Abstract
Integration of the human immunodeficiency virus type 1 (HIV-1) genome into the host chromosome is a vital step in the HIV life cycle. The highly conserved cytosine–adenine (CA) dinucleotide sequence immediately upstream of the cleavage site is crucial for integrase (IN) activity. As this viral enzyme has an important role early in the HIV-1 replication cycle, interference with the IN substrate has become an attractive strategy for therapeutic intervention. We demonstrated that a designed zinc finger protein (ZFP) fused to green fluorescent protein (GFP) targets the 2-long terminal repeat (2-LTR) circle junctions of HIV-1 DNA with nanomolar affinity. We report now that 2LTRZFP-GFP stably transduced into 293T cells interfered with the expression of vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped lentiviral red fluorescent protein (RFP), as shown by the suppression of RFP expression. We also used a third-generation lentiviral vector and pCEP4 expression vector to deliver the 2LTRZFP-GFP transgene into human T-lymphocytic cells, and a stable cell line for long-term expression studies was selected for HIV-1 challenge. HIV-1 integration and replication were inhibited as measured by Alu-gag real-time PCR and p24 antigen assay. In addition, the molecular activity of 2LTRZFP-GFP was evaluated in peripheral blood mononuclear cells. The results were confirmed by Alu-gag real-time PCR for integration interference. We suggest that the expression of 2LTRZFP-GFP limited viral integration on intracellular immunization, and that it has potential for use in HIV gene therapy in the future.
In this study, Sakkhachornphop and colleagues demonstrate that ex vivo transduction of human T lymphocytic cells with a lentiviral vector encoding a zinc finger protein that targets the 2-long terminal repeat (2-LTR) circle junctions of HIV-1 DNA leads to marked inhibition of HIV viral replication and integration as measured by p24 antigen assay and Alu-gag qPCR, respectively.
Introduction
Integration is a crucial step in the human immunodeficiency virus type 1 (HIV-1) replication cycle, as it ensures the production of viral progeny for the remaining life span of the host cell (Katz and Skalka, 1994). After double-stranded DNA is made from viral RNA by reverse transcriptase (RT), both terminal sequences of the viral DNA are then joined noncovalently and enter into the nucleus (Levy, 2007). The integrase (IN) enzyme removes the terminal guanine–thymine (GT) dinucleotide from each 3′-end long terminal repeat (LTR) of the viral genome to produce new 3′-hydroxyl ends (CA-3′OH) (Sherman and Fyfe, 1990; Bushman and Craigie, 1991), and the processed viral DNA is then inserted into the host chromosome (Vink and Plasterk, 1993). The conserved CA–TG dinucleotide pair at the viral DNA end is vital to maintain an integrative element at the ends of the viral DNA precursor (Whitcomb et al., 1990). Mutation, insertion, or deletion at the circle junction sequence in the virus results in the loss of integration into the host genome (Jurriaans et al., 1992). Integration also stabilizes the viral DNA against degradation, and the unintegrated viral DNA in an acutely infected cell is degraded within hours to days (Donehower and Varmus, 1984; Kim et al., 1989; Pauza, 1990; Barbosa et al., 1994; Pauza et al., 1994).
Highly active antiretroviral therapy (HAART), a combination therapy consisting of nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs), nonnucleoside reverse transcriptase inhibitors (NNRTIs), and protease inhibitors (PIs), is the therapeutic mainstay for HIV treatment (De Clercq, 2001; Flexner, 2007). HAART can dramatically reduce viral loads and increase CD4+ lymphocyte counts, which lead to improved health and longevity in HIV-infected individuals (Cavert et al., 1997; Palella et al., 1998). A new class of inhibitor targeting integrase is now in clinical use (Evering and Markowitz, 2008; Grant and Zolopa, 2008). However, resistance and toxicity have been reported (Charpentier et al., 2008; Delelis et al., 2009; Hicks et al., 2009; Mbisa et al., 2011). In addition, withdrawal of antiviral therapy results in the reemergence of the virus because latent viral particles remain in reservoir cells in lymphoid organs and in the peripheral blood (Schrager and D'Souza, 1998). Therefore, there is a need for a stable antiviral therapy such as stem cell gene therapy, which protects cells from viral infection, thereby reducing viral load without additional medical complications.
Gene therapy for HIV-1 infection has been continuously developing. Several strategies of gene therapy are based on inhibiting HIV-1 replication by interfering with the functions of HIV-1 RNAs or proteins. The interfering RNA strategies involve antisense RNAs (Cohli et al., 1994; Lu et al., 2004), ribozymes (Ramezani et al., 2002), RNA aptamers and decoys (Bohjanen et al., 1996), small interfering RNAs (siRNAs) (Lee et al., 2002), and short hairpin RNAs (shRNAs) (Lee et al., 2002). However, RNA approaches may be limited by escape mutations in the site-targeting regions, even without the changing of encoded protein. Many protein interference strategies involve trans-dominant negative mutants (Liem et al., 1993) and single-chain variable fragments (scFvs) to inhibit the function of intracellular target proteins in specific cellular compartments, such as IN (Levy-Mintz et al., 1996), reverse transcriptase (RT) (Shaheen et al., 1996), Rev (Vercruysse et al., 2010), gp120 (Marasco et al., 1993), and Gag p17 (Tewari et al., 2003). However, the reducing environment in the cytosol is an obstacle to the formation of disulfide bonds required for scFv function. Therefore, alternative scaffolds for conferring long-lasting protection against viral infection may circumvent the limitations of individual scFvs.
The Cys2His2 zinc finger protein (ZFP), a ubiquitous DNA-binding motif in humans, can be designed to target any DNA sequence specifically. Because of its excellent folding in cytoplasm and homing to the nucleus, this molecule would be a candidate framework for prolonged defense against HIV-1 challenge. Certain reports have manifested the interference with HIV-1 replication at the transcriptional level (Reynolds et al., 2003; Segal et al., 2004). Hence, our strategy relies on tracking noncovalent 2-LTR circle junctions, using a designed ZFP during the manipulation of HIV-DNA by IN. Therefore, the benefit of targeting the integrase recognition sequence at the 3′-end terminal part of HIV-1 LTR may be of use in future gene therapy.
We reported a designed Cys2His2 ZFP fused with green fluorescent protein (GFP) that targets the terminal region of the HIV-1 LTR (Sakkhachornphop et al., 2009). This ZFP, called 2LTRZFP-GFP, binds to the integrase recognition sequence at 2-LTR circle junctions with nanomolar affinity, which is similar to the affinity of HIV-1 IN (Bugreev et al., 2003; Deprez et al., 2004). The GFP reporter confirmed that 2LTRZFP-GFP folds correctly in the cytoplasm and homes to the nucleus (Sakkhachornphop et al., 2009). In this study, we investigated whether 2LTRZFP-GFP interferes with IN activity during the formation of HIV-1 DNA noncovalent 2-LTR circle junctions. We also demonstrated the inhibition of vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped lentiviral red fluorescent protein (RFP) integration in a stable 293T line expressing 2LTRZFP-GFP. Moreover, stable T cell lines expressing 2LTRZFP-GFP and peripheral blood mononuclear cells (PBMCs) were prepared for challenging with HIV-1NL4-3. 2LTRZFP-GFP successfully inhibited viral integration and replication as measured by Alu-gag qPCR and a p24 antigen assay, respectively. These findings indicate that viral integration can be inhibited by intracellular immunization with 2LTRZFP-GFP.
Materials and Methods
Construction of expression vectors
The 2LTRZFP-GFP and Aart-GFP gene fragments were amplified from pTriEx-4-2LTRZFP-GFP (as previously described; Sakkhachornphop et al., 2009) and pTriEx-4-Aart-GFP, respectively. Aart is a six-zinc finger protein designed to recognize a unique 18-bp target site not found in the human genome, and this gene was used as a control (Dreier et al., 2001). Both genes were cloned into pRRLSIN.cPPT.mPGK-GFP.WPRE, using the flanking XbaI and SalI sites, which led to the construction of pRRLSIN.cPPT.mPGK.2LTRZFP-GFP.WPRE and pRRLSIN.cPPT.mPGK.Aart-GFP.WPRE, and these fusion genes encoded N-terminal His6 and C-terminal GFP. These plasmids were driven by the murine phosphoglycerate kinase (mPGK) promoter. The ligation product was electrotransformed into competent Escherichia coli XL-1 Blue cells and plated on Luria–Bertani (LB) agar containing ampicillin (100 μg/ml). The plasmid minipreps were performed with a PureLink quick plasmid miniprep kit (Life Technologies, Carlsbad, CA). The sequences of the constructed plasmids were preliminarily analyzed by restriction digest with XbaI and SalI. PCR and DNA sequencing were performed to confirm the sequences. The pRRLSIN.cPPT.mPGK-RFP.WPRE construct was then prepared for the pseudotyped lentivirus challenge study. The system for production of the pseudotyped lentivirus was composed of the following components: (1) pMDLgag/polRRE in which the cytomegalovirus (CMV) immediate-early promoter drives the production of the gag/pol transcript containing the Rev-responsive element (RRE); (2) pRSV-Rev, which encodes HIV-1 Rev; (3) pMD.G, which encodes the envelope G protein of VSV; and (4) pRRLSIN.cPPT.mPGK-GFP.WPRE, in which an expression cassette for the transgene is flanked by the cis-acting sequences necessary for packaging, reverse transcription, and integration, including the 5′- and 3′-LTRs. The RFP gene fragment was amplified from pDsRed2-N1 (Clontech, Palo Alto, CA), and this gene was cloned in an analogous fashion to 2LTRZFP-GFP and Aart-GFP. The HIV-1 molecular clone used in this study was pNL4-3, which was obtained from the AIDS Research and Reference Reagent Program (National Institutes of Health, Bethesda, MD).
The 2LTRZFP-GFP and Aart-GFP gene fragments were also cloned into the pCEP4 vector (Life Technologies), using the flanking NheI and NotI sites to construct pCEP4-2LTRZFP-GFP or pCEP4-Aart-GFP, respectively. Expression was driven by the CMV immediate-early enhancer/promoter. The Epstein–Barr virus replication origin (oriP) and nuclear antigen (encoded by the EBNA-1 gene) were carried by this plasmid to permit extrachromosomal replication in mammalian cells. The ligation product was electrotransformed into competent E. coli XL-1 Blue cells and plated on LB agar containing ampicillin (100 μg/ml). Plasmid minipreps were performed with the PureLink quick plasmid miniprep kit. The plasmid sequences were preliminarily confirmed by digestion with NheI and NotI. PCR and DNA sequencing were performed for further sequence confirmation.
Cell cultures
The packaging cell line, 293T, was maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Grand Island, NY) containing penicillin (100 units/ml), streptomycin (100 μg/ml), 2 mM l-glutamine, and 10% fetal bovine serum (FBS) (HyClone, Cramlington, UK). CD4-positive human T-lymphocytic cell lines (SupT1 and stable lines susceptible to HIV-1 infection) were grown in RPMI 1640 medium supplemented with 10% FBS in a humidified atmosphere with 5% CO2 at 37°C.
Production of VSV-G-pseudotyped lentiviral particles
Third-generation pseudotyped lentiviral particles containing VSV-G were produced in 293T cells transiently cotransfected with four vectors. 293T cells (3.5×106 cells) were seeded into 10-cm dishes, and they were transfected using Lipofectamine and Plus reagent (Life Technologies). The amounts of DNA for transfection were as follows: 2.22 μg of pRRLSIN.cPPT.mPGK.2LTRZFP-GFP.WPRE, pRRLSIN.cPPT.mPGK.Aart-GFP.WPRE, or pRRLSIN.cPPT.mPGK-RFP.WPRE; 1.44 μg of the conditional packaging construct, pMDLgag/polRRE (for expression of gag and pol genes); 0.55 μg of the pRSV-Rev construct (for expression of rev cDNA); and 0.77 μg of the pMD.G construct (which encodes a heterologous envelope for VSV-G). After 5 hr, the transfection mixture was replaced with growth medium, and the cells were allowed to grow for 48 or 72 hr. Viruses were harvested from the culture supernatant, and they were filtered through a Millipore Millex-HA 0.45-μm filter unit, aliquoted, and frozen at –80°C. Viral culture supernatants were lysed with 0.2% Triton X-100, and a p24 antigen ELISA was performed with the Genscreen ULTRA HIV Ag-Ab assay (Bio-Rad, Marnes La Coquette, France). Viral load was determined with the COBAS AMPLICOR HIV-1 Monitor test (version 1.5; Roche Molecular Systems, Branchburg, NJ).
HIV-1 viral stocks
293T cells (3.5×106) were plated in 10-cm dishes. After approximately 12 hr, the cells were transfected with 5 μg of the pNL4-3 plasmid, using Lipofectamine and Plus reagent. After 5 hr, the transfection mixture was replaced with 10 ml of growth medium, and the cells were allowed to grow for 48 or 72 hr. The viruses were harvested from the culture supernatants, and they were filtered through a Millipore Millex-HA filter unit (0.45 μm), aliquoted, and frozen at –80°C. The p24 antigen ELISA was performed with the Genscreen ULTRA HIV Ag-Ab assay. The viral load was determined by COBAS AMPLICOR HIV-1 Monitor test (version 1.5).
Generation of stable lines expressing either 2LTRZFP-GFP or Aart-GFP by lentiviral gene transfer
293T cells (1×104) and SupT1 cells (1×105) were seeded in 24-well plates and incubated with 1 ml of VSV-G-pseudotyped lentiviral particles at 20 multiplicities of infection (MOI) in growth medium containing Polybrene (8 μg/ml; Sigma-Aldrich, St. Louis, MO). Spinoculation was performed by spinning the plates at 800×g at room temperature for 1.5 hr. The cells were then incubated in a humidified atmosphere with 5% CO2 at 37°C for 24 hr. After 24 hr, the culture supernatant was replaced with fresh growth medium, and the cells were maintained for 3 days. The efficiency of the stable transduction was determined on the basis of observations made by fluorescence microscopy and flow cytometry. A mixed population of transduced SupT1 cells was isolated by limiting dilution for single clone selection over the course of 3 weeks.
Generation of SupT1 cells stably expressing either 2LTRZFP-GFP or Aart-GFP by nonviral gene transfer
SupT1 cells (1×106) were electrotransfected with 5 μg of either pCEP4-2LTRZFP-GFP or pCEP4-Aart-GFP, using Nucleofector transfection reagent V (Amaxa Biosystems, Cologne, Germany), in accordance with the manufacturer's protocol for T-16 cells. The cells were maintained for 3 days. The efficiency of transfection was determined by fluorescence microscopy and flow cytometry. SupT1 cells that stably expressed the desired genes were continuously selected by limiting dilution in hygromycin B (500–1000 μg/ml; Life Technologies). The clones were maintained in hygromycin B (200 μg/ml) for long-term culture.
Preparation of primary T cells expressing either 2LTRZFP-GFP or Aart-GFP by electrotransfection
PBMCs were isolated by Ficoll-Hypaque centrifugation of buffy coats obtained from healthy blood donors. Cells were adjusted to 1×106 cells/ml in 5 ml of culture medium (RPMI 1640 medium supplemented with 10% FBS, interleukin-2 [10 U/ml], glutamine, and antibiotics) and stimulated with phytohemagglutinin (2 μg/ml; Sigma-Aldrich) in a humidified atmosphere with 5% CO2 at 37°C. After 24 hr of incubation, 2×106 cells were mixed with 5 μg of plasmid vector (pTriEx-4-2LTRZFP-GFP [Sakkhachornphop et al., 2009], pTriEx-4-Aart-GFP, or pTriEx-4-GFP) and GTporator electroporation solution (Protean, Budějovice, Czech Republic), which was provided by Watchara Kasinrerk (Division of Clinical Immunology, Department of Medical Technology, Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai, Thailand). The mix was electrotransfected, using program T-020 in the Nucleofector II device (Amaxa Biosystems). The transfected cells were further cultured for 24 hr, and the efficiency of transfection in each vector was measured by monitoring GFP expression by fluorescence microscopy and flow cytometry. The transfected PBMCs were subjected to HIV-1 challenge as described below.
Quantitation of integrated HIV-1 DNA (provirus) by Alu-gag qPCR assay
Two-step PCR amplification was performed according to a previously described protocol (O'Doherty et al., 2002; Agosto et al., 2007) with some modifications. Briefly, the first-round PCR was performed on extracted DNA from SupT1, SupT1 stable cell lines, and PBMCs expressing 2LTRZFP-GFP or Aart-GFP, using a High Pure PCR template preparation kit (Roche, Mannheim, Germany). The primer sequences used to detect HIV-1 integration were Alu forward, 5′-GCC TCC CAA AGT GCT GGG ATT ACA G-3′ (O'Doherty et al., 2002) and HIV Gag reverse, 5′-GTT CCT GCT ATG TCA CTT CC-3′ (Agosto et al., 2007). A Gag primer (GagREV_B, 5′-CGT TCT AGC TCC CTG CTT GCC CAT AC-3′) was designed to detect the background level of integrated transgenes resulting from lentiviral gene transfer. The reactions were performed in a volume of 25 μl containing 2.5×master mix (5 Prime, Gaithersburg, MD), 400 nM Alu forward primer, and 400 nM HIV Gag reverse or GagREV_B primer. The thermal cycler (MJ Mini thermal cycler and MiniOpticon real-time PCR system; Bio-Rad) was programmed to perform a 2-min hot start at 95°C followed by 40 cycles of denaturation at 95°C for 15 sec, annealing at 62°C for 15 sec, and extension at 72°C for 3.5 min.
The second-round RU5 kinetic PCR was performed with 10 μl of diluted (1:8) first-round amplicon. The primer sequences were R_FWD, 5′-TTA AGC CTC AAT AAA GCT TGC C-3′ and U5 _REV, 5′-GTT CGG GCG CCA CTG CTA GA-3′ (Liszewski et al., 2009). The RU5 molecular beacon probe, which was labeled at its 5′ terminus with the 6-carboxyfluorescein (FAM) reporter and at its 3′ terminus with the BlackBerry quencher (BBQ), had the following sequence: 5′-FAM-CCA GAG TCA CAC AAC AGA CGG GCA CA-BBQ-3′ (Liszewski et al., 2009). The reactions were performed in a final volume of 25 μl containing 2×DyNAmo probe qPCR master mix (Finnzymes, Espoo, Finland), 400 nM RU5 (R_FWD) primer, 400 nM RU5 (U5 _REV) primer, and 140 nM RU5 molecular beacon probe. The reactions were performed on an MJ Mini thermal cycler and MiniOpticon real-time PCR system with the following program: 20-sec hot start at 95°C followed by 50 cycles of denaturation at 95°C for 3 sec and annealing and extension at 63°C for 30 sec.
A primer–probe set designed to quantify the copy number of the cellular gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to adjust the amount of DNA in each qPCR assay. The GAPDH primer sequences were GAPDH_FWD, 5′-GAA GGT GAA GGT CGG AGT C-3′ and GAPDH_REV, 5′-GAA GAT GGT GAT GGG ATT TC-3′. The GAPDH molecular beacon probe was labeled at its 5′ terminus with an FAM reporter and at its 3′ terminus with BBQ. The GAPDH sequence was as follows: 5′-FAM-CAA GCT TCC CGT TCT CAG CCT-BBQ-3′. The reactions were performed in a final volume of 25 μl containing 2×DyNAmo probe qPCR master mix, 320 nM GAPDH_FWD primer, 320 nM GAPDH_REV primer, and 240 nM GAPDH molecular beacon probe. The reactions were performed on an MJ Mini thermal cycler and MiniOpticon real-time PCR system with the following program: 10-min hot start at 95°C followed by 45 cycles of denaturation at 95°C for 30 sec and annealing and extension at 63°C for 1 min.
Challenge of 2LTRZFP-GFP-expressing cells with VSV-G-pseudotyped lentiviral RFP
Two hundred thousand 293T cells or stably transduced 293T cells (2LTRZFP-GFP and Aart-GFP) were seeded into 6-well plates and then incubated overnight with diluted VSV-G-pseudotyped lentiviral RFP viral supernatant at 1 or 10 MOI in 3 ml of growth medium. The culture supernatant was then replaced with growth medium. Every 2 days after the challenge, the cells were subcultured at a 1:6 dilution. Flow cytometry was performed weekly by gating on green and red channels. The expression of RFP was analyzed in GFP-positive cells by gating 100,000 cells. For confocal imaging, the cells were seeded on 20-mm coverslips in a 6-well plate. The next day, the coverslips were washed with phosphate-buffered saline (PBS) and fixed in 4% formaldehyde for 15 min at room temperature followed by permeabilization with 0.1% Triton X-100 in PBS for 5 min. The cells were stained with 4′,6-diamidino-2-phenylindole (DAPI), and the slides were mounted with VECTASHIELD mounting medium (Vector Laboratories, Burlingame, CA) before microscopy analysis. The samples were viewed at ×100 magnification with a Zeiss LSM 700 confocal laser scanning microscope (Carl Zeiss MicroImaging, Jena, Germany), and the data were analyzed with ZEN 2010 software.
Flow cytometric analysis for CD4 expression
SupT1 cells and SupT1 cells stably expressing either 2LTRZFP-GFP or Aart-GFP were collected and washed three times with PBS. The Fc receptor on the cells (1×105 cells) was blocked by incubation with human AB serum on ice for 30 min. To 50 μl of blocked cells, 50 μl of purified anti-CD4 monoclonal antibody (mAb) (MT4-3, 20 μg/ml; kindly provided by W. Kasinrerk) in 1% bovine serum albumin (BSA)–PBS–NaN3 was added, and the samples were incubated on ice for 30 min. The cells were then washed twice with 1% BSA–PBS–NaN3 and resuspended in 20 μl of 1% BSA–PBS–NaN3. Subsequently, 25 μl of polyclonal rabbit anti-mouse immunoglobulins/RPE, rabbit F(ab′)2 (Dako, Denmark) was added to the samples, and the samples were incubated on ice for 30 min. Last, the cells were washed three times with 1% BSA–PBS–NaN3 and fixed with 1% paraformaldehyde in PBS. The fluorescence reactivity of the stained cells was analyzed by flow cytometry.
HIV-1 infection
SupT1 cells stably expressing either 2LTRZFP-GFP or Aart-GFP were maintained in growth medium for at least 4 weeks before HIV-1 infection. The cells were infected with cell-free HIV-1NL4-3 at 0, 1, and 5 MOI for 8 and 16 hr. The cells were then washed three times with prewarmed serum-free medium and suspended in growth medium. Every 2 days, the cells were split (1:2) to maintain a cell density of approximately 1×106 cells/ml, and the culture supernatants were collected for the HIV-1 p24 antigen assay (Genscreen ULTRA HIV Ag-Ab; Bio-Rad). The cell pellets were kept at –80°C until the determination of the inhibition of intracellular HIV-1 integration by Alu-gag qPCR. Cell viability was monitored by trypan blue exclusion staining.
For PBMCs, cells were maintained in culture medium with or without raltegravir at IC50 (10 nM) for 24 hr (Delelis et al., 2009). Then, 1×106 viable cells were challenged with HIV-1NL4-3 at 1 MOI for 16 hr, and washed three times with prewarmed medium and then cultured in growth medium. Cell viability was performed with PrestoBlue cell viability reagent (Life Technologies) at the challenge point. On day 2 after viral infection, the culture supernatants were substituted with fresh culture medium. The cell pellets were harvested on day 4 to determine the inhibition of intracellular HIV-1 integration by Alu-gag qPCR assay.
Results
Cloning of lentivirus-based vectors and nonviral vectors for stable cell line production
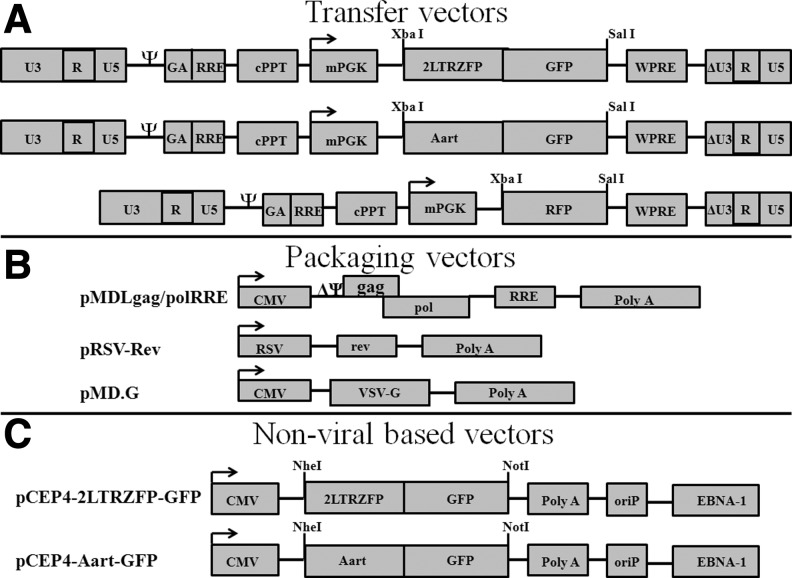
The XbaI and SalI sites in the pRRLSIN.cPPT.mPGK-GFP.WPRE vector were used as cloning sites for 2LTRZFP-GFP, Aart-GFP, and RFP to construct pRRLSIN.cPPT.mPGK.2LTRZFP-GFP.WPRE, pRRLSIN.cPPT.mPGK.Aart-GFP.WPRE, and pRRLSIN.cPPT.mPGK-RFP.WPRE, respectively. Expression from these transgenes was controlled by the mPGK promoter (Fig. 1A). The packaging vectors are schematically shown in Fig. 1B. The 2LTRZFP-GFP and Aart-GFP gene fragments were also cloned into the pCEP4 vector, and the flanking NheI and NotI sites were used to construct pCEP4-2LTRZFP-GFP and pCEP4-Aart-GFP to create N-terminal His6 and C-terminal GFP fusions. The expression of genes in these plasmids was driven by the CMV immediate-early enhancer/promoter (Fig. 1C). All plasmids were sequenced (data not shown).
FIG. 1.
Schematic drawing of the expression vectors for 2LTRZFP-GFP, Aart-GFP, and RFP. (A) The transfer vectors used in this study were derived from the third-generation, self-inactivating vector pRRLSIN.cPPT.mPGK-GFP.WPRE. The XbaI and SalI sites were used as the cloning sites for all transgenes. (B) The packaging vector pMDLgag/polRRE expresses the gag and pol transcript, intervening sequences, and polyadenylation site, and it is driven by the CMV promoter. pRSV-Rev expresses rev cDNA. pMD.G encodes a heterologous envelope to pseudotype the virus coding for VSV-G. (C) The nonviral based vector pCEP4 was used to construct pCEP4-2LTRZFP-GFP and pCEP4-Aart-GFP for stable cell line production. NheI and NotI were used as cloning sites for both transgenes. The EBV origin of replication (oriP) and nuclear antigen (EBNA-1) signal result in high-copy episomal replication in primate and canine cell lines (Yates et al., 1985; Reisman and Sugden, 1986). cPPT, central polypurine tract; GA, fragment of the HIV-1 gag gene; RRE, Rev-responsive element; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element.
Production and expression of VSV-G-pseudotyped lentiviruses containing 2LTRZFP-GFP, Aart-GFP, and RFP
VSV-G-pseudotyped lentiviral particles carrying the 2LTRZFP-GFP, Aart-GFP, and RFP genes were produced by cotransfection in human embryonic kidney 293T cells, using a four-component transient packaging vector system including (1) pMDLgag/polRRE, (2) pRSV-Rev, (3) pMD.G, and (4) pRRLSIN.cPPT.mPGK-GFP.WPRE with an expression cassette for 2LTRZFP-GFP, Aart-GFP, or RFP (Fig. 1A). High-titer viral stocks were harvested, followed by the measurement of p24 antigen and viral load. The results indicated the successful production of VSV-G-pseudotyped lentiviruses containing 2LTRZFP-GFP Aart-GFP, and RFP (data not shown).
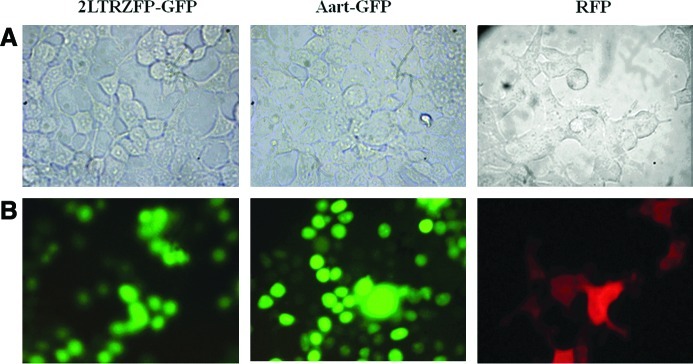
293T cells were transduced with VSV-G-pseudotyped lentivirus containing 2LTRZFP-GFP, Aart-GFP, or RFP. Both 2LTRZFP-GFP and Aart-GFP were observed predominantly in the nucleus when cells were observed by fluorescence microscopy, but RFP was homogeneously distributed throughout the cytoplasm and nucleus (Fig. 2). To determine the effects of the long-term expression of 2LTRZFP-GFP and Aart-GFP in 293T cells, stably transfected cells grown for 90 days were analyzed for GFP by flow cytometry. The percentages of cells containing 2LTRZFP-GFP and Aart-GFP in the stable 293T lines were 91 and 73%, respectively.
FIG. 2.
Expression of 2LTRZFP-GFP, Aart-GFP, and RFP in 293T cells. Stable 293T cell lines were created by transducing cells with pseudotyped lentivirus containing 2LTRZFP-GFP, Aart-GFP, and RFP. Expression was visualized by fluorescence microscopy. Each stable line was imaged at ×400 magnification in the same field. (A) Light-inverted channel. (B) Fluorescence channel. Color images available online at www.liebertonline.com/hum
2LTRZFP-GFP inhibits VSV-G-pseudotyped lentiviral RFP integration in 293T cells
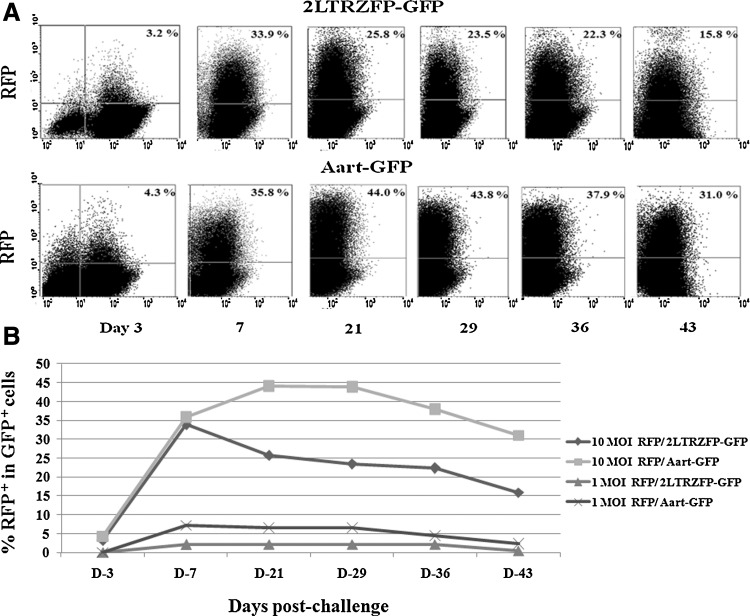
The inhibition of VSV-G-pseudotyped lentiviral RFP integration was studied in 293T cells stably transfected with either 2LTRZFP-GFP or Aart-GFP. Cells were subcultured every 2 days after challenge. The percentage of cells positive for RFP and GFP (Fig. 3A, upper right quadrants) was determined by flow cytometry to yield the proportion of GFP+ cells infected with VSV-G-pseudotyped RFP lentivirus in one round of infection. 2LTRZFP-GFP cells inoculated with VSV-G-pseudotyped RFP lentivirus at either 1 or 10 MOI had noticeably reduced RFP expression compared with expression in Aart-GFP control cells (Fig. 3B). RFP expression in both 2LTRZFP-GFP and Aart-GFP cells decreased during the experimental time course for up to 45 days postchallenge because of clonal variation and promoter silencing, which possibly influence the reduction of RFP intensity. Similar results were obtained by confocal microscopy. The cells were stained with DAPI to visualize the nuclei (Fig. 4).
FIG. 3.
Inhibition of VSV-G-pseudotyped lentiviral RFP expression by 2LTRZFP-GFP. 293T cells stably transfected with 2LTRZFP-GFP and Aart-GFP were challenged with VSV-G-pseudotyped lentiviral RFP at 1 and 10 MOI. (A) Comparison of inhibition of RFP expression in GFP+ cells in both stable lines infected with VSV-G-pseudotyped lentiviral RFP at 10 MOI was determined by flow cytometry on days 3, 7, 21, 29, 36, and 43 after challenge. Approximately 100,000 cells were gated per data point. The percentage of RFP expression in GFP+ cells is indicated in the upper right quadrant. (B) Comparison of inhibition of RFP expression in GFP+ cells in both stable lines infected with VSV-G-pseudotyped lentiviral RFP at 1 and 10 MOI. This graph is representative of two independent experiments.
FIG. 4.
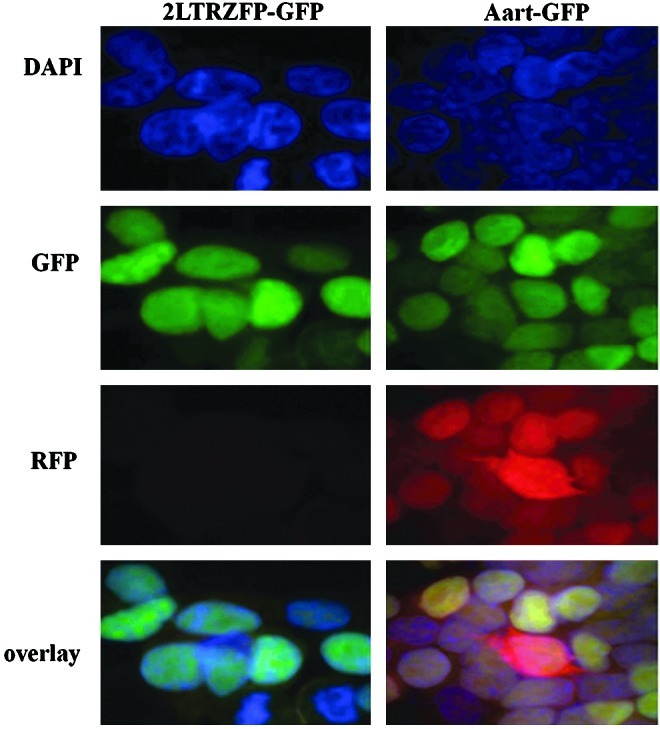
RFP expression in cells stably transfected with either 2LTRZFP-GFP or Aart-GFP. Cells that expressed 2LTRZFP-GFP or Aart-GFP were challenged with pseudotyped RFP lentivirus at 10 MOI. Twenty-nine days after challenge, the cells were stained with DAPI to observe the nuclei, and they were visualized in the same field by confocal microscopy in DAPI, GFP, and RFP channels. The bottom row is the overlay image. The original magnification was ×100. Color images available online at www.liebertonline.com/hum
2LTRZFP-GFP inhibits HIV-1 replication in virus-transduced SupT1 cell line
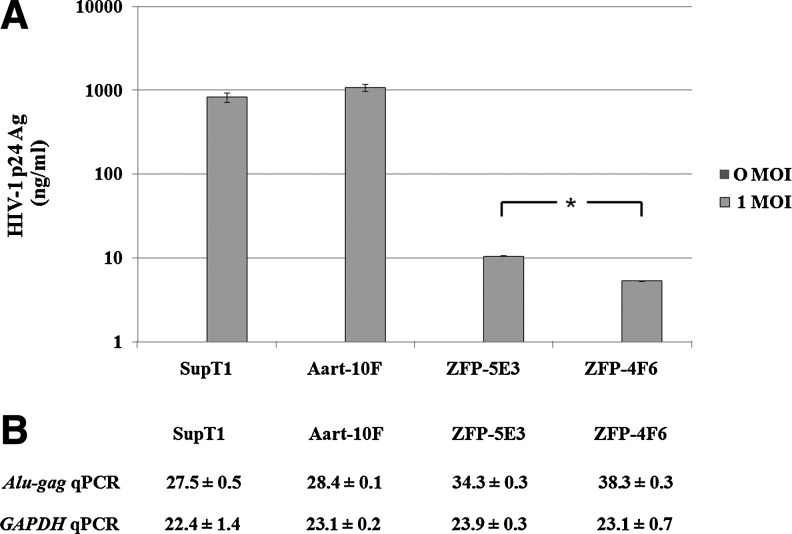
SupT1 and SupT1 cells transduced with either 2LTRZFP-GFP or Aart-GFP were analyzed for cell surface CD4 expression by staining the cells with purified anti-CD4 mAb. CD4 expression on the surface of all stable cells was greater than 97%. The efficiency of transduction, as represented by the percentage of GFP-positive cells, was 0, 97, 57, and 99%, whereas the mean fluorescence intensity (MFI) values were 2.54, 35.14, 16.23, and 36.30 for nontransduced SupT1 cells, Aart-10F stable clonal cells, ZFP-5E3 stable clonal cells, and ZFP-4F6 stable clonal cells, respectively. These cells were infected with HIV-1NL4-3 at 0 and 1 MOI. HIV-1 replication was monitored by quantifying the amount of p24 antigen released into the culture supernatants. At an MOI of 1 on day 13 after challenge, low levels of p24 antigen were observed in the supernatant of ZFP-4F6 cells, and a slightly higher level of HIV-1 p24 antigen was observed in the supernatant of ZFP-5E3 cells. However, the p24 level in ZFP-4F6 cells was significantly lower than in ZFP-5E3 cells (p<0.05, Mann–Whitney U test). Supernatants from controls (nontransfected SupT1 cells and Aart-10F-transfected cells) had high levels of p24 antigen (Fig. 5A).
FIG. 5.
Inhibition of HIV-1 replication in 2LTRZFP-GFP-transduced T-lymphoid cells. SupT1 cells were transduced with either 2LTRZFP-GFP or Aart-GFP, using third-generation lentiviral expression vectors. The following stable lines were propagated: Aart-10F, ZFP-5E3, and ZFP-4F6. These cells were infected with HIV-1NL4-3 at 0 and 1 MOI. (A) Comparison of HIV-1 p24 antigen in the culture supernatants of these cells on day 13 after challenge. The level of p24 antigen in the culture supernatants of ZFP-5E3 was significantly higher than in ZFP-4F6 (*p<0.05, Mann–Whitney U test). This graph is representative of two independent experiments. (B) Comparison of the mean Ct values±SD of Alu-gag qPCR and GAPDH qPCR of extracted genomic DNA from these clones on day 13 after challenge. The PCR assays were performed in triplicate.
A qPCR assay was performed on extracted genomic DNA from stably transduced cell lines that were infected with HIV-1NL4-3 at an MOI of 1 on day 13 after challenge. The mean cycle thresholds (Ct values)±standard deviation (SD) of Alu-gag qPCR of DNA extracted from SupT1, Aart-10F, ZFP-5E3, and ZFP-4F6 cells were 27.5±0.5, 28.4±0.1, 34.3±0.3, and 38.3±0.3, respectively, and the mean Ct values±SD of GAPDH qPCR of these clones were similar, with values of 22.4±1.4, 23.1±0.2, 23.9±0.3, and 23.1±0.7, respectively (Fig. 5B). These results indicated that the presence of ZFP-5E3 or ZFP-4F6 inhibited the integration process and that no inhibition of HIV-1 integration was observed in the control cells.
Inhibition of HIV-1 replication in nonvirally transduced SupT1 cells expressing 2LTRZFP-GFP
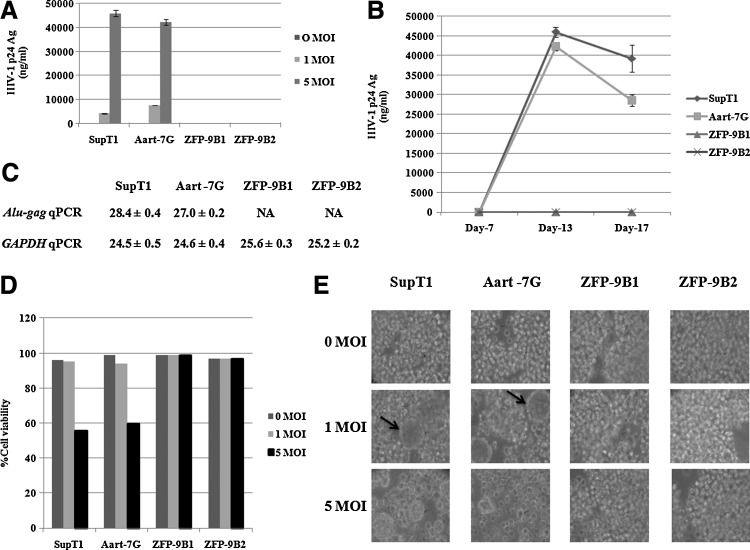
SupT1 cells were electrotransfected with the pCEP4-2LTRZFP-GFP and pCEP4-Aart-GFP vectors to produce stable lines. These cells were continuously selected by limiting dilution in hygromycin B. Flow cytometric analysis showed that the transfection efficiencies, based on the expression of GFP, were more than 90% with MFI values of 2.36, 52.08, 110.90, and 71.30 for nontransfected SupT1 cells and Aart-7G, ZFP-9B1, and ZFP-9B2 transfected SupT1 stable lines, respectively. In contrast, the cell surface expression levels of CD4 were greater than 90%. These cells were then infected with HIV-1NL4-3 at 0, 1, and 5 MOI. HIV-1 replication was determined by analysis of p24 antigen released into the culture supernatants. On day 13 after challenge, expression of ZFP-9B1 and ZFP-9B2 completely inhibited viral integration at both 1 and 5 MOI, and high levels of p24 antigen were observed in the supernatants of control SupT1 and Aart-7G cells (Fig. 6A). Protection due to expression of the ZFPs was observed throughout the experiment for up to 17 days postchallenge (Fig. 6B).
FIG. 6.
Inhibition of HIV-1 replication in SupT1 cells stably expressing 2LTRZFP-GFP. SupT1 cells were transfected with either the pCEP4-2LTRZFP-GFP or pCEP4-Aart-GFP vector. The following stable lines were produced by selection under hygromycin B treatment: Aart-7G, ZFP-9B1, and ZFP-9B2. These cells were infected with HIV-1NL4-3 at 0, 1, and 5 MOI. (A) Comparison of HIV-1 p24 antigen in supernatants from cells on day 13 after challenge. This graph is representative of two independent experiments. (B) Comparison of HIV-1 p24 antigen in supernatants from cells infected at 5 MOI on the indicated days after challenge. (C) Comparison of the mean Ct values±SD of Alu-gag qPCR and GAPDH qPCR of extracted genomic DNA from the indicated infected cells on day 13 after challenge. PCR assays were performed in triplicate. NA, below the detection threshold. (D) Cell viability was determined by trypan blue exclusion staining. (E) Inhibition of cytopathic effects by 2LTRZFP-GFP. The morphology of syncytial formations was imaged at ×400 magnification by inverted microscopy.
Genomic DNA was extracted from the cells on day 13 after challenge with HIV-1NL4-3 at an MOI of 5, and the relative amount of provirus was determined by the Alu-gag qPCR assay. The mean Ct values±SD of Alu-gag qPCR in nontransfected SupT1 cells and Aart-7G-transfected cells were 28.4±0.4 and 27.0±0.2, respectively, and no product was observed for cells transfected with either ZFP-9B1 or ZFP-9B2. The mean Ct values±SD of GAPDH qPCR of these clones were comparable (24.5±0.5, 24.6±0.4, 25.6±0.3, and 25.2±0.2, respectively) (Fig. 6C). These results confirmed that ZFP-9B1 and ZFP-9B2 completely inhibited HIV-1 integration. Cell viability was monitored by trypan blue exclusion staining. All cell lines at all MOIs on day 13 after challenge had almost 100% viability with the exception of nontransfected SupT1 cells and Aart-7G-transfected cells at an MOI of 5 (56 and 60% viability, respectively) (Fig. 6D). Syncytium formation, as indicated by arrows in Fig. 6E, resulting from cell–cell fusion mediated by viral infection, was observed in control SupT1 and Aart-7G cells, but syncytium formation was not observed in ZFP-expressing cells because of the inhibition of cytopathic effects by 2LTRZFP-GFP (Fig. 6E).
Inhibition of HIV-1 integration in PBMCs expressing 2LTRZFP-GFP
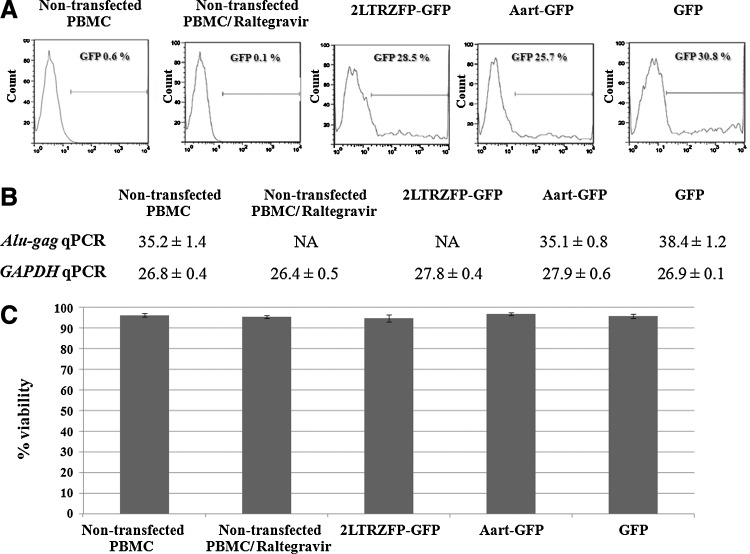
To reveal the potency of 2LTRZFP-GFP in a more physiological setting, PBMCs were electrotransfected with pTriEx-4-2LTRZFP-GFP, pTriEx-4-Aart-GFP, and pTriEx-4-GFP. Flow cytometric analysis of GFP expression was performed to monitor the efficiency of transfection with each vector. The results indicated that all transfected PBMCs showed a similar level of transfection, that is, approximately 26–31% of the PBMCs (Fig. 7A). The transfected PBMCs were challenged with HIV-1 and the relative amount of HIV-1 (integrated form) was determined by Alu-gag qPCR in extracted genomic DNA 4 days after infection. HIV-1 integration was inhibited in PBMCs transfected with 2LTRZFP-GFP, in contrast to other controls (Fig. 7B). The viability of all PBMC samples was comparable and close to 100% (Fig. 7C).
FIG. 7.
Inhibition of HIV-1 integration in PBMCs expressing 2LTRZFP-GFP. PBMCs were transiently transfected with pTriEx-4-2LTRZFP-GFP, pTriEx-4-Aart-GFP, and pTriEx-4-GFP. These cells were infected with HIV-1NL4-3 at 1 MOI. Raltegravir-treated cells were used as a control for inhibition of HIV integration. (A) Flow cytometric analysis of GFP expression, representing the efficiency of transfection. (B) Comparison of the mean Ct values±SD of Alu-gag qPCR and GAPDH qPCR of extracted genomic DNA from the indicated infected cells on day 4 after challenge. The PCR assays were performed in triplicate. NA, below the detection threshold. (C) Cell viability was determined with PrestoBlue cell viability reagent.
Discussion
In this study, we demonstrated that the expression of 2LTRZFP-GFP, in the nucleus of human T-lymphocytic cells, markedly inhibited viral replication and integration as measured by the p24 antigen assay and Alu-gag qPCR, respectively. The HIV-1 integrase recognition sequence is highly conserved and is processed by HIV-1 IN in the early phase of viral replication (LaFemina et al., 1991; Vink et al., 1991; van den Ent et al., 1994; Yoshinaga and Fujiwara, 1995; Katzman and Sudol, 1996; Balakrishnan and Jonsson, 1997). Although certain integrase inhibitors are currently being approved and used for the treatment of HIV-1-infected individuals in the clinic (Evering and Markowitz, 2008; Grant and Zolopa, 2008), remarkable resistance and toxicity have been identified (Charpentier et al., 2008; Delelis et al., 2009; Hicks et al., 2009; Mbisa et al., 2011). The efficacy of HAART has impressively lowered the viral count to undetectable levels in many HIV-1-infected individuals, which has resulted in a decreased mortality rate in these populations (Cavert et al., 1997; Palella et al., 1998). However, obvious eradication of the infection has failed because of the persistence of latent HIV-1 in resting memory CD4+ T cells (Han et al., 2004). Moreover, the emergence of multidrug-resistant viral strains in patients exposed to HAART regimens and the rapid spread of infection in developing countries constitute a significant challenge for AIDS treatment (Chen et al., 2007; Gupta and Pillay, 2007).
Numerous gene therapy strategies have been used to restrain HIV-1 replication by interfering with the functions of HIV-1 RNAs or proteins. Among the RNA approaches, one of them, an antisense RNA against the HIV-1 env gene, has demonstrated activity (Cohli et al., 1994; Lu et al., 2004). Rz1-9 is a multimeric hammerhead ribozyme targeting highly conserved sites in the env coding region of HIV-1 RNA (Ramezani et al., 2002). Tat-activated HIV-1 transcription has been inhibited by a small circular trans-activation response element (TAR) RNA decoy (Bohjanen et al., 1996), and small interfering RNAs (siRNAs) have been used to target HIV-1 rev transcripts in human cells (Lee et al., 2002). However, RNA strategies have been restricted by viral mutations in the targeted areas. The interfering protein strategies involve trans-dominant negative mutants (Liem et al., 1993) and single-chain antibodies (intrabodies) to inhibit the function of intracellular target proteins in specific cellular compartments such as IN (Levy-Mintz et al., 1996), reverse transcriptase (RT) (Shaheen et al., 1996), Rev (Vercruysse et al., 2010), gp120 (Marasco et al., 1993), and Gag p17 (Tewari et al., 2003). However, proper scFv folding requires disulfide bond formation, which makes the use of intrabodies difficult via gene therapy. Although gene therapy holds promise, a number of disadvantages must be overcome before it can be applied as an alternative therapy in HIV-infected individuals. The therapeutic advantage would rely on the ability and method by which the transgene inhibits HIV-1 replication, and on its interference with the life cycle of the virus. Inhibition of viral replication at the earliest step is believed to be optimal for preventing infection and would permit normal immune functions to be sustained. Therefore, preliminary studies are needed before these strategies may be assessed in clinical trials. A designed ZFP represents a promising strategy, in that it provides an ideal scaffold for superior folding in the cytoplasm and nuclear homing. Therefore, it could be an alternative scaffold for conferring long-lasting protection against viral infection.
Herein, 2LTRZFP-GFP, a previously described zinc finger protein construct that targets integrase recognition sequences at 2-LTR circle junctions of HIV-1 DNA, was stably expressed (Sakkhachornphop et al., 2009). This designed ZFP provided a steric barrier between IN and its substrate, blockading enzymatic processing by IN. Compared with cells expressing control ZFP, suppression of RFP expression was observed in 293T cells expressing 2LTRZFP-GFP that were inoculated with VSV-G-pseudotyped RFP lentivirus at both 1 and 10 MOI. To determine whether 2LTRZFP-GFP inhibited viral replication, we used a multiple-round infectious virus (HIV-1NL4-3) for the challenge. A third-generation lentiviral vector (Dull et al., 1998; Zufferey et al., 1998) was used to deliver the 2LTRZFP-GFP gene into a human T-lymphocytic cell line (SupT1). HIV-1 integration was inhibited in cells expressing 2LTRZFP-GFP but not in those expressing the control ZFP. However, the inhibition of HIV-1 integration depended on the amount of transgene expression, because the MFI of SupT1 cells stably transfected with pCEP4-based vector controlled by the CMV promoter was higher than that of stably transduced SupT1 cells with lentiviral transduction expressed under the control of the mPGK promoter. This evidence proved that the interference with HIV integration relied on the expression level of 2LTRZFP-GFP.
To improve safety and gene transfer efficiency, a third-generation lentiviral vector has been widely used for gene delivery into the host cell genome (Dull et al., 1998; Zufferey et al., 1998). Integration into certain sites can cause tumors by directly activating proto-oncogenes (Marchetti et al., 1995; Li et al., 2002). Therefore, gene delivery vehicles that direct therapeutic gene integration into “safe sites” within the human genome or that use technology based on a Sleeping Beauty transposon system have been developed (Tan et al., 2004; Tamhane and Akkina, 2008; Staunstrup et al., 2009; Su et al., 2009). In the present study, the pCEP4 vector was used for nonviral gene transfer. A complete inhibition of viral integration, as evidenced by undetectable levels of p24 antigen, was observed in the supernatants of cells expressing 2LTRZFP-GFP. This implied that the absence of p24 production was due to a blockade of viral integration, because replicable HIV was used.
We also demonstrated the potential application of 2LTRZFP-GFP as a gene therapeutic approach to confine HIV-1 integration in human PBMCs. The integration of HIV-1 DNA was absolutely disrupted in PBMCs expressing 2LTRZFP-GFP and raltegravir-treated cells analyzed by Alu-gag qPCR, whereas proviral DNA was observed in control groups. The inhibition efficiency of 2LTRZFP-GFP was comparable to that of a clinically used drug, that is, raltegravir. Regarding the function of 2LTRZFP-GFP proved in PBMCs, it indicates a potential application for HIV stem cell gene therapy.
In conclusion, our findings indicate that interference in the attachment of HIV integrase to 2-LTR junctions by designed ZFP appears to be highly effective in inhibiting HIV-1 integration without obvious toxicity to the host cells. As lymphoid organs and peripheral blood lymphocytes are reservoirs of latent virus (Schrager and D'Souza, 1998), we suggest the introduction of the 2LTRZFP gene into hematopoietic stem cells to achieve long-term protection (Tesio et al., 2008).
Acknowledgments
The authors thank Dr. Beatriz Gonzalez Alonso and Roberta Fuller for technical assistance, the Faculty of Allied Health Sciences at Chulalongkorn University in Bangkok for performing the confocal imaging experiments, and Dr. Bruce G. Weniger for editorial suggestions. This work was supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. program (grant number PHD/0112/2550), Fogarty AIDS International Research and Training Program, Johns Hopkins University, the National Center for Genetic Engineering and Biotechnology (BIOTEC) of the National Science and Technology Development Agency of Thailand, and U.S. National Institutes of Health grant GM065059 (C.F.B.). The authors thank the National Research University Project under Thailand's Office of the Higher Education Commission for partial financial support.
Author Disclosure Statement
The authors have no conflicts of interest to declare.
References
- Agosto L.M. Yu J.J. Dai J., et al. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology. 2007;368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan M. Jonsson C.B. Functional identification of nucleotides conferring substrate specificity to retroviral integrase reactions. J. Virol. 1997;71:1025–1035. doi: 10.1128/jvi.71.2.1025-1035.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa P. Charneau P. Dumey N., et al. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res. Hum. Retroviruses. 1994;10:53–59. doi: 10.1089/aid.1994.10.53. [DOI] [PubMed] [Google Scholar]
- Bohjanen P.R. Colvin R.A. Puttaraju M., et al. A small circular TAR RNA decoy specifically inhibits Tat-activated HIV-1 transcription. Nucleic Acids Res. 1996;24:3733–3738. doi: 10.1093/nar/24.19.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V. Baranova S. Zakharova O.D., et al. Dynamic, thermodynamic, and kinetic basis for recognition and transformation of DNA by human immunodeficiency virus type 1 integrase. Biochemistry. 2003;42:9235–9247. doi: 10.1021/bi0300480. [DOI] [PubMed] [Google Scholar]
- Bushman F.D. Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: Specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavert W. Notermans D.W. Staskus K., et al. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- Charpentier C. Karmochkine M. Laureillard D., et al. Drug resistance profiles for the HIV integrase gene in patients failing raltegravir salvage therapy. HIV Med. 2008;9:765–770. doi: 10.1111/j.1468-1293.2008.00628.x. [DOI] [PubMed] [Google Scholar]
- Chen L.F. Hoy J. Lewin S.R. Ten years of highly active antiretroviral therapy for HIV infection. Med. J. Aust. 2007;186:146–151. doi: 10.5694/j.1326-5377.2007.tb00839.x. [DOI] [PubMed] [Google Scholar]
- Cohli H. Fan B. Joshi R.L., et al. Inhibition of HIV-1 multiplication in a human CD4+ lymphocytic cell line expressing antisense and sense RNA molecules containing HIV-1 packaging signal and Rev response element(s) Antisense Res. Dev. 1994;4:19–26. doi: 10.1089/ard.1994.4.19. [DOI] [PubMed] [Google Scholar]
- De Clercq E. New developments in anti-HIV chemotherapy. Curr. Med. Chem. 2001;8:1543–1572. doi: 10.2174/0929867013371842. [DOI] [PubMed] [Google Scholar]
- Delelis O. Malet I. Na L., et al. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 2009;37:1193–1201. doi: 10.1093/nar/gkn1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez E. Barbe S. Kolaski M., et al. Mechanism of HIV-1 integrase inhibition by styrylquinoline derivatives in vitro. Mol. Pharmacol. 2004;65:85–98. doi: 10.1124/mol.65.1.85. [DOI] [PubMed] [Google Scholar]
- Donehower L.A. Varmus H.E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc. Natl. Acad. Sci. U.S.A. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier B. Beerli R.R. Segal D.J., et al. Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- Dull T. Zufferey R. Kelly M., et al. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evering T.H. Markowitz M. Raltegravir: An integrase inhibitor for HIV-1. Expert Opin. Investig. Drugs. 2008;17:413–422. doi: 10.1517/13543784.17.3.413. [DOI] [PubMed] [Google Scholar]
- Flexner C. HIV drug development: The next 25 years. Nat. Rev. Drug Discov. 2007;6:959–966. doi: 10.1038/nrd2336. [DOI] [PubMed] [Google Scholar]
- Grant P. Zolopa A. Integrase inhibitors: A clinical review of raltegravir and elvitegravir. J. HIV Ther. 2008;13:36–39. [PubMed] [Google Scholar]
- Gupta R.K. Pillay D. HIV resistance and the developing world. Int. J. Antimicrob. Agents. 2007;29:510–517. doi: 10.1016/j.ijantimicag.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Han Y. Lassen K. Monie D., et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks C. Gulick R.M. Roquebert B., et al. Raltegravir: The first HIV type 1 integrase inhibitor: Selection of the Q148R integrase inhibitor resistance mutation in a failing raltegravir containing regimen. Clin. Infect. Dis. 2009;48:931–939. doi: 10.1086/597290. [DOI] [PubMed] [Google Scholar]
- Jurriaans S. de Ronde A. Dekker J., et al. Analysis of human immunodeficiency virus type 1 LTR–LTR junctions in peripheral blood mononuclear cells of infected individuals. J. Gen. Virol. 1992;73:1537–1541. doi: 10.1099/0022-1317-73-6-1537. [DOI] [PubMed] [Google Scholar]
- Katz R.A. Skalka A.M. The retroviral enzymes. Annu. Rev. Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- Katzman M. Sudol M. Influence of subterminal viral DNA nucleotides on differential susceptibility to cleavage by human immunodeficiency virus type 1 and visna virus integrases. J. Virol. 1996;70:9069–9073. doi: 10.1128/jvi.70.12.9069-9073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y. Byrn R. Groopman J., et al. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: Evidence for differential gene expression. J. Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFemina R.L. Callahan P.L. Cordingley M.G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J. Virol. 1991;65:5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N.S. Dohjima T. Bauer G., et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Levy J.A. HIV and the Pathogenesis of AIDS. ASM Press; Washington, D.C.: 2007. Step involved in HIV:cell interaction and virus entry; p. 77. [Google Scholar]
- Levy-Mintz P. Duan L. Zhang H., et al. Intracellular expression of single-chain variable fragments to inhibit early stages of the viral life cycle by targeting human immunodeficiency virus type 1 integrase. J. Virol. 1996;70:8821–8832. doi: 10.1128/jvi.70.12.8821-8832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Li Z. Dullmann J. Schiedlmeier B., et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- Liem S.E. Ramezani A. Li X., et al. The development and testing of retroviral vectors expressing trans-dominant mutants of HIV-1 proteins to confer anti-HIV-1 resistance. Hum. Gene Ther. 1993;4:625–634. doi: 10.1089/hum.1993.4.5-625. [DOI] [PubMed] [Google Scholar]
- Liszewski M.K. Yu J.J. O'Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47:254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. Yu Q. Binder G.K., et al. Antisense-mediated inhibition of human immunodeficiency virus (HIV) replication by use of an HIV type 1-based vector results in severely attenuated mutants incapable of developing resistance. J. Virol. 2004;78:7079–7088. doi: 10.1128/JVI.78.13.7079-7088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco W.A. Haseltine W.A. Chen S.Y. Design, intracellular expression, and activity of a human anti-human immunodeficiency virus type 1 gp120 single-chain antibody. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7889–7893. doi: 10.1073/pnas.90.16.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A. Buttitta F. Miyazaki S., et al. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J. Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbisa J.L. Martin S.A. Cane P.A. Patterns of resistance development with integrase inhibitors in HIV. Infect. Drug Resist. 2011;4:65–76. doi: 10.2147/IDR.S7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty U. Swiggard W.J. Jeyakumar D., et al. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palella F.J., Jr. Delaney K.M. Moorman A.C., et al. HIV Outpatient Study Investigators. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- Pauza C.D. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- Pauza C. D. Trivedi P. McKechnie T. S., et al. 2-LTR circular viral DNA as a marker for human immunodeficiency virus type 1 infection in vivo. Virology. 1994;205:470–478. doi: 10.1006/viro.1994.1667. [DOI] [PubMed] [Google Scholar]
- Ramezani A. Ma X.Z. Nazari R., et al. Development and testing of retroviral vectors expressing multimeric hammerhead ribozymes targeted against all major clades of HIV-1. Front. Biosci. 2002;7:a29–a36. doi: 10.2741/ramezani. [DOI] [PubMed] [Google Scholar]
- Reisman D. Sugden B. Trans-activation of an Epstein–Barr viral transcriptional enhancer by the Epstein–Barr viral nuclear antigen 1. Mol. Cell. Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds L. Ullman C. Moore M., et al. Repression of the HIV-1 5′ LTR promoter and inhibition of HIV-1 replication by using engineered zinc-finger transcription factors. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1615–1620. doi: 10.1073/pnas.252770699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakkhachornphop S. Jiranusornkul S. Kodchakorn K., et al. Designed zinc finger protein interacting with the HIV-1 integrase recognition sequence at 2-LTR-circle junctions. Protein Sci. 2009;18:2219–2230. doi: 10.1002/pro.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrager L.K. D'Souza M.P. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA. 1998;280:67–71. doi: 10.1001/jama.280.1.67. [DOI] [PubMed] [Google Scholar]
- Segal D.J. Goncalves J. Eberhardy S., et al. Attenuation of HIV-1 replication in primary human cells with a designed zinc finger transcription factor. J. Biol. Chem. 2004;279:14509–14519. doi: 10.1074/jbc.M400349200. [DOI] [PubMed] [Google Scholar]
- Shaheen F. Duan L. Zhu M., et al. Targeting human immunodeficiency virus type 1 reverse transcriptase by intracellular expression of single-chain variable fragments to inhibit early stages of the viral life cycle. J. Virol. 1996;70:3392–3400. doi: 10.1128/jvi.70.6.3392-3400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P.A. Fyfe J.A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunstrup N. H. Moldt B. Mates L., et al. Hybrid lentivirus–transposon vectors with a random integration profile in human cells. Mol. Ther. 2009;17:1205–1214. doi: 10.1038/mt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su K. Wang D. Ye J., et al. Site-specific integration of retroviral DNA in human cells using fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc-finger protein E2C. Methods. 2009;47:269–276. doi: 10.1016/j.ymeth.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamhane M. Akkina R. Stable gene transfer of CCR5 and CXCR4 siRNAs by Sleeping Beauty transposon system to confer HIV-1 resistance. AIDS Res. Ther. 2008;5:16. doi: 10.1186/1742-6405-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W. Zhu K. Segal D.J., et al. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J. Virol. 2004;78:1301–1313. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesio M. Gammaitoni L. Gunetti M., et al. Sustained long-term engraftment and transgene expression of peripheral blood CD34+ cells transduced with third-generation lentiviral vectors. Stem Cells. 2008;26:1620–1627. doi: 10.1634/stemcells.2008-0161. [DOI] [PubMed] [Google Scholar]
- Tewari D. Notkins A.L. Zhou P. Inhibition of HIV-1 replication in primary human T cells transduced with an intracellular anti-HIV-1 p17 antibody gene. J. Gene Med. 2003;5:182–189. doi: 10.1002/jgm.336. [DOI] [PubMed] [Google Scholar]
- van den Ent F.M. Vink C. Plasterk R.H. DNA substrate requirements for different activities of the human immunodeficiency virus type 1 integrase protein. J. Virol. 1994;68:7825–7832. doi: 10.1128/jvi.68.12.7825-7832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse T. Pardon E. Vanstreels E., et al. An intrabody based on a llama single-domain antibody targeting the N-terminal α-helical multimerization domain of HIV-1 Rev prevents viral production. J. Biol. Chem. 2010;285:21768–21780. doi: 10.1074/jbc.M110.112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C. Plasterk R.H. The human immunodeficiency virus integrase protein. Trends Genet. 1993;9:433–438. doi: 10.1016/0168-9525(93)90107-s. [DOI] [PubMed] [Google Scholar]
- Vink C. van Gent D. C. Elgersma Y., et al. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb J.M. Kumar R. Hughes S.H. Sequence of the circle junction of human immunodeficiency virus type 1: Implications for reverse transcription and integration. J. Virol. 1990;64:4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J.L. Warren N. Sugden B. Stable replication of plasmids derived from Epstein–Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yoshinaga T. Fujiwara T. Different roles of bases within the integration signal sequence of human immunodeficiency virus type 1 in vitro. J. Virol. 1995;69:3233–3236. doi: 10.1128/jvi.69.5.3233-3236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R. Dull T. Mandel R.J., et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]