Abstract
For Serratia marcescens, secreted hemolysin/cytotoxin is not only secreted but also activated by an outer membrane protein. Excluding posttranslational processing by mass spectrometry, the conformation of active and inactive ShlA derivatives strongly differed in electrophoretic mobilities, gel permeation chromatography, sensitivity to trypsin, circular dichroism, and intrinsic fluorescence. We concluded that ShlB interacts with ShlA during secretion and imposes a conformational change in ShlA to form the active hemolysin.
Serratia marcescens synthesizes a hemolysin (2, 3-9, 13, 18, 25, 28) encoded by the shlA gene. It also acts as a cytotoxin on epithelial cells and fibroblasts, where it causes ATP depletion and potassium efflux (15). The ShlA toxin is secreted across the outer membrane by the ShlB protein, encoded by the shlB gene (7, 19, 24, 27, 30), thereby distinguishing this type of secretion from all other known secretion systems (3, 4). Homologous hemolysins are formed by Proteus mirabilis (33), Yersinia pestis (23), Haemophilus ducreyi (22, 32), and Edwardsiella tarda (10, 17, 31). In addition, the FHA filamentous hemagglutinin of Bordetella pertussis (34) and the HMWP1 and HMWP2 adhesins of nontypeable Haemophilus influenzae (1, 11) require a B component for cell surface exposure. This kind of secretion has been designated the two-partner secretion pathway to distinguish it from other protein secretion mechanisms and to emphasize its particular properties (20). A conserved region in these proteins (3), which is contained twice in S. marcescens ShlA (68-ANPNL and 109-NPNGIS), is important for ShlA activity; replacement of N-69 and N-111 by isoleucine abolishes the hemolytic activity (29).
In addition to secretion of ShlA, ShlB converts ShlA into a hemolysin. In the absence of ShlB, nonhemolytic ShlA (termed ShlA*) remains in the periplasm and displays at most 0.2% of the ShlB-dependent and -secreted hemolytic activity of ShlA (27).
Activation of ShlA* in vitro can be uncoupled from secretion. Crude extracts of cells that synthesize only ShlB activate ShlA* contained in crude extracts of cells that synthesize only ShlA* (21). Highly purified ShlB does not activate highly purified ShlA* unless phosphatidylethanolamine is added (16). ShlA* can also be converted to a hemolysin by addition of an ShlB-secreted and -activated N-terminal fragment of ShlA consisting of 255 residues (ShlA255) (14, 21). ShlA255 itself is nonhemolytic because it lacks the remaining, C-terminal part of ShlA (1,576 residues), which is required for pore formation in erythrocyte membranes (26, 28). ShlA255 also restores the hemolytic activity of ShlAΔ3-97, in which residues 3 to 97 of ShlA are deleted (29).
Results and Discussion.
The N-proximal 242-residue fragment of ShlA (ShlA242) complements full-length ShlA*, which in the absence of ShlB remains inactive in the periplasm (29). ShlA242 must be secreted by ShlB into the culture medium for complementation of ShlA*. This indicates that activation of ShlA by ShlB occurs in the ShlA242 fragment. Since full-length ShlA precipitates and is soluble in an active form only in 6 M urea, activation of ShlA by ShlB was studied with the experimentally more amenable ShlA242 fragment because of its better solubility and stability. Structural differences between active and inactive ShlA242 were examined using four constructs: (i) the secreted, active, N-terminal 242 amino acids plus six histidine residues at the C-terminal end (ShlA242-His6) and (ii) by mutated ShlB136ARSG-secreted but not activated ShlA242-His6, both isolated from the culture supernatant, (iii) ShlA242-His6 synthesized in cells lacking ShlB and therefore not secreted or activated (ShlA242*-His6), isolated from the periplasm, and (iv) ShlA242-His6 lacking a signal sequence, which therefore remained in the cytoplasm and formed inclusion bodies (C-ShlA242*-His6). All ShlA242-His6 derivatives contained an additional 33 amino acid residues between the His6 tag and ShlA242.
ShlA242-His6 was precipitated by (NH4)2SO4 from the culture supernatant and purified on a Ni2+-nitrilotriacetic acid (NTA) affinity column. Periplasmic ShlA242*-His6 was isolated from the (NH4)2SO4 precipitate of the periplasmic extract and purified by affinity column chromatography as described above. Cytoplasmic C-ShlA242*-His6 was isolated from the French press lysate of cells in the presence of 6 M urea and then purified by affinity column chromatography. The N-terminal sequence of all of the isolated derivatives started with AEIVAANGA (determined by Edman degradation), which is identical to the determined ShlA sequence (24). In addition, inactive ShlA242-His6, which is secreted but not activated by mutant ShlB136 (contains an insertion of the tetrapeptide ARSG between residues 136 and 137), was studied. Only ShlA242-His6 complemented ShlAΔ3-97 to an active hemolysin (82% lysed erythrocytes, compared to 0% for ShlAΔ3-97 alone). This result shows that the His tag and the additional amino acids in the linker region did not affect the complementing activity of ShlA242.
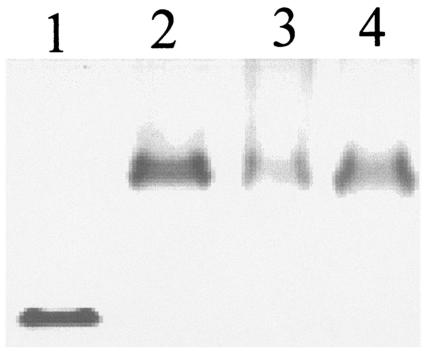
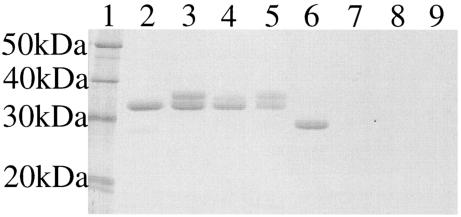
The electrophoretic mobility of active ShlA242-His6 in a native polyacrylamide gel, without addition of sodium dodecyl sulfate or mercaptoethanol and without heating, was much higher than that of the three inactive ShlA242-His6 derivatives isolated from the periplasm and cytoplasm and of an ShlA242-His6 derivative that was secreted but not activated by the mutant ShlB136 protein (35) (Fig. 1). Upon incubation with trypsin, ShlA242-His6 was truncated to approximately 90% of the nondegraded size, in contrast to the inactive ShlA242-His6 derivatives, which were completely degraded (Fig. 2). The differences between secreted and nonsecreted ShlA242-His6 indicated changes in the primary or secondary structures of cytoplasmic ShlA242-His6 during export across the cytoplasmic membrane, within the periplasm, during secretion across the outer membrane, or in the culture medium.
FIG. 1.
Native polyacrylamide gel electrophoresis of ShlA242-His6 derivatives. Lane 1, activated ShlA242-His6; lane 2, cytosolic C-ShlA242*-His6; lane 3, periplasmic ShlA242*-His6; lane 4, ShlA242*-His6 secreted by ShlB136.
FIG. 2.
Tryptic digest of ShlA242-His6 derivatives. Lane 1, molecular size marker; lane 2, secreted ShlA242-His6; lane 3, cytosolic C-ShlA242*-His6; lane 4, periplasmic ShlA242*-His6; lane 5, inactive secreted ShlA242*-His6; lanes 6 to 9, same as lanes 2 to 5, but samples were treated with trypsin.
The molecular masses of the ShlA242-His6 derivatives were estimated by gel permeation chromatography in the presence of 6 M urea to avoid aggregation and were found to be 41 kDa for active ShlA242-His6 and 64, 66, and 60 kDa for ShlA242*-His6, C-ShlA242-His6, and secreted but inactive ShlA242-His6, respectively. None of these values reflect the calculated molecular mass of 29 kDa. They suggest different shapes of the noncomplementing ShlA242 derivatives compared to complementing ShlA242 as a result of distinct conformations. The same chromatography with the buffer containing dithiothreitol did not much alter the chromatographic properties, excluding various disulfide bridges as causes of the distinct elution positions. Active ShlA242-His6 and inactive ShlA242*-His6 eluted at positions equivalent to proteins of 38 and 63 kDa, respectively (compared to 41 and 66 kDa under nonreducing conditions). The aberrant chromatographic behavior could also be the result of a covalent modification during the activation. Therefore, the molecular weights of the four ShlA242-His6 derivatives were determined by quadrupole electrospray mass spectrometry (12, 14). Each of the derivatives had a molecular weight of the calculated size (calculated value for ShlA242-His6, 29,069; determined value for active ShlA242-His6, 29,069, and for the inactive derivatives, 29,067, 29,067, and 29,066). This result excludes a covalent modification larger than the conversion of a carboxyl group to an amide. It is unlikely that such a small alteration would affect the properties of the ShlA242-His6 derivatives as profoundly as has been observed.
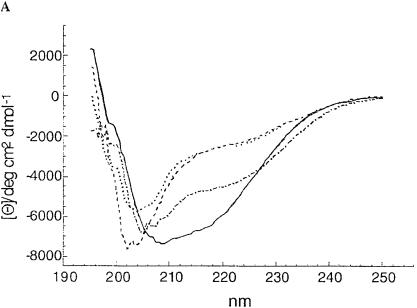
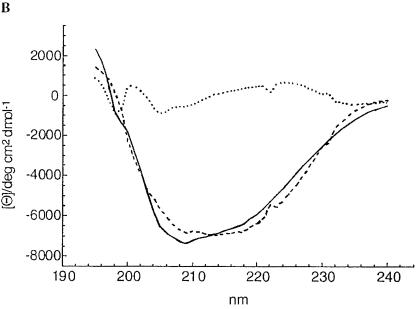
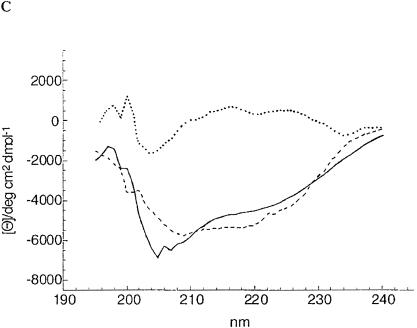
The altered electrophoretic mobility, the deviation of the size obtained by gel permeation chromatography from the calculated size, the exclusion of a covalent modification, and the trypsin sensitivity suggested that the inactive ShlA242*-His6 derivatives assume a conformation different from that of active ShlA242-His6. To evaluate conformational differences directly, the circular dichroism (CD) was determined. Profound differences between active ShlA242-His6 and the inactive ShlA242-His6 derivatives and smaller differences among the inactive ShlA242-His6 derivatives were found (Fig. 3). The spectra were compared with spectra of standard peptides to estimate the proportion of secondary-structure elements. Only the curves of ShlA242-His6 and ShlA242*-His6 were close enough to the calculated curves to allow a comparison. The two proteins strongly differed in their content of α-helix, β-turn, turn, and random structures (Table 1). The method provides only approximate values but is sufficiently accurate to reveal clear differences between the two proteins.
FIG. 3.
(A) CD spectra of secreted, active ShlA242-His6 (—), cytosolic C-ShlA242*-His6 (- - -), periplasmic ShlA242*-His6 ( · - · - · -), and inactive secreted ShlA242*-His6 ( · · · · · ). (B) Estimated CD spectrum of secreted, active ShlA242-His6 (—), calculated CD spectrum (- - -), and the difference between the two spectra ( · · · · · ). (C) estimated CD spectrum of periplasmic, inactive ShlA242*-His6 (—), calculated CD spectrum (- - -), and the difference between the two spectra ( · · · · · ).
TABLE 1.
Secondary structure elements of active ShlA242-His6 and inactive ShlA242*-His6
| Protein | Content (%)
|
|||
|---|---|---|---|---|
| α-Helix | β-Turn | Turn | Random | |
| ShlA242-His6 | 11.8 | 48.4 | 9.6 | 30.2 |
| ShlA242*-His6 | 14.3 | 31.3 | 17.4 | 37.0 |
Different conformations of active and inactive ShlA242-His6 derivatives were in addition revealed by fluorescence spectroscopy. The intensity and maximum intrinsic fluorescence depends on the surroundings of the aromatic amino acid residues and changes when different conformations provide different environments. Active ShlA242-His6 and inactive ShlA242-His6 were excited at 274 nm (excitation wavelength of tyrosine; ShlA242-His6 contains no tryptophan). The two proteins showed a similar emission maximum at 300 nm, but the fluorescence intensity at 300 nm of activated ShlA242-His6 was 1.5-fold higher than that of inactive C-ShlA242*-His6. The results imply a different conformation of active ShlA242-His6 in which the proportion of the tyrosine residues contained in a hydrophobic environment is greater than in C-ShlA242*-His6. Taking the results of this and our previous work together, we clearly demonstrate that ShlA is synthesized as an inactive protein and is activated through a conformational change triggered by ShlB. ShlB is a bifunctional protein involved in secretion and activation of ShlA.
Acknowledgments
We thank R. Süβmuth, Department of Organic Chemistry, University of Tübingen, for the N-terminal protein sequencing and mass spectrometry; A. Kapurniotu, Department of Biochemistry, University of Tübingen, for help with CD spectroscopy; and Karen A. Brune for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (BR 3110/2-1) and the Fonds der Chemischen Industrie.
Editor: J. T. Barbieri
REFERENCES
- 1.Barenkamp, S. J., and J. W. S. St. Geme. 1994. Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 62:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernheimer, A. W. 1988. Assay of hemolytic toxins. Methods Enzymol. 165:213-217. [DOI] [PubMed] [Google Scholar]
- 3.Braun, V., and R. Hertle. 1999. The family of Serratia and Proteus cytolysins, p. 349-361. In J. Alouf and J. H. Freer (ed.), The comprehensive source book of bacterial protein toxins. Academic Press, London, United Kingdom.
- 4.Braun, V., R. Schönherr, and S. Hobbie. 1993. Enterobacterial hemolysins: activation, secretion and pore formation. Trends Microbiol. 1:211-216. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., S. Hobbie, and R. Ondraczek. 1992. Serratia marcescens forms a new type of cytotoxin. FEMS Microbiol. Lett. 100:299-306. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., and T. Focareta. 1991. Pore-forming bacterial protein hemolysins (cytolysins). Crit. Rev. Microbiol. 18:115-158. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V., B. Neuss, Y. Ruan, E. Schiebel, H. Schöffler, and G. Jander. 1987. Identification of the Serratia marcescens hemolysin determinant by cloning into Escherichia coli. J. Bacteriol. 169:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, V., H. Gunther, B. Neuss, and C. Tautz. 1985. Hemolytic activity of Serratia marcescens. Arch. Microbiol. 141:371-376. [DOI] [PubMed] [Google Scholar]
- 9.Carbonell, G. V., and M. C. Vidotto. 1992. Virulence factors in Serratia marcescens: cell-bound hemolysin and aerobactin. J. Med. Biol. Res. 25:1-8. [PubMed] [Google Scholar]
- 10.Chen, J.-D., S.-Y. Lai, and S.-L. Huang. 1996. Molecular cloning, characterization, and sequencing of the hemolysin gene from Edwardsiella tarda. Arch. Microbiol. 165:9-17. [DOI] [PubMed] [Google Scholar]
- 11.Cope, L., R. Yongev, U. Müller-Eberhard, and E. C. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Covey, T. R., R. F. Bonner, B. I. Shushan, and J. Henion. 1988. The determination of protein, oligonucleotide and peptide molecular weights by ion-spray mass spectrometry. Rapid Commun. Mass Spectrom. 2:249-256. [DOI] [PubMed] [Google Scholar]
- 13.Hertle, R. 2000. Serratia type pore forming toxins. Curr. Protein Pept. Sci. 1:75-89. [DOI] [PubMed] [Google Scholar]
- 14.Hertle, R., R. Süssmuth, J. Jung, and V. Braun. 2000. Two step FPLC-purification of the Serratia marcescens hemolysin and peptide mapping with mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 737:13-23. [DOI] [PubMed] [Google Scholar]
- 15.Hertle, R., M. Hilger, S. Weingardt-Kocher, and I. Walev. 1999. Cytotoxic action of Serratia marcescens hemolysin on human epithelial cells. Infect. Immun. 67:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hertle, R., S. Brutsche, W. Groeger, S. Hobbie, W. Koch, U. Könninger, and V. Braun. 1997. Specific phosphatidylethanolamine dependence of Serratia marcescens cytotoxin activity. Mol. Microbiol. 26:853-865. [DOI] [PubMed] [Google Scholar]
- 17.Hirono, I., N. Tange, and T. Aoki. 1997. Iron-regulated haemolysin gene from Edwardsiella tarda. Mol. Microbiol. 24:851-856. [DOI] [PubMed] [Google Scholar]
- 18.König, W., Y. Faltin, J. Scheffer, H. Schöffler, and V. Braun. 1987. Role of cell-bound hemolysin as a pathogenicity factor for Serratia infections. Infect. Immun. 55:2554-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Könninger, U. W., S. Hobbie, R. Benz, and V. Braun. 1999. The haemolysin-secreting ShlB protein of the outer membrane of Serratia marcescens: determination of surface-exposed residues and formation of ion-permeable pores by ShlB mutants in artificial lipid bilayer membranes. Mol. Microbiol. 32:1212-1225. [DOI] [PubMed] [Google Scholar]
- 20.Locht, C., R. Antoine, and F. Jacob-Dubuisson. 2001. Bordetella pertussis, molecular pathogenesis under multiple aspects. Curr. Opin. Microbiol. 4:82-89. [DOI] [PubMed] [Google Scholar]
- 21.Ondraczek, R., S. Hobbie, and V. Braun. 1992. In vitro activation of the Serratia marcescens hemolysin through modification and complementation. J. Bacteriol. 174:5086-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer, K. L., and R. S. Munson, Jr.. 1995. Cloning and characterization of the genes encoding the haemolysin of Haemophilus ducreyi. Mol. Microbiol. 18:821-830. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, et al. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 24.Poole, K., E. Schiebel, and V. Braun. 1988. Molecular characterization of the hemolysin determinant of Serratia marcescens. J. Bacteriol. 170:3177-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan, Y., and V. Braun. 1990. Hemolysin as a marker for Serratia. Arch. Microbiol. 154:221-225. [DOI] [PubMed] [Google Scholar]
- 26.Schiebel, E., and V. Braun. 1989. Integration of the Serratia marcescens haemolysin into human erythrocyte membranes. Mol. Microbiol. 3:445-453. [DOI] [PubMed] [Google Scholar]
- 27.Schiebel, E., H. Schwarz, and V. Braun. 1989. Subcellular location and unique secretion of the hemolysin of Serratia marcescens. J. Biol. Chem. 264:16311-16320. [PubMed] [Google Scholar]
- 28.Schönherr, R., M. Hilger, S. Broer, R. Benz, and V. Braun. 1994. Interaction of Serratia marcescens hemolysin (ShlA) with artificial and erythrocyte membranes: demonstration of the formation of aqueous multistate channels. Eur. J. Biochem. 223:655-663. [DOI] [PubMed] [Google Scholar]
- 29.Schönherr, R., R. Tsolis, T. Focareta, and V. Braun. 1993. Amino acid replacements in the Serratia marcescens haemolysin ShIA define sites involved in activation and secretion. Mol. Microbiol. 9:1229-1237. [DOI] [PubMed] [Google Scholar]
- 30.Sieben, S., R. Hertle, J. Gumpert, and V. Braun. 1998. The Serratia marcescens hemolysin is secreted but not activated by stable protoplast-type L-forms of Proteus mirabilis. Arch. Microbiol. 170:236-242. [DOI] [PubMed] [Google Scholar]
- 31.Strauss, E. J., N. Ghori, and S. Falkow. 1997. An Edwardsiella tarda strain containing a mutation in a gene with homology to shlB and hpmB is defective for entry into epithelial cells in culture. Infect. Immun. 65:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Totten, P. A., D. V. Norn, and W. E. Stamm. 1995. Characterization of the hemolytic activity of Haemophilus ducreyi. Infect. Immun. 63:4409-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uphoff, T. S., and R. A. Welch. 1990. Nucleotide sequencing of the Proteus mirabilis calcium-independent hemolysin genes (hpmA and hpmB) reveals sequence similarity with the Serratia marcescens hemolysin genes (shlA and shlB). J. Bacteriol. 172:1206-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willems, R. J., C. Geuijen, H. G. J. van der Heide, G. Renauld, P. Bertin, W. M. R. van den Akker, C. Locht, and F. R. Mooi. 1994. Mutational analysis of the Bordetella pertussis fim/fha gene cluster: identification of a gene with sequence similarities to hemolysin accessory genes involved in export of FHA. Mol. Microbiol. 11:337-347. [DOI] [PubMed] [Google Scholar]
- 35.Yang, F.-L., S. Hobbie, and V. Braun. 2000. ShlB mutants of Serratia marcescens allow uncoupling of activation and secretion of the ShlA hemolysin. Int. J. Med. Microbiol. 290:529-538. [DOI] [PubMed] [Google Scholar]