Abstract
A comprehensive transcriptome assembly for pigeonpea has been developed by analyzing 128.9 million short Illumina GA IIx single end reads, 2.19 million single end FLX/454 reads, and 18 353 Sanger expressed sequenced tags from more than 16 genotypes. The resultant transcriptome assembly, referred to as CcTA v2, comprised 21 434 transcript assembly contigs (TACs) with an N50 of 1510 bp, the largest one being ∼8 kb. Of the 21 434 TACs, 16 622 (77.5%) could be mapped on to the soybean genome build 1.0.9 under fairly stringent alignment parameters. Based on knowledge of intron junctions, 10 009 primer pairs were designed from 5033 TACs for amplifying intron spanning regions (ISRs). By using in silico mapping of BAC-end-derived SSR loci of pigeonpea on the soybean genome as a reference, putative mapping positions at the chromosome level were predicted for 6284 ISR markers, covering all 11 pigeonpea chromosomes. A subset of 128 ISR markers were analyzed on a set of eight genotypes. While 116 markers were validated, 70 markers showed one to three alleles, with an average of 0.16 polymorphism information content (PIC) value. In summary, the CcTA v2 transcript assembly and ISR markers will serve as a useful resource to accelerate genetic research and breeding applications in pigeonpea.
Keywords: Cajanus cajan (L.), second-generation sequencing, transcriptome assembly, intron spanning region (ISR) markers
INTRODUCTION
Pigeonpea (Cajanus cajan (L.) Millspaugh) is an important food legume crop of tropical and subtropical regions of the world. It is a diploid (2n = 2x = 22), with moderate genome size of 858 Mbp (Greilhuber and Obermayer, 1998). The genus Cajanus comprises 32 species, most of which are found in India and Australia, and one is native to West Africa (Bohra et al., 2011a). Pigeonpea is grown in 4.67 Mha across the world and India is the world’s largest producer (van der Maesen, 1990). It is an important crop in south Asia, the Caribbean, and parts of Africa and South America. Pigeonpea is a vital source of protein (with 20–22% protein by dry weight), especially in vegetarian diets (Duke, 1981).
Key limitations to sustainable pigeonpea production are several abiotic stresses (e.g. drought, salinity and water-logging) and biotic stresses (e.g. Fusarium wilt (FW), sterility mosaic disease (SMD), and pod borer insects). Addressing these limitations is critical to meeting the demands of resource-poor people where pigeonpea is grown. Although there are continuing efforts for pigeonpea improvement through conventional breeding (Reddy et al., 1978; Saxena et al., 1983; Wanjari and Patel, 2003; Saxena, 2008), and molecular breeding has a great potential to enhance crop productivity (Varshney et al., 2010), limited availability of genomic resources coupled with a narrow genetic diversity in the cultivated gene pool have been serious bottlenecks to successful molecular breeding in pigeonpea (Varshney et al., 2009a).
Genomic resources like molecular markers, genetic maps, transcriptomic, or genome sequence data are prerequisites for undertaking molecular breeding in any crop. In the case of pigeonpea, efforts have been made only recently to develop some genomic resources. These include 88 860 bacterial artificial chromosome (BAC)-end sequences (BESs), 3072 BES-derived simple sequence repeat (SSR, or BES-SSR) markers, a 239 BES-SSR locus genetic map (Bohra et al., 2011a), 18 353 Sanger ESTs (Raju et al., 2010; unpublished), and 1.696 million FLX/454 reads (Dutta et al., 2011). Additionally, 494 353 FLX/454 reads and 128.9 million short Illumina GA IIx single end reads have also been generated (Dubey et al., 2011). The 494 353 FLX/454 reads along with the 10 817 Sanger ESTs available at the time were merged to generate a transcript assembly (CcTA v1) comprising 48 726 contigs (Dubey et al., 2011).
To improve the resources for pigeonpea genetics research and breeding applications, the present study was undertaken to develop a comprehensive transcriptome assembly based on a hybrid approach consisting of Sanger ESTs and mRNA-Seq data from two different next-generation sequencing platforms (Illumina GA IIx and FLX/454). This new assembly (CcTA v2) is available through the Legume Information System (LIS) website at http://cajca.comparative-legumes.org/data/lista_cajca-201012.tgz. These transcript assembly contigs (TACs) were aligned to the genome sequence of soybean (Glycine max), also in the Phaseoleae tribe and separated from Cajanus by about 20 Mya (Stefanovic et al., 2009). With the help of anchoring points between pigeonpea and soybean genomes, intron spanning region (ISR) markers were developed. Probable chromosomal assignments for more than 6000 ISR markers have been predicted for pigeonpea on the basis of location of the ISR markers in the soybean genome, and the locations of mapped BES-SSR loci in the pigeonpea genetic map that were used as anchor points between the pigeonpea and soybean genomes. Validation of a subset of ISR markers underlines the utility of these markers to enrich the existing pigeonpea genetic maps and identification of the quantitative trait loci (QTLs) for resistance/tolerance to biotic/abiotic stresses.
RESULTS
Development of an Improved Transcriptome Assembly
Four datasets, including 128.9 million Illumina GA IIx reads from 12 genotypes (Dataset I), 2.19 million FLX/454 reads from three genotypes (Datasets II and III), and 18 353 Sanger ESTs from six genotypes (Dataset IV) were processed to generate the transcriptome assembly CcTA v2 (Table 1). The transcriptome assembly comprises 21 434 TACs.
Table 1.
Details on Datasets Used for Developing Comprehensive Transcriptome Assembly (CcTA v2).
| Dataset/sequencing platform | Genotype | Tissues | Source | Number of transcript reads |
| Dataset I | ||||
| Illumina GA IIx | ICPL 332 | SMD-challenged leaves | ICRISAT/NCGR | 16 361 115 |
| Illumina GA IIx | ICPW 94 | FW-challenged roots | ICRISAT/NCGR | 15 828 791 |
| Illumina GA IIx | ICPL 99050 | FW-challenged roots | ICRISAT/NCGR | 13 498 156 |
| Illumina GA IIx | ICP 7035 | SMD-challenged leaves | ICRISAT/NCGR | 13 223 516 |
| Illumina GA IIx | ICPB 2049 | FW-challenged roots | ICRISAT/NCGR | 11 494 670 |
| Illumina GA IIx | BSMR 736 | SMD-challenged leaves | ICRISAT/NCGR | 11 065 219 |
| Illumina GA IIx | ICP 28 | FW-challenged roots | ICRISAT/NCGR | 9 721 562 |
| Illumina GA IIx | ICPL 20096 | SMD-challenged leaves | ICRISAT/NCGR | 9 507 797 |
| Illumina GA IIx | ICPL 87091 | FW-challenged roots | ICRISAT/NCGR | 8 977 567 |
| Illumina GA IIx | TAT 10 | SMD-challenged leaves | ICRISAT/NCGR | 7 932 691 |
| Illumina GA IIx | TTB 7 | SMD-challenged leaves | ICRISAT/NCGR | 4 122 216 |
| Illumina GA IIx | Asha (ICPL 87119) | FW-challenged roots | ICRISAT/NCGR | 7 182 619 |
| Dataset II | ||||
| FLX/454 | Asha (ICPL 87119) | Pooled RNA from 4 tissues | NRCPB | 906 300 |
| FLX/454 | UPAS 120 | Pooled RNA from 4 tissues | NRCPB | 790 424 |
| Dataset III | ||||
| FLX/454 | PusaAgeti | Pooled RNA from 31 tissues | ICRISAT | 494 353 |
| Dataset IV | ||||
| Sanger | ICPL 20102 | FW-challenged roots | ICRISAT | 3168 |
| Sanger | ICP 2376 | FW-challenged roots | ICRISAT | 2880 |
| Sanger | TTB 7 | SMD-challenged leaves | ICRISAT | 1920 |
| Sanger | ICP 7035 | SMD-challenged leaves | ICRISAT | 1920 |
| Sanger | Asha (ICPL 87119) and UPAS 120 | Root and shoot | NCBI dbEST | 8465 |
SMD, sterility mosaic disease; FW, Fusarium wilt, ICRISAT, International Crops Research Institute for the Semi-Arid Tropics; NCGR, National Center for Genome Resources; NRCPB, National Research Center on Plant Biotechnology; NCBI, National Centre for Biotechnology Information.
When the datasets above were analyzed individually in earlier studies, a wide range of TAC counts were reported: 4557 contigs from 9888 Sanger ESTs (Raju et al., 2010) and 43 324 contigs from 1.696 million FLX/454 reads (Dutta et al., 2011). The CcTA v1 (Dubey et al., 2011), assembled from 494 353 454/FLX reads and 10 817 Sanger ESTs, produced an assembly of 48 726 contigs. The transcriptome assembly in this study, referred to as CcTA v2, has numerous improved characteristics (Table 2). For instance, the CcTA v2 has a total of 21 434 TACs, with N50 of 1510 bp, while the CcTA v1 included 48 726 contigs, with N50 length of only 285 bp (Dubey et al., 2011). The largest TAC in CcTA v2 is 7909 bp, which is almost four times larger than that in the CcTA v1, with 2067 bp.
Table 2.
Comparative Analysis of Four Assemblies.
| CcTA v2 (this study) | CcTA v1 (Dubey et al., 2011) | Dutta et al. (2011) | Raju et al. (2010) | |
| Sequence data used | 128.9 million Illumina short reads + 2.19 million FLX/454 reads + 18,353 Sanger ESTs | 494 353 FLX/454 reads and 10 817 Sanger ESTs | 1.696 million FLX/454 reads | 9888 Sanger ESTs |
| Number of genotypes providing sequence data | >16 | 5 | 2 | 4 |
| Program(s) used for assembly | ABySS and miraEST | CAP3 | Lasergene SeqMan Pro™ v8.0.12 | CAP3 |
| Total number of transcript assembly contigs | 21 434 | 48 726 | 43 324 | 4557 |
| N50 (bp) | 1510 | 287 | 1222 | 701 |
| Largest contig (bp) | 7909 | 2067 | 7783 | 3430 |
| Shortest contig (bp) | 85 | 52 | 32 | 46 |
To check for microbial contamination, if any, all 21 434 TACs of the transcriptome assembly (CcTA v2) were BLASTed against NCBI’s bacterial genomes database. Only 49 TACs had significant hits to bacterial genomes. These 49 TACs were further BLASTed against NCBI’s EST_others database (non-human, non-mouse). While 45 TACs had hit with pigeonpea cDNA libraries or other plant cDNAs, only four TACs (lista_cajca-201012 TACs# 9664, 18316, 21003, and 21371) still showed hit with microbial ESTs. However, three of these TACs (lista_cajca-201012 TACs# 9664, 18316, and 21371) could be mapped to the soybean genome at >90% sequence similarity and 80% coverage of query length. Therefore, these TACs also could be considered legitimate transcript contigs. The remaining one TAC (lista_cajca-201012 TAC# 21003) could also be mapped to soybean genome at a lower stringency, namely >85% sequence similarity and >50% coverage of query length. Therefore, the developed transcriptome assembly seems to be of high quality without any microbial contamination.
Comparison of CcTA v2 with the Soybean Genome
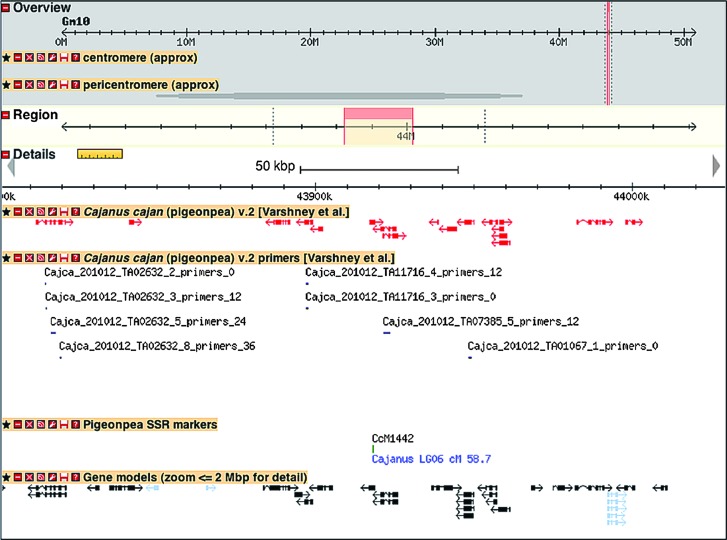
All TACs of CcTA v2 were aligned to the soybean genome (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v7.0/Gmax/) using the HMM-based alignment program Exonerate 2.2.0 (Slater and Birney, 2005), to investigate gene coverage and gene structures of pigeonpea. Using the alignment criteria of 80% identity and 50% coverage, of the 21 434 TACs, 16 622 (77.5%) could be aligned. These alignments can be visualized in the soybean genome browser, using the track ‘Cajanus cajan (pigeonpea) v.2’, at http://soybase.org/gb2/gbrowse/gmax1.01. It is encouraging to note that 27 490 predicted soybean genes have matches, and that the overall distribution of soybean gene models covered by the CcTA v2 is relatively even across all 20 soybean chromosomes, ranging from 50 to 73% of predicted soybean genes per chromosome with pigeonpea matches (Table 3). Further, 9863 (59.3%) out of 16 622 CcTA v2 TACs had two good matches with the soybean genome, consistent with the recent genome duplication in soybean with respect to pigeonpea, while an additional 20% had three or four good matches, as might be expected given the ancient whole-genome duplication event in pappilionoids ∼59 Mya (Schmutz et al., 2010). Details of the number of pigeonpea TACs mapped onto soybean at a given number of times are given in Supplemental Table 1. Synteny and commonalities between pigeonpea TACs and soybean genes can be visualized genome-wide in soybean Gbrowse (http://soybase.org/gb2/gbrowse/gmax1.01) (Figure 1).
Table 3.
Mapping of Pigeonpea Transcriptome Assembly TACs onto Soybean Genome Build 1.0.9.
| Soybean (Gm) chromosome | Total unique CcTA v2 TACs hits | Genes covered on each Gm chromosome | Total number of genes on each Gm chromosome | Percent of genes covered on each Gm chromosome |
| Gm01 | 1558 | 1200 | 2023 | 59.30 |
| Gm02 | 1985 | 1582 | 2652 | 59.70 |
| Gm03 | 1524 | 1244 | 2196 | 56.60 |
| Gm04 | 1656 | 1293 | 2088 | 61.90 |
| Gm05 | 1738 | 1556 | 2147 | 72.50 |
| Gm06 | 1960 | 1324 | 2686 | 49.30 |
| Gm07 | 1791 | 1378 | 2298 | 60.00 |
| Gm08 | 2392 | 1930 | 3130 | 61.70 |
| Gm09 | 1693 | 1324 | 2305 | 57.40 |
| Gm10 | 1861 | 1497 | 2479 | 60.40 |
| Gm11 | 1839 | 1455 | 2304 | 63.20 |
| Gm12 | 1453 | 1212 | 2016 | 60.10 |
| Gm13 | 2418 | 1893 | 3119 | 60.70 |
| Gm14 | 1359 | 1061 | 1825 | 58.10 |
| Gm15 | 1690 | 1288 | 2250 | 57.20 |
| Gm16 | 1172 | 999 | 1803 | 55.40 |
| Gm17 | 1757 | 1351 | 2252 | 60.00 |
| Gm18 | 1738 | 1374 | 2452 | 56.00 |
| Gm19 | 1608 | 1300 | 2177 | 59.70 |
| Gm20 | 1504 | 1229 | 1975 | 62.20 |
| Total | 27 490 | 46 177 | 59.50 |
Figure 1.
A Sample View of Pigeonpea TACs, Markers, and Candidate ISR Markers onto Soybean Genome Sequence.
This image is from the SoyBase GBrowse viewer for soybean, at http://soybase.org/gb2/gbrowse, shows 200 kb from Gm10, starting from position 43 820 000. At a zoom level >2 Mbp, a heat map showing transcript density will be displayed. Assemblies for all 20 Gm chromosomes are displayed alongside regions of sequence homology in Cc. Red: there was at least one additional reported CcTA v2 alignment. Green: there were no other reported alignments.
Development of ISR Markers
The alignment of the CcTA v2 transcriptome assembly with the soybean genome predicted 10 009 intron spanning regions (ISRs), for a total of 5033 TACs. The alignments and primer sets can be viewed on the SoyBase genome browser at http://bit.ly/Cajca_ISR3, and are available for download at http://cajca.comparative-legumes.org/data/contrib/cajanus_cajan_v2_primers.txt.gz. A minimum of one and a maximum of 19 ISRs were designed against each of the matched soybean gene (varying based on the number of introns in a gene and the ability of the primer prediction software to identify low-copy ISR markers across the introns). The largest number of genes with ISR markers (1516) contained two markers.
Selection of Genome-Wide Set of ISR Markers
In addition to the identification of ISR markers in pigeonpea, approximate mapping positions for a portion of these markers were predicted based on syntenic regions between pigeonpea and soybean genomes, using mapped BES-SSR loci in the pigeonpea genetic map as anchor points. In this context, 239 pigeonpea BES-SSR loci (Bohra et al., 2011a), mapped onto the genome sequence of soybean, were used for identifying the syntenic regions between the pigeonpea and soybean genomes. Of these 239 BES-SSR loci, 93 showed probable synteny in soybean chromosomes, and were used to identify putative linkage groups for 6284 pigeonpea ISR markers.
This method produced putative linkage group assignments for all 11 of the pigeonpea linkage groups. The strongest associations were for Gm10 and CcLG02, and for Gm14 and CcLG10; for these associations, there were 12 and 10 markers in syntenic regions, respectively; and putative placements for 888 and 738 ISRs on CcLG02 and CcLG10, respectively. Details of correspondences of other ISR markers between soybean chromosome and expected pigeonpea CcLGs are given in Supplemental Table 2.
ISR Marker Polymorphism
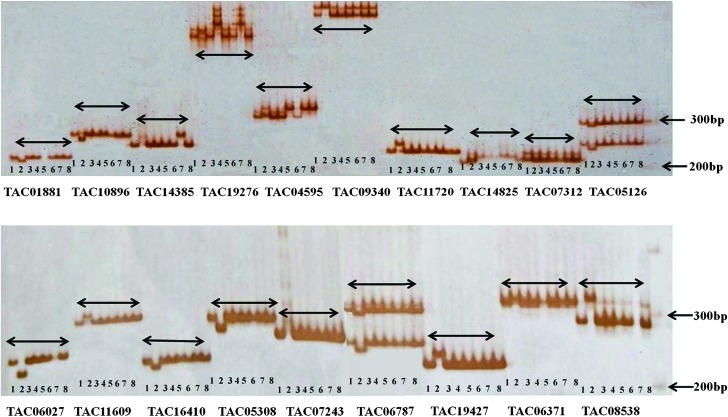
Primer pairs were designed and synthesized for a total of 128 ISR markers. All these primer pairs were screened for amplification of DNA from two pigeonpea genotypes, namely ICP 28 and the popular variety Asha (ICPL 87119). This analysis identified a set of 116 markers (90.6%) with scorable amplicons. These 128 ISR markers corresponded to 10 pigeonpea linkage groups, generally distributed evenly—as assessed relative to the syntenic regions on soybean chromosomes. Screening of these 116 ISRs on eight pigeonpea genotypes including seven cultivated (ICPL 332, ICPL 99050, ICPB 2049, ICP 28, ICPL 20096, ICPL 87091, and ICPL 87119) and one wild (ICPW 94) showed length polymorphism (two to three alleles) (Figure 2) with 70 (54.6%) markers (Table 4 and Supplemental Table 3). The polymorphism information content (PIC) value for the polymorphic markers ranged from 0.19 to 1.00, with an average of 0.16.
Figure 2.
Amplification Pattern of ISR Markers on MDE Gel.
From left to right lanes 1–8 for each marker: ICPL 332, ICPW 94, ICPL 99050, ICPB 2049, ICP 28, ICPL 20096, ICPL 87091, and ICPL 87119. Arrows indicate ladder bp fragments.
Table 4.
Distribution of ISR Markers on Pigeonpea Linkage Groups and Their Polymorphism Status.
| Pigeonpea LG | Total ISR markers showing mapping positions | Markers selected for analysis | Markers amplified | Markers showing polymorphism |
| CcLG01 | 333 | 10 | 8 | 5 |
| CcLG02 | 1778 | 20 | 17 | 12 |
| CcLG03 | 699 | 17 | 16 | 9 |
| CcLG04 | 763 | 10 | 7 | 6 |
| CcLG05 | 27 | – | – | – |
| CcLG06 | 257 | 8 | 8 | 6 |
| CcLG07 | 478 | 15 | 15 | 11 |
| CcLG08 | 394 | 5 | 5 | 4 |
| CcLG09 | 281 | 15 | 15 | 6 |
| CcLG10 | 1011 | 16 | 14 | 5 |
| CcLG11 | 263 | 12 | 11 | 6 |
| Total | 6284 | 128 | 116 | 70 |
With an objective of identification of SNPs and indels with ISR markers, as an example, sequence data were generated for eight pigeonpea genotypes with six ISR markers (TAC12160, TAC08538, TAC10495, TAC04354, TAC18853, and TAC07243). After trimming low-quality sequences at both ends, sequence data were aligned and analyzed for occurrence of SNPs and indels in the eight genotypes. While six markers showed 27 SNPs and two ISR markers (TAC08538 and TAC07243) showed two indels between the wild species, ICPW 94 and seven cultivated species (ICPL 332, ICPL 99050, ICPB 2049, ICP 28, ICPL 20096, ICPL 87091, and ICPL 87119) (Supplemental Table 4 and Supplemental Figure 1). Indels observed in the cases of TAC08538 and TAC07243 markers were confirmed on mutation detection enhancement (MDE) gel as mentioned above (Figure 2).
DISCUSSION
In pigeonpea, conventional breeding to date has proceeded without the support of molecular methods. Limited use of germplasm over the course of pigeonpea domestication has also resulted in a very narrow genetic base (Yang et al., 2006). As a result, pigeonpea genetic improvement programs have made comparatively little progress and hence have faced problems in addressing key constrains to crop production, including a range of abiotic stresses (e.g. drought, salinity, water-logging) and biotic stresses (e.g. Fusarium wilt, sterility mosaic disease, Helicoverpa armigera). Only during last five years some efforts have been made to develop genomic resources such as SSRs, ESTs, genetic, maps and transcriptome assemblies in pigeonpea (Varshney et al., 2009a, 2010). The available genetic maps, especially based on intra-specific mapping populations, do not have a good marker density (Bohra et al., 2011b; Gnanesh et al., 2011).
This study reports a comprehensive transcriptome assembly that is based on 131 million sequence reads coming from a range of tissues of more than 16 different pigeonpea genotypes. We employed two assembly programs in order to take advantage of the characteristics of the constituent sequences. All Illumina GA IIx reads were assembled into contigs with the ABySS program, and FLX/454 reads were assembled with the miraEST program. Contigs generated for Illumina GA IIx and FLX/454 reads were assembled together with Sanger ESTs using the miraEST program. Therefore, the developed assembly is a hybrid assembly. Hybrid assemblies are considered superior over pure assembly based on sequence data coming from one sequencing platform, as weaknesses from single sequencing platforms may be compensated by different characteristics of sequences from other platforms (Schatz et al., 2010; Garg et al., 2011).
The assembly described in this study is coming from 31 tissues representing a range of development and growth stages as well as challenged by different stresses. Thus, this assembly (CcTA v2) can be considered the most comprehensive transcriptome assembly of pigeonpea. The completeness and quality of this assembly can be assessed by comparing it with other earlier assemblies (Raju et al., 2010; Dubey et al., 2011; Dutta et al., 2011). The average TAC length as well as N50 of TAC in the CcTA v2 is much better than the CcTA v1. It is important to mention here that earlier assemblies were developed based on CAP3 (Dubey et al., 2011; Raju et al., 2010) and Lasergene SeqMan Pro™ v8.0.12 (Dutta et al., 2011) programs, while the present assembly has been developed using two powerful assembly programs that can accommodate large amounts of next-generation sequence: ABySS and miraEST.
The transcriptome assembly developed here can be used for a variety of applications to advance genetics research and breeding applications in pigeonpea. For instance, this assembly can provide the information about gene content and function, for identification of candidate genes, for development of molecular markers such as SSRs, SNPs, etc. (Varshney et al., 2009b). The majority of these applications have already been explored and a preview of such applications for the pigeonpea transcriptome was presented in our earlier studies (Dubey et al., 2011; Dutta et al., 2011). Therefore, these topics are not being discussed in this study. We have demonstrated one major application of the transcriptome assembly in the development of genome-wide marker datasets for enriching the genetic map of pigeonpea, using a comparative genomics approach that employs the soybean genome sequence and the BES-SSR loci-based genetic map of pigeonpea (Bohra et al., 2011a). For instance, comparison of the CcTA v2 with soybean genome identified the homologs for 77.5% of the pigeonpea TACs, and covering 27 490 genes in the soybean genome. Of these, the majority of TACs (9863/16 622) mapped twice against soybean genome—as expected, because of the ∼13-Mya genome duplication in soybean. Occurrence of more than two hits in the soybean genome for a given pigeonpea TAC is also not surprising, as ∼75% of soybean genes were found in multiple copies, due to older genome duplications in soybean’s history (Schmutz et al., 2010). Based on exon–intron boundaries, 10 009 primer pairs, designed for 5033 TACs, can be used for checking the length or sequence polymorphism between the parental genotypes of mapping populations. To shortlist a set of markers that can be mapped across the pigeonpea genome, 177 anchor points were identified between the soybean and pigeonpea genomes, with the genetically mapped pigeonpea BES-SSR loci being placed by sequence homology onto the soybean genome sequence. Based on this information, 6284 ISR markers were identified that have putative chromosomal placements in the pigeonpea genome. A subset (128) of these markers was further analyzed for length (indel) polymorphism in eight parental genotypes of mapping populations segregating for three important traits, FW, SMD, and pod borer that are significant for pigeonpea improvement.
While 90.6% (116) markers provided scorable amplicons, 54.6% (70) markers showed polymorphism with two to three alleles in the genotypes analyzed on MDE gel. Utility of single-strand confirmation polymorphism-based SNPs and indels has been shown earlier in several species like common bean (Galeano et al., 2009), pearl millet (Thudi et al., 2010), etc. Although the majority of the markers showed polymorphism between ICPW 94 (C. scrabaeoides) and other genotype(s) of cultivated species (C. cajan), only three markers showed length polymorphism between the parental combinations of cultivated species. These results are not unexpected, as very low levels of polymorphism in some other cultivated species have been observed in several earlier studies (Yang et al., 2006; Varshney et al., 2009a; Raju et al., 2010; Varshney et al., 2010; Bohra et al., 2011a, 2011b; Dubey et al., 2011; Dutta et al., 2011; Gnanesh et al., 2011). On the other hand, sequence analysis of amplicons generated for six ISR markers showed their utility for identification of SNPs and indels at sequence level. Two common markers used for length as well as sequence polymorphism also confirmed the length polymorphism on MDE gel at sequence level. In brief, the validation results with some markers for detection of polymorphisms on MDE gel and sequence level underline the importance of developed resource of ISR markers. These markers should be useful for genetic mapping and trait mapping in breeding programs to develop the superior pigeonpea varieties with enhanced crop productivity.
In conclusion, the present study demonstrated a high-quality comprehensive transcriptome assembly of the important legume crop pigeonpea using Sanger and second-generation sequencing (FLX/454 and Illumina GA IIx) technologies. The results deliver novel information for future genetic studies in pigeonpea and provide a robust transcriptome assembly. The identification of syntenic regions between the pigeonpea and the sequence of a related phaseolid legume, soybean, provides greater insight into the gene content of pigeonpea. For the ISR markers identified, their putative mapping positions and parental polymorphism information will be a useful resource for molecular breeding programs to develop elite pigeonpea cultivars.
METHODS
Sequence Datasets
The following four datasets were used for defining the transcriptome assembly: (1) 128.9 million Illumina GA IIx short single end reads (1x36-nt) generated from 12 genotypes at ICRISAT and NCGR (Dubey et al., 2011; unpublished), referred to as Dataset I; (2) 1.696 million FLX/454 reads generated from the genotypes ‘Asha’ and ‘UPAS 120’ at NRCPB (Dutta et al., 2011), referred to as Dataset II; (3) 494 353 FLX/454 reads generated from ‘PusaAgeti’ at ICRISAT and J. Craig Venter Institute (JCVI) (Dubey et al., 2011), referred to as Dataset III; and (4) 18 353 vector-trimmed Sanger ESTs downloaded from dbEST (www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html) (the majority of which were generated at ICRISAT (Raju et al., 2010) and NRCPB, referred to as Dataset IV) (see Table 1).
Sequence Assembly
Sequence datasets, as mentioned above, were assembled using the programs ABySS (Simpson et al., 2009) and miraEST (Chevreux et al., 2004), using the following three steps. In the first step, all Illumina GA IIx reads (Dataset I) from 12 genotypes were pooled and assembled together using ABySS. In the second step, FLX/454 reads from three genotypes (Datasets II and III) were trimmed of adapter sequences and assembled individually using the miraEST assembler. Subsequently, the pooled Illumina GA IIx (step 1 by ABySS) and FLX/454 (step 2 by miraEST) assemblies were merged with vector-trimmed Sanger ESTs of Dataset IV using the miraEST program. Both programs were run with the default settings, except for the following parameters: for ABySS, scaffolding ‘on’ at the paired end stage; and for miraEST: number of threads was seven and these options specified as ‘no’: Load straindata, Enforce presence of qualities, Extra gap penalty, and Wants quality file. In order to decrease runtime, number of processors used was seven. Since we were interested in a consensus assembly, the ‘Load straindata’ option was turned off. During the second stage of the assembly in which FLX/454 and Sanger ESTs were merged, there were no quality scores for the synthetic ESTs. Therefore, ‘Enforce presence of qualities’ and ‘Wants quality file’ options were specified to ‘no’. By turning off ‘Extra gap penalty’, we avoided penalizing gaps during the smith waterman alignment, especially since FLX/454 data are known to have homopolymer errors.
In order to check for microbial contamination, BLAST of all 21 434 CcTA v2 contigs was carried out against NCBI’s microbial genomes database (www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html). The TACs of CcTA v2 having significant hits to bacterial genomes have been later on run against NCBI’s EST_others database (non-human, non-mouse) to check whether they could be mapped to other plant genomes.
Mapping of the Pigeonpea Transcriptome Assembly onto Soybean Genome
All TACs of the CcTA v2 assembly were aligned to soybean genome (Schmutz et al., 2010) build 1.0.9 using Exonerate 2.2.0 (Slater and Birney, 2005), with parameters and flags ‘percent 25’ (to report only alignments over 25% of the maximum score attainable by each query) and ‘refine region’ (to perform an exhaustive alignment over the region in which the heuristic alignment was found). Alignments were filtered to require at least 80% alignment identity and 50% query coverage. If this resulted in more than 12 matches for a given sequence, the sequence was considered repetitive, and all matches were discarded.
Mapping of BES-SSR Loci of Pigeonpea onto the Soybean Genetic Map
All genetically mapped BES-SSR loci onto the pigeonpea genetic map (Bohra et al., 2011a) were anchored to the soybean genome using BLASTN (Altschul et al., 1997) of the corresponding BESs (with maximum E-value 1e-8), followed by manual selection for best hits with matches up to two homologous soybean regions.
Identification of Intron Spanning Region (ISR) Markers
Alignment results of pigeonpea TACs with the soybean genome were analyzed for identification of flanking intron junctions. The Exonerate alignment of the TACs, in Exonerate ‘vulgar’ (Verbose Useful Labelled Gapped Alignment Report) output format, was used to identify intron junctions in the TAC fasta sequences. These junctions were used to design the primer pairs using Primer3 (Rosen and Skaletsky, 2000) and BatchPrimer3 (You et al., 2008). Primer pairs were remapped to the soybean genome (to evaluate for repetitive sequences) using e-PCR (Schuler, 1997), with parameters ‘-n3 -g1 -t3 -m400 -d50-1000’. These parameters have the following effects: ‘-n3’ allows up to three mismatches per primer; ‘-g1’ allows up to one gap per primer; ‘-t3’ specifies output in tabular format; ‘-m400’ specifies an allowable margin for the product of 400 bases; and ‘-d50-1000’ specifies the default PCR product size range. Primer pairs with more than two alignments at these parameters were discarded.
Putative approximate mapping positions for the identified ISR markers were imputed based on anchoring points between pigeonpea and soybean genetic maps using BES-SSR loci of pigeonpea. Where there are two or more pigeonpea SSR markers with proximity in both pigeonpea and soybean (i.e. with nearby cM values in pigeonpea and nearby nucleotide positions in soybean chromosome pseudomolecules), tentative pigeonpea linkage groups (CcLGs) were assigned for ISR candidate markers occurring between the neighboring pigeonpea SSR markers.
ISR Analysis
Polymerase chain reactions (PCRs) for amplification of ISR loci were performed on eight pigeonpea genotypes (seven cultivated and one wild species) in a 5-μl reaction volume as described by Gujaria et al. (2011). Amplified products were denatured and separation was undertaken on MDE gel electrophoresis as described earlier (Thudi et al., 2010).
Allele Re-Sequencing and SNP Detection
For detection of SNP or indel polymorphism in the case of ISR markers, PCR products for eight pigeonpea genotypes using six ISR markers were sequenced in both directions using Sanger sequencing methodology. Sequence data analysis and SNP identification among the selected genotypes were carried out as described in our earlier study (Gujaria et al., 2011).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
The authors thank the CGIAR Generation Challenge Programme (GCP), Mexico (G.D.M., R.K.V., and N.K.S.) and the Indian Council of Agricultural Research (ICAR), India (N.K.S. and R.K.V.) for sponsoring this research. No conflict of interest declared.
Supplementary Material
Acknowledgments
The authors are thankful to Anuja Dubey and Rachit Saxena for their help extended throughout this study.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohra A, et al. Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea, (Cajanus spp.) BMC Plant Biol. 2011a;11:56. doi: 10.1186/1471-2229-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohra A, Saxena RK, Gnanesh BN, Saxena KB, Byregowda M, Rathore A, Kavi Kishor PB, Cook DR, Varshney RK. An intra-specific consensus genetic map of pigeonpea (Cajanus cajan (L.) Millspaugh) derived from six mapping populations. 2011b doi: 10.1007/s00122-012-1916-5. BMC Genomics (submitted revised version) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey A, et al. Defining the transcriptome assembly and its use for genome dynamics and transcriptome profiling studies in pigeonpea (Cajanus cajan L.) DNA Res. 2011;18:153–164. doi: 10.1093/dnares/dsr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke JA. Handbook of Legumes of World Economic Importance. New York: Plenum Press; 1981. p. p. 345. [Google Scholar]
- Dutta S, et al. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea (Cajanus cajan (L.) Millspaugh) BMC Plant Biol. 2011;11:17. doi: 10.1186/1471-2229-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano CH, Fernández AC, Gómez M, Blair MW. Single strand conformation polymorphism based SNP and Indel markers for genetic mapping and synteny analysis of common bean (Phaseolus vulgaris L.) BMC Genomics. 2009;10:629. doi: 10.1186/1471-2164-10-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Patel RK, Jhanwar S, Priya P, Bhattacharjee A, Yadav G, Bhatia S, Chattopadhyay D, Tyagi AK, Jain M. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol. 2011;156:1661–1678. doi: 10.1104/pp.111.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanesh BN, Bohra A, Sharma M, Byregowda M, Pandey S, Wesley V, Saxena RK, Saxena KB, Kavi Kishor PB, Varshney RK. Genetic mapping and quantitative trait locus analysis of resistance to sterility mosaic disease in pigeonpea (Cajanus cajan (L.) Millsp.) Field Crops Res. 2011;123:53–61. [Google Scholar]
- Greilhuber J, Obermayer R. Genome size variation in Cajanus cajan (Fabaceae): a reconsideration. Plant Syst. Evol. 1998;212:135–141. [Google Scholar]
- Gujaria N, et al. Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.) Theor. Appl. Genet. 2011;122:1577–1589. doi: 10.1007/s00122-011-1556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju NL, Gananesh BN, Lekha P, Jayashree B, Pande S, Hiremath PJ, Byregowda M, Singh NK, Varshney RK. The first set of EST resource for gene discovery and marker development in pigeonpea (Cajanus cajan L.) BMC Plant Biol. 2010;10:45. doi: 10.1186/1471-2229-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BVS, Green JM, Bise SS. Genetic male sterility in pigeonpea. Crop Sci. 1978;18:362–364. [Google Scholar]
- Rosen S, Skaletsky HJ. Primer 3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Saxena KB. Genetic improvement of pigeonpea: a review. Tropical Plant Biol. 2008;1:159–178. [Google Scholar]
- Saxena KB, Wallis ES, Byth DE. A new gene for male sterility in pigeonpea (Cajanus cajan (L.) Millsp) Heredity. 1983;51:419–421. [Google Scholar]
- Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second generation sequencing. Genome Res. 2010;20:1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schuler GD. Sequence mapping by electronic PCR. Genome Res. 1997;7:541–750. doi: 10.1101/gr.7.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Pfeil BE, Palmer JD, Doyle JJ. Relationships among phaseolid legumes based on sequences from eight chloroplast regions. Syst. Bot. 2009;34:115–125. [Google Scholar]
- Thudi M, Senthilvel S, Bottley A, Hash CT, Reddy AR, Feltus AF, Paterson AH, Hoisington DA, Varshney RK. A comparative assessment of the utility of PCR-based marker systems in pearl millet. Euphytica. 2010;174:253–260. [Google Scholar]
- van der Maesen LJG. Pigeonpea: origin, history, evolution and taxonomy. In: Nene YL, Hall SD, Sheila VK, editors. Pigeonpea. Wallingford: CAB International; 1990. pp. 15–46. [Google Scholar]
- Varshney RK, Close TJ, Singh NK, Hoisington DA, Cook DR. Orphan legume crops enter the genomics era. Curr. Opin. Plant Biol. 2009a;12:202–210. doi: 10.1016/j.pbi.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Varshney RK, et al. Pigeonpea genomics initiative (PGI): an international effort to improve crop productivity of pigeonpea (Cajanus cajan L.) Mol. Breeding. 2010;26:393–408. doi: 10.1007/s11032-009-9327-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Nayak SN, May GD, Jackson SA. Next generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009b;27:522–530. doi: 10.1016/j.tibtech.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Wanjari KB, Patel MC. Fertility restorers isolated from germplasm for cytoplasmic male sterility in pigeonpea. PKV Res. J. 2003;27:111–113. [Google Scholar]
- Yang S, Pang W, Ash G, Harper J, Carling J, Wenzl P, Huttner E, Zong X, Kilian A. A Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Theor. Appl. Genet. 2006;113:585–595. doi: 10.1007/s00122-006-0317-z. [DOI] [PubMed] [Google Scholar]
- You FM, Huo N, Gu YQ, Luo M, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD. BatchPrimer3: a high throughput web application for PCR and sequencing primer designing. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.