Abstract
Narrow spectrum antimicrobial activity has been designed to reduce the expression of two essential genes, one coding for the protein subunit of RNase P (C5 protein) and one for gyrase (gyrase A). In both cases, external guide sequences (EGS) have been designed to complex with either mRNA. Using the EGS technology, the level of microbial viability is reduced to less than 10% of the wild-type strain. The EGSs are additive when used together and depend on the number of nucleotides paired when attacking gyrase A mRNA. In the case of gyrase A, three nucleotides unpaired out of a 15-mer EGS still favor complete inhibition by the EGS but five unpaired nucleotides do not.
Keywords: RNase P, tRNA processing
Current antimicrobial drugs inhibit bacteria primarily by targeting essential proteins (or protein-mediated processes) conserved among many bacterial species (1). Accordingly, medical and agricultural antimicrobials not only treat pathogenic bacterial infections, but also affect commensal bacteria. This broad spectrum of activity creates side effects for individual patients (2, 3) and also exerts selective pressure for the emergence, spread, and interspecies transfer of resistance (4, 5). Here, we report antimicrobial activity mediated by using techniques of external guide sequences (EGSs) that enhance RNase P-mediated mRNA cleavage (6, 7). We inhibited bacterial growth by reducing the level of expression for two different essential proteins that exist in fewer than 1,500 copies per Escherichia coli, RNase P C5 protein (8), and gyrase A (9).
RNase P catalyzes tRNA processing (10). The tRNA products of these cleavages result in important components of the protein synthetic mechanisms. This enzyme contains a catalytic RNA subunit (M1 RNA) and a protein subunit (C5 protein) in E. coli. Gyrase A catalyzes chromosomal DNA supercoiling during replication (11), is the molecular target of quinolone antimicrobials (12), and can mediate quinolone resistance (13–15). Eukaryotes also express essential RNase P subunits (10) and gyrase (16) enzymes (the later a target for antineoplastic agents), but they are quite distinct from their bacterial homologs in RNA and protein sequence (17) as well as in some aspects of enzymatic reaction details (10, 16).
E. coli EGSs were based on species-specific gyrase A sequences that are not present in Salmonella typhimurium, but which encode identical gyrase A protein sequences in both species (15, 18), and the C5 protein subunit of RNase P. This system provides a foundation for general strategies to attack bacteria and a tool for the inducible disruption of bacterial gene products. It also suggests mechanisms for a degree of exquisitely narrow spectrum antimicrobial activity, discriminating between and selectively inhibiting even closely related species of bacteria based on fine differences in bacterial coding sequences.
The inhibition of bacterial growth via EGS-targeted technology exhibits species specificity and dose–response. The cleavage is independent of certain mutations of up to three bases in the gyrase A EGS complementary sequence.
Materials and Methods
Bacterial Strains, Plasmids, and EGSs.
EGS oligonucleotides were designed to include a region of complementarity to target mRNA followed by a 3′-ACCA terminal sequence. This strategy will allow formation of a duplex molecule recognized as a substrate by endogenous RNase P with resultant cleavage of target mRNA (6). The individual EGS oligonucleotide sequences are listed in Figs. 1 and 2.
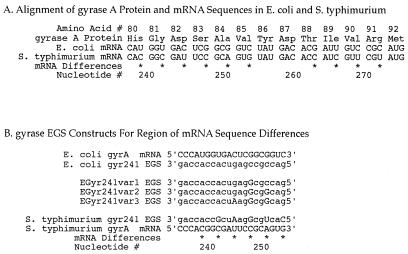
Figure 1.
EGS and target mRNA sequences. Nucleotide number 1 occurs at the mRNA start codon. All of the EGSs have a 3′-acca tail, important for recognition and cleavage of target mRNA in an EGS:mRNA duplex by endogenous RNase P. The remainder of the EGS oligonucleotide is designed to be complementary to its mRNA target and to guide target mRNA cleavage at the nucleotide specified (e.g., E. coli C5 mRNA is cleaved at nucleotide 32 with E. coli C5 EGS 32). (A) C5, the protein subunit of prokaryotic RNaseP, is shown with two mRNA sequences, both of which encode functional C5 protein. Synthetic C5 mRNA has been reengineered for increased recombinant protein synthesis, via codon substitution. The nucleotides that differ in synthetic C5 as compared with E. coli C5 mRNA are starred and result in the EGSs against synthetic C5 mRNA having several base pair mismatches with corresponding E. coli mRNA segments. Although differing at their start codons (GUG for E. coli C5 and AUG for synthetic C5: both coding for M) the two resultant C5 protein sequences are identical (MVKLAFPRELRLLTPSQFTF, for mRNA shown here). (B) The targeted section of EGyrA mRNA, with three EGS oligonucleotides.
Figure 2.
Gyrase A variation between E. coli and S. typhimurium. One portion of the gyrase A molecule was studied in particular detail (see text). This portion of gyrase mRNA was used as a target for EGSs designed to assess EGSs' species specificity and nucleotide-level specificity. (A) The gyrase A protein has identical amino acid sequence in E. coli and S. typhimurium over this region. However, due to codon use variation, these two species have several differences in their respective mRNAs encoding gyrase A (*). Numbering is per EGyr sequence of Wang (16). (B) EGS oligonucleotides designed to target cleavage of this portion of mRNA at nucleotide 241. EGSs all include a 3′-acca tail and an oligonucleotide region complementary to their mRNA target Here, complemetarity is designed to be imperfect in some constructs. EGSs shown here include: EGyr241 EGS (against EGyr mRNA) and SGyr241 EGS (against SGyr mRNA); as well as three variants of the EGyr241 EGS, which differ from this EGS by step-wise introductions of one, two, or three nucleotide changes (EGyr241var1, var2, or var3 EGSs, respectively). Changes compared to EGyr241 are indicated in uppercase letters.
E. coli BL21 (DE3) and EGS expression vector plasmid construction steps are described in detail elsewhere (19). For the vector that expresses four EGSs, the complete two EGS cassette (including T7 promoter and terminator) was excised by digestion with EcoRI and HindIII, treated with Klenow polymerase, gel-purified, and then introduced by blunt end ligation into the Klenow-treated HindIII site just 3′ to the complete cassette of another two EGS vector. One negative control vector (no T7 promoter) was produced from a single EGS vector by removal of the T7 promoter, single EGS, hammerhead, and part of the T7 terminator. Constructs were verified by sequencing.
Inducible EGSs are under the control of the lac operator and, therefore, are regulated by isopropyl β-d-thiogalactoside (IPTG). Technical details of this construction are reported elsewhere (7).
Assessing Bacterial Viability.
The impact of EGS expression on bacterial viability in vivo was assessed by using quantitative plate colony counts of E. coli liquid cultures. E. coli transformants containing plasmid vectors encoding the inducible expression of various EGS oligonucleotides were each grown in LB carbenicillin liquid culture (LB medium supplemented with 50 μg/ml carbenicillin) from a single colony picked from a fresh, overnight LB ampicillin (LBa) agar plate culture. Liquid cultures were grown in side arm flasks in a 37°C shaker until entering log phase. On entry into log phase, liquid cultures received a fresh aliquot of carbenicillin stock (for an additional 50 μg/ml final concentration) and were split into two culture tubes. These split cultures were incubated in a 37°C shaker in parallel: one tube with the inducer, IPTG (from single-use frozen stock aliquots), for 2 mM final concentration for EGS production and one tube without. At various times after growth in liquid culture, sample aliquots were removed and equally serially diluted, and spread on LB and LBa (100 μg/ml) agar plates for overnight growth at 37°C.
Plasmid loss in a given culture was assessed by the presence of many fewer colonies on LBa plates than on LB plates. Every culture was plated in quadruplicate, and an inhibition index (see below) was calculated to quantitate impact on bacterial viability after EGS induction by IPTG.
Assays of mRNA Cleavage in Vitro.
Assays of target RNA cleavage in vitro by recombinant RNase P guided by EGS oligonucleotides were performed as detailed (20). The assays in vitro were performed by using reaction mixes incubated for 15 min at 37°C. Target mRNAs encoded either the E. coli C5 protein component of RNase P or a synthetic C5 protein (described below) and were produced by transcription in vitro from plasmid cDNA. These plasmids were linearized by EcoRI or HindIII digestion of the E. coli C5 or synthetic C5 cDNA plasmids, pFHC1008, and pBSC5, respectively. The resultant E. coli C5 mRNA of 535 nt and synthetic C5 mRNA of 280 nt were radiolabeled via incorporation of [32P-α]-GTP during transcription in vitro.
Results
Cleavage of Target mRNA in Vitro.
Two mRNA target molecules that would disrupt cell function when cleaved were examined: the mRNAs encoding protein subunits of gyrase A or RNase P. For gyrase A mRNA the region around nucleotide 240 was selected for detailed study because of four special features. The protein segment encoded by this region of mRNA includes the proposed active site of the gyrase molecule (21) is also the quinolone antimicrobial binding site (22) and bestows quinolone resistance after well defined mutations (14, 15). This region of gyrase A mRNA is also notable (see Figs. 1 and 2) for encoding identical protein sequences in E. coli and S. typhimurium, despite containing extensive RNA differences between these two closely related enteric bacteria, due to codon use variability (15, 18). This property might allow for specific disruption of gyrase protein production in E. coli but not in S. typhimurium, via mRNA-specific discrimination and cleavage.
Transcription in vitro and subsequent T1 RNase digestion of target mRNA revealed several accessible guanine residues in single-stranded regions of mRNA for potential EGS binding. These single-stranded regions included nucleotides −2 to 16 and 32–48 in the mRNA encoding the C5 protein subunit of RNase P, and nucleotides 211–217, 223–224, and 241–244 in the mRNA encoding gyrase A (both numbered with start codons beginning as nucleotide 1).
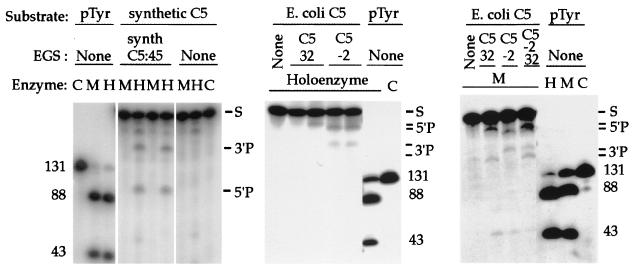
Two forms of C5 mRNA are tested as cleavage targets by their respective complementary EGSs: synthetic C5 (used as a negative control in experiments in vivo) and naturally occurring E. coli C5. In each case, target mRNA was cleaved by RNase P after mixture with complementary EGSs. The sizes of the mRNA cleavage products were consistent with single-site cleavage of mRNA targets, occurring at the intended site of EGS hybridization, as predicted from past studies of EGS-directed mRNA cleavage in prokaryotic systems. The C5:−2 and the C5:+32 both directed RNase P-mediated cleavage of the E. coli C5 mRNA target, but these two EGSs did not exhibit cooperative activity beyond the sum of their individual effects (Fig. 3, lane C5: −2/32).
Figure 3.
EGS-directed cleavage of RNA substrate targets in vitro. The components of each reaction mixture are listed above each lane. Target substrates for cleavage include: two preparations of mRNA encoding the C5 protein, one naturally occurring E. coli C5 mRNA (E. coli C5), and one synthetic E. coli C5 mRNA reengineered via conservative codon substitutions (Fig. 1; synthetic C5); as well as precursor tRNA (pTyr). EGSs include an oligonucleotide with sequences complementary to synthetic C5 mRNA, predicted to guide mRNA cleavage at nucleotide 45 (synth C5 EGS:45), and oligonucleotides complementary to naturally occurring E. coli C5 mRNA, predicted to guide mRNA cleavage at nucleotides −2 (C5 EGS:−2) or 32 (C5 EGS:32). Total reaction volumes: 10 μl. RNase P holoenzyme (20 nM M1 RNA and 200 nM C5 protein) was incubated in binding buffer (20 mM Hepes⋅KOH, pH 8/400 mM NH4OAc/10 mM Mg(OAc)2/5% glycerol). M1 RNA (200 nM) was incubated in 50 mM Tris, pH 7.5/100 mM NH4Cl/100 mM MgCl2/4% polyethylene glycol. The enzymes were incubated for 15 min in the presence of mRNA substrate (10 nM) and EGS RNAs (50 nM). When the same EGS is listed for more than one otherwise identical enzyme lane, the molar concentration ratio of EGS to target increases from 10× to 50× going rightward. Reference cleavage of precursor to tRNATyr (pTyr) provides size markers, and the target mRNA substrates (S) and cleavage reaction 3′ and 5′ products are indicated. C, no addition other than substrate (control); H, holoenzyme added; M, M1 RNA added.
Decreased Bacterial Viability After EGS Induction.
The quantitative culture plates from a representative experiment in vivo are shown in Fig. 4. This figure includes transformants that each express two different inducible EGS oligonucleotides, targeting different portions of a single mRNA molecule. Decreased colony counts are observed after induction of EGS expression in the two transformants each encoding two EGS molecules against E. coli gyrase A (EGyrA241/211, targeting cleavage of EGyrA mRNA at nucleotides 241 and 211; and EGyrA241/223, targeting cleavage at nucleotides 241 and 223), as well as the transformant with two EGSs against the E. coli C5 subunit of RNase P (EC5 32/−2, targeting E. coli C5 mRNA cleavage at nucleotides 32 and −2). In each case, there was greater inhibition of bacterial viability at 25 h after induction than at 8 h.
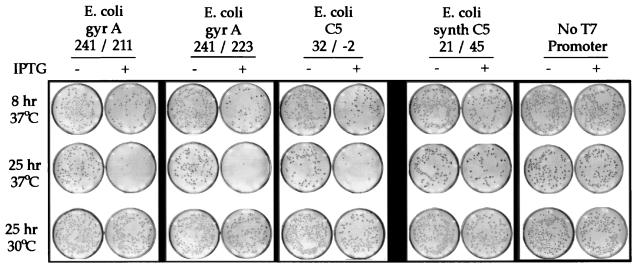
Figure 4.
Growth of bacteria of E. coli transformants with EGS constructs. Liquid cultures were each inoculated with a single E. coli colony and grown until entering log phase. In log phase, each culture was split and either received IPTG to induce expression of EGS oligonucleotides or was not induced. These split cultures then were incubated in parallel for the number of hours shown, at the temperature shown. The E. coli transformants contained plasmids encoding various EGS constructs designed to cleave target mRNA at the nucleotides listed (e.g., EGyrA 241/211 EGS is designed to cleave gyrase A mRNA at nucleotides 241 and 211). Negative controls included E. coli with EGS designed to cleave synthetic (non-naturally occurring) mRNA for C5 at nucleotides 21 and 45; and an E. coli with an EGS expression vector lacking its promoter and EGS encoding segments. The original plates were electronically scanned, and images were printed by laser printer.
Two negative controls were used. One was a transformant with deletion of the inducible promoter and EGS encoding portions of its EGS expression vector. This promoterless construct consistently exhibited little, if any, impact on viability after induction. The other negative control was one of a set of transformants able to express EGS oligonucleotides designed to be noncomplementary to the target mRNAs encoding EGyrA or RNase P C5. For C5, this included two EGSs complementary to mRNA reengineered via codon substitution for expressing functional E. coli synthetic C5 protein (23). This EGS pair against synthetic C5 is noncomplementary to naturally occurring E. coli C5 mRNA at 13 of 30 nt over the regions of EGS targeting (see Fig. 1). For EGyrA, negative controls included EGS constructs complementary to the gyrase A mRNA of S. typhimurium, which differs in sequence from EGyrA mRNA (see Fig. 2) at 6 of 16 nt starting at nucleotide 240. These negative controls also showed little effect on cell viability after induction of EGS expression.
The impact of EGS induction on cell viability was quantitated by counting colonies for a given transformant and then calculating its inhibition index. This inhibition index is defined as the ratio of colonies growing without EGS induction versus with EGS induction for a given transformant such that an inhibition index of 5 represents a 5-fold decrease in growth after EGS induction. The colony counts, and calculated inhibition index values, for another experiment using the constructs each encoding two EGSs against either EGyrA mRNA or C5 mRNA are shown in Table 1. In these data, the colony counts for sets of plated transformants with versus without EGS induction are listed for 5 and 24 h after induction. For the T7 promoter and EGS-deficient construct, addition of the EGS-inducing agent (IPTG) has little effect as indicated by an inhibition index near 1 (i.e., the colony counts with and without EGS induction are nearly equal). By contrast, whereas the C5 32/−2 has only a mild (1.5-fold) effect on growth at 5 h, it progresses to a 15-fold reduction in colony counts with EGS induction at 24 h. As in Fig. 4, the constructs against EGyrA (including here an EGS targeting gyrase A mRNA at 242) also inhibit E. coli viability.
Table 1.
Summary viability of E. coli
| 5 h
|
24 h
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| +
IPTG
|
No IPTG
|
Inhibition index | +
IPTG
|
No IPTG
|
Inhibition index | |||||
| LBa | LB | LBa | LB | LBa | LB | LBa | LB | |||
| No T7 promoter | 452 | 428 | 447 | 463 | 1.0 ± 0.1 | 114 | 143 | 188 | 154 | 1.4 ± 0.1 |
| 424 | 406 | 356 | 472 | 120 | 112 | 170 | 165 | |||
| E. coli C5 32/−2 | 176 | 137 | 278 | 295 | 1.5 ± 0.2 | 5 | 4 | 86 | 79 | 15 ± 2.0 |
| 157 | 319 | 282 | 283 | 4 | 8 | 84 | 81 | |||
| EGyr241/211 | 61 | 71 | 250 | 290 | 4.2 ± 0.2 | 1 | 3 | 62 | 43 | 12 ± 6.0 |
| 69 | 58 | 251 | 281 | 5 | 8 | 48 | 35 | |||
| EGyr241/223 | 78 | 71 | 302 | 309 | 3.8 ± 0.5 | 4 | 7 | 37 | 43 | 7.0 ± 0.5 |
| 62 | 110 | 301 | 284 | 7 | 4 | 53 | 34 | |||
| EGyr242/211 | 184 | 136 | 346 | 310 | 1.9 ± 0.2 | 10 | 13 | 65 | 59 | 5.6 ± 0.3 |
| 193 | 159 | 307 | 298 | 11 | 7 | 51 | 59 | |||
| EGyr242/223 | 149 | 117 | 281 | 286 | 2.2 ± 0.2 | 7 | 4 | 74 | 48 | 14 ± 2.0 |
| 136 | 108 | 263 | 265 | 4 | 4 | 70 | 73 | |||
Colony counts of individual plates are listed for quantitative plate cultures from a single assay in vivo performed as described in Materials and Methods. The E. coli transformant panel included a negative control (no T7 promoter) described in Fig. 4 and text. Other transformants encoded dual EGS constructs against either E. coli C5 (E. coli C5 32/−2) or EGyr A (EGyr241/211, etc.), named according to the EGS nomenclature described in Fig. 1 and the text. Quantitative plating was performed at two times: 5 and 24 h after liquid cultures were split for parallel incubation either with induction (+ IPTG) or without induction (no IPTG) for EGS expression. Duplicate plates are listed for both LB and LBa agar plates. An inhibition index is calculated (see text) from the ratio of the colony counts of a given transformant without EGS induction versus with EGS induction, such that an inhibition index of 1 corresponds with no decrease in colony counts after EGS induction, and an inhibition index of 5 corresponds with a 5-fold decrease in colonies with EGS induction as compared to without EGS induction.
Overall (see Table 2), quantitative plating studies of E. coli transformants that each contain two EGSs demonstrated a greater than 10-fold decrease in viable colony counts after induction of EGS production. This effect was observed for double-EGS constructs complementary to either of the targets we investigated: RNase P C5 protein or gyrase A.
Table 2.
Inhibition index values
| Experiment | Index values
|
|||
|---|---|---|---|---|
| 4–8 h | 5–8 h | 21 h | 24 h | |
| Time course | ||||
| No T7 promoter | 1.2 | 1.3 | ||
| E. coli C5 32/−2 | 2.7 | 13.0 | ||
| EGyr241/211 | 4.0 | 17.0 | ||
| EGyr241/223 | 4.3 | 14.0 | ||
| Synth C5 21/45 | 2.4 | 2.6 | ||
| Dose effects | ||||
| No T7 promoter | 1.6 | |||
| EGyr241 | 6.9 | |||
| EGyr241/223 | 10.0 | |||
| EGyr241/223 C5 32/−2 | 26.0 | |||
| No T7 promoter | 1.7 | |||
| E. coli C5 32/−2 | 5.6 | |||
| E. coli 32/32 | 7.9 | |||
| Nucleotide level specificity | ||||
| No T7 promoter | 1.3 | |||
| EGyr241 | 7.4 | |||
| EGyr241 var1 | 14.0 | |||
| EGyr241 var2 | 6.3 | |||
| EGyr241 var3 | 6.3 | |||
| SGyr241 | 1.3 | |||
| Species specificity | ||||
| No T7 promoter | 1.4 | 1.1 | ||
| EGyr241/211 | 2.0 | 23.0 | ||
| EGyr241/223 | 2.1 | 7.1 | ||
| SGyr241/EGyr211 | 1.7 | 0.9 | ||
| SGyr241/EGyr223 | 1.6 | 1.2 | ||
Summary of multiple assays in vivo, listed as inhibition index values from quantitative plate cultures, as described in Table 1. Results represent two separate experiments done on different days. Time course: Panel from assays performed early (5–8 h) or late (24 h) after cultures were split and grown in parallel either with or without IPTG to induce EGS expression. Negative control transformants include no T7 promoter, and synthC5 21/45. Dose effects: Comparison between inhibition of growth of transformants containing vectors expressing one, two, or four EGSs (EGyr241, EGyr241/223, and EGyr241/223 C532/−2, respectively). Also comparison between transformants with vectors encoding two EGSs, either different (C5 32/−2) or identical (C5 32/32) in their EGS sequences. Nucleotide level specificity: Assessment of effects of various EGSs directed against the mRNA for gyrase A, designed to cleave position 241. Bacterial species specificity was addressed with EGyr241 and SGyr241, as explained in Fig. 2 and text. Tolerance of nucleotide mismatches was assessed with EGyr241var1, 2 and 3 constructs, as per Fig. 1. Species specificity: Extension of EGyr241 and SGyr241 studies to dual EGS vectors, expressing EGSs against either E. coli or S. typhimurium gyrase mRNA sequences.
We investigated the kinetics of growth inhibition at different culture temperatures designed to alter replication rates. Using a liquid culture temperature of 30°C rather than 37°C resulted in complete loss of inhibitory effects after addition of IPTG (see Fig. 4).
Additive Effects of Multiple EGSs.
To further assess dose dependence of growth inhibition effects after induction of EGSs, vectors were constructed expressing one, two, or four EGSs against E. coli C5 and/or gyrase A. As shown in Table 2, as the number of EGSs expressed in a transformant was doubled, effects of EGS induction in vivo also approximately doubled. In the EGyr system, a single EGS construct inhibits growth approximately half as much as does a construct with that EGS plus another EGS directed against another portion of gyrase A mRNA (with inhibition indices of 6.9 and 10.4, respectively).
There are roughly equal effects on growth by double-EGS constructs against E. coli C5, whether they encode two EGSs that are identical (C5 32/32) or different (C5 32/−2). This finding suggests that, in vivo, two EGSs directed against two different portions of a target mRNA do not achieve synergy beyond effects from merely doubling expression of a single EGS. This observation is consistent with cleavage results in vitro with EGSs C5:−2, C5:32, and C5:−2 plus C5:32 in Fig. 3, where cooperativity in target cleavage guided by EGSs C5:−2 used together with C5:32 is not seen.
We also assessed effects of concurrent targeting of both mRNAs. Using a construct encoding four EGSs, composed of the two double-EGS cassettes for EGyr241/223 and EC5 32/−2, we observed our maximal inhibition of E. coli viability, with an inhibition index of about 26, as well as subjective signs of poor viability including colony morphologic changes, poor growth in general, and frequent plasmid loss. Quantitatively, the roughly doubled effects of the four-EGS construct as compared with either of its two precursor double-EGS constructs indicates an additive rather than a multiplicative effect of targeting both E. coli C5 and EGyrA mRNAs concurrently.
The additive effect observed with increased copies of EGSs appears EGS-dependent given the lack of growth inhibition by negative control EGS constructs, which differ only in their EGS sequences from other, inhibitory, single and double-EGS constructs [see Table 2; in particular, S. typhimurium gyrase (SGyr)241, SGyr241/223, SGyr241/211, and synthC5 21/45].
Nucleotide Sequence Level Specificity.
Past work showed that a single base-pair mismatch between an EGS and target mRNA did not prevent RNase P-mediated cleavage in vivo (19). To further define the mismatch tolerance of the system presented here, we produced variants of one of our single-EGS constructs against EGyr (EGyr241). Three different variant EGSs were derived from EGyr241, by the stepwise introduction of one, two, or three nucleotide changes. In an effort to disrupt EGS binding with its mRNA target, these changes were introduced into the midregion of the predicted EGS-mRNA duplex, using purine substitutions to maximize steric effects, and avoiding the creation of potential guanine-uracil base pairs to minimize wobbling. These changes are in positions G246→A, C249→G, and C252→G. In addition, an EGS (SGyr241) was made which is complementary to the mRNA in S. typhimurium corresponding to the EGyr mRNA targeted by EGyr241. This SGyr241 EGS varies from the EGyr241 EGS at 5 of the 15 nt that are predicted to bind target mRNA in the EGS-target mRNA duplex.
Of the EGyr241 variants containing one (EGyrVar1), two (EGyrVar2), or three (EGyrVar3) successive nucleotide changes, all were able to inhibit E. coli growth after EGS induction, with inhibition indices very similar to that seen with EGyr241 (see Table 2). For this EGS-mRNA pair, as many as three mismatches between EGS and mRNA targets are tolerated without disruption of observed in vivo effects. By contrast, the SGyr241 EGS, differing from EGyr241 by 5 nt, was quite distinct from EGyr241 in vivo, in that SGyr241 did not inhibit E. coli viability.
Species-Specific Targeting of E. coli Gyrase A mRNA.
Given that there is a section of gyrase A in E. coli and S. typhimurium in which the amino acid sequence is the same but the respective mRNA sequences differ, the EGS against this portion of SGyr (SGyr241) was further examined.
SGyr241 was substituted for EGyr241 in the double-EGS constructs against EGyrA described above (EGyr241/211 and EGyr241/223). The resulting vectors encode S. typhimurium-specific SGyr241 EGS in tandem with EGSs directed against gyrase A mRNA sequences (near nucleotides 211 and 223) shared by E. coli and S. typhimurium. As seen for SGyr241 as compared with EGyr241 in single-EGS vectors (see Table 2), these dual-EGS constructs containing the S. typhimurium-specific SGyr241 (SGyr241/EGyr211 and SGyr241/EGyr223) did not inhibit E. coli growth, whereas dual-EGS constructs containing EGyr241 did here, by 7- to 20-fold (see Table 2).
Discussion
This study extends previous work using EGS techniques (6, 7) in bacteria (7, 8, 19) to inactivate essential gene products. The ability to concurrently target multiple gene products for inactivation provides one strategy for combating antimicrobial resistance that arises from a single-step mutation of one gene product. Even in our inducible system for gene inactivation, we encountered some of these difficulties from dynamically inactivating essential genes: particularly effective EGS constructs made transformants more difficult to grow and tended to lose their plasmid vectors more rapidly and often than did the negative control EGS constructs. We suspect this result represents leaky read-through from our promoter even without the addition of IPTG for induction.
When microbial viability was assessed at a lower liquid culture temperature (30ο C) or assayed relatively early after EGS induction in liquid culture (up to 6 h), no loss of viability is found compared with wild type. We propose that the dynamics of the EGS effects on viability depend on both rates of production of EGSs and the depletion of targets by cell fission. In fact, neither the lower temperature nor the early time samples give the cells adequate time to dilute out through cell division the preformed essential gene products to a level sufficiently low to affect viability.
We roughly double effects on bacterial viability when we add a second distinct EGS directed against a second portion of a single mRNA target. Compared with a single EGS, we quadruple effects when we use four distinct EGSs targeting two different mRNAs: two EGSs against gyrase A and two EGSs against RNase P.
Other, more qualitative, observations also suggest that specific EGS expression disrupts bacterial viability. Loss of the EGS vector plasmid (manifest by loss of ampicillin resistance carried by the vector plasmid, as detected by greater colony numbers on LB as compared with LBa plates) occurred most often with plasmids encoding the EGSs most inhibitory to bacterial growth and never from the transformant that was deficient for T7 promoter and EGS (data not shown). In addition, transformants containing four EGS constructs in tandem, which had the highest inhibition index values of all our constructs (see above), exhibited smaller colony size when grown on agar plates that were impregnated with IPTG than when grown on plates not containing IPTG. These transformants were also more difficult to grow in general than the transformants containing EGS constructs with lower inhibition index values.
At the level of nucleotide specificity we found that (for the EGS targeting EGyr mRNA for cleavage at position 241) one, two, or three unpaired nucleotides in a 15-mer EGS did not interfere with EGS impact on viability, whereas five unpaired nucleotides did. The location of the unpaired nucleotides is important because three contiguous unpaired bases might very well disallow the RNase P-mediated effects. Our results, however, suggest that an EGS could still function despite several point mutations between it and the bacterial target mRNA, depending precisely on the sequence of the unpaired bases.
Targeting mRNA as a tool for antimicrobial development takes advantage of microbial genomic database advances (24) well suited for rapid identification of candidate mRNA sequences. We were particularly interested in mRNA sequences that differed between species, but which encoded identical proteins, such as for gyrase A. We designed EGS molecules to be specific for either E. coli or S. typhimurium gyrase mRNA and demonstrate that E. coli mRNA for gyrase A can be targeted for in vivo effects by EGSs complementary to E. coli mRNA sequence, but not by EGSs complementary to S. typhimurium mRNA sequence. This system raises the potential for using species-specific EGSs to target mRNA differences between species. Currently, even so-called narrow spectrum antimicrobials inhibit many species beyond the relevant pathogen (25), thereby allowing the creation of selective pressure on other organisms for the emergence and spread of antimicrobial resistance (5).
Acknowledgments
We thank Dr. J. Wang for plasmids and the members of our laboratory for helpful discussions. This research was conducted while J.M. was a Pfizer Postdoctoral Fellow. This work was supported by a National Institutes of Health research fellow training program (NIH T32 AI07210–17) and National Institutes of Health Grant GM19422 (to S.A.).
Abbreviations
- EGS
external guide sequence
- IPTG
isopropyl β-d-thiogalactoside
- EGyr
E. coli gyrase
- SGyr
S. typhimurium gyrase
- LBa
LB ampicillin
References
- 1.Jacobs R F, Schutze G E, Young R A, Kearns G L, James L P. In: Principles and Practice of Pediatric Infectious Diseases. Long S S, Pickering L K, Prober C G, editors. New York: Churchill Livingstone; 1997. pp. 1604–1662. [Google Scholar]
- 2.Baltimore R S, Jenson H B. In: Pediatric Infectious Diseases: Principles and Practice. Jenson H B, Baltimore RS, editors. East Norwalk, CT: Appleton and Lange; 1995. pp. 7–12. [Google Scholar]
- 3.Walsh C. Nature (London) 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 4.Neu H C. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 5.Cohen M L. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 6.Forster A C, Altman S. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 7.Guerrier-Takada C, Altman S. Methods Enzymol. 1999;313:442–456. doi: 10.1016/s0076-6879(00)13028-9. [DOI] [PubMed] [Google Scholar]
- 8.Guerrier-Takada C, Li Y, Altman S. Proc Natl Acad Sci USA. 1995;92:11115–11119. doi: 10.1073/pnas.92.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Ames G F. Proc Natl Acad Sci USA. 1988;85:8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman S, Kirsebom L. In: The RNA World. Gesteland R F, Cech T R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 351–380. [Google Scholar]
- 11.Levine C, Hiasa H, Marians K J. Biochim Biophys Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 12.Hooper D C, Wolfson J S. In: Quinolone Antimicrobial Agents. Hooper D C, Wolfson J S, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 53–75. [Google Scholar]
- 13.Cullen M E, Wyke A W, Kuroda R, Fisher L M. Antimicrob Agents Chemother. 1989;33:886–894. doi: 10.1128/aac.33.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griggs D J, Gensberg K, Piddock L J. Antimicrob Agents Chemother. 1996;40:1009–1013. doi: 10.1128/aac.40.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 17.Wyckoff E, Natalie D, Nolan J M, Lee M, Hsieh T. J Mol Biol. 1989;205:1–13. doi: 10.1016/0022-2836(89)90361-6. [DOI] [PubMed] [Google Scholar]
- 18.Swanberg S L, Wang J C. J Mol Biol. 1987;197:729–736. doi: 10.1016/0022-2836(87)90479-7. [DOI] [PubMed] [Google Scholar]
- 19.Guerrier-Takada C, Salavati R, Altman S. Proc Natl Acad Sci USA. 1997;94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Guerrier-Takada C, Altman S. Proc Natl Acad Sci USA. 1992;89:3185–3189. doi: 10.1073/pnas.89.8.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida H, Nakamura M, Bogaki M, Ito H, Kojima T, Hattori H, Nakamura S. Antimicrob Agents Chemother. 1993;37:839–845. doi: 10.1128/aac.37.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willmott C J, Maxwell A. Antimicrob Agents Chemother. 1993;37:126–127. doi: 10.1128/aac.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vioque A, Arnez J, Altman S. J Mol Biol. 1988;202:835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- 24.Fraser C M, Eisen J A, Salzberg S L. Nature (London) 2000;406:799–803. doi: 10.1038/35021244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert D N, Moellering R C, Sande M A. The Sanford Guide to Antimicrobial Therapy 1998. Vienna, VA: Antimicrobial Therapy; 1998. pp. 52–54. [Google Scholar]