Abstract
We report the cocrystal structures of a computationally designed and experimentally optimized retro-aldol enzyme with covalently bound substrate analogs. The structure with covalently bound substrate analog is similar but not identical to the design model, with an RMSD over the active site residues and equivalent substrate atoms of 1.4Å. As in the design model, the binding pocket orients the substrate through hydrophobic interactions with the naphthyl moiety such that the oxygen atoms analogous to the carbinolamine and β-hydroxyl oxygens are positioned near a network of bound waters. However, there are differences between the design model and the structure: the orientation of the naphthyl group and the conformation of the catalytic lysine are slightly different; the bound water network appears to be more extensive; and the bound substrate analog exhibits more conformational heterogeneity than in typical native enzyme-inhibitor complexes. Alanine scanning of the active site residues shows that both the catalytic lysine and the residues around the binding pocket for the substrate naphthyl group make critical contributions to catalysis. Mutating the set of water-coordinating residues also significantly reduces catalytic activity. The crystal structure of the enzyme with a smaller substrate analogue that lacks the naphthyl rings shows the catalytic lysine to be more flexible than in the naphthyl substrate complex; increased preorganization of the active site would likely improve catalysis. The covalently bound complex structures and mutagenesis data highlight strengths and weaknesses of the de novo enzyme design strategy.
Introduction
Computational enzyme design approaches have the potential to produce new enzymatic catalysts for many chemical reactions. However, computational design is still in its infancy. While structures of apo-enzymes have been solved for several designed enzymes1; 2, there are no structures to date of designed enzymes with covalently bound substrate analogs.
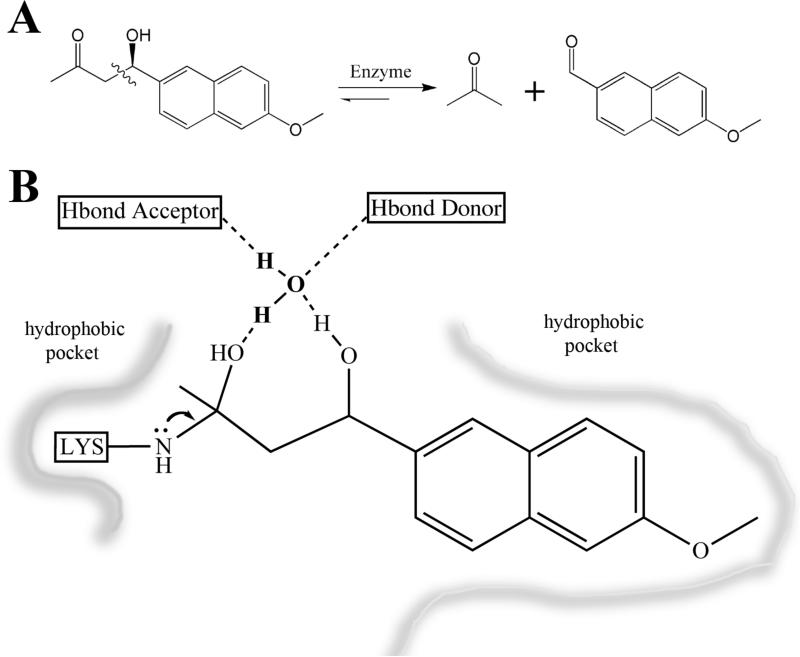
As described previously1, a two stage computational approach was used to design enzymes catalyzing the retro-aldol reaction shown in Figure 1. In the first step, ideal active site “theozymes” were generated that consist of superimposed carbinolamine intermediate and bond-breaking transition state models surrounded by a catalytic lysine residue which forms a Schiff base with the substrate and hydrogen bonding residues to position a water molecule near the carbinolamine oxygen. Locations in a set of scaffold proteins where one of these ideal active sites could be created were identified using the RosettaMatch algorithm3. In the second step, the residues within 8 Å around the model ligand were optimized to maximize the binding of the intermediate/transition state models, the packing around the Schiff-base lysine residue, and nonpolar packing around the substrate napthyl group.
Figure 1.
Retro-aldol reaction and active site description. A. The schematic of the retroaldolase reaction. The product is fluorescent (⌊ex = 330 nm, ⌊em = 452 nm)28B. The minimal active site used in the design calculations. The Schiff base forming lysine serves as an electron sink to promote bond cleavage. The bridging water is positioned by hydrogen bonding residues to allow proton shuttling on and off the substrate and product.
In this study, we focus on one of the most active of the designs identified in our previous study, RA341 which has a kcat/KM of 0.11 M-1s-1, comparable to computationally designed catalysts 2; 4; 5 for other reactions and in the range of previous peptide and catalytic antibody aldol catalysts6; 7; 8; 9; 10; 11; 12. To identify shortcomings in the design calculations and guide improvement of the design methodology, we first used mutagenesis and screening to optimize the active site residues and to identify positions that were suboptimal in the original design. Next, we solved the crystal structure of the optimized enzyme covalently bound to substrate analogs both with and without the naphthyl group that was modeled in the design process. The contributions of the residues in the active site to catalysis were probed by alanine scanning mutagenesis, and the contributions of the sequence changes that arose during optimization were investigated by reversion mutations. The structures and mutational analyses illustrate how the designed and optimized residues influence catalysis, and reveal areas for improvement in computational design methodology.
Results
Design and optimization of the retro-aldol enzyme RA34
The RA34 design was created using RosettaMatch 1 to identify a location on the TIM barrel indole-3-glycerolphosphate synthase (1lbf) scaffold 13 where the ideal active site schematically illustrated in Figure 1B could be recapitulated. Rosetta design 14 calculations were subsequently carried out to optimize substrate/transition state binding. In total, the design calculations introduced 13 mutations from the original 1lbf scaffold. The final design model contains (i) a catalytic lysine residue, (ii) surrounding hydrophobic residues to hold the lysine in place and lower its pKa8, (iii) several neighboring polar residues to stabilize the carbinolamine reaction intermediate via water mediated interactions15, and (iv) a hydrophobic pocket designed to bind and orient the substrate naphthyl group. As reported previously 1, the activity of the purified RA34 protein for the retro-aldol cleavage of (±)-methodol was well above background, but the catalytic parameters were quite low: kcat = 0.7 × 10-4 s-1 and kcat/KM = 0.11 M-1s-1.
To investigate the extent the active site could be optimized and positions in which the computational design was suboptimal, we screened amino acid sequence variants at 19 positions around the active site (supplementary Figure S1). 12 residues were mutated into all other amino acids and 7 residues participating primarily in packing interactions were mutated to other hydrophobic residues. We started from a variant, Y51T, found serendipitously to increase RA34 activity by about 2-fold. Sequence changes were introduced via Kunkel mutagenesis using degenerate oligonucleotides. After transformation into BL21(DE3) cells, individual constructs were expressed and retroaldolase activity was measured in the clarified lysates. We began by screening single amino acid substitutions at each position (supplementary Table S1). Subsequently, variants that displayed at least 2-fold improvements in activity over the original design were combined to select the best possible combination, termed RA34.2 (supplementary Figure S1). Additional mutations, found previously to increase the solubility of the scaffold (Olga Kheronsky, personal communication), were made outside of the active site, producing the variant RA34.3. An additional round of screening single mutations was performed, and the top 3 active variants (RA34.4, RA34.5, and RA34.6) were identified and purified. Of these, RA34.6 had the highest activity, with a kcat of 3.6 × 10-4s-1, and a KM of 30 μM and a kcat/KM of ~12 M-1s-1 (Table 1). Further single mutations of RA34.6 yielded less than 2-fold improvement.
Table 1.
Steady-state enzyme activity
| KM (μM) | kcat (s-1) | kcat/KM (M-1 s-1) | |||
|---|---|---|---|---|---|
| RA34 optimization | |||||
| RA34 original design | 620 | 0.7E-04 | 0.11 ± 0.01 | 1.1 E+04 | 0.009 |
| RA34 Y51T | 626 | 1.22E-04 | 0.19 ± 0.01 | 1.9 E+04 | 0.016 |
| RA34.6 | 30 | 3.60E-04 | 12 ± 1 | 5.54E+04 | 1 |
| Ala mutation scan | |||||
| K159A | 1696 | 6.63E-07 | (3.9±0.4)E-04 | 1.02E+02 | 3.3E-05 |
| C83A | 102 | 3.29E-04 | 3.2 ± 1.5 | 5.06E+04 | 0.3 |
| S210A | 41 | 1.53E-04 | 3.7 ± 1.2 | 2.35E+04 | 0.32 |
| W8A | 34 | 6.65E-05 | 2.0 ± 0.7 | 1.02E+04 | 0.18 |
| P57A | 136 | 1.37E-04 | 1.0 ± 0.2 | 2.11E+04 | 0.09 |
| F112A | 25 | 5.82E-05 | 2.3 ± 0.3 | 8.95E+03 | 0.19 |
| I157A | 39 | 1.72E-05 | 0.4 ± 0.2 | 2.65E+03 | 0.04 |
| T51A | 55 | 2.77E-04 | 5.0 ± 1.1 | 4.26E+04 | 0.44 |
| C180A | 31 | 2.62E-04 | 8.5 ± 0.4 | 4.03E+04 | 0.72 |
| T211A | 45 | 4.19E-04 | 9.3 ± 2.4 | 6.45E+04 | 0.83 |
| M53A | 98 | 1.54E-04 | 1.6 ± 0.5 | 2.37E+04 | 0.14 |
| W58A | 143 | 1.19E-04 | 0.8 ± 0.04 | 1.83E+04 | 0.07 |
| W184A | 102 | 1.18E-04 | 1.2 ± 0.2 | 1.82E+04 | 0.1 |
| S81A | 32 | 2.69E-04 | 8.4 ± 0.6 | 4.14E+04 | 0.7 |
| S181A | 42 | 3.67E-04 | 8.7 ± 1.6 | 5.65E+04 | 0.75 |
| S231A | 27 | 1.21E-04 | 4.5 ± 0.8 | 1.86E+04 | 0.39 |
| L108A | 40 | 1.53E-04 | 3.8 ± 0.6 | 2.35E+04 | 0.33 |
| I133A | 37 | 1.75E-04 | 4.7 ± 0.4 | 2.69E+04 | 0.39 |
| I233A | 62 | 1.93E-04 | 3.1 ± 0.6 | 2.97E+04 | 0.27 |
| Reversion mutants | |||||
| C83T | 107 | 2.34E-04 | 2.2 ± 1.3 | 3.60E+04 | 0.21 |
| I233G | 110 | 8.54E-05 | 0.8 ± 0.6 | 1.31E+04 | 0.08 |
| T211Y | 82 | 2.33E-04 | 2.8 ± 1.8 | 3.58E+04 | 0.27 |
| C180V | 58 | 2.58E-04 | 4.4 ± 1.7 | 3.97E+04 | 0.4 |
| I157L | 70 | 3.08E-04 | 4.4 ± 2.6 | 4.74E+04 | 0.41 |
| P131A | 88 | 2.98E-04 | 3.4 ± 1.9 | 4.58E+04 | 0.31 |
| Water network mutants | |||||
| T51L | 69 | 6.05E-05 | 0.88 ± 0.02 | 9.31E+03 | 0.073 |
| T51I | 65 | 4.54E-05 | 0.70 ± 001 | 6.98E+03 | 0.058 |
| S81V | 47 | 7.24E-05 | 1.55 ± 0.21 | 1.11E+04 | 0.13 |
| T51V/S81A | 50 | 2.91E-05 | 0.59 ± 0.08 | 4.47E+03 | 0.049 |
| S210A/S231A | 66 | 1.41E-05 | 0.21 ± 0.01 | 2.17E+03 | 0.018 |
| T51V/S81A/S210A/S231A | 96 | 1.30E-06 | 0.014 ± 0.003 | 2.00E+02 | 0.0012 |
In total, the seven mutations around the active site in RA34.6 relative to the original designed enzyme RA34 (Y51T, T83C, A131P, L157I, V180C, Y211T, and G233I) resulted in a 100-fold increase in kcat/KM: from kcat/KM=0.11 M-1s-1 to kcat/KM =12 M-1s-1. Although the kcat/KM value of RA34.6 is still low compared to most native aldolases16; 17, which are in the range of 105 M-1s-1, it represents a 5 × 106-fold acceleration over the second-order rate constant for the same reaction catalyzed by the lysine analog butylamine at pH 7.5 18. The rate acceleration of RA34.6 relative to the first-order nonenzymatic reaction in solution (kcat/kuncat = 5.5×104) exceeds those of all but the best of the aldolase catalytic antibodies and peptides that have been developed6; 7; 8; 9; 10; 11; 12 based on the enamine mechanism of natural enzymes19; 20. The rate acceleration of RA34.6 exceeds that of computationally designed enzymes2; 4; 5 and is comparable to that of designs after improvement by directed evolution21; 22.
The positions and identities of the substitutions found during optimization in the original Rosetta design model suggest that they could increase activity by improving packing around the catalytic lysine and the substrate naphthyl group as well as by altering the hydrogen-bonding network with substrate oxygens. To evaluate these possibilities, we performed structural studies of RA34.6.
Crystal structures of RA34.6
Previous attempts at solving the structure of the original RA34 design were unsuccessful1, but we were able to crystallize and determine the structures at 2.1-2.4Å (Table 2) of the improved RA34.6 protein in the absence of added inhibitors or substrate analogues, and of complexes of RA34.6 both with a mechanism-based inhibitor (1-(6- methoxy-2-naphthalenyl)-1,3-butanedione) and a substrate analogue that lacks the naphthyl rings (4-hydroxy-4-methyl-2-pentanone). The initial phases were determined via molecular replacement, using the coordinates of indole-3-glycerolphosphate synthase (RCSB PDB entry 1a53, whose protein sequence is identical to 1lbf) as a search model. In the original search model and corresponding starting model used for rebuilding, all residues that were subjected to computational redesign and subsequent mutagenesis were converted to alanine and then rebuilt manually into initial electron density maps to avoid modeling bias that might arise from the original computational design produced by Rosetta.
Table 2.
Crystallization, data collection, structure determination, and refinement
| Cross-linked Inhibitor | Cross-linked 4h4m2p | Apo | |
|---|---|---|---|
| Data Collection | |||
| Space group | P3(1) 2 1 | P3(1) 2 1 | P3(1) 2 1 |
| Cell dimensions | 62.68 62.68 123.68 90 90 120 | 61.92 61.92 121.24 90 90 120 | 62.59 62.59 123.70 90 90 120 |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 |
| Resolution (Å) | 50 - 2.09 (2.16 - 2.09) | 50 - 2.4 (2.49 - 2.4) | 50 - 2.1 (2.16 - 2.1) |
| Observations | 110,823 | 176,391 | 248,913 |
| Unique reflections | 17,318 | 11,053 | 17,270 |
| Data coverage (%) | 99.0 (95.2) | 99.1 (99.7) | 96.8 (87.8) |
| Redundancy | 2.9 | 3.9 | 5.0 |
| Rlin | 3.7 (15.3) | 3.9 (29.0) | 7.8 (34.0) |
| I/σI | 22.0(6.9) | 21.1 (5.8) | 16.3 (5.1) |
| Mean FOM | 0.80 | 0.74 | 0.74 |
| Refinement | |||
| Resolution range (Å) | 19.47 - 2.09 | 53.6 - 2.4 | 50 - 2.1 |
| Reflections | 16,229 | 10,320 | 15,688 |
| Completeness | 99.35% | 98.22% | 96.28% |
| Total atoms | |||
| Protein | 1979 | 2036 | 1994 |
| Water | 61 | 60 | 82 |
| Ligand | 17 | 7 | |
| Ion (SO4) | 10 | 5 | 15 |
| Rworking set | 21.1% | 22.4% | 22.3% |
| Rfree | 26.0% | 26.8% | 29.0% |
| Rmsd | |||
| Bonds (Å) | 0.008 | 0.012 | 0.022 |
| Angles | 1.028 | 1.4 | 2.0 |
| Ramachandran Plot | |||
| Most favored | 92.1% | 93.3% | 90.7% |
| Additional allowed | 6.2% | 6.7% | 8.4% |
| Generously allowed | 0.4% | 0% | 2% |
| Disallowed | 1.3% | 0% | 0% |
Crystals were grown via vapor diffusion. Equal amounts of protein at 5mg/ml in 100mM NaCl, 25mM Tris pH7.5 were mixed with well solutions consisting of 2M ammonium sulfate, 4% peg400, 100mM Na Acetate pH 5.5. Data were collected at -180°C in cryoprotectant buffer composed of 2.1M ammonium sulfate, 5% peg400, .2M sodium acetate, 500mM sodium chloride on a Rigaku Micromax 7HF with a Saturn 944+. Data were indexed, integrated, and scaled using HKL2000 package . Molecular replacement and refinement were performed with the Phaser and Refmac modules of CCP4i. Coot was used for model building.
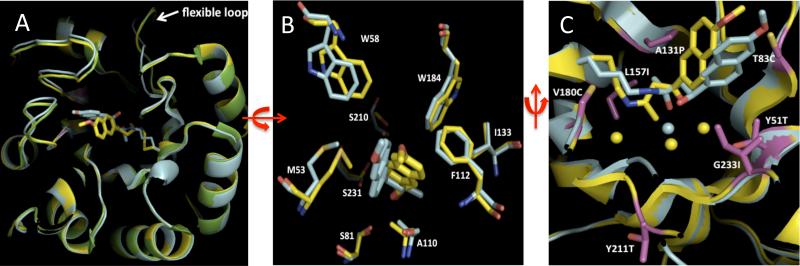
The structure of RA34.6 apo protein was determined to 2.1 Å resolution (Figure 2 and Table 2). Overall, the crystal structure is very similar to the RA34 design model with a RMSD of 0.4 Å over the C〈 atoms (the RMSD relative to the original structure of 1lbf is also approximately 0.4 Å). The most pronounced structural discrepancy is in a flexible loop (residue numbers 210-216) (Figure 3A), which may be associated with the Y211T mutation. Density around the catalytic lysine suggests that more than one rotamer may be populated. Significant additional poorly ordered density emanates from the amino group of the catalytic lysine and extends into the binding site (Figure 2A), which suggests Schiff base formation with an unknown compound from the protein preparation. Mass spectrometry of small crystals of the apo protein showed the molecular weight to be 128 Da larger than expected; the adduct likely formed during crystallization as freshly purified RA34.6 samples have the expected molecular weight.
Figure 2.
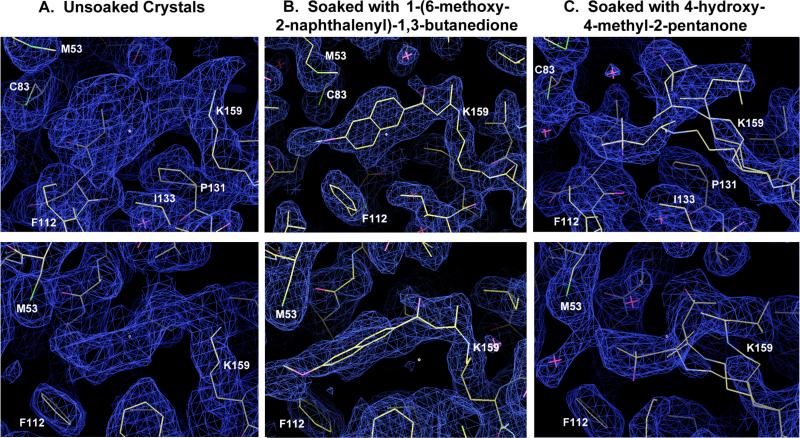
Unbiased composite omit maps calculated after building and refinement of protein side chains. The atoms and models corresponding to the bound substrate and product analogs were not modeled or used in any way for phase calculations prior to generation of these maps. The three maps are all shown at two similar angles (viewed from the side of the binding pocket and from the top) for each crystal and data set. The contour level of panel A (corresponding to the unsoaked crystals of freshly purified enzyme) is 1.2 sigma; the contour levels of panels B and C (corresponding to soaks with substrate and product analogues) is 2 sigma. The ring system of the bound naphthalene group of the substrate analog (panel B) shown clear signs of mobility in the plane of the rings (i.e. up and down in the upper portion of panel B); the connectivity and rotameric modeling of the chemical linkage to the active site lysine (K159) looks clear. Inverting the stereochemistry of this linkage results in clear positive difference density where the carbonyl oxygen is modeled.
Figure 3.
Comparison of the structure of RA34.6 to the original design model. A. Overlay of apo RA34.6 (green), bound RA34.6 (yellow) and the original design model RA34 (cyan) 1. The backbone conformations of the apo and holo RA34.6 are nearly identical with a C〈 RMSD of 0.15 Å. The C〈 RMSD of bound RA34.6 to the original design model of RA34 is 0.4 Å. The flexible loop in the highest discrepancy region, amino acid numbers 210-216, is highlighted. B. Active site comparison of cocrystal structure of RA34.6 (yellow) and the design model (cyan) after superimposing all C〈 atoms of the structures. The “superimposable” active site includes carbinolamine intermediate, covalently bound Lys159, hydrogen-binding residues (Ser231, Ser210 and Ser81) and other hydrophobic residues contacting the substrate (Phe112, Ala110, Trp184, Met53, Trp58 and Ile133). The heavy atom RMSD of this superimposable active site is 1.4Å between the original design and the crystal structure of the evolved variant. C. Comparison of the water-mediated interactions in the active site in the RA34 model (cyan) with putative water positions modeled in the RA34.6 substrate analog complex structure (yellow). Only the sidechains of the crystal structure are shown for simplicity. Optimized substitutions (from RA34 to RA34.6) in the active site are highlighted in purple. The modeled and crystallographic bound water molecules are shown as spheres. The Protein Data Bank entry code of apo and holo RA34.6 are 3O6Y and 3NL8, respectively. The RA34 design model was reported previously 1.
To better understand the interactions of RA34.6 with substrate, we next determined the structure of RA34.6 with the covalently bound diketone substrate analog (1-(6-methoxy-2-naphthalenyl)-1,3-butanedione) at 2.1 Å resolution (Figure 2B). The bound diketone was modeled in the final stage of model building and was fitted into density that was clearly observable in completely unbiased composite omit electron density maps (Figure 2). Electron density emanating from the catalytic lysine is consistent with the mechanism-based inhibitor, and differs in its shape, size and strength from that observed in the “apo” structure (Figure 2A). Though the density that we assign to the bound diketone analog exhibits considerable positional heterogeneity in the pocket (Figure 2B), the chemical linkage to the lysine is unambiguous, and the overall shape of the aromatic planar bicyclic naphthalene ring is well defined.
The RA34.6 structure with bound inhibitor was overall similar to the original RA34 design model, with an 〈-carbon RMSD of 1.4 Å over the active site atoms including the catalytic lysine, the three polar residues designed to position bound water molecules, and six surrounding apolar residues which pack on the napthyl group and buttress the catalytic lysine (Figure 3). Detailed comparison of the RA34.6 structure to the design model reveals several interesting differences. First, the catalytic lysine, K159, appears to have more than one conformation in the structure, but is also better packed by non-polar groups than in original RA34 design model. The two mutations introduced by optimization that contribute to better packing are L157I, which packs underneath the lysine and A131P, which alters the lysine backbone conformation and may stabilize a slightly bent conformation of the lysine side chain (supplementary Figure S3).
The second difference we observed between the RA34.6 structure and the RA34 design model was that the naphthyl group of the substrate rotates and adopts an orientation similar to but different than that in the model. There is also a slight translational movement out of the hydrophobic pocket (Figure 3A and 3B).
The third intriguing feature of the structure is a network of several putative water molecules near the oxygens of the substrate analog, within interaction distance to four polar residues at the bottom of the pocket (Figure 4C). In the design model of RA34, we introduced hydrogen bonds from Tyr51 and Ser81 to a single water molecule to facilitate the reaction and help shuffle the proton generated during the reaction. The RA34.6 structure, however, may indicate a more extensive water network with hydrogen bonds to at least three waters from four residues (Thr51, Ser81, Ser210, and Ser231).
Figure 4.
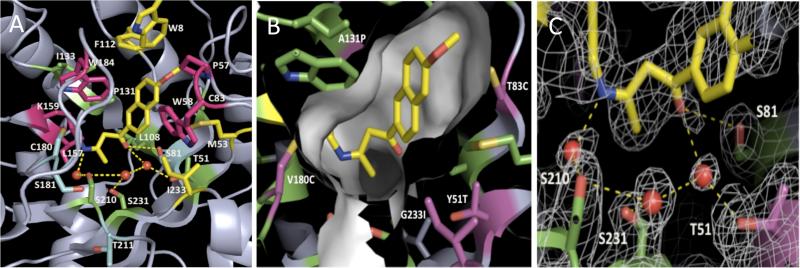
Contributions of active site residues to catalysis. A. RA34.6 active site alanine scanning results. Residues which when mutated to alanine decrease activity more than 8-fold, 4-fold, 2-fold and less than 2-fold are shown in red, yellow, green and blue, respectively. The ligand is in yellow. B. The RA34.6 binding pocket; green side chains are from the original design, purple side chains were those changed during the optimization procedure. C. The electron density map of the active site of RA34.6 with bound substrate shows the three putative waters and the interacting residues around the active site.
A fourth notable feature of the structure is the tight packing of the naphthyl ring by the active site residues (Figure 4B). The substitutions G233I, T83C, and A131P introduced by optimization contribute to this tight packing. To determine the importance of the naphthyl ring packing, we solved the structure of RA34 complexed with a smaller substrate lacking the naphthyl rings (4-hydroxy-4-methyl-2-pentanone) to 2.4 Å resolution. In a prior study of designed retroaldolase RA61, this compound showed substantially reduced activity as a substrate, consistent with an important role of hydrophobic interactions of the enzyme with the naphthyl rings18. The electron density emanating from the catalytic lysine is again consistent with the size of the ligand, in this case considerably smaller than the unidentified compound present in the apo structure.
The electron density shows multiple conformations of the smaller substrate analog bound to the catalytic lysine within the active site; the substrate without the napthyl group is less well ordered than the naphthyl-containing substrate (Figure 2C). This observation provides further indication that interactions between the designed active site pocket and the naphthyl rings help position the substrate for catalysis.
Active site mutagenesis
To probe the roles of individual sidechains in these four active site features, we carried out a series of mutagenesis studies around the active site (Table 1).
First, we mutated all the active site residues in RA34.6 to alanine (Figure 4A and Table 1 Ala mutation scan). Mutation of the catalytic lysine to alanine reduces activity by > 104-fold, consistent with the critical role of the residue in Schiff base formation. Besides the crucial lysine, the largest decreases in activity upon alanine mutation were observed for large non-polar side chains that pack against the naphthyl ring and catalytic lysine in the structure. These interactions contribute both to substrate binding and to positioning of the substrate and catalytic lysine in orientations appropriate for catalysis, consistent with the crystal structure observations.
Second, we mutated residues that were changed during the optimization back to their identities in the original design to further understand how the active site optimization increased activity (Table 1 Reversion mutants). Reverting mutations P131A, G233I and T83C, which pack around the naphthyl ring of the substrate (Fig 4B), decreased kcat/KM by 3-fold, 13-fold and 5-fold, respectively. Reversion of L157I and V180C, which surround the catalytic lysine (Figure 4B), reduced activity 2-3 fold. Though the active site changes near the lysine could in principle affect activity by perturbing the lysine pKa, RA34 and RA34.6 have similar pKa values (7.2-7.3) under subsaturating conditions (supplementary Figure S2), and given the Brønsted slope for the reaction, small changes in pKa are not expected to have large impacts on reactivity18. Therefore, it is likely that the increased packing around the catalytic lysine increases activity by properly positioning the amine group for catalysis.
We also carried out single and combined mutations to evaluate the importance of the putative network of water molecules and interacting side chains (Figure 4C and Table 1 Water network mutants). Mutation of these side chains individually to alanine (T51A, S81A, S210A, S231A) had small effects on activity (1-4 fold) (Table 1-Ala scan), but mutation of two together (S210A/S231A) showed a larger effect (>50 fold) (Table 1-Water network). Mutations of the three residues simultaneously to alanine and Thr51 to valine led to a >800-fold loss in activity. Individual mutations to hydrophobic residues that can exclude water (T51L, T51I, S81V) resulted in much larger reductions in kcat(10 fold or greater) compared to Ala mutants (Table 1-Water network). These results suggest that a water network formed by these residues may make critical contributions to catalysis.
Overall, these results support the role of the designed lysine in forming the Schiff base intermediate during the reaction. They further suggest the importance of active site side chains both in positioning the substrate within the active site pocket and in surrounding active site water molecules that can facilitate proton transfers in the catalytic cycle.
Discussion
The catalytic machinery of RA34.6 revealed by the crystal structures is similar in broad outline to the design model. The catalytic lysine is in a hydrophobic pocket and forms a Schiff base with the substrate, the naphthyl ring is packed by hydrophobic residues, and the hydroxyl group of the carbinolamine (not present in the inhibitor structure) is likely to interact with water molecules coordinated with surrounding residues. However, the structures also illustrate limitations of the RA34.6 active site. For example, the naphthyl ring system is well ordered in one dimension (Figure 2B, lower panel), but appears to exhibit considerable motion in the plane of the ring (Figure 2B, upper panel). This structural variation suggest that the substrate is not bound as precisely as the corresponding group in native aldolase enzyme-substrate complex structures15; 23; 24; 25. The observation of an unidentified endogenous compound in the apo protein structure suggests that the binding pocket acts as a somewhat non-specific hydrophobic binding chamber. The flexibility of the catalytic lysine suggested by the multiple conformers in the crystal structure is also likely to compromise catalytic activity. Finally, the water network that appears to be visible in electron density maps with the bound inhibitor differs from the modeled water molecule in the original design. In the design model a water molecule was positioned to interact with the substrate oxygens and potentially facilitate proton transfer chemistry; in the structure there appears to be a more extensive group of waters that may serve a similar function. These results suggest that the identities of the side chains surrounding the water network are important, but the specific hydrogen bonding solvent network in the design model is not recapitulated in the experimental structures. A future direction for improvement of activity would be to incorporate polar interactions that allow the enzyme to directly interact with the carbinolamine and ®-alcohol group.
The RA34.6 catalyst is the product of computational design followed by experimental active site optimization. The starting computationally designed RA34 catalyst has a kcat/KM of 0.11 M-1s-1, and a kcat 104-fold over the background kuncat. Improvement of this activity by active site mutagenesis reduced the KM 20 fold and increased kcat by 5 fold, for an overall 100-fold improvement of kcat/KM in RA34.6. Improved catalysts could be achieved both by improving the methodology along the lines described in the previous paragraph, and by more extensive directed evolution. Continued efforts to develop catalysts with increased activities should provide insights into the features responsible for the remarkably high activities of native enzymes.
In summary, computational design is still at its early stage: key catalytic residues can be placed appropriately and hydrophobic pockets complementary in shape to the substrate can be created, but the finer details of polar networks are likely very challenging to control precisely. Progress on this front will require more accurate models of the delicate tradeoff between favorable hydrogen bonding interactions and the cost of desolvation. More precise recapitulation of the designed binding mode would also be facilitated by substrates with more binding “handles” (asymmetrically disposed hydrogen bonding groups, for example) than the relatively small, non-polar substrates used in our retroaldolase and Kemp eliminase design efforts. The crystal structures and mutational analysis suggest several routes by which higher activities could potentially be achieved either by further computational design or directed evolution—more extensive packing could better position the catalytic lysine and napthyl group, and replacement of some of the water mediated interactions with general base interactions, as in native enzymes, could facilitate substrate positioning and proton transfers.
Materials and Methods
Saturation mutagenesis for active site optimization and cell lysates preparation
Saturation mutagenesis was performed at each active site position using Kunkel mutagenesis with degenerate oligonucleotides 26. The resulting mutants were transformed into E. coli (BL21 DE3) cells, and individual clones were grown and screened for increased catalytic activity using a cell lysate assay. For initial screening, we used 3ml cultures of mutant enzymes because the activity signal was low, whereas 1 mL cultures could be used for later variants with higher activity. Cultures of the mutants were grown in 96 well plates at 37°C in LB with 25 mg/mL Kan until OD ~ 0.6. Expression was then induced by the addition of IPTG to a concentration of 1.0 mM. The cells were then grown 4 h at 37°C before being pelleted by centrifugation and removing the supernatant media by aspiration. The pellets were frozen at -20°C overnight. The cells were lysed with 5 cycles of dry ice/ethanol bath for 2 minutes and then 5 minutes in 10°C water. The pellet was then resuspended in 250 μL 25 mM HEPES, 100 mM NaCl, pH 7.5, and incubated on ice for 15 minutes before centrifugation. 180 μL of supernatant was transferred for activity assay.
Protein purification for steady-state kinetics assays
Proteins were expressed in BL21-star (DE3) cells using auto-induction media for 8 hours at 37°C followed by 24 hours at 18°C. The cells were sonicated and purified over Qiagen Ni-NTA resin. After elution the proteins were dialyzed at least three times into a 100-fold excess of buffer: 25 mM HEPES, 100 mM NaCl at pH 7.5 or by desalting column (Sephadex G-25, HiPrep 26/10 desalting column from GE Healthcare) to remove the imidazole. The proteins are then collected, flash frozen and stored at -80; or diluted for the activity assay.
Catalytic assays for cell lysate and purified proteins
For assay of cell lysates, 145μl cell lysate supernatant or diluted purified protein (10μM) in buffer (25 mM HEPES, 100 mM NaCl at pH 7.5) was pipetted into each well of a 96 Well Black Flat Bottom Polystyrene Non Binding Surface Microplates (Corning, Lowell, MA). Then 5 μL of 10.0 mM 4-hydroxy-4-(6-methoxy-2-naphthyl)-2-butanone 27 in CH3CN was than added and the enzyme activity was monitored at room temperature by fluorescence in a Spectramax M5e (Molecular Devices, Sunnyvale, CA) with λex = 330 nm (9 nm bandwidth) and λem = 452 nm (15 nm badwidth) with an additional filter at 435 nm for increased noise reduction. Quartz cuvette measurements were used to verify the plate measurements. The reactions were controlled for evaporation and were generally stable up to 4-5 hours with minimal evaporation- though most kinetic measurements were done within the first hour.
Assays for retro-aldol cleavage of (±)-methodol with purified protein27, were performed at room temperature by following product fluorescence as for cell lysates. Measurements were performed in triplicate and averaged. Steady state kinetic parameters kcat and KM were derived by fitting initial rates to the Michaelis-Menten equation.
Crystallization
RA34.6 was overexpressed from E. coli strain BL21 (RIL) as a non-cleavable C-terminal 6x His-tagged protein construct using the pET-29b vector (Novagen), and purified via affinity chromatography against Talon metal-chelate resin (Clontech).
The diketone inhibitor (1-(6-methoxy-2-naphthalenyl)-1,3-butanedione was prepared by oxidation of methodol28 with Dess-Martin periodinane29 (see supplemental material). For the bound inhibitor structures, RA34.6 (42 μM, 1ml) plus 50 μL 10 mM diketone inhibitor, final concentration ~500 μM) was incubated for 30 mins; then, 10μL Na(CN)BH3 (5M in 1M NaOH stock, final concentration ~50 mM) was added and incubated another 30 mins. The pH of the reaction is about 7.5. Afterwards, an aliquot was taken for mass spectrometry to observe the extent of covalent modification. Finally the rest of the sample was purified by gel filtration column and used to set up the crystallization tray.
For the 4-hydroxy-4-methyl-2-pentanone structure: RA34.6 (47.7 μM, 1ml) plus 50 μL 10mM 4-hydroxy-4-methyl-2-pentanone (~500 μM) was incubated at room temperature for 30mins, after which 10 μL Na(CN)BH3 (5 M in 1 M NaOH stock, final concentration ~50 mM) was added and incubated another 30 minutes. An aliquot was taken for mass spectrometry to determine the extent of covalent modification (about 50% was derivatized judging by MS). Gel filtration was used to clean the protein sample, which was subsequently used to set up the crystal tray.
Purified proteins were each concentrated to approximately 5 mg/mL in 100 mM NaCl, 25 mM Tris, pH 7.5, by centrifugation against low molecular weight cut-off sieves (Centricon) and then screened for crystallization conditions using a commercial sparse matrix screen (Nextal Classic Suite; Qiagen). Crystals were grown by equilibration of the protein against a reservoir containing 2 M ammonium sulfate, 4% PEG400, 100 mM sodium acetate pH 5.5. The crystals were frozen in a cryo buffer of 2.1 M ammonium sulfate, 5% PEG400, 0.2 M sodium acetate, and 500 mM NaCl. For the structure with the inhibitor, the crystals were exposed to the inhibitor prior to freezing.
Molecular replacement was performed using the PDB file (1a53) from the RCSB database30, corresponding to the parental protein prior to computational redesign, with all sidechains and extended peptide regions that were subjected to redesign deleted. The designed active site was rebuilt from the density (Figure 1). All stages of molecular replacement, model building, and refinement were performed using programs from the CCP4 computational suite 31 (PHASER32, COOT33 and REFMAC34).
For apo RA34.6 protein, crystals were grown via vapor diffusion. Equal amounts of protein at 5 mg/ml in 100 mM NaCl 25 mM Tris pH 7.5 were mixed with well solutions consisting of 2 M ammonium sulfate, 4% PEG400, 100 mM sodium acetate pH 5.5. Data were collected at -180°C in cryoprotectant buffer composed of 2.1 M ammonium sulfate, 5% PEG400, 0.2 M sodium acetate, 500 mM sodium chloride on a Rigaku Micromax 7HF with a Saturn 944+. Data were indexed, integrated, and scaled using HKL2000 package. Molecular replacement and refinement were performed with the Phaser and Refmac modules of CCP4i. Coot was used for model building.
Subsequent incubation and cross-linking of the purified enzyme with the naphthyl substrate analog or with the acetone product analog resulted in features of electron density that were consistent with each compound, with each being present at higher occupancy than the unidentified ligand in the unsoaked crystals.
Supplementary Material
Acknowledgements
We thank Dan Herschlag for helpful discussions and comments on an earlier version of the manuscript, Arshiya Quadri for help with protein purification, and DARPA, HHMI and the Schweizerischer Nationalfonds for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS:
Coordinates and structure factors have been deposited in the Protein Data Bank with RCSD PDB accession codes 3O6Y, 3NL8 and 3NXF.
References
- 1.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, 3rd, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–91. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–5. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 3.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Rothlisberger D, Baker D. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–94. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolon DN, Mayo SL. Enzyme-like proteins by computational design. Proc Natl Acad Sci U S A. 2001;98:14274–9. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St Clair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–13. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reymond J-L, Chen Y. Catalytic, enantioselective aldol reaction using antibodies against a quaternary ammonium ion with a primary amine cofactor. Tetrahedron Letters. 1995;36:2575–2578. [Google Scholar]
- 7.Wagner J, Lerner RA, Barbas CF., 3rd Efficient aldolase catalytic antibodies that use the enamine mechanism of natural enzymes. Science. 1995;270:1797–800. doi: 10.1126/science.270.5243.1797. [DOI] [PubMed] [Google Scholar]
- 8.Barbas CF, 3rd, Heine A, Zhong G, Hoffmann T, Gramatikova S, Bjornestedt R, List B, Anderson J, Stura EA, Wilson IA, Lerner RA. Immune versus natural selection: antibody aldolases with enzymic rates but broader scope. Science. 1997;278:2085–92. doi: 10.1126/science.278.5346.2085. [DOI] [PubMed] [Google Scholar]
- 9.Hilvert D. Critical analysis of antibody catalysis. Annu Rev Biochem. 2000;69:751–93. doi: 10.1146/annurev.biochem.69.1.751. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann T, Zhong G, List B, Shabat D, Anderson J, Gramatikova S, Lerner RA, Barbas CF. Aldolase Antibodies of Remarkable Scope. Journal of the American Chemical Society. 1998;120:2768–2779. [Google Scholar]
- 11.Muller MM, Windsor MA, Pomerantz WC, Gellman SH, Hilvert D. A rationally designed aldolase foldamer. Angew Chem Int Ed Engl. 2009;48:922–5. doi: 10.1002/anie.200804996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnsson K, Allemann RK, Widmer H, Benner SA. Synthesis, structure and activity of artificial, rationally designed catalytic polypeptides. Nature. 1993;365:530–2. doi: 10.1038/365530a0. [DOI] [PubMed] [Google Scholar]
- 13.Hennig M, Darimont BD, Jansonius JN, Kirschner K. The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product. J Mol Biol. 2002;319:757–66. doi: 10.1016/S0022-2836(02)00378-9. [DOI] [PubMed] [Google Scholar]
- 14.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–8. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 15.Fullerton SW, Griffiths JS, Merkel AB, Cheriyan M, Wymer NJ, Hutchins MJ, Fierke CA, Toone EJ, Naismith JH. Mechanism of the Class I KDPG aldolase. Bioorg Med Chem. 2006;14:3002–10. doi: 10.1016/j.bmc.2005.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Acc Chem Res. 2001;34:938–45. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 17.Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50:4402–10. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 18.Lassila JK, Baker D, Herschlag D. Origins of catalysis by computationally designed retroaldolase enzymes. Proc Natl Acad Sci U S A. 2010;107:4937–42. doi: 10.1073/pnas.0913638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu X, Tanaka F, Lerner RA, Barbas CF, 3rd, Wilson IA. Direct observation of an enamine intermediate in amine catalysis. J Am Chem Soc. 2009;131:18206–7. doi: 10.1021/ja907271a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka F, Fuller R, Barbas CF., 3rd Development of small designer aldolase enzymes: catalytic activity, folding, and substrate specificity. Biochemistry. 2005;44:7583–92. doi: 10.1021/bi050216j. [DOI] [PubMed] [Google Scholar]
- 21.Khersonsky O, Rothlisberger D, Dym O, Albeck S, Jackson CJ, Baker D, Tawfik DS. Evolutionary optimization of computationally designed enzymes: Kemp eliminases of the KE07 series. J Mol Biol. 2010;396:1025–42. doi: 10.1016/j.jmb.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 22.Khersonsky O, Rothlisberger D, Wollacott AM, Murphy P, Dym O, Albeck S, Kiss G, Houk KN, Baker D, Tawfik DS. Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution. Journal of molecular biology. 2011;407:391–412. doi: 10.1016/j.jmb.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorentzen E, Siebers B, Hensel R, Pohl E. Mechanism of the Schiff base forming fructose-1,6-bisphosphate aldolase: structural analysis of reaction intermediates. Biochemistry. 2005;44:4222–9. doi: 10.1021/bi048192o. [DOI] [PubMed] [Google Scholar]
- 24.Heine A, DeSantis G, Luz JG, Mitchell M, Wong CH, Wilson IA. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science. 2001;294:369–74. doi: 10.1126/science.1063601. [DOI] [PubMed] [Google Scholar]
- 25.Choi KH, Shi J, Hopkins CE, Tolan DR, Allen KN. Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate. Biochemistry. 2001;40:13868–75. doi: 10.1021/bi0114877. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985;82:488–92. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka F, Fuller R, Shim H, Lerner RA, Barbas CF., 3rd Evolution of aldolase antibodies in vitro: correlation of catalytic activity and reaction-based selection. Journal of molecular biology. 2004;335:1007–18. doi: 10.1016/j.jmb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 28.List B, Barbas CF, 3rd, Lerner RA. Aldol sensors for the rapid generation of tunable fluorescence by antibody catalysis. Proc Natl Acad Sci U S A. 1998;95:15351–5. doi: 10.1073/pnas.95.26.15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dess DB, Martin JC. Readily accessible 12-I-5 oxidant for the conversion of primary and secondary alcohols to aldehydes and ketones. The Journal of Organic Chemistry. 1983;48:4155–4156. [Google Scholar]
- 30.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–95. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.