The plant hormone salicylic acid (SA) plays a prominent role in modulating plant immune responses against diverse pathogens. SA also influences other physiological processes in plants, such as senescence-associated gene expression, basal thermogenesis, and seed germination (Vlot et al., 2009). Because of the critical role of SA in regulating plant immunity, growth, and development, there has been immense research about SA, which has resulted in the discovery of numerous plant genes involved in SA biosynthesis or signal transduction. One of the most notable findings was the identification of NPR1 (non-expressor of pathogenesis protein 1) (Cao et al., 1994; Delaney et al., 1995), a gene that encodes a master regulatory protein of SA-dependent defense responses and is a transcriptional co-activator of the TGA clade of bZIP transcription factors (transcription factors that contain basic region/leucine zipper motif). NPR1 exists in at least two forms in the cell. When the SA level is low in the cell (e.g. in the absence of pathogen infection), NPR1 is localized in the cytoplasm as an oligomer through intermolecular disulphide bonds. However, when the SA level is high (e.g. after pathogen infection), redox changes in the cytosol trigger the reduction of disulphide bonds, and monomeric NPR1 enters the nucleus and functions as a transcriptional co-activator at the target gene promoter (Mou et al., 2003). The npr1 mutant could not initiate the SA-associated global transcriptional response program and is defective in all major SA-dependent defense responses, suggesting a central role in SA signal transduction (Cao et al., 1994; Delaney et al., 1995). Despite these impressive advances, however, one of the most fundamental questions—the identity of the SA receptor—has remained unanswered.
There have been serious efforts to identify SA receptors in the past, using biochemical approaches. These efforts led to the identification of four tobacco SA-binding proteins (SABPs), including a catalase, a methyl salicylate esterase (SABP2), a cytoplasmic ascorbate peroxidase, and a chloroplastic carbonic anhydrase (Vlot et al., 2009). For example, SABP2 shows a very high affinity for SA; it uses methyl salicylate (MeSA) as a substrate and converts it to SA. Catalase and ascorbate peroxidase may be involved in redox changes after SA exposure, which could result in the breakdown of NPR1 oligomers into monomers, thereby facilitating NPR1 translocation into the nucleus (Tripathi et al., 2010). Although these SABPs are involved in mediating some aspects of SA metabolism or action, genetic evidence for them functioning as SA receptors is lacking. In particular, none is required for SA-mediated global defense gene expression, which is a hallmark function of NPR1. As such, it was widely believed that the true SA receptors are yet to be discovered. Using different ligand–receptor binding methods, two research groups now report that NPR1 or NPR1-related proteins, NPR3 and NPR4, are the long-sought-after SA receptors in Arabidopsis. Whereas Wu et al. provided evidence, using a special equilibrium dialysis ligand binding method, that NPR1 itself is a SA receptor, Fu et al. found that NPR1 does not bind to SA directly in conventional ligand binding assays, but two NPR1-related proteins, NPR3 and NPR4, bind to SA and function as SA receptors. In this Research Highlight, we will focus on the work of Fu and colleagues.
After failing to detect SA binding to NPR1 in a conventional ligand–receptor binding assay, Fu and colleagues carried out experiments to further understand the dynamic turnover of the NPR1 protein in the cell. These authors followed an intriguing observation in their previous research. NPR1 contains an N-terminal Broad-Complex, Tramtrack and Bric-a-brac/Pox virus and Zinc finger (BTB/POZ) domain, a central ankryin repeat domain, and a C-terminal region with transactivation activity. Some BTB domain-containing proteins interact with Cullin 3 (CUL3) E3 ligase and mediate substrate recognition, ubiquitination, and subsequent degradation by the 26S proteasome. Fu et al. examined the possibility of NPR1 functioning as an adaptor of a CUL3 E3 ligase to degrade a substrate protein (presumably a negative regulator of SA-dependent defenses). Unexpectedly, their previous study showed that the NPR1 protein itself is degraded by the proteasome. In this study, they wanted to identify the adaptor proteins of the CUL3 E3 ligase that target NPR1 for degradation. They thought that NPR1 paralogs NPR3 and NPR4 might be such adaptors because both contain the BTB domain and, more importantly, the npr3npr4 double mutant exhibits enhanced disease resistance, a phenotype that is opposite to that of the npr1 mutant. Indeed, Fu and colleagues found that NPR1 protein levels were higher in the npr4 and npr3npr4 mutants in comparison with that in wild-type plants. A series of further experiments convincingly show that NPR4 and NPR3 bind to both NPR1 and CUL3, and act as CUL3 adaptors for the degradation of NPR1 (Fu et al., 2012). Furthermore, degradation of NPR1 through the proteasome was observed in wild-type Arabidopsis plants, but not in the npr3 npr4 double mutant.
Major advances in recent years have firmly established a new paradigm in plant hormone receptor biology: plant hormones, such as auxin and jasmonate, directly promote the physical interaction between E3 ubiquitin ligases and their substrate proteins. In other words, E3 ligases and substrate proteins function as co-receptors for sensing auxin and jasmonate (Santner and Estelle, 2009). Amazingly, this paradigm also seems to be true for SA. Fu and colleagues found that not only SA, but also 2,6-dichloroisonicotinic acid (INA), a widely used biologically active analogue of SA, promote the NPR1–NPR3 interaction in yeast two-hybrid (Y2H) assays. In contrast, SA disrupted the interaction between NPR1 and NPR4. The authors verified the opposing effect of SA on NPR1–NPR3 and NPR1–NPR4 interactions using in vitro pull-down assays.
Fu and colleagues went on to determine SA-binding affinities for NPR3 and NPR4 using conventional ligand–receptor binding assays. They found that SA binds to NPR3 and NPR4 with vastly different binding affinities. The dissociation constant (Kd) for NPR4 was very low (46.2 ± 2.35 nM), whereas the Kd value for NPR3 was very high and could not be estimated accurately, but may be ~1000 nM. Furthermore, SA seems to bind to NPR3 and NPR4 at more than one site.
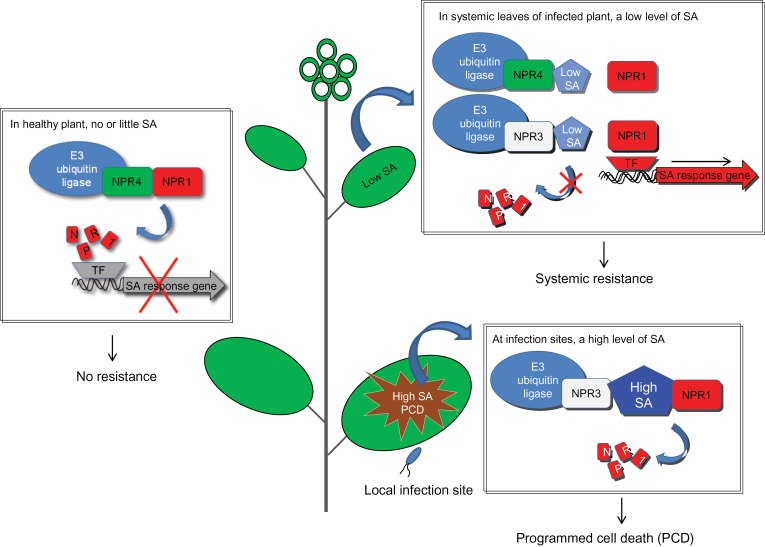
Because NPR3 and NPR4 promote degradation of NPR1, which is a positive regulator of SA-dependent defenses, an important question arises: What is the logic for these two proteins to function as SA receptors and then to remove NPR1? Fu and colleagues addressed this question. Their data show that SA has dynamic roles in pathogen-infected (local) and uninfected (systemic) regions of the same plant. In local tissue, infection by avirulent pathogens results in the accumulation of SA to a very high level and the development of defense-associated programmed cell death (PCD) whereas, in systemic tissue, SA accumulates to a lower level, which is sufficient to activate pathogen resistance without triggering PCD. In fact, PCD in pathogen-infected tissue is highly local, with clear borders. These authors found that NPR1–green fluorescent protein (GFP) fusion protein was markedly reduced inside the local tissue that undergoes PCD, and that the level was highest in plant cells surrounding the PCD lesion. Furthermore, Fu and colleagues found that the npr3 npr4 mutant, which accumulates NPR1 to a high level, failed to undergo PCD in local tissue upon infection with an avirulent bacterium, suggesting that NPR1 is an inhibitor of PCD and must be removed for PCD to proceed in local tissue. In systemic tissue, the npr3 npr4 double mutant has elevated basal resistance, presumably due to the elevated level of NPR1. However, the npr3 npr4 double mutant lost the capacity to further increase systemic resistance after local inoculation of avirulent bacterium, consistently with the conclusion that NPR3 and/or NPR4 are receptors that sense SA in systemic tissue.
Based on their findings, Fu and colleagues hypothesized that, in healthy plants with only a basal level of SA, NPR4, as part of the CUL3–NPR4 ubiquitin ligase, interacts with NPR1 to remove the NPR1 protein, thereby preventing the activation of energy-consuming defense responses (Figure 1). When plants are under pathogen attack, SA levels increase both in infected tissue and other parts of plants, with the highest concentration at the infection site. In infected tissue, a high concentration of SA promotes interaction of NPR3 with NPR1 to mediate degradation of NPR1, leading to PCD at the site of attack. In uninfected parts of the plant (e.g. uninfected leaves) in which SA accumulates at lower levels compared to the infection site, NPR1–NPR3 and NPR1–NPR4 interactions are both weakened, resulting in accumulation of NPR1, inhibition of PCD, expression of defense genes, and establishment of systemic acquired resistance.
Figure 1.
A Hypothetical Model for NPR3 and NPR4 Functioning as SA Sensors in Plants.
Identification of NPR3 and NPR4 as SA receptors has solved a major puzzle in plant science, and is expected to have a long-lasting impact on future research on SA signaling. However, this is likely an exciting beginning, not the end of SA receptor biology. Much is to be learned regarding the interplay of NPR3 and NPR4 receptors and other SABPs (including possibly NPR1; Wu et al., 2012) in living cells. It remains an open question whether BTB domain-containing NPR1, 3, and 4, as part of the E3 ubiquitin ligases, target negative regulators of SA-dependent defenses. Finally, crystal structure analyses of NPR3 and NPR4 receptors would be the next crucial step to further unravel the binding sites and exact SA-sensing mechanisms of these receptors.
Funding
This research was supported by the Howard Hughes Medical Institute (HHMI), the Gordon and Betty Moore Foundation (GBMF), the US National Institutes of Health, and the US Department of Energy (Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science; #DE-FG02-91ER20021). S.Y.H. is an HHMI-GBMF Investigator. No conflict of interest declared.
References
- Cao H.,, Bowling S.A.,, Gordon A.S.,, Dong X., (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 6, 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T.P.,, Friedrich L.,, Rylas J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl Acad. Sci. U S A. 92, 6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 486, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z.,, Fan W.,, Dong X., (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 113, 935–944 [DOI] [PubMed] [Google Scholar]
- Santner A.,, Estelle M., (2009). Recent advances and emerging trends in plant hormone signaling. Nature. 459, 1071–1078 [DOI] [PubMed] [Google Scholar]
- Tripathi D.,, Jiang Y.L.,, Kumar D., (2010). SABP2, a methyl salicylate esterase is required for the systemic acquired resistance induced by acibenzolar-S-methyl in plants. FEBS Lett. 584, 3458–3463 [DOI] [PubMed] [Google Scholar]
- Vlot A.C.,, Dempsey D.A.,, Klessig D.F., (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206 [DOI] [PubMed] [Google Scholar]
- Wu Y.,, Zhang D.,, Chu J.Y.,, Boyle P.,, Wang Y.,, Brindle I.D.,, De Luca V.,, Després C. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Reports. 1, 639–647 [DOI] [PubMed] [Google Scholar]