection (P = 0.08), mainly because of a wide variation of IgG responses in the mice. The diminished IgG response trend in CD4D mice would be similar to previous observations on the antibody response following a viral infection (35). Nevertheless, the antibody response did not correlate with clearance. For example, on days 13 and 15 after a single infection, both B6 and CD4D mice made similar amounts of E. chaffeensis-specific antibody. However, the majority of the B6 mice cleared infection around this time, while the CD4D mice remained infected, suggesting that humoral immunity may not be essential for resolving infections. This would be consistent with the observation that adoptive transfer of E. chaffeensis-specific antibody could prevent the death of E. chaffeensis-infected SCID mice but is unable to mediate clearance (29, 47), probably because it removes cell-free bacteria but is ineffective in attacking intracellular organisms that continue to replicate (28). B6 mice generated specific antibodies against E. chaffeensis antigens that CD4D mice did not. Therefore, it may be possible that very specific antibodies are generated in the presence of CD4+ T cells that contribute to host resistance. Moreover, our data do not preclude a role for antibody in critical functions such as phagocytosis, antibody-dependent cellular cytotoxicity, or complement activation. Additional experiments will be needed to determine the importance of these processes during E. chaffeensis infection.
The lack of an IgG antibody response in MHCII knockout mice is consistent with our previous work (20) and that with E. ruminantium (7) and the concept that helper T cells are required for isotype switching (21). Moreover, it is not surprising that helper T cells not only activate macrophages to mediate bacterial clearance (20) but also contribute to the more robust antibody responses seen in the wild-type mice after a second injection of the bacteria in our study.
In conclusion, the results from this study support the hypothesis that CD4+ T cells impact the course of E. chaffeensis infection. However, CD4+ T-cell help can be complemented by other types of helper T cells, because CD4 knockout mice also can cure an infection. Therefore, CD4+ T cells are not absolutely necessary for E. chaffeensis resistance, but the most effective host response is accomplished only with the CD4+ T cells.
Acknowledgments
This study was supported by the National Institutes of Health grants AI50785, AI55052, RR17686, and RR16475; NASA grant NAG2-1274; and KSU Agricultural Experimental Station Animal Health funds section 1433 grant 4-81321.
Editor: J. T. Barbieri
Footnotes
Kansas Agricultural Experiment Station publication no. 04-021-J.
REFERENCES
- 1.Andrew, H. R., and R. A. Norval. 1989. The carrier status of sheep, cattle and African buffalo recovered from heartwater. Vet. Parasitol. 34:261-266. [DOI] [PubMed] [Google Scholar]
- 2.Barry, M., and R. C. Bleackley. 2002. Cytotoxic T lymphocytes: all roads lead to death. Nat. Rev. Immunol. 2:401-409. [DOI] [PubMed] [Google Scholar]
- 3.Beharka, A. A., J. W. Armstrong, and S. K. Chapes. 1998. Macrophage cell lines derived from major histocompatibility complex II-negative mice. In Vitro Cell Dev. Biol. Anim. 34:499-507. [DOI] [PubMed] [Google Scholar]
- 4.Breitschwerdt, E. B., B. C. Hegarty, and S. I. Hancock. 1998. Sequential evaluation of dogs naturally infected with Ehrlichia canis, Ehrlichia chaffeensis, Ehrlichia equi, Ehrlichia ewingii, or Bartonella vinsonii. J. Clin. Microbiol. 36:2645-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, W. C., T. C. McGuire, W. Mwangi, K. A. Kegerreis, H. Macmillan, H. A. Lewin, and G. H. Palmer. 2002. Major histocompatibility complex class II DR-restricted memory CD4+ T lymphocytes recognize conserved immunodominant epitopes of Anaplasma marginale major surface protein 1a. Infect. Immun. 70:5521-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunner, K. T., J. Mauel, J. C. Cerottini, and B. Chapuis. 1968. Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology 14:181-196. [PMC free article] [PubMed] [Google Scholar]
- 7.Byrom, B., A. F. Barbet, M. Obwolo, and S. M. Mahan. 2000. CD8+ T cell knockout mice are less susceptible to Cowdria ruminantium infection than athymic, CD4+ T cell knockout, and normal C57BL/6 mice. Vet. Parasitol. 93:159-172. [DOI] [PubMed] [Google Scholar]
- 8.Casciotti, L., K. H. Ely, M. E. Williams, and I. A. Khan. 2002. CD8+-T-cell immunity against Toxoplasma gondii can be induced but not maintained in mice lacking conventional CD4+ T cells. Infect. Immun. 70:434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, S. M., V. L. Popov, H. M. Feng, and D. H. Walker. 1996. Analysis and ultrastructural localization of Ehrlichia chaffeensis proteins with monoclonal antibodies. Am. J. Trop. Med. Hyg. 54:405-412. [DOI] [PubMed] [Google Scholar]
- 10.Cowley, S. C., and K. L. Elkins. 2003. Multiple T cell subsets control Francisella tularensis LVS intracellular growth without stimulation through macrophage interferon gamma receptors. J. Exp. Med. 198:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson, J. E., B. E. Anderson, D. B. Fishbein, C. Y. Sanchez, C. Y. Goldsmith, K. H. Wilson, and C. W. Duntley. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson, J. E., K. L. Biggie, C. K. Warner, K. Cookson, S. Jenkins, J. F. Levine, and J. G. Olson. 1996. Polymerase chain reaction evidence of Ehrlichia chaffeensis, an etiologic agent of human ehrlichiosis, in dogs from southeast Virginia. Am. J. Vet. Res. 57:1175-1179. [PubMed] [Google Scholar]
- 13.Dawson, J. E., J. E. Childs, K. L. Biggie, C. Moore, D. Stallknecht, J. Shaddock, J. Bouseman, E. Hofmeister, and J. G. Olson. 1994. White-tailed deer as a potential reservoir of Ehrlichia spp. J. Wildl. Dis. 30:162-168. [DOI] [PubMed] [Google Scholar]
- 14.D'Orazio, S. E., D. G. Halme, H. L. Ploegh, and M. N. Starnbach. 2003. Class Ia MHC-deficient BALB/c mice generate CD8+ T cell-mediated protective immunity against Listeria monocytogenes infection. J. Immunol. 171:291-298. [DOI] [PubMed] [Google Scholar]
- 15.Dugan, V. G., S. E. Little, D. E. Stallknecht, and A. D. Beall. 2000. Natural infection of domestic goats with Ehrlichia chaffeensis. J. Clin. Microbiol. 38:448-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumler, J. S., W. L. Sutker, and D. H. Walker. 1993. Persistent infection with Ehrlichia chaffeensis. Clin. Infect. Dis. 17:903-905. [DOI] [PubMed] [Google Scholar]
- 17.Felek, S., A. Unver, R. W. Stich, and Y. Rikihisa. 2003. Sensitive detection of Ehrlichia chaffeensis in cell culture, blood, and tick specimens by reverse transcription-PCR. J. Clin. Microbiol. 39:460-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finelli, A., K. M. Kerksiek, S. E. Allen, N. Marshall, R. Mercado, I. Pilip, D. H. Busch, and E. G. Pamer. 1999. MHC class I restricted T cell responses to Listeria monocytogenes, an intracellular bacterial pathogen. Immunol. Res. 19:211-223. [DOI] [PubMed] [Google Scholar]
- 19.Fishbein, D., L. Sawyer, C. Holland, E. Hayes, W. Okoroanyanwu, B. Williams, R. Sikes, M. Ristic, and J. McDade. 1987. Unexplained febrile illnesses after exposure to ticks: infection with an Ehrlichia? JAMA 257:3100-3104. [PubMed] [Google Scholar]
- 20.Ganta, R. R., M. J. Wilkerson, C. Cheng, A. M. Rokey, and S. K. Chapes. 2002. Persistent Ehrlichia chaffeensis infection occurs in the absence of functional major histocompatibility complex class II genes. Infect. Immun. 70:380-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grusby, M. J., R. S. Johnson, V. E. Papaioannou, and L. H. Glimcher. 1991. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science 253:1417-1420. [DOI] [PubMed] [Google Scholar]
- 22.Hart, M. L., D. A. Mosier, and S. K. Chapes. 2003. Toll-like receptor 4-positive macrophages protect mice from Pasteurella pneumotropica-induced pneumonia. Infect. Immun. 71:663-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huerkamp, M. J. 2003. It's in the bag: easy and medically sound rodent gas anesthesia introduction. Tech. Talk 5:3. [Google Scholar]
- 24.Janssen, E. M., E. E. Lemmens, T. Wolfe, U. Christen, M. G. von Herrath, and S. P. Schoenberger. 2003. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 421:852-856. [DOI] [PubMed] [Google Scholar]
- 25.Kocan, A. A., G. C. Levesque, L. C. Whitworth, G. L. Murphy, S. A. Ewing, and R. W. Barker. 2000. Naturally occurring Ehrlichia chaffeensis infection in coyotes from Oklahoma. Emerg. Infect. Dis. 6:477-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordick, S. K., E. B. Breitschwerdt, B. C. Hegarty, K. L. Southwick, C. M. Colitz, S. I. Hancock, J. M. Bradley, R. Rumbough, J. T. McPherson, and J. N. MacCormack. 1999. Coinfection with multiple tick-borne pathogens in a Walker Hound kennel in North Carolina. J. Clin. Microbiol. 37:2631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwok, L. Y., S. Lutjen, S. Soltek, D. Soldati, D. Busch, M. Deckert, and D. Schluter. 2003. The induction and kinetics of antigen-specific CD8 T cells are defined by the stage specificity and compartmentalization of the antigen in murine toxoplasmosis. J. Immunol. 170:1949-1957. [DOI] [PubMed] [Google Scholar]
- 28.Li, J. S., and G. M. Winslow. 2003. Survival, replication, and antibody susceptibility of Ehrlichia chaffeensis outside of host cells. Infect. Immun. 71:4229-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J. S., E. Yager, M. Reilly, C. Freeman, G. R. Reddy, A. A. Reilly, F. K. Chu, and G. M. Winslow. 2001. Outer membrane protein-specific monoclonal antibodies protect SCID mice from fatal infection by the obligate intracellular bacterial pathogen Ehrlichia chaffeensis. J. Immunol. 166:1855-1862. [DOI] [PubMed] [Google Scholar]
- 30.Maeda, K., N. Markowitz, R. C. Hawley, M. Ristic, D. Cox, and J. E. McDade. 1987. Human infection with Ehrlichia canis, a leukocytic rickettsia. N. Engl. J. Med. 316:853-856. [DOI] [PubMed] [Google Scholar]
- 31.Mercado, R., S. Vijh, S. E. Allen, K. Kerksiek, I. M. Pilip, and E. G. Pamer. 2000. Early programming of T cell populations responding to bacterial infection. J. Immunol. 165:6833-6839. [DOI] [PubMed] [Google Scholar]
- 32.Mwangi, D. M., D. J. McKeever, J. K. Nyanjui, A. F. Barbet, and S. M. Mahan. 2002. Immunisation of cattle against heartwater by infection with Cowdria ruminantium elicits T lymphocytes that recognise major antigenic proteins 1 and 2 of the agent. Vet. Immunol. Immunopathol. 85:23-32. [DOI] [PubMed] [Google Scholar]
- 33.Paddock, C. D., D. P. Suchard, K. L. Grumbach, W. K. Hadley, R. L. Kerschmann, N. W. Abbey, J. E. Dawson, B. E. Anderson, K. G. Sims, J. S. Dumler, and B. G. Herndier. 1993. Brief report: fatal seronegative ehrlichiosis in a patient with HIV infection. N. Engl. J. Med. 329:1164-1167. [DOI] [PubMed] [Google Scholar]
- 34.Paddock, C. D., J. W. Sumner, G. M. Shore, D. C. Bartley, R. C. Elie, J. G. McQuade, C. R. Martin, C. S. Goldsmith, and J. E. Childs. 1997. Isolation and characterization of Ehrlichia chaffeensis strains from patients with fatal ehrlichiosis. J. Clin. Microbiol. 35:2496-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahemtulla, A., T. M. Kundig, A. Narendran, M. F. Bachmann, M. Julius, C. J. Paige, P. S. Ohashi, R. M. Zinkernagel, and T. W. Mak. 1994. Class II major histocompatibility complex-restricted T cell function in CD4-deficient mice. Eur. J. Immunol. 24:2213-2218. [DOI] [PubMed] [Google Scholar]
- 36.Reardon, M. J., and K. R. Pierce. 1981. Acute experimental canine ehrlichiosis. I. Sequential reaction of the hemic and lymphoreticular systems. Vet. Pathol. 18:48-61. [DOI] [PubMed] [Google Scholar]
- 37.Schwacha, M. G., C. P. Schneider, and I. H. Chaudry. 2002. Differential expression and tissue compartmentalization of the inflammatory response following thermal injury. Cytokine 17:266-274. [DOI] [PubMed] [Google Scholar]
- 38.Shedlock, D. J., J. K. Whitmire, J. Tan, A. S. MacDonald, R. Ahmed, and H. Shen. 2003. Role of CD4 T cell help and costimulation in CD8 T cell responses during Listeria monocytogenes infection. J. Immunol. 170:2053-2063. [DOI] [PubMed] [Google Scholar]
- 39.Stenger, S. 2001. Cytolytic T cells in the immune response to Mycobacterium tuberculosis. Scand. J. Infect. Dis. 33:483-487. [DOI] [PubMed] [Google Scholar]
- 40.Sun, J. C., and M. J. Bevan. 2003. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300:339-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Telford, S. R., and J. E. Dawson. 1996. Persistent infection of C3H/HeJ mice by Ehrlichia chaffeensis. Vet. Microbiol. 52:103-112. [DOI] [PubMed] [Google Scholar]
- 42.Thimme, R., S. Wieland, C. Steiger, J. Ghrayeb, K. A. Reimann, R. H. Purcell, and F. V. Chisari. 2003. CD8+ T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 77:68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Totte, P., I. Esteves, N. Gunter, D. Martinez, and A. Bensaida. 2002. Evaluation of several flow cytometric assays for the analysis of T-cell responses in goats. Cytometry 49:49-55. [DOI] [PubMed] [Google Scholar]
- 44.Walker, D. H., J. P. Olano, and H. M. Feng. 2001. Critical role of cytotoxic T lymphocytes in immune clearance of rickettsial infection. Infect. Immun. 69:1841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wijburg, O. L., N. van Rooijen, and R. A. Strugnell. 2002. Induction of CD8+ T lymphocytes by Salmonella typhimurium is independent of Salmonella pathogenicity island 1-mediated host cell death. J. Immunol. 169:3275-3283. [DOI] [PubMed] [Google Scholar]
- 46.Winslow, G. M., E. Yager, K. Shilo, D. N. Collins, and F. K. Chu. 1998. Infection of the laboratory mouse with the intracellular pathogen Ehrlichia chaffeensis. Infect. Immun. 66:3892-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winslow, G. M., E. Yager, K. Shilo, E. Volk, A. Reilly, and F. K. Chu. 2000. Antibody-mediated elimination of the obligate intracellular bacterial pathogen Ehrlichia chaffeensis during active infection. Infect. Immun. 68:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright, A. D., and S. K. Chapes. 1999. LPS sensitivity in recombinant mice lacking functional alleles at MHCII, Lps, and Nramp1 genes. J. Endotoxin Res. 5:297-305. [Google Scholar]
- 49.Zaugg, J. L., D. Stiller, M. E. Croan, and S. D. Lincoln. 1986. Transmission of Anaplasma marginale Theiler by males of Dermacentor andersoni Stiles fed on an Idaho field infected, chronic carrier cow. Am. J. Vet. Res. 47:2269-2271. [PubMed] [Google Scholar]