Abstract
Intimin is the primary adhesin of Escherichia coli O157:H7, the most common infectious cause of bloody diarrhea in the United States and the leading cause of acute kidney failure in children who develop hemolytic uremic syndrome. Cattle are the primary reservoir of E. coli O157:H7. Indeed, most cases of E. coli O157:H7 infection in the United States occur after ingestion of contaminated undercooked hamburger or produce that had contact with bovine manure. Because intimin is required for persistent colonization of neonatal calves and adult cattle, we hypothesized that an intimin-based vaccination strategy in calves would reduce colonization of cattle with E. coli O157:H7. To test this concept in a small-animal model, we developed transgenic tobacco plant cells that express the carboxy-terminal host cell-binding domain of E. coli O157:H7 intimin. Mice were either immunized intraperitoneally with intimin expressed from the plant cells, fed transgenic plant cells, or both. Here we show that these mice generated an intimin-specific mucosal immune response when primed parenterally and then boosted orally and also exhibited a reduced duration of E. coli O157:H7 fecal shedding after challenge.
Escherichia coli O157:H7 is the most common cause of bloody diarrhea, or hemorrhagic colitis, in the United States, with an estimated incidence of 73,480 cases per annum (7, 34). Moreover, hemolytic uremic syndrome, a sequela of E. coli O157:H7 infection, is the most frequent basis for acute kidney failure in U.S. children (7). These organisms are typically transmitted directly or indirectly from infected cattle to humans. Both beef and dairy cattle can be sporadically and asymptomatically colonized with E. coli O157:H7 and shed the bacteria, which can survive in broad ecological niches beyond the bovine gastrointestinal tract, into the environment in their feces (10, 17, 25, 28). Moreover, contacts with the farming environment and livestock density are major risk factors for human infection and disease caused by E. coli O157:H7. Many of the foods implicated in human disease are of bovine origin or are food or water that have come into contact with contaminated meat or bovine fecal material (14, 43).
A number of investigators have concluded that a decrease in the amount of E. coli O157:H7 shed as well as in the number of cattle that excrete the serotype could cause a significant reduction in the prevalence of the bacteria in cattle and the farm environment. The hypothesis that vaccination of cattle or treatment of the animals with an agent to diminish the level of colonization and shedding of E. coli O157:H7 could potentially lead to a decline in the incidence of human E. coli O157:H7-related disease (17, 22, 47) was suggested by the findings from a stochastic simulation model designed by Jordan et al. (22). Based on this idea, B. Finlay's group, our laboratory, and others have begun to design and/or test E. coli O157:H7 vaccine protocols for use in cattle. Indeed, Finlay and colleagues have initiated field studies of E. coli O157:H7 secreted products as a subcutaneously administered bovine vaccine [B. Finlay, Abstr. 5th Int. Symp. “Shiga toxin (verocytotoxin)-producing Escherichia coli infections,” abstr. p. 23, 2003; R. Moxley, D. Smith, T. Klopfenstein, G. Erickson, J. Folmer, C. Macken, S. Hinkley, A. Potter, and B. Finlay, Abstr. 5th Int. Symp. “Shiga toxin (verocytotoxin)-producing Escherichia coli infections,” abstr. p. 23, 2003].
We selected a different immunogen, expression system, and route of administration for proof of concept studies to assess, in a small-animal model, the feasibility of an E. coli O157:H7 vaccine for cattle. For our vaccine candidate, we chose intimin, an outer membrane protein of E. coli O157:H7 that is required for attaching and effacing lesion formation as well as for bacterial adherence to mammalian cells and the intestinal mucosa of calves, piglets, and ferrets (8, 21, 32, 46). Intimin is the product of the eae (E. coli attach and efface) gene, which is contained within an approximately 43-kb pathogenicity island called the locus of enterocyte effacement (23, 24, 36). The carboxy-terminal portion of intimin binds the bacterium-encoded translocated intimin receptor (Tir) and a host cell receptor, nucleolin, to mediate intimate attachment of the bacteria to the eucaryotic cell surface (11, 12, 38). The reasons we consider intimin an attractive candidate for an E. coli O157:H7 antitransmission vaccine for cattle are based on both in vitro and in vivo studies. Specifically, members of our laboratory previously found that antibodies against the carboxy-terminal third of the molecule block adherence of wild-type E. coli O157:H7 to HEp-2 cells (13, 33). In addition, our laboratory, with E. Dean-Nystrom's group, showed that colostrum from pigs immunized with intimin isolated from E. coli O157:H7 contains anti-intimin antibodies that can protect suckling piglets from colonization with E. coli O157:H7 (13). These tissue culture experiments and passive transfer studies suggest that antibodies specific to intimin play an important role in blocking adherence of the bacterium to host cells and can protect the host from E. coli O157:H7-mediated disease.
For delivery of intimin as a vaccine for cattle, we sought an oral inoculation system to facilitate induction of mucosal antibodies and for ease of administration. Therefore, we elected to use a transgenic plant cell system for intimin expression, with the ultimate goal of moving the antigen into whole-plant expression and delivery systems. Transgenic plants offer the flexibility to function as low-cost, efficient, and practical vaccine antigen oral delivery systems to stimulate mucosal immunity or to boost and shift initial immunity to a mucosal antibody response (27, 44). Indeed, transgenic plants have already been used as successful vaccine antigen production and delivery systems (18, 29, 30, 40). The vaccine antigens expressed by plants include hepatitis B surface antigen (26, 31, 42), enterotoxigenic E. coli heat-labile toxin B subunit (LT-B) (18, 27, 30, 40), Norwalk virus capsid protein (29, 41), cholera toxin B subunit (2, 3), and many others. For this study, we created carboxy-terminal third of intimin-expressing plant cells and evaluated the capacity of this transgenic material to induce adherence-blocking antibodies and to reduce levels and/or time of E. coli O157:H7 fecal shedding in a mouse model of intimin-dependent colonization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are summarized in Tables 1 and 2, respectively. Enterohemorrhagic E. coli (EHEC) O157:H7 strain 86-24 (15) was isolated in 1986 from a patient in Seattle, Wash., and was kindly provided by Phil Tarr. EHEC O157:H7 strain 86-24 that expresses the green fluorescent protein arabinose-inducible plasmid p166 was used to visualize bacteria in the HEp-2 cell adherence assay, as previously described (13, 38). A streptomycin-resistant derivative of strain 86-24 (called 86-24 Strr [35]) and its isogenic, intimin-negative mutant strain 86-24 eaeΔ10 (called 86-24 Strr eaeΔ10 [32]) were used to distinguish EHEC O157:H7 from other fecal bacteria during the colonization assay by plating on sorbitol-MacConkey agar with 100 μg of streptomycin per ml. In addition, several sorbitol-negative colonies from different mice at each collection time point were tested by slide agglutination with O157 antiserum (Difco Laboratories, Sparks, Md.) to confirm that the colonies were of the O157 serogroup. All mice were free of Strr, O157 agglutination-positive colonies prior to challenge. Agrobacterium tumefaciens strain EHA105, plasmids with plant-specific expression components (pBTI210.3, pBTI210.4, and pIBT210), and the binary vector pGPTV-Kan were provided by the Boyce Thompson Institute for Plant Research (Cornell University, Ithaca, N.Y.). All E. coli strains and clones were grown in Luria-Bertani (LB) broth or on LB agar. Prior to plant cell transformation, Agrobacterium strains were grown in YM broth (0.4 g of yeast extract, 10 g of mannitol, 0.1 g of NaCl, 0.2 g of MgSO4 · 7H2O, 0.5 g of K2HPO4 per liter). Antibiotics were added as needed for selection at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; carbenicillin, 100 μg/ml. Plasmid DNA was isolated by the Miniprep procedure (Qiagen, Valencia, Calif.). Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.), and restriction enzyme-digested DNA fragments were purified by agarose gel electrophoresis and eluted from the gel with Geneclean spin columns and reagents (Bio 101, Carlsbad, Calif.). T4 DNA ligase was purchased from U.S. Biochemicals (Cleveland, Ohio). PCRs were done with AmpliTaq (Roche, Branchburg, N.J.) and Pfu Turbo (Stratagene, La Jolla, Calif.) polymerases in an MJ Research Minicycler (Watertown, Mass.). Sequencing was performed with the ABI Big Dye sequencing kit (Applied Biosystems, Inc., Foster City, Calif.). Products were separated and analyzed by the Biomedical Instrumentation Center at Uniformed Services University of the Health Sciences with Applied Biosystems sequencer model 377 or 3100. Sequence results were compared to published sequences by use of the Wisconsin Sequence Analysis package from the Genetics Computer Group, Inc. (Madison, Wis.).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| 86-24 | E. coli O157:H7, eae positive | 15 |
| 86-24 GFP | E. coli O157:H7(p166) | 38 |
| 86-24 Strr | E. coli O157:H7, Strr | 35 |
| 86-24 eaeΔ10 Strr | E. coli O157:H7, eae negative, Strr | 32 |
| DH5α | E. coli cloning host, recA | 16 |
| XL-1 Blue | E. coli cloning host, recA lacIq | 6; Stratagene |
| A. tumefaciens EHA105 | Plant transformation strain | 20 |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Intimin plasmids | ||
| pMW103 | Truncated eae (bp 1959-2804) in pQE31 (Qiagen) | 13 |
| pNR37 | eae from pMW103, plant optimized | This study |
| Plant expression vectors | ||
| pIBT210 | CaMV 35S promoter, TEV 5′ UTR, Vsp 3′ UTR | 18 |
| pBTI210.3 | CaMV 35S promoter, TMVΩ 5′ UTR, Vsp 3′ UTR | Hugh Mason, while at Boyce Thompson Institute for Plant Research |
| pBTI210.4 | CaMV 35S promoter, TEV 5′ UTR, VspB 3′ UTR, with plant-specific signal peptide sequence | Hugh Mason, while at Boyce Thompson Institute for Plant Research |
| pGPTV-Kan | Binary vector, Kanr | 4 |
| Plant-specific intimin expression vectors | ||
| pNR49 | eae from pNR37 with signal peptide sequence from pBTI210.4 | This study |
| pNR50 | eae from pNR37 | This study |
Optimization of eae (nucleotides 1959 to 2804) for plant expression.
Thirteen independent changes were made to the nucleotide sequence of the intimin gene from pMW103 with the QuikChange site-directed mutagenesis kit (Stratagene). The seven sets of oligonucleotides listed in Table 3 were designed according to the protocol listed in the QuikChange manual. The initial mutagenesis reaction was done with 50 ng of pMW103 and 125 ng of each primer from primer set 1 in a total volume of 50 μl. In addition, 1 μl of Pfu Turbo polymerase was added to each reaction tube immediately prior to placing the sample tubes into the Minicycler. The samples were subjected to a 30-s hot start at 95°C and then 12 cycles of 95°C for 30 s, 55°C for 60 s, and 68°C for 12 min. After the cycles were completed, each reaction mixture was digested with DpnI, and 1 to 5 μl of the digested reaction was transformed into the supercompetent cells provided with the kit according to the protocol outlined in the QuikChange manual. The transformed cells were plated on LB agar with ampicillin. Several ampicillin-resistant colonies were selected, and 3 ml of overnight LB broth culture was prepared from each colony. Bacteria were then harvested by centrifugation, and plasmid DNA was purified from the pelleted organisms. The eae sequences were amplified by PCR from the plasmids, and the PCR products were then eluted from an agarose gel and sequenced. Clones for which the sequence of the mutated, PCR-amplified eae gene had been confirmed were used in the next round of mutagenesis with the next set of mutagenesis primers. The final plant-optimized intimin clone from pMW103 (13) that contained all 13 nucleotide changes was called pNR37. The intimin protein encoded by this gene has an amino-terminal histidine tag and consists of the carboxy-terminal 261 amino acids of intimin from E. coli O157:H7 (Int261).
TABLE 3.
Oligonucleotide sets for eae (nucleotides 1959 to 2804) plant expression optimization by QuikChange site-directed mutagenesis
| Oligonucleotide | Sequencea |
|---|---|
| 1-forward | GGGTCAGCCAGTTAACAACCAATCCGTTACATTCTCAACAAACTTTGG |
| 1-reverse | CCAAAGTTTGTTGAGAATGTAACGGATTGGTTGTTAACTGGCTGACCC |
| 2-forward | GGCTGCAATATGGTCAATTTAAACTGAAAGCAAGCGG |
| 2-reverse | CCGCTTGCTTTCAGTTTAAATTGACCATATTGCAGCC |
| 3-forward | GGGAAAGTCACTTTGAATGGTAAAGGCTCTGTCGTAATTAAAGCC |
| 3-reverse | GGCTTTAATTACGACAGAGCCTTTACCATTCAAAGTGACTTTCCC |
| 4-forward | GGTGATAAGCAAACAGTAAGTTACACTATCAAGGCACCGTCG |
| 4-reverse | CGACGGTGCCTTGATAGTGTAACTTACTGTTTGCTTATCACC |
| 5-forward | GCTATGTCCATTTGCAAAAACTTGTTACCATCCACACAGACGG |
| 5-reverse | CCGTCTGTGTGGATGGTAACAAGTTTTTGCAAATGGACATAGC |
| 6-forward | GGTATTGTCAGATATCTATGACTCATGGGGGGCTGCAAACAAGTATAGCC |
| 6-reverse | GGCTATACTTGTTTGCAGCCCCCCATGAGTCATAGATATCTGACAATACC |
| 7-forward | GGATTAAACAGACATCTAGTGAGCAACGTTCTGGAGTATCAAGC |
| 7-reverse | GCTTGATACTCCAGAACGTTGCTCACTAGATGTCTGTTTAATCC |
Sites of mutagenesis are indicated in bold.
Plant expression plasmid construction.
The nucleotide sequence for the VspA signal peptide was amplified by PCR from pHB306 (37). A fragment that contained the nucleotide sequences for the tobacco etch virus (TEV) 5′ untranslated region (UTR) and the VspA signal peptide (encodes MAMKVLVFFVATILVAWGA) was ligated into the XhoI and SacI sites of pIBT210 (18) to make pBTI210.4. Plasmid pBTI210.4 codes for a plant-specific signal peptide 5′ to a SacI site, a cauliflower mosaic virus (CaMV) 35S promoter, the TEV 5′ UTR, and a VspB 3′ flank. The gene for Int261 was amplified by PCR from pNR37 to include flanking SacI sites; this fragment was cloned into the SacI site of pBTI210.4. The EcoRI-HindIII fragment of the resultant plasmid was then cloned into pGPTV-Kan (4) to create pNR49.
Plasmid pBTI210.3 was created by digestion of pHB211.1 (37) with NcoI. This restriction enzyme-digested DNA was 3′ end-filled with Klenow polymerase and then further digested with XhoI and mung bean nuclease. The resultant blunt-ended vector fragment was ligated to generate pHB211. The HindIII-NcoI fragment of pHB211 that contained the sequences for the CaMV 35S promoter fused to the TMV 5′ UTR was ligated into pIBT210 to create pBTI210.3. Site-directed mutagenesis was used to introduce an NcoI site 5′ of the histidine-tagged Int261 gene from pNR37. The plasmid was cut with NcoI and KpnI, and the fragment that contained the Int261 gene was cloned into pBTI210.3. The EcoRI-HindIII fragment of the resultant plasmid was cloned into pGPTV-Kan to create pNR50.
NT-1 cell culture and transformation.
Nicotiana tabacum cv. Bright Yellow 2 (NT-1) cells (1) were grown at 25°C on a rotary shaker (150 rpm) in 40 ml of NT medium [Murashige minimal organics medium (Invitrogen Life Technologies, Carlsbad, Calif.), 30 g of sucrose per liter, 3 μM thiamine, 0.58 mM myoinositol, 1.3 mM KH2PO4, 1 μM 2,4-dichlorophenoxyacetic acid (Sigma, St. Louis, Mo.), 2.5 mM 2(N-morpholino)ethanesulfonic acid, pH 5.7]. These cells were subcultured (1 part cells to 19 parts NT medium) every 7 days. To facilitate transformation, binary vectors of interest were introduced into A. tumefaciens EHA105 by electroporation. Agrobacterium strains that contained the construct of interest were used to transform NT-1 cells as previously described (1). The NT-1 cells were kindly provided by the Boyce Thompson Institute for Plant Research.
Immunoblot analysis of Int261 expressed in plant cells.
A small amount of plant cell material (∼0.03 g), resuspended in an extraction buffer (25 mM sodium phosphate [pH 6.6], 100 mM NaCl, 10 mM EDTA, 0.1% Triton X-100, 10 μg of leupeptin per ml, and 50 mM sodium ascorbate), was disrupted by sonication (25-s pulse; 5 s on, 10 s off). The lysed material was clarified by centrifugation (12,000 × g, 3 min, 4°C), and proteins in the extract were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins in the Tris-glycine-sodium dodecyl sulfate-polyacrylamide gels were transferred onto nitrocellulose and probed with anti-intimin polyclonal antibodies. Antigen amounts were quantified by comparing the Western blot band intensity to that of a known concentration of control purified Int261 (∼35 kDa) from pMW103 (13) by use of the NIH Image (v. 1.61) program (38).
Mouse feeding assay.
NT-1 cells or transgenic NT-1 cell clones that expressed Int261 were grown in 40-ml suspension cultures to confluence (about 7 days). The cells were pelleted by gentle centrifugation, and culture medium was removed with a pipette. Five grams of NT-1 cell material was divided into aliquots in small, individual plastic weigh dishes, and 0.5 g of sucrose was added to each sample. A 7.5-μg dose of purified cholera toxin (CT) (Sigma) was also added to appropriate samples to serve as an oral adjuvant. The addition of this dose of CT did not appear to have any detrimental effects on the mice (i.e., loose stool or diarrhea). Female BALB/c mice of 16 to 18 g were made to fast overnight before they were allowed to eat the plant material ad libitum. All of the plant material was usually consumed within 8 h. The mice were bled by a tail vein cut, and fecal pellets were collected both prior to and after feeding. Blood samples were allowed to clot at room temperature for 15 min and centrifuged at 10,000 × g for 10 min to pellet red blood cells. A second, 2-min centrifugation step was used to further clarify each serum sample. These clarified samples were then stored at −20°C until assayed. Fecal samples were frozen, lyophilized overnight, and then homogenized in phosphate-buffered saline (PBS), pH 7.2 (for every 15 pellets, 0.8 ml of PBS was used). The resuspended fecal material was centrifuged for 5 min, and each supernatant was then transferred to a clean tube and stored at −20°C until assayed. Mice immunized intraperitoneally (i.p.) with purified His-tagged Int261 (from pMW103 [13]) plus TiterMax (TiterMax USA Inc., Norcross, Ga.) (20 μg on day 0 and 10 μg on days 7 and 14) served as the positive control.
ELISA.
For measurement of anti-intimin antibodies, an enzyme-linked immunosorbent assay (ELISA) was developed. For that purpose, U-bottomed 96-well microtiter plates were coated with 50 ng of purified histidine-tagged Int261 from pMW103 (13) per well. Serum samples were diluted from a titer of 1:50 to 1:781,250 and fecal pellet extracts were diluted from 1:50 to 1:3,200 and incubated overnight at 4°C. The plates were washed, and goat anti-mouse immunoglobulin G (IgG) (1:3,000) or goat anti-mouse IgA (1:3,000) conjugated to alkaline phosphatase was added to the appropriate plates. The plates were then incubated at room temperature for 1 h. The antigen-antibody reactions were detected colorimetrically by incubation with tetramethylbenzidine peroxidase substrate (Bio-Rad) for 15 min followed immediately by measurement of the absorbance at 600 nm with an ELx800 microtiter plate reader (Bio-Tek Instruments Inc., Winooski, Vt.). The titer was defined as the reciprocal of the highest dilution of serum or fecal extract that gave an absorbance reading above both background and preimmune levels. Mice were considered responders if they had detectable titers in serum or fecal material at the last collection point. Mice with no titer or that lost titer by the last collection point were called nonresponders. The geometric mean of the responders' titers was used to determine the mean response per group.
E. coli O157:H7 colonization of mice.
Female BALB/c mice of 16 to 18 g (Charles River Laboratories, Inc.) were made to fast overnight and fed a total inoculum of 108 to 109 CFU of E. coli O157:H7 strain 86-24 Strr or 86-24 Strr eaeΔ10 in each of two doses administered 4 h apart. Fecal pellets were collected daily from individual mice and weighed, and serial dilutions of fecal pellet homogenates were prepared in PBS. These homogenates were plated on sorbitol-MacConkey agar (45) with 100 μg of streptomycin per ml to determine the CFU per gram of feces and the duration of colonization.
Statistical methods.
Determination of the mean duration of colonization and statistical comparison of the duration of colonization among mouse groups were done with the SPSS 11.0 statistical package. The log-rank test was used to determine the statistical significance of the difference in duration of colonization among the mouse groups.
RESULTS
Construction of NT-1 plant cells that express the carboxy-terminal third of intimin.
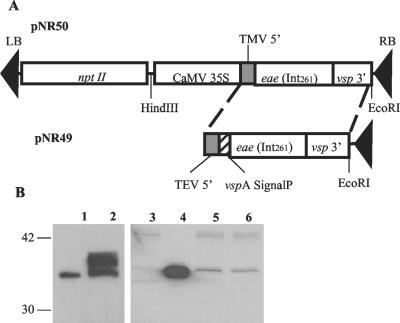
We began by generating intimin-expressing transgenic plant cells. For this purpose, we constructed two expression vectors (pNR49 and pNR50) with the eae gene from pMW103 (13) that we had optimized for expression in plants (Fig. 1A). The intimin protein encoded by this gene has an amino-terminal histidine tag and consists of the carboxy-terminal 261 amino acids of intimin from E. coli O157:H7 (Int261). The carboxy terminus of intimin is the cell-binding domain (11, 12). A. tumefaciens EHA105 was used to transfer the DNA for Int261 expression from the binary vectors into the nicotine-free tobacco cell line NT-1 (1, 20). Kanamycin-resistant calli were selected, and Int261 expression was assayed by Western blot analysis (Fig. 1B).
FIG. 1.
Expression of Int261 in NT-1 cells. (A) The Int261 plant transformation vectors pNR50 and pNR49 are comprised of an Int261 expression cassette with a CaMV 35S promoter and a neomycin phosphotransferase II expression cassette for detection of kanamycin resistance in successfully transformed clones. A VspA signal peptide was included in pNR49 to increase protein expression. (B) Immunoblot analysis of Int261 expressed with or without the signal peptide. Int261 (∼35 kDa) purified by nickel affinity chromatography from pMW103 (13) (∼75 ng) served as the positive control (lanes 1 and 4). NT-1 cells transformed with pGPTV-Kan vector only did not express Int261 (lane 3). NT-1 cells transformed with pNR49 (CSP20) expressed several intimin-specific bands (lane 2). NT-1 cells transformed with pNR50 (C34) from both plate (lane 5) and broth (lane 6) cultures expressed a single intimin-specific band (∼35 kDa).
Two independent transgenic NT-1 cell clones were used for the experiments described below. Clone 20 (CSP20) was created by transformation of NT-1 cells with pNR49. This clone expressed Int261 with a plant-specific signal peptide (∼35 kDa) and produced 10 to 13 μg of Int261 per g of total plant material. Clone 34 (C34) was generated by transformation of NT-1 cells with pNR50. This clone expressed Int261 without a signal peptide and produced ∼3 μg of Int261 (∼35 kDa) per g of total plant material (Fig. 1B).
Glycosylation of Int261 expressed with a plant signal peptide.
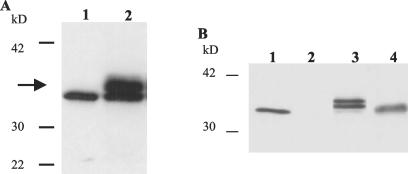
Although CSP20 expressed four times more antigen than C34, CSP20 accumulated higher-molecular-weight bands over time in culture (Fig. 2A). Moreover, this plant material, when fed to mice, elicited an antibody response (as assessed by ELISA) that recognized Int261 purified from that clone, but not from bacterial cells. In addition, mice fed CSP20 were colonized at the same level and for the same duration as mice fed plant cells alone when challenged with wild-type E. coli O157:H7 strain 86-24 (data not shown). Based on these findings, we speculated that the higher-molecular-weight bands noted in Fig. 2A might represent a glycosylated form of Int261 that was recognized by the murine host differently than the protein made by bacteria. To test this theory, Int261 from CSP20 was purified by nickel affinity chromatography and then treated with trifluoromethanesulfonic acid (TFMS) (GlycoFree deglycosylation kit from Glyco, Inc). TFMS nonselectively removes both N- and O-linked glycans from glycoproteins without altering the primary protein structure. The appearance of a band with a molecular mass consistent with that of control Int261 (∼35 kDa, purified from pMW103 [13] by nickel affinity chromatography) after treatment with TFMS strongly suggested that Int261 from CSP20 was in fact glycosylated (Fig. 2B). We noted that the Int261 sequence contains two potential asparagine-linked glycosylation sites (NQS and NTS) that could be modified by glycosylation in this endoplasmic reticulum-targeted form. The two higher-molecular-weight bands observed for Int261 from CSP20 could result from glycosylation of either one or both of these N-linked sites. Since this probable glycosylation of Int261 in CSP20 adversely affected the immune response to this antigen, we elected to proceed with the C34 clone that expressed Int261 without the addition of a signal peptide and in an apparently unglycosylated state (Fig. 1B).
FIG. 2.
Immunoblot analysis of Int261 from NT-1 cell clone CSP20. (A) The higher-molecular-weight, intimin-specific bands (arrow) accumulated as the culture aged. Lane 1, control Int261 purified from pMW103 (13); lane 2, Int261 from CSP20 in continuous culture for over 6 months. (B) Int261 from CSP20 treated with TFMS. Int261 purified by Ni affinity chromatography from pMW103 (13) shows the expected size of approximately 35 kDa (lane 1). Plant cell sonic extracts from NT-1 cells alone did not produce Int261 (lane 2). Int261 from CSP20 was purified by nickel affinity chromatography and showed two higher-molecular-weight intimin-specific bands compared to the control (lane 3). When material from lane 3 was treated with TFMS, a single band of approximately 35 kDa appeared that was consistent with the size of the control Int261 band (lane 4).
Generation of intimin-specific serum and fecal antibodies.
We tested several different vaccination strategies with C34 in mice. The vaccine protocols and the serum (IgG) and fecal (IgA and IgG) ELISA results are summarized in Table 4. To maximize the immune response to orally administered Int261, we tried a priming-boosting protocol (groups E and F) similar to that used to elicit an enhanced immune response to the B subunit pentamers of E. coli LT-B (27). Five of ten mice primed by injection of purified Int261 from C34 and then fed nontransgenic plant material developed an intimin-specific fecal antibody response. These results suggest that even a single inoculation with Int261 from C34 can elicit a mucosal antibody response to intimin. Seven of ten mice primed by injection of Int261 from C34 and then boosted by feeding with C34 (with CT as an oral adjuvant) produced an intimin-specific fecal IgA response. The Fisher's exact test was used to compare percentages of responders, and we found that both groups E and F had a significantly higher number of fecal IgA responders than the negative control group, B (P = 0.033 and 0.003, respectively). There was no significant difference in percentages of responders between groups E or F and group A (P = 0.175 and 1.000, respectively). In addition, 3 of 10 mice developed an IgG response in serum, and serum samples from these mice blocked adherence of wild-type E. coli O157:H7 to HEp-2 cells compared to preimmune serum controls (data not shown).
TABLE 4.
Immunogenicity in BALB/c mice of NT-1 cells that express Int261
| Group, immunization protocol | Vaccination method | IgG titer (no. of responders/total) in seruma | Titer (no. of responders/total) in fecesa |
|---|---|---|---|
| A, i.p. Int261 from pMW103 | 20 μg of Int261 plus TiterMax i.p. on day 0, 10 μg of Int261 plus TiterMax i.p. on days 10 and 20 | 300,000 (10/10)b | 560 (8/10) (IgA only)c |
| B, Fed NT-1 cells only | 5 g of NT-1 cells fed on days 0, 7, and 14 | <50 (0/10) | <50 (0/10) |
| C, Fed C34 cells only | 5 g of C34 with 15 μg of Int261 fed on days 0, 7, and 14 | <50 (0/10) | 340 (4/10) (IgG and IgA) |
| D, Fed C34 cells with CTd | 5 g of C34 with 15 μg of Int261 plus CT fed on days 0, 7, and 14 | <50 (0/10) | 280 (6/10) (IgG and IgA) |
| E, i.p. Int261 from C34, fed NT-1 cells only | ∼15 μg of Int261 from C34 plus TiterMax i.p. on day 0; 5 g of NT-1 cells fed on days 7 and 14 | 50 (1/10) | 150 (5/10) (IgA only) |
| F, i.p. Int261 from C34, fed C34 cells with CT | ∼15 μg of Int261 from C34 plus TiterMax i.p. on day 0; 5 g of C34 cells with 15 μg of Int261 plus CT fed on days 7 and 14 | 80 (3/10) | 180 (7/10) (IgA only) |
Serum and fecal pellet extracts were collected throughout the experiment and were evaluated by ELISA for the presence of antibodies against Int261. The titer was defined as the geometric mean of the reciprocal of the highest dilution that gave an absorbance (A600) above both preimmune and background levels. Responders were defined as mice that had a detectable titer at the final sample collection point (7 days after the final feeding or injection).
Significantly higher IgG titer in serum and number of responders than other groups (P < 0.0001, two-sided t test; P < 0.003, Fisher's exact test).
IgA titer in feces was significantly different than that for groups E and F (P = 0.019 and 0.013, respectively [two-sided t test]). The number of fecal IgA responders was significantly different from that for groups E and F (P = 0.175 and 1.000, respectively [Fisher's exact test]).
CT, cholera toxin (7.5 μg; Sigma).
Colonization of mice by E. coli O157:H7 is dependent on intimin.
We determined that E. coli O157:H7 strain 86-24 Strr (35) reproducibly colonized female BALB/c mice at modest levels for approximately 10 days (mean, 6.4 to 13.4 days for unimmunized mice). Furthermore, in an experiment limited to 7 days, E. coli O157:H7 86-24 Strr colonized BALB/c mice a mean of 6.4 days, whereas the intimin-negative mutant 86-24 Strr eaeΔ10 showed a statistically significant reduction in the duration of colonization (mean, 3.2 days; P < 0.005; log-rank test).
Challenge of vaccinated mice with E. coli O157:H7.
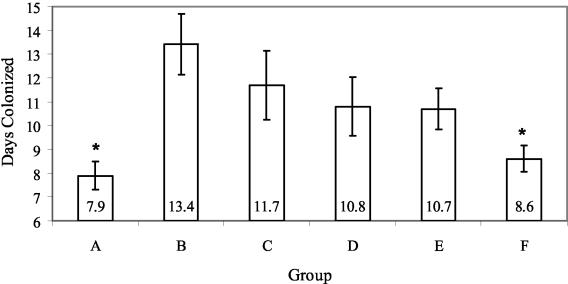
With this intimin-dependent mouse model of E. coli O157:H7 colonization, we orally infected the same groups of immunized mice described in Table 4 with strain 86-24 Strr. We found that mice primed with an i.p. injection of Int261 from C34 and then fed homologous C34 plant material with CT showed a statistically significant (P < 0.002; log-rank test) reduction in the duration of colonization (as measured by fecal shedding) compared to the negative control mice that were fed nontransgenic NT-1 cells only (Fig. 3). The mice that were primed and boosted with Int261 from transgenic plant material also exhibited a statistically significant decrease in the duration of bacterial colonization compared to mice that were only immunized i.p. with Int261 from C34 and then boosted with nontransgenic plant material (P = 0.033; log-rank test). These results suggest that the feeding of transgenic, Int261-expressing plant material plays a significant role in boosting the immune response to Int261 and in reducing the duration of bacterial colonization. The positive control mice, injected i.p. three times with highly purified Int261 from pMW103 (13), also displayed a statistically significant reduction in the duration of colonization versus the negative control (P < 0.002; log-rank test). There was no difference in the duration of shedding between the positive control immunization method and the priming-boosting protocol with oral delivery of transgenic plant material that expressed Int261 as a boost. This last observation suggests that immunization with a plant-based oral vaccine can induce an immune booster response sufficient to provide protection from a challenge with wild-type E. coli O157:H7.
FIG. 3.
Duration of E. coli O157:H7 colonization in mice immunized with Int261. The same groups of mice as those shown in Table 4 were challenged with wild-type E. coli O157:H7. Mice from both groups A and F (*) showed a statistically significant (P < 0.002; log-rank test) reduction in the duration of colonization compared to group B. No other groups demonstrated a statistically significant reduction compared to group B.
DISCUSSION
Many pathogens infect or invade via mucosal surfaces, so the capacity of plant-based vaccines to induce mucosal immunity is a great advantage. Plant cells act as a natural microencapsulation system to protect the vaccine antigens from being degraded in the upper digestive tract before they can reach the gut-associated lymphoid tissue. Recent studies suggest that plant-based oral vaccines can significantly boost mucosal immune responses primed by parenteral injection (27, 44). Parenteral priming of the immune system may allow the gut-associated lymphoid tissue to react successfully to the small amounts of antigen delivered during oral immunization and thus decrease the possibility of inducing oral immunotolerance to plant-based and other orally delivered vaccine antigens. Our studies support these recent findings by demonstrating that parenteral priming of mice with intimin purified from transgenic plant cells can assist in the development of an intimin-specific fecal immune response when these mice are subsequently boosted with oral feeding of the same intimin-expressing transgenic plant material. Furthermore, in this study mice that were parenterally primed and then given an oral booster showed a statistically significant decrease in the duration of colonization by wild-type E. coli O157:H7 upon challenge (P < 0.002; log-rank test). Mice immunized entirely by oral feeding did exhibit a reduction in the duration of colonization versus unimmunized mice, but the reduction was not statistically significant. These results suggest that a combination of vaccination strategies with a vaccine antigen produced in and delivered by transgenic plants can function in inducing beneficial, specific immune responses. Further studies are needed to determine whether an oral-only immunization route would suffice if the antigen levels in the plant material or the number of doses delivered were increased so as to induce an immune response sufficient to result in a reduction in the duration of bacterial colonization. Such approaches might eliminate the requirement for priming by parenteral injection of intimin.
Our discovery that nonglycosylated bacterial proteins can be glycosylated in transgenic plant systems highlights an issue previously not reported in other studies of vaccine antigen expression in transgenic plants. We found that the glycosylation of Int261 expressed in NT-1 cells adversely affected the immunogenicity of the molecule and resulted in an aberrant immune response that was unable to block adherence of E. coli O157:H7. Although the addition of a plant-specific signal peptide increased the vaccine antigen expression levels in the plant cells, the signal peptide may have caused the protein to be glycosylated in the plant expression system by trafficking the newly expressed protein directly into the endoplasmic reticulum, where it would be more accessible to the glycosylation pathways in the Golgi apparatus. The removal of the plant-specific signal peptide from the Int261 plant expression constructs resulted in decreased protein expression but appeared to produce an unglycosylated protein. Our experience emphasizes one of the challenges involved in the successful production of bacterial vaccine antigens in transgenic plant systems.
Our finding that these transgenic cells synthesize Int261 in a conformation that is immunologically equivalent to the native protein underscores the feasibility of transgenic plant-based systems for expression and delivery of oral vaccines. For further vaccine development, we plan to express a signal sequence-free Int261 construct in a whole plant, perhaps by use of recently developed chloroplast transformation and expression models (19) that maximize protein expression and minimize protein modification (such as glycosylation). The selected plant system will likely be alfalfa because it is compatible with oral administration to cattle and is amenable to both nuclear and chloroplast transformation methods. Debate still exists as to whether feedlot food type, holding pen cleanliness, or water quality management would offer the benefit of reducing EHEC O157:H7 colonization levels in cattle (9, 28, 39). One might hypothesize that removing or reducing environmental sources of EHEC O157:H7 infection, such as soiled bedding or contaminated water sources, would reduce the prevalence of EHEC O157:H7 in cattle. Unfortunately, these hypotheses have not been adequately tested. However, the notion that reduction of sources of EHEC O157:H7 infection and contamination would reduce not only the prevalence of EHEC O157:H7 in cattle, but also the risk of human infection through other environmental sources such as drinking or recreational water and fruits and vegetables, has strong intuitive appeal. We believe that the use of transgenic plants for the production and delivery of an EHEC O157:H7 vaccine for cattle, alone or in conjunction with other potential control methods (5), could lead to a significant decrease in the level of colonization or percentage of cattle infected with pathogenic E. coli O157:H7. Such a reduction will likely translate into a decline in beef carcass and environmental contamination by E. coli O157:H7 and a decrease in transmission of the bacterium to humans.
Acknowledgments
We gratefully acknowledge Brian Maloney, Stephen Darnell, and Humberto Carvalho for technical assistance and Susan Rasmussen for her critical evaluation of the manuscript. We are also appreciative of the kind gift of purified bacterially expressed intimin provided by James Sinclair for this study. Additionally, we thank the Boyce Thompson Institute for the training N.J. received in plant cell transformation methods.
This work was supported by National Institutes of Health grant AI 20148-20 (A.O.B.), U.S. Department of Agriculture grant 97-35201-4578 (A.O.B.), an American Meat Institute Foundation grant (A.O.B.), and the Defense Advanced Research Projects Agency grant N65236-98-1-5411 (H.S.M.).
Editor: J. T. Barbieri
REFERENCES
- 1.An, G. 1985. High-efficiency transformation of cultured tobacco cells. Plant Physiol. 79:568-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, T., D. K. Chong, and W. H. Langridge. 1998. Efficacy of a food plant-based oral cholera toxin B subunit vaccine. Nat. Biotechnol. 16:292-297. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa, T., D. K. Chong, J. L. Merritt, and W. H. Langridge. 1997. Expression of cholera toxin B subunit oligomers in transgenic potato plants. Transgenic Res. 6:403-413. [DOI] [PubMed] [Google Scholar]
- 4.Becker, D., E. Kemper, J. Schell, and R. Masterson. 1992. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20:1195-1197. [DOI] [PubMed] [Google Scholar]
- 5.Brashears, M. M., M. L. Galyean, G. H. Loneragan, J. E. Mann, and K. Killinger-Mann. 2003. Prevalence of Escherichia coli O157:H7 and performance by beef feedlot cattle given Lactobacillus direct-fed microbials. J. Food Prot. 66:748-754. [DOI] [PubMed] [Google Scholar]
- 6.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-378. [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1994. Addressing emerging infectious disease threats: a prevention strategy for the United States (executive summary). Morb. Mortal. Wkly. Rep. 43(RR-5):1-18. [PubMed] [Google Scholar]
- 8.Dean-Nystrom, E. A., B. T. Bosworth, H. W. Moon, and A. D. O'Brien. 1998. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect. Immun. 66:4560-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diez-Gonzalez, F., T. R. Callaway, M. G. Kizoulis, and J. B. Russell. 1998. Grain feeding and the dissemination of acid-resistant Escherichia coli from cattle. Science 281:1666-1668. [DOI] [PubMed] [Google Scholar]
- 10.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankel, G., D. C. Candy, P. Everest, and G. Dougan. 1994. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect. Immun. 62:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel, G., D. C. Candy, E. Fabiani, J. Adu-Bobie, S. Gil, M. Novakova, A. D. Phillips, and G. Dougan. 1995. Molecular characterization of a carboxy-terminal eukaryotic-cell-binding domain of intimin from enteropathogenic Escherichia coli. Infect. Immun. 63:4323-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gansheroff, L. J., M. R. Wachtel, and A. D. O'Brien. 1999. Decreased adherence of enterohemorrhagic Escherichia coli to HEp-2 cells in the presence of antibodies that recognize the C-terminal region of intimin. Infect. Immun. 67:6409-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin, P. M. 1998. Epidemiology of Shiga toxin-producing Escherichia coli infections in humans in the United States, p. 15-22. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 15.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, D. D., T. E. Besser, D. H. Rice, and P. I. Tarr. 1998. Ecology of Escherichia coli O157:H7 in cattle and impact of management practices, p. 85-91. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 18.Haq, T. A., H. S. Mason, J. D. Clements, and C. J. Arntzen. 1995. Oral immunization with a recombinant bacterial antigen produced in transgenic plants. Science 268:714-716. [DOI] [PubMed] [Google Scholar]
- 19.Heifetz, P. B., and A. M. Tuttle. 2001. Protein expression in plastids. Curr. Opin. Plant Biol. 4:157-161. [DOI] [PubMed] [Google Scholar]
- 20.Hood, E. E., S. B. Gelvin, L. S. Melchers, and A. Hoekma. 1993. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2:208-218. [Google Scholar]
- 21.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan, D., S. A. McEwen, A. M. Lammerding, W. B. McNab, and J. B. Wilson. 1999. Pre-slaughter control of Escherichia coli O157 in beef cattle: a simulation study. Prev. Vet. Med. 41:55-74. [DOI] [PubMed] [Google Scholar]
- 23.Kaper, J. B., S. Elliott, V. Sperandio, N. T. Perna, G. F. Mayhew, and F. R. Blattner. 1998. Attaching-and-effacing intestinal histopathology and the locus of enterocyte effacement, p. 163-182. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 24.Kaper, J. B., L. J. Gansheroff, M. R. Wachtel, and A. D. O'Brien. 1998. Intimin-mediated adherence of Shiga toxin-producing Escherichia coli and attaching-and-effacing pathogens, p. 148-156. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 25.Keen, J. E., and R. O. Elder. 2002. Isolation of Shiga-toxigenic Escherichia coli O157 from hide surfaces and the oral cavity of finished beef feedlot cattle. J. Am. Vet. Med. Assoc. 220:756-763. [DOI] [PubMed] [Google Scholar]
- 26.Kong, Q., L. Richter, Y. F. Yang, C. J. Arntzen, H. S. Mason, and Y. Thanavala. 2001. Oral immunization with hepatitis B surface antigen expressed in transgenic plants. Proc. Natl. Acad. Sci. USA 98:11539-11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauterslager, T. G., D. E. Florack, T. J. van der Wal, J. W. Molthoff, J. P. Langeveld, D. Bosch, W. J. Boersma, and L. A. Hilgers. 2001. Oral immunisation of naive and primed animals with transgenic potato tubers expressing LT-B. Vaccine 19:2749-2755. [DOI] [PubMed] [Google Scholar]
- 28.LeJeune, J. T., T. E. Besser, and D. D. Hancock. 2001. Cattle water troughs as reservoirs of Escherichia coli O157. Appl. Environ. Microbiol. 67:3053-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason, H. S., J. M. Ball, J. J. Shi, X. Jiang, M. K. Estes, and C. J. Arntzen. 1996. Expression of Norwalk virus capsid protein in transgenic tobacco and potato and its oral immunogenicity in mice. Proc. Natl. Acad. Sci. USA 93:5335-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason, H. S., T. A. Haq, J. D. Clements, and C. J. Arntzen. 1998. Edible vaccine protects mice against Escherichia coli heat-labile enterotoxin (LT): potatoes expressing a synthetic LT-B gene. Vaccine 16:1336-1343. [DOI] [PubMed] [Google Scholar]
- 31.Mason, H. S., D. M. Lam, and C. J. Arntzen. 1992. Expression of hepatitis B surface antigen in transgenic plants. Proc. Natl. Acad. Sci. USA 89:11745-11749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKee, M. L., A. R. Melton-Celsa, R. A. Moxley, D. H. Francis, and A. D. O'Brien. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect. Immun. 63:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKee, M. L., and A. D. O'Brien. 1996. Truncated enterohemorrhagic Escherichia coli (EHEC) O157:H7 intimin (EaeA) fusion proteins promote adherence of EHEC strains to HEp-2 cells. Infect. Immun. 64:2225-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melton-Celsa, A. R., J. E. Rogers, C. K. Schmitt, S. C. Darnell, and A. D. O'Brien. 1998. Virulence of Shiga toxin-producing Escherichia coli (STEC) in orally-infected mice correlates with the type of toxin produced by the infecting strain. Jpn. J. Med. Sci. Biol. 51(Suppl.):S108-S114. [DOI] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. F. Mayhew, G. Pósfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richter, L. J., Y. Thanavala, C. J. Arntzen, and H. S. Mason. 2000. Production of hepatitis B surface antigen in transgenic plants for oral immunization. Nat. Biotechnol. 18:1167-1171. [DOI] [PubMed] [Google Scholar]
- 38.Sinclair, J. F., and A. D. O'Brien. 2002. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-gamma of enterohemorrhagic Escherichia coli O157:H7. J. Biol. Chem. 277:2876-2885. [DOI] [PubMed] [Google Scholar]
- 39.Smith, D., M. Blackford, S. Younts, R. Moxley, J. Gray, L. Hungerford, T. Milton, and T. Klopfenstein. 2001. Ecological relationships between the prevalence of cattle shedding Escherichia coli O157:H7 and characteristics of the cattle or conditions of the feedlot pen. J. Food Prot. 64:1899-1903. [DOI] [PubMed] [Google Scholar]
- 40.Tacket, C. O., H. S. Mason, G. Losonsky, J. D. Clements, M. M. Levine, and C. J. Arntzen. 1998. Immunogenicity in humans of a recombinant bacterial antigen delivered in a transgenic potato. Nat. Med. 4:607-609. [DOI] [PubMed] [Google Scholar]
- 41.Tacket, C. O., H. S. Mason, G. Losonsky, M. K. Estes, M. M. Levine, and C. J. Arntzen. 2000. Human immune responses to a novel Norwalk virus vaccine delivered in transgenic potatoes. J. Infect. Dis. 182:302-305. [DOI] [PubMed] [Google Scholar]
- 42.Thanavala, Y., Y. F. Yang, P. Lyons, H. S. Mason, and C. Arntzen. 1995. Immunogenicity of transgenic plant-derived hepatitis B surface antigen. Proc. Natl. Acad. Sci. USA 92:3358-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valcour, J. E., P. Michel, S. A. McEwen, and J. B. Wilson. 2002. Associations between indicators of livestock farming intensity and incidence of human Shiga toxin-producing Escherichia coli infection. Emerg. Infect. Dis. 8:252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Heijden, P. J., A. T. Bianchi, B. A. Bokhout, M. Dol, J. W. Scholten, and W. Stok. 1989. Quantification of antigen-specific antibody-secreting cells in the small intestine and other lymphoid organs of mice after oral booster immunization. Immunology 66:404-409. [PMC free article] [PubMed] [Google Scholar]
- 45.Wells, J. G., B. R. Davis, I. K. Wachsmuth, L. W. Riley, R. S. Remis, R. Sokolow, and G. K. Morris. 1983. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J. Clin. Microbiol. 18:512-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woods, J. B., C. K. Schmitt, S. C. Darnell, K. C. Meysick, and A. D. O'Brien. 2002. Ferrets as a model system for renal disease secondary to intestinal infection with Escherichia coli O157:H7 and other Shiga toxin-producing E. coli. J. Infect. Dis. 185:550-554. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, T., M. P. Doyle, B. G. Harmon, C. A. Brown, P. O. Mueller, and A. H. Parks. 1998. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J. Clin. Microbiol. 36:641-647. [DOI] [PMC free article] [PubMed] [Google Scholar]