Abstract
Mapping the functionality of GTPases through small molecule inhibitors represents an underexplored area in large part due to the lack of suitable compounds. Here we report on the small chemical molecule 2-(benzoylcarbamothioylamino)-5,5-dimethyl-4,7-dihydrothieno[2,3-c]pyran-3-carboxylic acid (PubChem CID 1067700) as an inhibitor of nucleotide binding by Ras-related GTPases. The mechanism of action of this pan-GTPase inhibitor was characterized in the context of the Rab7 GTPase as there are no known inhibitors of Rab GTPases. Bead-based flow cytometry established that CID 1067700 has significant inhibitory potency on Rab7 nucleotide binding with nanomolar inhibitor (Ki) values and an inhibitory response of ≥97% for BODIPY-GTP and BODIPY-GDP binding. Other tested GTPases exhibited significantly lower responses. The compound behaves as a competitive inhibitor of Rab7 nucleotide binding based on both equilibrium binding and dissociation assays. Molecular docking analyses are compatible with CID 1067700 fitting into the nucleotide binding pocket of the GTP-conformer of Rab7. On the GDP-conformer, the molecule has greater solvent exposure and significantly less protein interaction relative to GDP, offering a molecular rationale for the experimental results. Structural features pertinent to CID 1067700 inhibitory activity have been identified through initial structure activity analyses and identified a molecular scaffold that may serve in the generation of more selective probes for Rab7 and other GTPases. Taken together, our study has identified the first competitive GTPase inhibitor and demonstrated the potential utility of the compound for dissecting the enzymology of the Rab7 GTPase as well as serving as a model for other small molecular weight GTPase inhibitors.
Keywords: Rab, Rho, Rac, Cdc42 and Ras GTPases; chemical biology; drug discovery; therapeutics; fluorescent GTP and GDP; enzyme kinetics
The Ras superfamily of GTPases, which includes Arf, Rho, Ras and Rab GTPase subfamilies, regulate a broad range of cellular processes ranging from membrane trafficking to the control of cell proliferation (1–3). Alteration of small GTPase functions is a hallmark of genetic and sporadic human diseases, making GTPase family members attractive though understudied targets (2, 4, 5). In a cellular context, GTPases are molecular regulators that switch between GDP (inactive) and GTP (active) bound conformational states and are temporally and spatially regulated (6). The conformational switch is key to the regulation of cell physiology and is therefore closely controlled by regulatory proteins. Among such regulators, guanosine exchange factors (GEFs) catalyze release of bound GDP to allow for GTP binding and activation in association with organelle membranes (7, 8). Inactivation is mediated by GTPase activating proteins (GAPs) that facilitate GTP-hydrolysis by the membrane localized small GTPase proteins and lead to cytosolic translocation of the now inactive GTPase (7, 8). To date, inhibitors of small GTPase function have focused on blocking membrane recruitment, through inhibition of protein prenylation or inhibition of regulatory protein interactions (2, 9, 10). Because of the broad cellular importance of lipid modifications for protein function, the former approach has not yielded the selectivity required for many applications, though the pursuit of more specific prenylation inhibitors remains under active investigation (11–14). The inhibition of regulatory protein interactions holds promise for being more specific, with the best-characterized example provided by Brefeldin-A mediated inhibition of Arf GEF activity (15). Rho-family GEF inhibitors have been identified through screening and rationale based design, leading to a handful of Rho-family selective inhibitors (16–19). Despite the therapeutic success and selectivity of competitive nucleotide binding inhibitors for targeting tyrosine kinases, no similar inhibitors have been identified for GTPases and there are no small molecules reported to date targeting the Rab-family of GTPases, in spite of their cellular importance in cancer and genetic diseases (2, 20).
Among the Rab GTPases, Rab7 occupies a critical nexus in the endocytic pathway where it governs cellular processes that are potential candidates for small molecule modulation. Such small molecules could have utility both for dissecting the mechanistic details of processes governed by Rab7, as well as for the future development of targeted therapies for diseases whose treatment may benefit from the modulation of specific GTPase activities (2, 4). Rab7 and its effectors are central to the regulation of growth factor signaling and receptor down regulation through lysosomal degradation or exosome release (21–26). Increased receptor downregulation may be pertinent to reducing hyperactive signaling in cancers where expression of epidermal growth factor receptor is elevated and/or constitutively active and where Rab7 is overexpressed (27, 28). Autophagy is central to cellular homeostasis, wherein Rab7 facilitates delivery of autophagosomes to lysosomes (29, 30). Imbalances in autophagy are associated with immune dysregulation, cancer, and neurodegeneration (31–33). Disease causing point mutants of Rab7 alter its nucleotide binding characteristics and lead to the peripheral neuropathy Charcot-Marie-Tooth (CMT) type 2B (34–36). CMT2B mutant Rab7 protein expression leads to altered neurite outgrowth and prolonged nerve growth factor receptor signaling from endosomes (23, 37). The highlighted examples demonstrate the importance of Rab7 in fundamental cellular processes and illustrate the potential for targeted manipulation of Rab7 function to manage human disease. For example, upregulation or enhancement of wild-type Rab7 activity could be beneficial in autosomal dominant neuropathy such as Charcot-Marie-Tooth disease, while downregulation or functional inhibition of Rab7 may be a benefit in some cancers. Thus, Rab7 presents a good candidate for exploring how small molecules can modulate its functional characteristics in vitro and in cell based systems.
Here we report on Rab7 inhibition by a sulfur based small molecule, 2-(benzoylcarbamothioylamino)-5,5-dimethyl-4,7-dihydrothieno[2,3-c]pyran-3-carboxylic acid. The molecule was identified through high-throughput screening (HTS) using a novel bead-based flow cytometry assay (38, 39). This is a simple, easy to implement experimental approach which is not susceptible to the underestimation of equilibrium dissociation (Kd) values seen with classical radioactive ligand binding assays, unless nucleotide free protein is used, which however tends to be unstable (40). We characterized the effects of the small molecule on Rab7 interaction with fluorescently linked nucleotides (BODIPY-GTP and BODIPY-GDP). We also present evidence on the structural activity relationships of a small cohort of analogs that identify the critical structural features responsible for the GTPase inhibitory activity of CID 1067700.
Results
Identification of CID 1067700 as an inhibitor of nucleotide binding by Ras-related GTPases
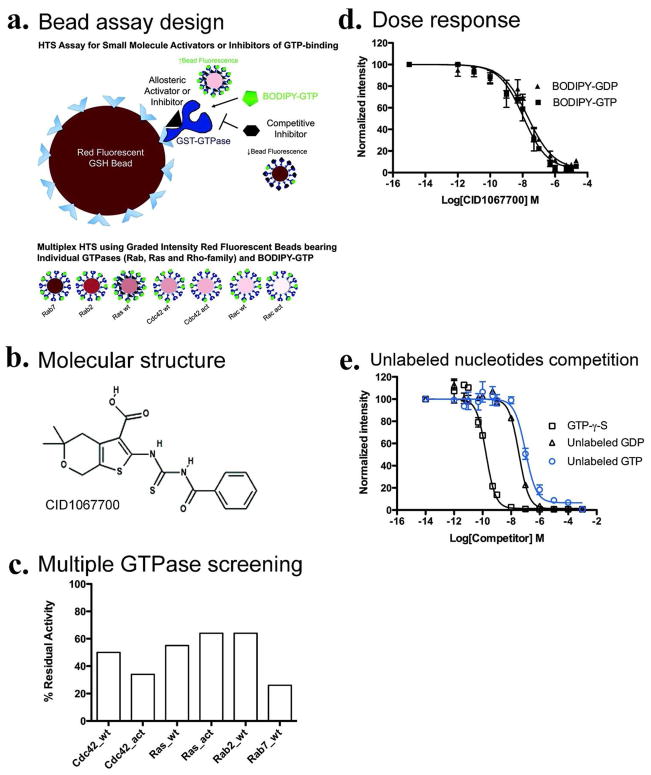
CID 1067700 was identified as an inhibitor of BODIPY-GTP binding using a high throughput screen that we described previously in the context of an allosteric Rho GTPase inhibitor (41). Briefly, six sets of beads (each with a unique red fluorescence intensity) were individually coated with six representative GST-tagged Ras-related GTPases (Cdc42 wt, Rac1wt, Rac1Q61L, Rab2 wt, Rab7wt, and H-Ras wt) (Fig. 1a). The individually conjugated beads were then assayed in the presence of individual compounds in the Molecular Libraries Small Molecule Repository using HyperCyt flow cytometry to identify molecules that altered the binding of fluorescent nucleotide to Ras-family GTPases (42). CID 1067700 (Fig. 1b) was identified as a hit that decreased fluorescent nucleotide (BODIPY-GTP) binding in the primary screen and was confirmed in secondary, multiplex dose response assays to have an EC50 of 20–500 nM and at least 40% inhibitory activity against all tested GTPases (Fig. 1c). CID 1067700 was the only compound identified in the screen of ~ 300, 000 small molecules to have significant inhibitory activity against the Rab GTPases (Rab2 and Rab7). Because of its pronounced inhibitory effect (>80%) on Rab7 and the absence of any known small molecule inhibitors for Rab-family GTPases, we further characterized the mechanism of CID 1067700 inhibition using Rab7 as a model protein.
Figure 1. Identification of CID 1067700 as a pan GTPase inhibitor using high-throughput screening (HTS) on small GTPases.
(a) Schematic diagram of the bead based assay used to measure BODIPY-linked guanosine triphosphate (GTP) binding by flow cytometry to glutathione-S-transferase (GST)–GTPase chimeras immobilized on GSH-beads. For HTS, beads of varying red fluorescence intensities were used as identifiers for individual protein-conjugated bead sets. The assay is sensitive to both increases and decreases in bead associated fluorescence and was used to identify both activators and inhibitors in a single screen. (b) Chemical structure of CID 1067700 an inhibitor of nucleotide binding with (c) Activity against multiple GTPases measured as residual nucleotide binding activity (BODIPY-GTP, 100 nM) in the presence of increasing concentrations of compound. (d) Nanomolar concentrations of CID 1067700 inhibit Rab7wt protein nucleotide binding; BODIPY-GTP (1 nM, filled squares) and BODIPY-GDP (0.4 nM, filled triangles). (e) Unlabeled nucleotides (GTP, GDP and GTP-γ-S) effectively competes BODIPY-GTP (100 nM) binding.
Dose dependent inhibition of Rab7 nucleotide binding by CID 1067700
Single-plex dose response measurements were used to determine the inhibitory response and potency of CID 1067700 on nucleotide binding by Rab7. The BODIPY-GTP and BODIPY-GDP concentrations in the assay were fixed at or below the previously determined equilibrium dissociation constants (Kd=100 nM for BODIPY-GTP; Kd=40 nM for BODIPY-GDP) for the wild-type (wt) Rab7 protein (38). Increasing CID 1067700 concentrations resulted in strong inhibition of binding of the BODIPY-linked nucleotides, providing respective EC50 values of 11.22 ± 1.34 nM for BODIPY-GTP and 20.96 ± 1.34 nM for BODIPY-GDP (Fig. 1d) and calculated Ki values of 12.89 nM and 19.70 nM respectively (Table 1 and 2). The maximal inhibitory response was ≥97% for both nucleotides. These results collectively demonstrate high efficacy and potency of CID 1067700 with respect to inhibiting Rab7 nucleotide binding. Maximum inhibition of BODIPY-linked nucleotide binding by CID 1067700 occurred at ~1 μM. The slight differences between the deduced EC50 values using BODIPY-GTP or BODIPY-GDP may be attributed to higher affinity of Rab7 for GDP than for GTP (38, 43). For comparative purposes, we also assayed the competition between BODIPY-linked nucleotides and unlabeled GTP, GDP and GTP-γ-S nucleotides (Fig. 1e). Consistent with our expectations, the unlabeled nucleotides effectively competed with BODIPY-GTP to ≈100 %. This confirmed that the fluorescently linked nucleotides indeed bind the nucleotide binding pocket of Rab7 and, together with our published work (38, 39), confirm the suitability of the assay for determining the mechanism of small molecule action.
Table 1.
| Nucleotide: | Rab7wt | Rab7Q67L | Rab7T22N | |
|---|---|---|---|---|
|
| ||||
| GTP* | GDP* | GTP* | GDP* | |
| a Bmax (BODIPY/bead): Control | 2110.00 ± 42.88 | 2651.00 ± 57.56 | 2660.00 ± 83.51 | 3275.00 ± 76.61 |
| Bmax (BODIPY/bead): Treated | 2250.00 ± 55.75 | 2779.00 ± 57.58 | 3372.00 ± 187.40 | 3515.00 ± 148.30 |
| b EC50 (nM): Control | 90.89 ± 7.19 | 51.57 ± 4.97 | 220.50 ± 22.30 | 106.70 ± 9.420 |
| EC50 (nM): Treated | 258.40 ± 19.79 | 140.4 ± 10.42 | 654.40 ± 86.32 | 294.50 ± 37.15 |
| c Ki (nM) | 12.89 (95 % confidence interval: 8.440 to 19.690) | |||
| c Ki (nM) | 19.70 (95 % confidence interval: 13.370 to 29.004) | |||
| Equilibrium binding off rates | ||||
| d BODIPY-GTP: | ||||
| CID1067700 competitor (K−1 sec−1): 2.21e-2 ± 1.30e-3 | ||||
| Unlabeled GDP competitor (K−1 sec−1): 3.27e-2 ± 1.70e-3 | ||||
| e BODIPY-GDP: | ||||
| CID1067700 competitor (K−1 sec−1): 1.02e-3 ± 3.28e-5 | ||||
| Unlabeled GDP competitor (K−1 sec−1): 1.60e-3 ± 2.92e-5 | ||||
Maximal number of bound BODIPY-nucleotide molecules per bead (Bmax) and
Cytometry detector output was converted to BODIPY-Nucleotide/bead using standard fluorescein beads from Invitrogen after correcting for the non-specific binding
Off rates were calculated using two phase exponential to fit BODIPY-GTP kinetic data obtained under equilibrium conditions.
Off rates were calculated using equation; to fit BODIPY-GDP kinetic data obtained under equilibrium conditions.
The samples labeled control, were treated with 1 % DMSO final while those labeled treated were subjected to CID1067700 treatment.
Table 2.

| ||||||
|---|---|---|---|---|---|---|
| PubChem SID | PubChem CID | R1 | R2 | R3 | L | Ki (μM) Rab7 |
| 57578339 | 1067700 | H | Me | Me |
|
0.013 |
| 99381128 | 46916263 | H | Me | H |

|
0.021 |
| 99381129 | 46916266 | H | H | H |

|
0.065 |
| 99381130 | 46916265 | H | Me | Me |

|
0.050 |
| 99381117 | 740871 | H | Me | Me |

|
Inactive |
| 99381118 | 1251121 | Me | Me | Me |

|
Inactive |
| 99381127 | 1280844 | H | Me | Me |
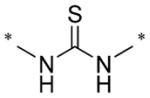
|
Inactive |
To test the mechanism of CID 1067700 inhibition, equilibrium nucleotide binding assays were used. We first tested the inhibition of Rab7wt by CID 1067700 under conditions where CID 1067700 concentration was held fixed while increasing the concentration of the fluorescent nucleotide competitor (Fig. 2a–d). The concentrations of CID 1067700 used were equivalent to at least 10 times the deduced EC50 for CID 1067700 that was measured against the two fluorescent nucleotides (Fig. 1d). The inhibitory effect of CID 1067700 was most pronounced at lower nucleotide concentrations, resulting in an overall rightward shift of the dose response curves (Fig. 2b, d). Significantly, both control (DMSO only) and inhibitor (CID 1067700-treated) curves had statistically similar Bmax values that correspond to the number of binding sites on Rab7 (Fig. 2a, c). Thus, at high concentrations, guanine nucleotides (GTP or GDP) out compete the CID 1067700 compound for the binding pocket, accounting for the statistically similar Bmax values. On the contrary, there was ~3-fold increase in the observed EC50 for both nucleotides in the presence of CID 1067700, indicating competition by the inhibitor for the nucleotide binding site. Statistically similar Bmax values ruled out inhibition by CID 1067700 via non-competitive mechanism. Inhibitor constants (Ki) calculated using the Cheng-Prusoff equation (44) showed that within statistically acceptable experimental limits, CID 1067700 is slightly better at inhibiting BODIPY-GTP binding relative to BODIPY-GDP binding (Table 1). The difference is however not significant given that the deduced Ki values overlap within the confidence interval of the measurements (Table 1). Analysis of the constitutively active Rab7Q67L and dominant negative Rab7T22N mutants, that mimic the GTP-bound and GDP-bound conformers of Rab7, respectively, showed that both were similarly inhibited by CID 1067700 (Fig. 2e–h). The composite data suggest that the CID 1067700 compound and the nucleotide most likely compete for the nucleotide binding pocket of Rab7.
Figure 2. CID 1067700 competitively inhibits BODIPY-linked nucleotide binding by wildtype and mutant forms of Rab7.
(a–b) CID 1067700 (100 nM) does not alter Rab7wt Bmax for BODIPY-GTP, but does alter apparent EC50 for BODIPY-GTP (filled squares); observed as a rightward shift of log plot of BODIPY-GTP binding by Rab7wt. (c–d) CID 1067700 (200 nM) does not alter Rab7wt Bmax for BODIPY-GDP; but does alter apparent EC50 for BODIPY-GDP (filled triangles); observed as rightward shift of log plot of BODIPY-GDP binding by Rab7wt. (e–f) CID 1067700 (100 nM) does not alter constitutively active Rab7Q67L mutant Bmax for BODIPY-GTP, but does alter apparent EC50 for BODIPY-GTP (filled squares); observed as a rightward shift of log plot of BODIPY-GTP binding by Rab7Q67L. (g–h) CID 1067700 (200 nM) does not alter Rab7T22N Bmax for BODIPY-GDP; but does alter apparent EC50 for BODIPY-GDP (filled triangles); observed as rightward shift of log plot of BODIPY-GDP binding by Rab7T22N. In all experiments, equilibrium binding reactions performed in 1% DMSO (final) served as the controls (open circles) and fluorescence attributed to non-specific nucleotide binding subtracted accordingly.
CID 1067700 equilibrates quickly and does not affect nucleotide release by Rab7
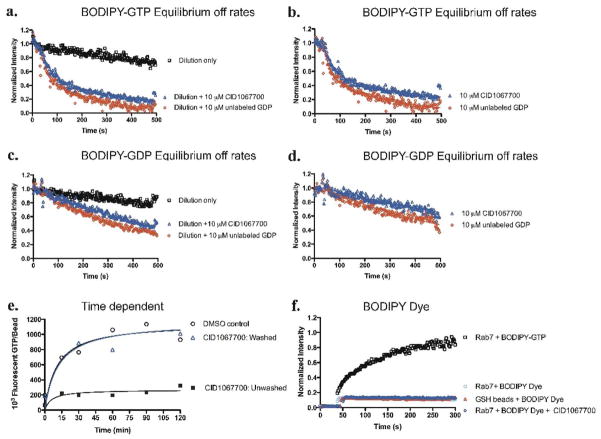
To further confirm the mode of Rab7 nucleotide inhibition by CID 1067700, we analyzed the fluorescent nucleotide dissociation rates from Rab7 in the presence of CID 1067700. We analyzed bound nucleotide dissociation by first pre-binding BODIPY-GTP or BODIPY–GDP to GST-tagged Rab7wt protein to equilibrium and then assaying for the loss of fluorescence over time occasioned by the release of bound nucleotide (Fig. 3a–d) at room temperature. As a competitor, we used CID 1067700 (10 μM) or unlabeled GDP (10 μM). Dissociation rate constants (Koff) were calculated from the data fit to exponential functions using Prism software. Koff values were statistically similar for BODIPY-GTP or BODIPY-GDP irrespective of whether unlabeled GDP or CID 1067700 was used as the competitor (Fig. 3a–d and Table 1). Comparatively, the observed slower off rate of BODIPY-GDP relative to that of BODIPY-GTP is consistent with the significant difference in Rab7 affinity for BODIPY-GDP relative to BODIPY-GTP. The time dependence of Rab7 inhibition of BODIPY-GTP binding by CID 1067700 demonstrated fast binding kinetics (maximal inhibition observed within 2 min) and there was no further time dependent increase in the inhibition of BODIPY-GTP binding (Fig. 3e). CID 1067700 could also be completely washed off following pre-incubation and restored BODIPY-GTP binding to DMSO control levels (Fig. 3e). The data indicate that covalent interaction between the acyl thiourea group of CID 1067700 and Rab7 was not a factor in its inhibitory properties. Independent chemical stability experiments performed with CID 1067700 and excess glutathione provided a third line of evidence that CID 1067700 remained unchanged and did not form covalent conjugates or other products when monitored by liquid chromatography mass spectrometry (LCMS) over 72 h. Together, these findings confirm that the CID 1067700 equilibrates rapidly with Rab7 and has no effect on the rate of release of bound BODIPY-GTP or BODIPY-GDP by Rab7, further substantiating the competitive binding of the CID 1067700 to the nucleotide pocket of Rab7.
Figure 3. CID 1067700 has no effect on the rate of release of bound BODIPY-linked nucleotide by wild type Rab7 under equilibrium binding conditions.
(a) Rab7 was pre-incubated with BODIPY-GTP (100 nM) for 2 h 15 min at 4°C, conditions that allow nucleotide binding to equilibrium. Dissociation assays were initiated by dilution +/− the addition of either CID 1067700 (10 μM) or unlabeled GDP (10 μM) and decrease in fluorescence due to nucleotide dissociation was measured in real time. (b) Two-phase exponential analysis of a, normalized by division of dilution only values (c). Rab7 was preincubated with BODIPY-GDP (40 nM) for 2 h 15 min at 4°C as for (a). Dissociation assays were initiated by dilution +/− CID 1067700 (10 μM) or unlabeled GDP (10 μM) and decrease in fluorescence due to nucleotide dissociation was measured in real time (d) Single phase exponential analysis of c, normalized by division of dilution only values (e) CID 1067700 exhibits fast binding kinetics and is reversible. Equal concentrations (140 nM) of Rab7 and CID 1067700 used against 100 nM BODIPY-GTP. CID 1067700 pre-incubated with Rab7 and either not washed (closed boxes) or washed (open blue triangles) and (f). Rab7 specifically binds guanine nucleotide moiety on BODIPY-GTP (100 nM, blue open boxes) without additional interaction with the BODIPY fluorescent moeity. 100 nM BODIPY dye was used in this control experiment and exhibited only background binding.
A potential caveat of using fluorescent nucleotide derivatives in our assays is the possibility that the small molecule might compete a putative interaction between Rab7 and the BODIPY moiety of the labeled nucleotide or might directly quench the fluorophore. Based on the ability of unlabeled nucleotides to compete 100% for BODIPY-GTP binding (Fig. 1e), we considered anomalies due to the BODIPY moiety binding to Rab7 unlikely. Nevertheless, we tested for both scenarios using unconjugated BODIPY dye. As expected, we did not observe any significant interaction between bead bound Rab7 and unconjugated BODIPY, nor was there any interaction between the BODIPY dye and ‘naked’ glutathione (GSH) beads (Fig. 3f). There was also no interaction between the CID 1067700 small molecule and BODIPY fluorescent moiety of the dye (Fig. 3f). The controls further confirmed that the inhibitory effect associated with CID 1067700 is a direct result of its interaction with Rab7 and ruled out possible indirect inhibitory effects of CID 1067700 via fluorescence quenching.
Molecular docking of CID 1067700 on Rab7wt GDP and GTP bound crystal structures shows optimal binding to the nucleotide binding pocket of the GTP-conformer
The molecular docking of CID 1067700 to the nucleotide binding site of Rab7 as predicted by the experimental data was examined using molecular docking OpenEye Fred docking software (Fig. 4a–h). Docking of CID 1067700 on the GDP- and GTP-conformers of Rab7 revealed that the molecule fills the nucleotide binding pocket of both conformers in a manner that mimics the normal nucleotide binding (Fig. 4a–d). However, the interaction map revealed (Figure 4e-h) that the compound had fewer potential interactions with the GDP-conformer where the binding pocket is more exposed to solvent (Fig. 4e–f), which may explain why CID 1067700 has a slightly lower potency against BODIPY-GDP bound Rab7 than BODIPY-GTP bound Rab7. The good alignment between CID 1067700 and GNP where the 4,7-dihydro-4H-thieno[2,3-c]pyran ring of CID 1067700 has the same orientation as the guanine ring is the basis for our model of the predicted binding of CID 1067700 to the nucleotide binding pocket of Rab7. To confirm the most likely binding mode, however, a crystal structure determination of CID 1067700 complexed to Rab7 will be required.
Figure 4. CID 1067700 docks optimally in the nucleotide binding pocket of Rab7 in the GTP-bound conformation.
CID 1067700 docked in the nucleotide binding site of Rab7 in the (a–b) GDP-bound (PDB 1VG9) and (c–d) (GNP)- bound (PDB 1VG8) conformations. Molecular docking carried out using Fred docking software. Both GTP or GDP and CID 1067700 are shown simultaneously docked in the pocket for purposes of comparing their orientations in the pocket. Interaction maps for (e–f) Rab7-GDP vs. CID 1067700 and (g–h) Rab7-GNP vs. CID 1067700 illustrate differences in number and sites of interaction.
Structure activity analyses identify critical determinants for competitive inhibition
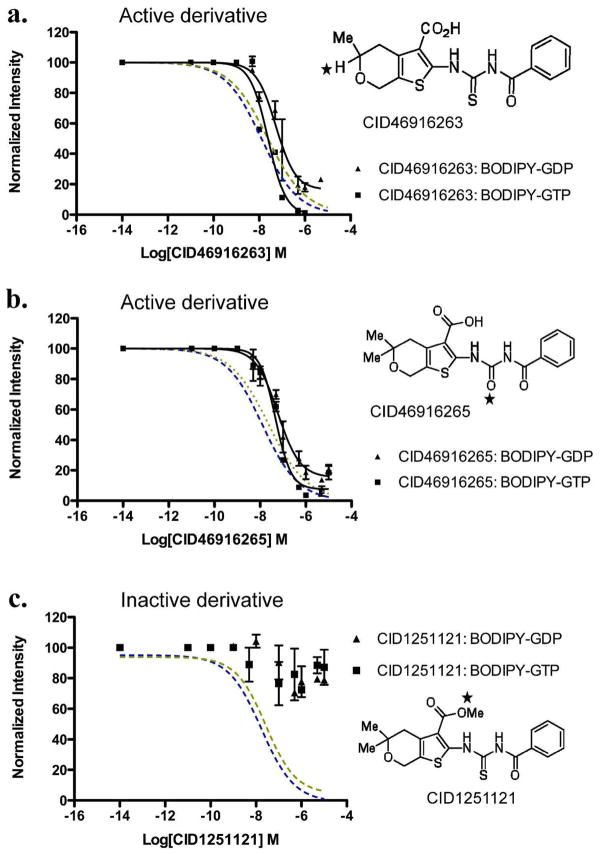
To determine structure activity relationships (SAR), thirty-nine variants of the parent compound were evaluated. This preliminary SAR set was obtained through synthetic effort (32 compounds) or commercial acquisition (7 compounds). The parent scaffold, represented by CID 1067700, was modified in any of the four major regions to afford the initial analog collection. The structural alterations focused on esterification of the carboxylic acid (R1), geminal substitution changes on the fused pyran ring (R2, R3), mutation of the ring-fused pyran to an N-methylated ring-fused piperidine, revision of the N-acyl thiourea linker (L), or alteration of the phenyl substituent by ortho-, meta-, or para-substitution or replacement with aliphatic or heterocylic moieties. Of the structural changes surveyed with R1-R3 and L, only three of the compounds retained activity in the intial multiplex GTPase screen with all other derivatives in that series being inactive (Table 2, select compounds shown). Two of the active derivatives and one inactive derivative were assayed in single-plex dose response assays and EC50 determined against Rab7 (Fig. 5a–c). Esterification of the carboxylic acid moiety resulted in loss of nucleotide binding inhibition (Fig. 5c and Table 2), a finding that supports the suggested participation of this group in hydrogen bonding (Fig. 4). Removal of one or both of the gem-dimethyl groups from the parental thiophenylpyran structure (R2–R3) resulted in a 10- or 20-fold loss in potency respectively when compared across multiple GTPases (Table 2 and data not shown). The effect on potency observed with the gem dimethyl substitution suggests an advantageous lipophilic ligand-binding site interaction that is lost upon substituent removal.
Figure 5. Structure activity relationships identify importance of linker and R-groups in inhibitory activity of CID 1067700.
Blue and red dotted lines represent inhibitory dose response curves of the parent CID 1067700 compound against BODIPY-GTP (1 nM) and BODIPY-GDP (0.4 nM), respectively. (a) Nanomolar concentrations of CID 46916263 derivative with only single methyl replacement on the pyran group and intact carbamothioylamino linker inhibits Rab7wt protein nucleotide binding; BODIPY-GTP (1 nM, filled squares) and BODIPY-GDP (0.40 nM, filled triangles). (b) Nanomolar concentrations of CID 46916265 derivative with only alteration of the thiourea moiety of the carbamothioylamino linker inhibits Rab7wt protein nucleotide binding; BODIPY-GTP (1nM, filled squares) and BODIPY-GDP (0.4 nM, filled triangles) and (c) Nanomolar concentrations of CID 1251121 derivative with alteration of the carboxylic acid group does not inhibit nucleotide binding by Rab7wt protein.
Alteration of the N-acyl thiourea linker further attenuated potency as demonstrated by the complete loss of activity for the amide CID 740871 (Table 2). While further SAR work is required to attribute specific linker functionality with preferred binding interactions, these initial results indicate that the orientation of tethered groups, and the length and hydrogen-bonding character of the linker region are critical to maintaining a beneficial activity profile. Generally, of the 2- or 3- or 4-substituted phenyl rings that were tested, 2-substituted phenyl rings were the least active (data not shown). Regardless of the electron distribution of the substituent installed on the ring, none was more beneficial than the parent structure, which possessed an unsubstituted phenyl ring. Replacement of the phenyl ring with other heterocycles such as 2-furyl or 2- thiophene gave analogs with activity profiles analogous to the parent hit. The 2-thiophene analog possessed a slightly better potency profile across all GTPases tested, and no changes in selectivity were noted. A non-aromatic cyclohexyl substituent was also surveyed without benefit.
Discussion
In the present study we identify CID 1067700 as a small molecule that inhibits nucleotide (BODIPY-GTP and BODIPY-GDP) binding by Rab7 with deduced inhibitor constant (Ki) value in the nanomolar range (12.89 to 19.70 nM) and a maximal efficacy of nucleotide binding inhibition of ≥97% for both BODIPY-linked nucleotides. Kinetic and equilibrium nucleotide binding studies demonstrate that the parent compound (CID 1067700) acts as a competitive inhibitor and as predicted by molecular docking, exhibits a good fit to the nucleotide binding pockets of both the GTP- and GDP-bound conformations of Rab7. By assaying initial on rate kinetics of Rab7 nucleotide binding in the presence of CID 1067700, we noted a decrease in the number of available BODIPY-GTP and BODIPY-GDP binding sites on Rab7 across the entire nucleotide concentration range indicating that CID 1067700 was competing with BODIPY-linked nucleotides for the binding sites on Rab7 (data not shown). Based on equilibrium nucleotide binding kinetics and dissociation measurements, we showed that CID 1067700 equilibrates quickly with Rab7 and does not affect bound nucleotide dissociation rates further supporting a competitive mechanism. Structure activity relationships demonstrate a dependence on the hydrophobic interactions with the substituted ring-fused pyran, the hydrogen bonding capability of the carboxylic acid, and the integrity of an extended N-acyl thiourea linker to properly extend and orient the tethered aryl functionality. Although our assays focused on characterizing the small molecule on Rab7, CID 1067700 also exhibits inhibitory activity on other small GTPases. To the best of our knowledge CID 1067700 is the first example of a competitive guanine nucleotide binding inhibitor characterized for any member of the Ras-super family of GTPases.
CID 1067700 was found to inhibit nucleotide binding by both the Rab7Q67L and Rab7T22N mutants that are known to be constitutively in the GTP and GDP bound states, respectively (45). Although the Q67L mutation lies in the nucleotide binding pocket of Rab7, the actual location of the Gln67 residue in the pocket is far removed from all predicted contacts with CID 1067700. From analysis of the crystal structure, amino acid residue Thr22 is involved in interactions with the Mg+2 cofactor which is required for guanine nucleotide binding. Although our small molecule inhibitor binds to the same pocket, molecular docking studies suggest that Mg+2 is not essential for its binding and therefore, it is not surprising that there was no impact of the Rab7T22N mutant on CID 1067700 inhibition. Thus, the compound could have the utility against relevant human disease mutants of Rab7 such as those responsible for Charcot-Marie-Tooth type 2B (CMT2B) disease.
The identification of CID 1067700 as a competitive inhibitor of Rab7 nucleotide binding presents an exciting and novel route to characterize the Rab family of proteins. Most small molecules reported to be active against low molecular GTPases are allosteric inhibitors and have activities that are restricted to the Rho-family GTPases based on inhibition of specific Rho-family GEF or effector protein interactions with Cdc42, Rac1 and RhoA. CID 10677000 is the first inhibitor with demonstrated activity against any member of the Rab subfamily of GTPases. Even though small GTPases share a common nucleotide binding fold, CID 1067700 had significantly less activity against other Ras-superfamily GTPases and even against another Rab family member (Rab2). Thus, the identification of CID 1067700, together with our analyses of structure activity relationships, suggest that it may be possible with further development to prepare more specific analogs that target individual subsets of GTPases analogous to nucleotide binding inhibitors of kinases (46).
The identification of small molecule inhibitors for GTPases has unique advantages when compared to traditional approaches such as conditional knock out or RNA interference that have been the primary means to perturb Rab GTPase functionality. In functional studies, small molecules can be applied to cells to rapidly and reversibly inhibit molecular targets of interest. This is exemplified by Brefeldin A, an allosteric inhibitor of Arf GTPases (15) and inhibitors of phosphoinositide 3-kinases that have been shown to inhibit specific kinase isoforms (46). Inhibitors of nucleotide binding may also have utility for inhibiting protein-protein interactions. Rab7 function in regulating endocytosis depends on numerous protein-protein interaction partners including hVps39 (25, 26), TBC1D15 (47), RILP (48, 49), ORPIL (50, 51), Rabring7 (52) and the alpha proteasome subunit XAPC7 (26, 53). Susceptibility of these Rab7-protein interactions may be useful for designing new disease interventions. Use of CID 1067700 may also be useful for dissecting the order of Rab7 binding to a multimeric complex and/or the importance of nucleotide bound status for stability of the protein complex involving Rab7. Taken together, our findings present CID 1067700 as a novel competitive inhibitor of small GTPase nucleotide binding that has potential for dissecting protein function. It may serve as a springboard for further development of families of compounds selective for individual GTPases.
Methods
Reagents
Reagents used in this study were obtained from Sigma (St. Louis, MO, USA) unless otherwise indicated. Sephadex G-25, glutathione (GSH) Sepharose 4B, and Superdex peptide beads (13 μm with an exclusion limit of 7 kDa) were from Amersham Biosciences. BODIPY (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene or dipyrromethene boron difluoride) nucleotide analogues (BODIPY-GTP; 2′-(or 3′)-O-[N-(2-aminoethyl)urethane] G-35778 and BODIPY-GDP; 2′-(or-3′)-O-[N-(2-aminoethyl)urethane], G-22360) and BODIPY dye were from Invitrogen Molecular Probes (Carlsbad, CA, USA). Concentrations of BODIPY-linked nucleotides were based on absorbance measurements and an extinction coefficient value of 80,000 M−1cm−1. 2-(benzoylcarbamothioylamino)-5,5-dimethyl-4,7-dihydrothieno[2,3-c]pyran-3-carboxylic acid with compound identification number; CID 1067700 was from ChemDiv Inc (San Diego, CA, USA).
Expression and purification of GST-Rab7
Recombinant GST–Rab7 was expressed in E. coli BL21(DE3) cells. Cultures were grown at 37 °C to a bacterial density of 0.5–0.7 absorbance units measured at 595 nm and protein expression induced by transfer to room temperature and addition of 0.2 mM isopropyl-beta-D-1-thiogalactopyranoside (IPTG) for 16–18 h to maximize yield of properly folded active fusion protein. Purification of GST-Rab7 was performed according to standard procedures as previously described (38).
Synthesis of glutathione (GSH) beads for flow cytometry assay
High GSH density beads used for flow cytometry were synthesized by loading Superdex peptide beads with GSH as previously reported (38, 39).
Immobilization of Rab7 on GSH beads for flow cytometry
All nucleotide binding measurements were performed in the Hepes buffer (30 mM Hepes, pH 7.5, 20 mM NaCl and 100 mM KCl) supplemented with 1 mM EDTA, 1 mM dithiothreitol (DTT) and 0.1 % bovine serum albumin (BSA). A BectonDickinson FACScan flow cytometer with a 488-nm excitation Argon (Ar) laser and standard detection optics was used for all assays (38). Purified GST-Rab7 protein was incubated in 96-well plates at 4 °C overnight with 105 GSH beads in a total volume of 100 μL of Hepes buffer supplemented with 1 mM EDTA and 1 mM dithiothreitol (DTT) added fresh. Unbound protein was removed by centrifugation twice at 800 g followed by resuspension in fresh buffer.
Dose response or competition binding assays
Dose response assays involving CID 1067700 were conducted in two different modes. In the first case, 2×103 GSH beads loaded with GST-Rab7 were incubated with a fixed concentration of nucleotide (BODIPY-GDP or BODIPY-GTP) for 1h in the presence of 1% DMSO (final) or increasing concentrations of CID 1067700 (10−6–20) μM in 1% DMSO and a total volume of 20 μL on a 96-well plate. CID 1067700 was added from a stock solution of CID 1067700 (10 mM) prepared in 100% DMSO and stored in single use aliquots at −20 °C. In the second mode, 2×103 GSH beads loaded with GST-Rab7 protein were incubated for 2 h at 4 °C with varying concentrations of BODIPY-linked guanine nucleotide (0–2) μM in a total volume of 20 μL on a 96-well plate in the presence of either 1 % DMSO or a fixed CID 1067700 concentration (0.1–0.2) μM. For measurements on the flow cytometer, samples were transferred to a tube suitable (Product No. 352008, BD Bioscience, San Jose, California, U.S.A.) for flow cytometry and were diluted at least 10-fold in Hepes buffer as before. This dilution step was necessary to ensure discrimination between bead-associated fluorescence and background fluorescence of soluble proteins as well as ensured sufficient sample volume for the measurement. Similarly, competition assays involving regular unlabeled nucleotides (GTP, GDP and GTP-γ-S) were conducted by incubating GST-Rab7 with a fixed concentration of BODIPY-GTP (100 nM) against increasing concentrations (10−6–1000) μM of unlabeled nucleotide in a total volume of 20 μL on a 96-well plate for 2 hr. Final fluorescence measurements were then obtained as before.
Nucleotide binding and dissociation kinetics assays
A two-pronged approach was employed to assay how CID 1067700 affects kinetics of nucleotide binding by Rab7. Nucleotide dissociation under equilibrium conditions was accomplished by first loading GST-Rab7 with fixed concentrations of BODIPY-GTP (100 nM) or BODIPY-GDP (40 nM) for 2hr at 4 °C followed by initiating release of bound nucleotide with either unlabeled GDP (10 μM) or CID 1067700 (10 μM). As a negative control, GST-Rab7 was preloaded with unlabelled GDP (1 mM) for 2hr 15 min at 4 °C prior to adding BODIPY-GTP or BODIPY–GDP at the above concentrations for a further 2hr 15 min and then similarly initiating release of bound BODIPY-linked nucleotide with unlabeled GDP (10 μM) to obtain fluorescence for non-specific binding under equilibrium binding conditions. This low level bead associated fluorescence was subtracted from specific binding to obtain accurate measurement of the off rates. Data normalization was performed by dividing values from competitor treated experimental set by those obtained from dilution only set. Time dependent nucleotide binding by Rab7 in the presence of CID 1067700 was carried out using BODIPY-GTP in the presence of either 1 % DMSO (final) or 1 μM CID 1067700. Either was added to glutathione (GSH) bead mobilized GST-Rab7 and the fluorescence of bound BODIPY-GTP monitored as a function of time up to the equilibrium point of 2 h.
Molecular docking of CID 1067700 on Rab7
Docking calculations were performed using OpenEye Fred (Fred, version 2.2.5, OpenEye Scientific Software, Inc., Santa Fe, NM, USA, www.eyesopen.com, 2010) docking software. Rab7 crystal structures (PDB code: 1T91 GDP-conformer, 1VG8 GNP-conformer) were used to dock CID 1067700 on Rab7 for the wild type. Chemgauss3 scoring function was used to evaluate ligand binding potential. Docking simulations provide only a qualitative assessment of binding probability of our ligand to Rab7 and should be examined with care however the results indicate that CID 1067700 can potentially bind to Rab7 nucleotide binding site and no steric constraints were observed.
Data analyses
All data processing and analyses employed GraphPad Prism software (GraphPad Software). For kinetic experiments, raw data acquired were first processed using IDLE query software (obtained from University of New Mexico Center for Molecular Discovery, UNM CMD) before further analysis using GraphPad Prism. All experiments are representative of at least three independent trials.
Synthesis of CID 1067700 derivatives for structure activity analyses
General experimental and analytical details
1H and 13C NMR spectra were recorded on a Bruker AM 400 spectrometers (operating at 400 and 101 MHz) in CDCl3 with 0.03% tetramethylsilane (TMS) or DMSO-d6 as an internal standard. The chemical shifts (δ) reported are given in parts per million (ppm) and the coupling constants (J) are in Hertz (Hz). The spin multiplicities are reported as s = singlet, br. s = broad singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublet and m = multiplet. The LCMS analysis was performed on an Agilent 1200 RRL chromatograph with photodiode array UV detection and an Agilent 6224 TOF mass spectrometer. The chromatographic method utilized the following parameters: a Waters Acquity BEH C-18 2.1 × 50 mm, 1.7 μm column; UV detection wavelength = 214 nm; flow rate = 0.4 ml/min; gradient = 5–100% acetonitrile over 3 minutes with a hold of 0.8 minutes at 100% acetonitrile; the aqueous mobile phase contained 0.15% ammonium hydroxide (v/v). The mass spectrometer utilized the following parameters: an Agilent multimode source which simultaneously acquires ESI+/APCI+; a reference mass solution consisting of purine and hexakis (1H, 1H, 3H-tetrafluoropropoxy) phosphazine; and a solvent consisting of 90:10:0.1 MeOH:water:formic acid which was introduced to the LC flow prior to the source to assist ionization. Melting points were determined on a Stanford Research Systems OptiMelt apparatus. Synthesized analogs were prepared by the general route depicted in Figure 6. Stepwise procedures for all synthesized analogs and intermediates are described. Analytical characterization is provided for all tested commercial and locally synthesized compounds itemized in this manuscript.
Figure 6. General scheme used for synthesis of CID 1067700 analogs.
Reagents and conditions: a) morpholine sulfur, EtOH, 50°C; b) PhCONCS or PhCONCO, THF, 50°C; c) TFA, CH2Cl2, rt.

tert-Butyl 2-amino-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate
Following a standard procedure, tetrahydro-4H-pyran-4-one (0.28 mL, 3.0 mmol, 1 equiv.), tert-butyl cyanoacetate (0.47 mL, 3.3 mmol, 1.1 equiv.), sulfur (0.106 g, 3.30 mmol, 1.1 equiv.), and morpholine (0.39 mL, 4.5 mmol, 1.5 equiv.) were dissolved in EtOH (9 mL) and stirred at 50 °C1 for 16 h. The solvent was removed and water (7 mL) was added. The product was extracted with EtOAc (2 × 7 mL) and purified by preparatory HPLC (0–20% EtOAC:hexanes). The product was isolated as a clear, pale yellow oil (0.77 g, quantitative yield). 1H NMR (400 MHz, CDCl3) δ 5.98 (s, 2H), 4.55 (t, J = 2.0 Hz, 2H), 3.90 (t, J = 5.6 Hz, 2H), 2.79 (ddd, J = 7.6, 3.8, 2.0 Hz, 2H), 1.54 (s, 9H). 13C NMR (101 MHz, CDCl3) δ 165.38, 161.65, 130.33, 114.60, 106.76, 80.37, 65.20, 64.66, 28.57, 27.94.

tert-Butyl 2-(3-benzoylthioureido)-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate
tert-Butyl 2-amino-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate (0.099 g, 0.39 mmol, 1 equiv.) and PhCONCS (0.06 mL, 0.4 mmol, 1 equiv.) were dissolved in THF (2 mL) and stirred at 50 °C for 16 h. The solvent was removed and the product was purified by preparatory HPLC (0–30% EtOAC:hexanes). The product was isolated as a white solid (0.131 g, 80% yield). 1H NMR (400 MHz, CDCl3) δ 14.75 (s, 1H), 9.21 (s, 1H), 7.92 (dd, J = 5.2, 3.3 Hz, 2H), 7.64–7.57 (m, 1H), 7.54–7.46 (m, 2H), 4.71 (s, 2H), 3.94 (t, J = 5.6 Hz, 2H), 2.90 (t, J = 5.6 Hz, 2H), 1.63 (s, 9H). 13C NMR (101 MHz, CDCl3) δ 173.72, 165.23, 164.54, 147.15, 133.48, 131.81, 129.36, 129.04, 127.77, 125.77, 117.76, 82.36, 65.20, 64.71, 28.51, 27.38.

2-(3-benzoylthioureido)-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylic acid (SID 99381129, CID 46916266)
To a solution of tert-Butyl 2-(3-benzoylthioureido)-5,7-dihydro-4H-thieno[ 2,3-c]pyran-3-carboxylate (0.104 g, 0.25 mmol, 1 equiv.) in DCM (10 mL) was added TFA (2 mL, 26 mmol, 100 equiv.), and the reaction was stirred at rt for 16 h. The solvent was removed and the product was triturated with 1:1 Et2O/hexanes (25 mL) and filtered. The product was purified by mass-directed reverse-phase chromatography and isolated as a white solid (3 mg, 3% yield). 1H NMR (400 MHz, CDCl3) δ 14.92 (br s, 1H), 9.02–9.09 (m, 1H), 7.83–7.91 (m, 1H), 7.55–7.66 (m, 2H), 7.41–7.50 (m, 2H), 7.19–7.23 (m, 1H), 4.69–4.76 (m, 2H), 3.92–3.98 (m, 2H), 2.88–3.03 (m, 2H). LCMS retention time: 2.955 min; LCMS purity at 215 nm = 100%. HRMS m/z calculated for C16H15N2O4S2 [M++1]: 363.0468, found 363.0464.

tert-Butyl 2-amino-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate
A mixture of 2,2-dimethyldihydro-2H-pyran-4(3H)-one (0.300 g, 2.34 mmol, 1 eq), tert-butyl cyanoacetate (0.37 mL, 2.57 mmol, 1.1 eq), sulfur (0.083 g, 2.57 mmol, 1.1 eq), morpholine (0.30 mL, 3.51 mmol, 1.5 eq), and ethanol (7 mL) was heated at 50 °C for 16 h. The reaction mixture was then filtered, and the filter cake washed with ethyl acetate (20 mL). Purification by silica gel chromatography (0–20% EtOAc:Hex ramp over 20 min) afforded the desired product tert-butyl 2-amino-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate as a yellow solid (0.631 g, 2.23 mmol, 95 % yield). 1H NMR (400 MHz, CDCl3) δ 5.88 (s, 2H), 4.52 (t, J = 1.9, 2H), 2.64 (t, J = 1.9, 2H), 1.53 (s, 9H), 1.26 (s, 6H).

tert-Butyl 2-(3-benzoylureido)-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate
tert-Butyl 2-amino-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate (0.116 g, 0.41 mmol, 1 equiv.) and PhCONCO (0.06 mL, 0.5 mmol, 1.2 equiv.) were dissolved in THF (2 mL) and stirred at 50 °C for 16 h. The solvent was removed and the product was purified by preparatory HPLC (0–40% EtOAC:hexanes). The product was isolated as a white solid (0.105 g, 59% yield). 1H NMR (400 MHz, CDCl3) δ 13.26 (s, 1H), 9.81 (s, 1H), 8.13 (d, J = 7.0 Hz, 2H), 7.65 (d, J = 6.7 Hz, 3H), 4.72 (s, 2H), 2.75 (s, 2H), 1.64 (s, 9H), 1.32 (s, 6H).

2-(3-Benzoylureido)-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylic acid (SID 99381130, CID 46916265)
To a solution of tert-butyl 2-(3-benzoylureido)-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate (0.105 g, 0.24 mmol, 1 equiv.) in DCM (10 mL) was added TFA (0.5 mL, 6.5 mmol, 27 equiv.), and the reaction was stirred at rt for 5 days. The solvent was removed under reduced pressure and the product was purified by preparatory HPLC (0–5% MeOH/DCM), followed by mass-directed reverse-phase chromatography and isolated as a white solid (6 mg, 7% yield). 1H NMR (400 MHz, DMSO) δ 13.11 (s, 1H), 11.39 (s, 1H), 8.05–7.99 (m, 2H), 7.69–7.63 (m, 1H), 7.55 (t, J = 7.7 Hz, 2H), 4.62 (s, 2H), 2.71 (s, 2H), 1.22 (s, 6H). LCMS retention time: 2.872 min; LCMS purity at 215 nm = 100%. HRMS m/z calculated for C18H19N2O5S [M++1]: 375.1009, found 375.1013.
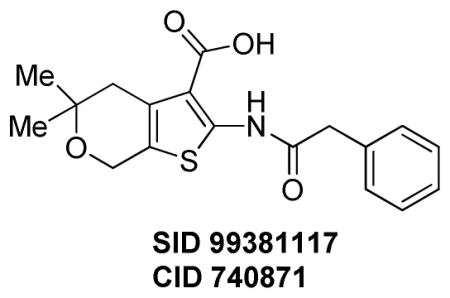
5,5-Dimethyl-2-(2-phenylacetamido)-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylic acid (SID 99381117, CID 740871)
5,5-Dimethyl-2-(2-phenylacetamido)-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylic acid was purchased from ChemBridge (CAS 303966-15-6) and purified by mass-directed reverse-phase chromatography and isolated as a white solid (3 mg, 100% purity). 1H NMR (400 MHz, CDCl3) δ 10.88 (s, 1H), 7.43–7.37 (m, 2H), 7.33 (dd, J = 6.7, 4.3 Hz, 3H), 4.72 (s, 2H), 3.85 (s, 2H), 2.85 (s, 2H), 1.35 (s, 6H). LCMS retention time: 3.045 min; LCMS purity at 215 nm = 100%. HRMS m/z calculated for C18H20NO4S [M++1]: 346.1108, found 346.1110.

Methyl 2-(3-benzoylthioureido)-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate (SID 99381118, CID 1251121)
To a combined mixture of methyl 2-amino-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate (ChemBridge, CAS 713111-73-0, 31 mg, 0.13 mmol, 1 equiv.) and PhCONCS (0.02 mL, 0.15 mmol, 1.2 equiv.) was added THF (0.7 mL). The resulting solution was stirred at 50 °C for 3 days. The solvent was then removed under reduced pressure and the crude product was triturated with hexanes, filtered, and rinsed with hexanes (4 × 5 mL). The product was further purified by preparatory HPLC (0–50% EtOAC:hexanes) and isolated as a white solid (38 mg, 72% yield). 1H NMR (400 MHz, CDCl3) δ 14.83 (s, 1H), 9.14 (s, 1H), 7.98 (dd, J = 8.4, 1.2 Hz, 2H), 7.72–7.63 (m, 1H), 7.56 (dd, J = 10.5, 4.8 Hz, 2H), 4.77 (t, J = 1.5 Hz, 2H), 4.04 (s, 3H), 2.85 (s, 2H), 1.35 (s, 6H). LCMS retention time: 3.508 min; LCMS purity at 215 nm = 96%. HRMS m/z calculated for C19H21N2O4S2 [M++1]: 405.0937, found 405.0942.

2-(3-Benzoylthioureido)-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylic acid (SID 85747738/CID 1067700)
2-(3-benzoylthioureido)-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylic acid was purchased from Chemdiv Inc. (CAS 314042-01-8) and purified by mass-directed reverse-phase chromatography to yield a white solid (6 mg). 1H NMR (400 MHz, DMSO) δ 14.84 (s, 1H), 13.39 (s, 1H), 11.81 (s, 1H), 7.98 (dd, J = 1.2, 8.4, 2H), 7.74–7.62 (m, 1H), 7.55 (t, J = 7.7, 2H), 4.67 (s, 2H), 2.75 (s, 2H), 1.24 (s, 6H). 13C NMR (101 MHz, DMSO) δ 174.43, 166.86, 165.35, 146.72, 133.08, 132.06, 129.10, 128.75, 128.41, 124.09, 116.50, 70.14, 58.82, 37.27, 26.19. LCMS retention time: 1.871 min; LCMS purity at 214 nm = 92.8%. HRMS m/z calculated for C18H19N2O4S2 [M++1]: 391.0781, found 391.0777.

2-(3-Benzoylthioureido)-5-methyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylic acid (SID 9931128/CID 46916263)
To tert-butyl 2-(3-benzoylthioureido)-5-methyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate (0.028 g, 0.065 mmol, 1 eq.) was added a 40% v/v solution of trifluoroacetic acid in dichloromethane (2.5 mL). After stirring the solution for 4 hours at rt, the solvent was evaporated in vacuo, and the residue purified by reverse-phase chromatography to yield the product as a white semi-solid (0.011 g, 0.029 mmol, 46% yield). 1H NMR (400 MHz, Acetone) δ 8.07 (d, J = 7.4, 2H), 7.68 (t, J = 7.0, 1H), 7.56 (t, J = 7.5, 2H), 4.76 (q, J = 14.7, 2H), 3.82–3.68 (m, 1H), 3.05 (d, 1H), 1.47 (d, J = 6.5, 1H), 1.32 (d, J = 6.1, 3H). 13C NMR (126 MHz, Acetone) δ 175.37, 166.92, 165.37, 161.89, 148.65, 134.12, 133.28, 131.39, 129.62, 129.28, 126.49, 71.61, 65.11, 35.05, 21.82. LCMS retention time: 3.095 min; Purity at 214 nm = 93.3%. HRMS m/z calculated for C17H17N2O4S2 [M+ + 1] 377.0624, found 377.0622.
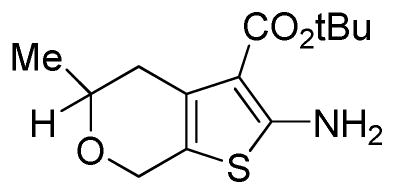
tert-Butyl 2-amino-5-methyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate
A mixture of 2-methyldihydro-2H-pyran-4(3H)-one (0.320 g, 2.80 mmol, 1 eq), tert-butyl cyanoacetate (0.44 mL, 3.08 mmol, 1.1 eq), sulfur (0.099 g, 3.08 mmol, 1.1 eq), morpholine (0.36 mL, 4.21 mmol, 1.5 eq), and ethanol (7 mL) was heated at 50 °C for 16 hours. The reaction mixture was then filtered, and the filter cake washed with ethyl acetate (20 mL). Purification by silica gel chromatography (0–30% EtOAc:Hex ramp over 20 min) afforded the desired product tert-butyl 2-amino-5-methyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate as a pale yellow solid (0.745 g, 2.77 mmol, 99 % yield). 1H NMR (400 MHz, CDCl3) δ 5.90 (s, 2H), 4.71–4.49 (m, 2H), 3.82–3.65 (m, 1H), 2.86–2.73 (m, 1H), 2.52–2.37 (m, 1H), 1.52 (d, J = 1.4, 9H), 1.31 (dd, J = 2.1, 6.2, 3H).

tert-Butyl 2-(3-benzoylthioureido)-5-methyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate
To a solution of tert-butyl 2-amino-5-methyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate (0.104 g, 0.386 mmol, 1 eq) in dry ethanol (1 mL) under argon was added benzoyl isothiocyanate (0.15 mL, 1.08 mmol, 2.8 eq). The mixture was heated at reflux for 16 hours. The solvent was evaporated in vacuo. Purification by reverse-phase chromatography (10–100% CH3CN: Water ramp over 20 min) afforded the desired product tert-butyl 2-(3-benzoylthioureido)-5-methyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate as a pale yellow oil (0.057 g, 0.132 mmol, 34 % yield). 1H NMR (400 MHz, DMSO) δ 14.72 (s, 1H), 11.89 (s, 1H), 8.06–7.95 (m, 2H), 7.69 (t, J = 7.4, 1H), 7.57 (t, J = 7.7, 2H), 4.78 (d, J = 14.8, 1H), 4.66 (d, J = 15.0, 1H), 3.80–3.63 (m, 1H), 2.93 (d, J = 16.7, 1H), 2.50–2.44 (m, 1H), 1.59 (s, 9H), 1.30 (d, J = 6.1, 3H).

tert-Butyl 5,5-dimethyl-2-(3-phenylthioureido)-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate
To a solution of tert-butyl 2-amino-5,5-dimethyl-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate (0.100 g, 0.353 mmol, 1 eq) in dry tetrahyrofuran (5 mL) under argon was added phenyl isothiocyanate (0.05 mL, 0.388 mmol, 1.1 eq). The mixture was heated at reflux for 16 hours. The solvent was evaporated in vacuo. Purification by reverse-phase chromatography (0–100% CH3CN: Water ramp over 20 min) afforded the desired product tertbutyl 5,5-dimethyl-2-(3-phenylthioureido)-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylate as a yellow solid (0.045 g, 0.108 mmol, 30 % yield). 1H NMR (400 MHz, DMSO) δ 11.92 (s, 1H), 11.05 (s, 1H), 7.50 (d, J = 7.6, 2H), 7.42 (t, J = 7.9, 2H), 7.26 (t, J = 7.3, 1H), 4.61 (s, 2H), 2.65 (s, 2H), 1.53 (s, 9H), 1.23 (s, 6H).
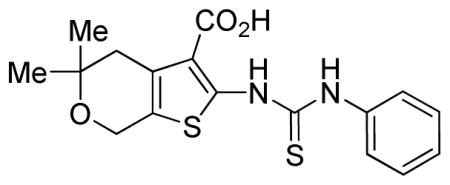
5,5-Dimethyl-2-(3-phenylthioureido)-5,7-dihydro-4H-thieno[2,3-c]pyran-3-carboxylic acid (SID 99381127/CID 1280844)
To 5,5-dimethyl-2-(3-phenylthioureido)-5,7-dihydro-4H-thieno[ 2,3-c]pyran-3-carboxylic acid (0.037 g, 0.088 mmol, 1 eq.) was added a 40% v/v solution of trifluoroacetic acid in dichloromethane (2.5 mL). After stirring the solution for 4 hours at rt, the solvent was evaporated in vacuo and the residue purified by reverse-phase LCMS to yield the product as a white solid (0.020 g, 0.065 mmol, 63% yield). 1H NMR (400 MHz, Acetone) δ 7.97 (d, J = 7.7, 2H), 7.88 (t, J = 7.7, 2H), 7.73 (t, J = 6.9, 1H), 4.32 (s, 2H), 3.11 (s, 2H), 1.73 (s, 6H). LCMS retention time: 3.113 min; Purity at 214 nm = 100%. HRMS m/z calculated for C17H19N2O3S2 [M+ + 1] 363.0832, found 363.0829.
Acknowledgments
This work is generously supported by the National Science Foundation (MCB0956027), National Institutes of Health (R03MH081231 and R21NS066435) to AWN, and P30CA1181000, U54MH074425 and U54MH084690 to LAS and TO. The University of Kansas Specialized Chemistry Center gratefully acknowledges the National Institutes of Health, MLPCN, for financial support (U54 HG005031). We appreciate critical reading of the manuscript by Dr. Roger Goody (MPI, Dortmund, Germany). We thank J. Kelly and L. Thal for administrative support. We also thank E. Romero and P. Simons for technical support. Small molecule screening was performed in the NMMLSC/UNMCMD and follow up flow cytometry assays were conducted in the Flow Cytometry Shared Resource Center supported by the University of New Mexico Health Sciences Center and the University of New Mexico Cancer Center.
References
- 1.East MP, Kahn RA. Models for the functions of Arf GAPs. Semin Cell Dev Biol. 2010;22:3–9. doi: 10.1016/j.semcdb.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agola J, Jim P, Ward H, Basuray S, Wandinger-Ness A. Rab GTPases as regulators of endocytosis, targets of disease and therapeutic opportunities. Clin Genet. 2011;80:305–318. doi: 10.1111/j.1399-0004.2011.01724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 4.Mitra S, Cheng KW, Mills GB. Rab GTPases implicated in inherited and acquired disorders. Semin Cell Dev Biol. 2011;22:57–68. doi: 10.1016/j.semcdb.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal. 2010;8:23. doi: 10.1186/1478-811X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant BJ, Gorfe AA, McCammon JA. Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLoS Comput Biol. 2009;5:e1000325. doi: 10.1371/journal.pcbi.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebti SM, Hamilton AD. Farnesyltransferase and geranylgeranyltransferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 10.Sousa SF, Fernandes PA, Ramos MJ. Farnesyltransferase inhibitors: a detailed chemical view on an elusive biological problem. Curr Med Chem. 2008;15:1478–1492. doi: 10.2174/092986708784638825. [DOI] [PubMed] [Google Scholar]
- 11.Sane KM, Mynderse M, Lalonde DT, Dean IS, Wojtkowiak JW, Fouad F, Borch RF, Reiners JJJ, Gibbs RA, Mattingly RR. A novel geranylgeranyl transferase inhibitor in combination with lovastatin inhibits proliferation and induces autophagy in STS-26T MPNST cells. J Pharmacol Exp Ther. 2010;333:23–33. doi: 10.1124/jpet.109.160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machida S, Kato N, Harada K, Ohkanda J. Bivalent Inhibitors for Disrupting Protein Surface-Substrate Interactions and for Dual Inhibition of Protein Prenyltransferases. J Am Chem Soc. 2011;133:958–963. doi: 10.1021/ja1086112. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher S, Keaney EP, Cummings CG, Blaskovich MA, Hast MA, Glenn MP, Chang SY, Bucher CJ, Floyd RJ, Katt WP, Gelb MH, Van Voorhis WC, Beese LS, Sebti SM, Hamilton AD. Structure-based design and synthesis of potent, ethylenediamine-based, mammalian farnesyltransferase inhibitors as anticancer agents. J Med Chem. 2010;53:6867–6888. doi: 10.1021/jm1001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna CE, Kashemirov BA, Blazewska KM, Mallard-Favier I, Stewart CA, Rojas J, Lundy MW, Ebetino FH, Baron RA, Dunford JE, Kirsten ML, Seabra MC, Bala JL, Marma MS, Rogers MJ, Coxon FP. Synthesis, chiral high performance liquid chromatographic resolution and enantio-specific activity of a potent new geranylgeranyl transferase inhibitor, 2-hydroxy-3-imidazo[1,2-a]pyridin-3-yl-2-phosphonopropionic acid. J Med Chem. 2010;53:3454–3464. doi: 10.1021/jm900232u. [DOI] [PubMed] [Google Scholar]
- 15.Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner RD. Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67:601–616. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- 16.Nassar N, Cancelas J, Zheng J, Williams DA, Zheng Y. Structure-function based design of small molecule inhibitors targeting Rho family GTPases. Curr Top Med Chem. 2006;6:1109–1116. doi: 10.2174/156802606777812095. [DOI] [PubMed] [Google Scholar]
- 17.Pelish HE, Peterson JR, Salvarezza SB, Rodriguez-Boulan E, Chen JL, Stamnes M, Macia E, Feng Y, Shair MD, Kirchhausen T. Secramine inhibits Cdc42-dependent functions in cells and Cdc42 activation in vitro. Nat Chem Biol. 2006;2:39–46. doi: 10.1038/nchembio751. [DOI] [PubMed] [Google Scholar]
- 18.Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- 19.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartmann JT, Haap M, Kopp HG, Lipp HP. Tyrosine Kinase Inhibitors - A Review on Pharmacology, Metabolism and Side Effects. Curr Drug Metab. 2009 doi: 10.2174/138920009788897975. [DOI] [PubMed] [Google Scholar]
- 21.Progida C, Malerod L, Stuffers S, Brech A, Bucci C, Stenmark H. RILP is required for the proper morphology and function of late endosomes. J Cell Sci. 2007;120:3729–3737. doi: 10.1242/jcs.017301. [DOI] [PubMed] [Google Scholar]
- 22.Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009;284:12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.BasuRay S, Mukherjee S, Romero E, Wilson MC, Wandinger-Ness A. Rab7 mutants associated with Charcot-Marie-Tooth disease exhibit enhanced NGFstimulated signaling. PLoS One. 2010;5:e15351. doi: 10.1371/journal.pone.0015351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Gassart A, Geminard C, Hoekstra D, Vidal M. Exosome secretion: the art of reutilizing nonrecycled proteins? Traffic. 2004;5:896–903. doi: 10.1111/j.1600-0854.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- 25.Cao C, Laporte J, Backer JM, Wandinger-Ness A, Stein MP. Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic. 2007;8:1052–1067. doi: 10.1111/j.1600-0854.2007.00586.x. [DOI] [PubMed] [Google Scholar]
- 26.Dong J, Chen W, Welford A, Wandinger-Ness A. The proteasome alpha-subunit XAPC7 interacts specifically with Rab7 and late endosomes. J Biol Chem. 2004;279:21334–21342. doi: 10.1074/jbc.M401022200. [DOI] [PubMed] [Google Scholar]
- 27.Croizet-Berger K, Daumerie C, Couvreur M, Courtoy PJ, van den Hove MF. The endocytic catalysts, Rab5a and Rab7, are tandem regulators of thyroid hormone production. Proc Natl Acad Sci U S A. 2002;99:8277–8282. doi: 10.1073/pnas.122187699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roepstorff K, Grovdal L, Grandal M, Lerdrup M, van Deurs B. Endocytic downregulation of ErbB receptors: mechanisms and relevance in cancer. Histochem Cell Biol. 2008;129:563–578. doi: 10.1007/s00418-008-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 30.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deretic V. Autophagy in infection. Curr Opin Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon JM, FitzPatrick D, Schmedding E, De Vriendt E, Jacobs A, Van Gerwen V, Wagner K, Hartung HP, Timmerman V. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003;72:722–727. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spinosa MR, Progida C, De Luca A, Colucci AM, Alifano P, Bucci C. Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease. J Neurosci. 2008;28:1640–1648. doi: 10.1523/JNEUROSCI.3677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cogli L, Piro F, Bucci C. Rab7 and the CMT2B disease. Biochem Soc Trans. 2009;37:1027–1031. doi: 10.1042/BST0371027. [DOI] [PubMed] [Google Scholar]
- 37.Cogli L, Progida C, Lecci R, Bramato R, Kruttgen A, Bucci C. CMT2B-associated Rab7 mutants inhibit neurite outgrowth. Acta Neuropathol. 2010;120:491–501. doi: 10.1007/s00401-010-0696-8. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz SL, Tessema M, Buranda T, Pylypenko O, Rak A, Simons PC, Surviladze Z, Sklar LA, Wandinger-Ness A. Flow cytometry for real-time measurement of guanine nucleotide binding and exchange by Ras-like GTPases. Anal Biochem. 2008;381:258–266. doi: 10.1016/j.ab.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tessema M, Simons PC, Cimino DF, Sanchez L, Waller A, Posner RG, Wandinger-Ness A, Prossnitz ER, Sklar LA. Glutathione-S-transferase-green fluorescent protein fusion protein reveals slow dissociation from high site density beads and measures free GSH. Cytometry A. 2006;69:326–334. doi: 10.1002/cyto.a.20259. [DOI] [PubMed] [Google Scholar]
- 40.Goody RS, Frech M, Wittinghofer A. Affinity of guanine nucleotide binding proteins for their ligands: facts and artefacts. Trends Biochem Sci. 1991;16:327–328. doi: 10.1016/0968-0004(91)90134-h. [DOI] [PubMed] [Google Scholar]
- 41.Surviladze Z, Waller A, Wu Y, Romero E, Edwards BS, Wandinger-Ness A, Sklar LA. Identification of a small GTPase inhibitor using a high-throughput flow cytometry bead-based multiplex assay. J Biomol Screen. 2010;15:10–20. doi: 10.1177/1087057109352240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramirez S, Aiken CT, Andrzejewski B, Sklar LA, Edwards BS. High-throughput flow cytometry: validation in microvolume bioassays. Cytometry A. 2003;53:55–65. doi: 10.1002/cyto.a.10035. [DOI] [PubMed] [Google Scholar]
- 43.Simon I, Zerial M, Goody RS. Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J Biol Chem. 1996;271:20470–20478. doi: 10.1074/jbc.271.34.20470. [DOI] [PubMed] [Google Scholar]
- 44.Giraldo J, Serra J, Roche D, Rovira X. Assessing receptor affinity for inverse agonists: Schild and Cheng-Prusoff methods revisited. Curr Drug Targets. 2007;8:197–202. doi: 10.2174/138945007779315687. [DOI] [PubMed] [Google Scholar]
- 45.Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nature Reviews Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang XM, Walsh B, Mitchell CA, Rowe T. TBC domain family, member 15 is a novel mammalian Rab GTPase-activating protein with substrate preference for Rab7. Biochem Biophys Res Commun. 2005;335:154–161. doi: 10.1016/j.bbrc.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 48.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 50.Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizuno K, Kitamura A, Sasaki T. Rabring7, a novel Rab7 target protein with a RING finger motif. Mol Biol Cell. 2003;14:3741–3752. doi: 10.1091/mbc.E02-08-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee S, Dong J, Heincelman C, Lenhart M, Welford A, Wandinger-Ness A. Functional analyses and interaction of the XAPC7 proteasome subunit with Rab7. Methods Enzymol. 2005;403:650–663. doi: 10.1016/S0076-6879(05)03056-9. [DOI] [PubMed] [Google Scholar]