Abstract
The posttranslational modification of eukaryotic intracellular proteins by O-linked N-acetylglucosamine (O-GlcNAc) monosaccharides is essential for cell viability, yet its precise functional roles are largely unknown. O-GlcNAc transferase utilizes UDP-GlcNAc, the end product of hexosamine biosynthesis, to catalyze this modification. The availability of UDP-GlcNAc correlates with glycosylation levels of intracellular proteins as well as with transcriptional levels of some genes. Meanwhile, transcription factors and RNA polymerase II can be modified by O-GlcNAc. A linkage between transcription factor O-GlcNAcylation and transcriptional regulation therefore has been postulated. Here, we show that O-GlcNAcylation of a chimeric transcriptional activator containing the second activation domain of Sp1 decreases its transcriptional activity both in an in vitro transcription system and in living cells, which is in concert with our observation that O-GlcNAcylation of Sp1 activation domain blocks its in vitro and in vivo interactions with other Sp1 molecules and TATA-binding protein-associated factor II 110. Furthermore, overexpression of O-GlcNAc transferase specifically inhibits transcriptional activation by native Sp1 in cells. Thus, our studies provide direct evidence that O-GlcNAcylation of transcription factors is involved in transcriptional regulation.
A broad variety of cytoplasmic and nuclear proteins in eukaryotic cells are modified by O-linked N-acetylglucosamine (O-GlcNAc) monosaccharides at serine and threonine residues (1). This posttranslational modification, termed O-GlcNAcylation, is catalyzed by O-GlcNAc transferase (OGT), an enzyme recently shown to be necessary for cell survival (2). UDP-GlcNAc, which is synthesized de novo from glucose via the hexosamine biosynthetic pathway serves as the substrate for protein O-GlcNAcylation. The availability of UDP-GlcNAc correlates with glycosylation levels of intracellular proteins (3, 4) as well as with transcriptional levels of some genes (5–9). However, how glucose flux through the hexosamine pathway regulates gene transcription remains elusive. Based on the observation that many transcription factors are modified by O-GlcNAc (10, 11), most often in the transcriptional activation domain (10, 11), the intriguing hypothesis has been raised that O-GlcNAcylation of transcription factors could regulate gene transcription in response to glucose flux (8–10, 12). Because O-GlcNAcylation and phosphorylation often occur reciprocally in transcription factors (1, 9, 13), it has been difficult to elucidate the direct effects of O-GlcNAc on transcriptional regulation. Our finding that a region in the activation domain of the transcription factor Sp1 is exclusively subject to O-GlcNAcylation, but not to phosphorylation, allows us to assess the functional roles of O-GlcNAc in transcription directly (12).
The first transcription factor shown to bear the O-GlcNAc modification was Sp1 (10), a ubiquitous transcription factor that plays a vital role in the control of TATA-less housekeeping gene transcription (14). The N-terminal portion of the molecule contains two glutamine-rich activation domains, each associated with a serine-/threonine-rich region, whereas the C-terminal region contains the zinc-finger DNA-binding domain (15). The second glutamine-rich activation domain has been shown to be involved both in the homomultimerization of Sp1 (16) and in the interaction with the general transcription factor (TFIID) via TATA-binding protein-associated factor II 110 (TAFII110) (17, 18). The homomultimerization is required for synergistic activation of transcription by Sp1 (19) whereas the interaction with TFIID via TAFII110 is required for the effect of Sp1 on DNA polymerase II-dependent transcription. These protein–protein interactions appear to be dependent on the glutamine/hydrophobic patches within this domain of Sp1 (18), a motif that is conserved in the activation domains of other transcription factors such as CREB and VP16 (20, 21). Furthermore, we have identified a dominant O-GlcNAcylation site in the C-terminal region of this domain (12). Based on these characteristics of this glutamine-rich activation domain of Sp1, we developed a model Sp1 peptide, termed SpE, which spans this domain and in which we could control the O-GlcNAc state either by mutagenesis or by use of different expression systems. Using an in vitro pull-down assay, we showed that the interaction of SpE with either the full-length Sp1 or TAFII110 was inhibited dramatically by O-GlcNAcylation of the SpE peptide, suggesting that the O-GlcNAc residue interrupted the hydrophobic interactions between the SpE peptide and its partners (12). However, in the same study, we were unable to detect an effect of the glycosylation site mutation on the ability of the SpE segment of Sp1 to activate transcription in vivo in HeLa cells. Thus, the significance of the in vitro behavior of the SpE in the pull-down reactions to transcription remained unclear.
This study was designed to determine the role of O-GlcNAcylation in Sp1-driven transcription. Using an in vitro transcription system, we showed that O-GlcNAcylation of the SpE model peptide markedly inhibited the ability of this peptide to activate transcription. However, in HeLa and HepG2 cells, we confirmed that SpE-mediated transcription was not influenced by a mutation that prevented O-GlcNAcylation of the dominant modification site. However, we could detect a significant difference in transcriptional competence of the glycosylation-site mutant over the wild-type SpE in pancreatic β cells that naturally express high levels of OGT (3, 22) and also in HeLa and HepG2 cells that were transfected with an OGT expression vector. Indeed, even the full-length Sp1-driven transcription could be repressed by overexpression of OGT. Together, these results suggest that O-GlcNAcylation of Sp1 represses Sp1-mediated transcription. We postulate that inhibition of the hydrophobic interactions by O-GlcNAcylation of the Sp1 activation domain must occur to prevent untimely, ectopic, and nonspecific protein—protein interactions between transcription factors before their proper assembly on the cognate DNA template.
Materials and Methods
Plasmids and Recombinant Vaccinia Viruses.
The cDNA encoding amino acids 424–521 of human Sp1 (SpE) was amplified by PCR, fused in-frame with the cDNA encoding Gal4 (1), and subcloned into pGEX-2T (Amersham Pharmacia) for the expression of the glutathione S-transferase (GST)-Gal4-SpE fusion in Escherichia coli. The cDNAs encoding GST-Gal4-SpE, GST-SpE, and the full-length Sp1 were subcloned into pTM3, and recombinant vaccinia viruses were generated as described (23). The cDNA encoding Gal4 (1–94) was fused in-frame with the cDNA for SpE and subcloned into pECE for transient transfection. The serine residue in SpE that corresponds to serine 484 in Sp1 was converted to an alanine by site-directed mutagenesis. The cDNA encoding rat OGT was subcloned into pcDNA3 (Invitrogen).
Protein Expression and Purification.
GST fusions were expressed in E. coli and in mammalian BSC40 cells and were purified as described (12, 22). Proteins bound to glutathione-Sepharose 4B (Amersham Pharmacia) were either eluted with 30 mM glutathione or cleaved with 10 units of thrombin per mg of fusion protein. Proteins were quantified by scanning Coomassie blue-stained SDS/PAGE minigels.
Galactosyltransferase Labeling.
GST-SpE and GST-SpE(S) produced in BSC40 cells were labeled with autogalactosylated bovine milk galactosyltransferase according to the method of Holt and Hart (24).
Coprecipitation Assay.
The full-length Sp1 and GST-SpE or GST-SpE(S) were coexpressed in BSC40 cells for 24 h by using recombinant vaccinia viruses. Cells were extracted with lysis buffer (20 mM Hepes, pH 7.9/400 mM NaCl/0.5% Nonidet P-40/3 mM MgCl2/0.1 mM EDTA/0.1 mM EGTA/10% glycerol/1 mM DTT/1 mM PMSF/2 μg/ml leupeptin/2 μg/ml aprotinin), clarified by centrifugation, and incubated with glutathione-Sepharose beads at 4°C for 3 h. Then, the beads were washed in the lysis buffer. Precipitated proteins were subjected to immunoblot analysis with anti-Sp1 antiserum.
Mass Spectroscopy.
Fusion proteins were passed through an immobilized trypsin cartridge column (PerSeptive Biosystems, Framingham, MA) for digestion. The tryptic digests were analyzed by matrix-assisted laser desorption ionization/time of flight mass spectroscopy as described (12).
In Vitro Transcription.
Transcription reactions were performed in HeLa nuclear extract according to the manufacturer's protocol (Promega) with the following modifications: GTP was substituted by 0.1 mM 3′-O-methyl-GTP, and the indicated amounts of the fusion proteins were added into the reaction mixtures (25 μl) that contained 0.1 pmol of template DNA and 1,000 units of RNase T1 (Ambion, Austin, TX).
Analysis of Gal4-SpE Glycosylation Coupled to Transcription.
DNA templates were biotinylated by incorporating biotin-16-dUTP (Boehringer Mannheim) at the 3′ end with a terminal deoxynucleotidyltransferase and were immobilized on Dynabeads–280 Streptavidin according to the manufacturer's protocol (Dynal). In vitro transcription reactions (100 μl) were performed for 50 min. Then, immobilized DNA templates were separated from nuclear extract and washed sequentially with buffer D (20 mM Hepes, pH 7.9/100 mM KCl/0.5 mM DTT/0.2 mM EDTA/20% glycerol) and with digestion buffer (0.1 M Tris⋅Cl, pH 8.0/1 mM CaCl2). GST-Gal4-SpE proteins bound to immobilized templates were released by tryptic digestion and were subjected to mass spectroscopy. GST-Gal4-SpE proteins soluble in nuclear extract were affinity-purified on glutathione-Sepharose beads, eluted with reduced glutathione, then subjected to tryptic digestion and mass spectroscopy. A fraction (1/100 vol) of each sample was saved at each step for immunoblot analysis using anti-GST antibody (Sigma).
Transient Transfection Assays.
HIT-T15, HeLa, and HepG2 cells were transiently transfected by electroporation as described (12), and luciferase activities were assayed 48 h later. Transfection efficiencies were normalized by using a cotransfected β-galactosidase plasmid. Each transfection was done in duplicate or triplicate and repeated three times.
Immunoblot and Northern Blot Analyses.
Immunoblotting was performed as described (23). For Northern blotting, total RNA was isolated by acid guanidinium thiocyanate–phenol/chloroform extraction from HIT-T15, HeLa, and HepG2 cells, resolved on a formaldehyde-agarose gel, transferred onto a nylon membrane, and then probed with 32P-labeled rat OGT cDNA. The blot was reprobed with β-actin sequence for data normalization.
Results
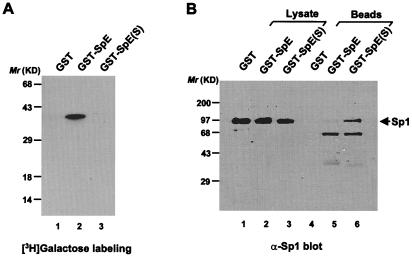
Using in vitro binding studies, we previously found that O-GlcNAcylation of a model peptide spanning the second activation domain of Sp1, termed SpE, blocked physical interactions of the peptide with full-length Sp1 and TAFII110 (12). To determine whether the protein-interaction properties of this peptide in vivo also are dependent on the O-GlcNAc state, we coexpressed GST-SpE or its serine 484-to-alanine mutant [GST-SpE(S)] and the full-length Sp1 in BSC40 cells by using recombinant vaccinia virus. Galactosyltransferase labeling showed that wild-type GST-SpE was modified by O-GlcNAc but that the glycosylation-site mutant was not (Fig. 1A), confirming our earlier finding that serine 484 is the dominant glycosylation site in SpE. As a result, the unglycosylated mutant GST-SpE(S) coprecipitated Sp1 efficiently, whereas the glycosylated form of the peptide was much less efficient, demonstrating that O-GlcNAc in SpE disrupts its interaction with the full-length Sp1 both in vitro (12) and in vivo (Fig. 1B).
Figure 1.
O-GlcNAc blocks interaction between SpE and the full-length Sp1 in vivo. (A) Approximately equivalent amounts (0.5 μg) of GST fusion proteins were subjected to in vitro [3H]galactose labeling by galactosyltransferase. (B) The full-length Sp1 coprecipitated with the wild-type and mutant GST-SpE was analyzed by immunoblot with anti-Sp1 antiserum. One percent of whole-cell lysates were analyzed as well.
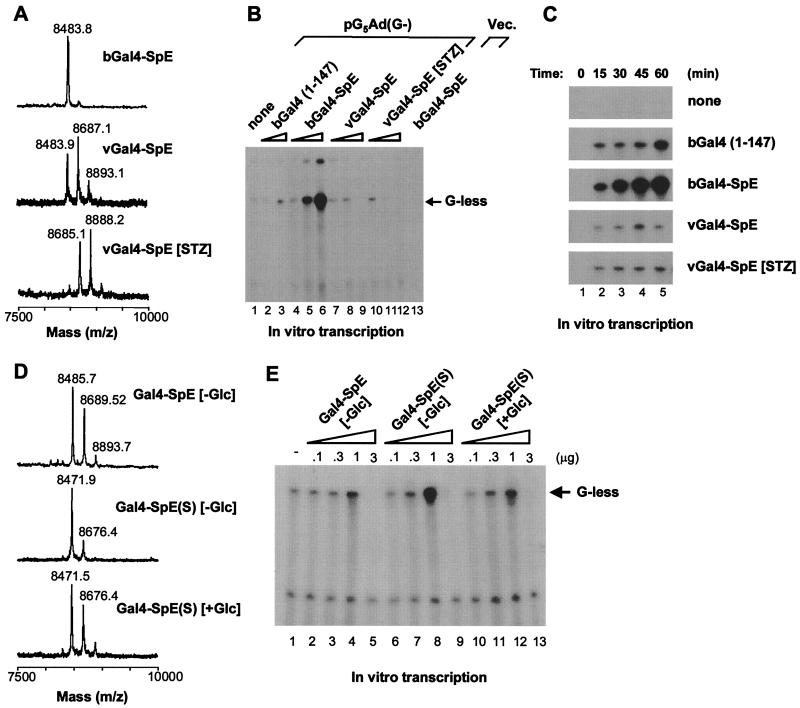
To determine whether the blockade of protein interactions by O-GlcNAc affects SpE transactivation capability, SpE was fused to the Gal4 DNA-binding domain to generate a chimeric activator, Gal4-SpE. Mass spectroscopic analysis showed that the fusion protein expressed in bacteria lacked glycosylation and that the protein produced in mammalian cells using recombinant vaccinia virus carried predominantly a single O-GlcNAc in the SpE region (Fig. 2A). Streptozotocin (STZ) is a GlcNAc analog that selectively inhibits O-GlcNAc-selective N-acetyl-β-d-glucosaminidase (O-GlcNAcase), the enzyme that catalyzes the removal of O-GlcNAc from proteins (25), resulting in the hyperglycosylation of cellular proteins (3, 22, 26). Indeed, the SpE protein expressed in STZ-treated mammalian cells mainly contained two O-GlcNAc moieties (Fig. 2A), suggesting the presence of at least a second O-GlcNAcylation site in the SpE region. In contrast, no phosphorylation sites were found in this region. These chimeras, with diverse glycosylation states, were tested for competency to stimulate transcription in in vitro transcription reactions with a DNA template containing five tandem Gal4-binding sites upstream of the minimal adenovirus major later promoter and a G-less transcription cassette, pG5Ad(G−). All chimeras stimulated transcription in a dose-dependent manner; however, the unglycosylated form was markedly more competent than the glycosylated forms in stimulating transcription in vitro (Fig. 2B). Interestingly, the addition of 3 μg of the glycosylated proteins to the reactions resulted in the squelching of transcription, whereas the same dose of the unglycosylated protein stimulated transcription to a maximal extent. The time course study was consistent with the dose-response analysis, demonstrating at each time point the greater activity of the unglycosylated protein in the transcription assay (Fig. 2C). These results indicate that O-GlcNAc inhibits SpE-mediated transcriptional activation, probably as a result of the reduced affinity of this Sp1 activation domain for the other proteins involved in transcriptional activation.
Figure 2.
Transcriptional activation by Gal4-SpE chimeras with diverse glycosylation status. (A) Mass spectra of tryptic digests of Gal4-SpE fusions. The GST fusions were produced in bacteria (bGal4-SpE) and in BSC40 cells infected with recombinant vaccinia virus (vGal4-SpE). Some infected cells were treated with streptozotocin (vGal4-SpE [STZ]). (B) Dose-response of Gal4-SpE fusions in activating transcription in vitro. Gal4-SpE proteins (0.3, 1.0, and 3.0 μg) were added to reactions with linearized DNA template pG5Ad(G−). Gal4 (1) (0.3 and 3.0 μg) was added to reactions as a control (lanes 2 and 3). bGal4-SpE (1.0 μg) was added to a reaction with an empty vector (Vec.) (lane 13). (C) Time course of Gal4-SpE-activated transcription in vitro. One microgram of each fusion was used. (D) Mass spectra of tryptic digests of the wild-type and mutant Gal4-SpE, which were expressed in BSC40 cells grown either in glucose-free ([−Glc]) or glucose-containing ([+Glc]) medium. (E) Dose-dependent transcriptional activation by the wild-type and mutant Gal4-SpE. Fusion proteins (0.1, 0.3, 1.0, and 3.0 μg) were added to transcription reactions with the linearized DNA template pG5Ad(G−).
The Gal4-SpE protein produced in bacteria may differ from that expressed in mammalian cells not only in glycosylation status, but also in the conformation or other posttranslational modifications. To allow expression of the chimeric activator in various glycosylation states by using the same vaccinia expression system, we made use of the mutant, Gal4-SpE(S), in which the dominant glycosylation site was eliminated by the replacement of serine 484 with alanine. The mutant protein, Gal4-SpE(S), essentially was unglycosylated when expressed in glucose-starved mammalian cells (Fig. 2D), and this form of the protein was more active at stimulating transcription in vitro than the glycosylated wild-type form of the protein (Fig. 2E). However, when Gal4-SpE(S) was expressed in cells exposed to glucose, it was glycosylated at a site distinct from the dominant glycosylation site and to a level comparable to that of the wild-type protein synthesized in glucose-starved cells (Fig. 2D). Glycosylation at this distinct site resulted in the same attenuation of transcriptional activity as did glycosylation at the dominant site (Fig. 2E). The observations suggest a tight correlation between the glycosylation status of Gal4-SpE and its ability to stimulate transcription in vitro. However, the attenuation of transcriptional activation does not appear to depend strictly on the exact residue modified by O-GlcNAc. This result also suggests that OGT recognizes a conformation in its substrate rather than specific consensus modification sites.
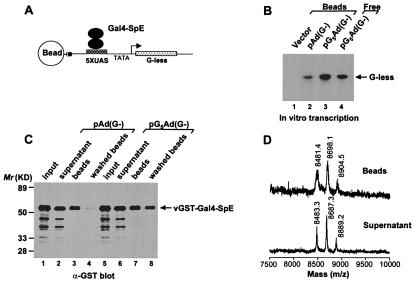
To determine whether Gal4-SpE is deglycosylated during the in vitro transcription process, we analyzed the glycosylation status of the protein that was either bound to DNA template or free in reaction buffer. To isolate DNA-bound proteins, the DNA template was immobilized on magnetic beads (Fig. 3A). This immobilized template was fully capable of supporting in vitro transcription that could be stimulated to some extent by the glycosylated Gal4-SpE (Fig. 3B). Immunoblot analysis confirmed that the Gal4-SpE protein was bound specifically to the Gal4 consensus elements in the immobilized template (Fig. 3C). After the in vitro transcription reaction, the glycosylation state of Gal4-SpE was unaltered whether bound to the template or free in the nuclear extract (Fig. 3D). Although O-GlcNAcase has been found in the nucleus (25), we detected very little enzyme activity in the transcriptionally competent nuclear extract (data not shown). Furthermore, the amount of Gal4-SpE substrate may have overwhelmed the capacity of the enzyme. The failure of the glycosylated form of Gal4-SpE to be converted to the unglycosylated form in the in vitro system might account for the relatively weak transcriptional activity of the glycoprotein.
Figure 3.
GST-Gal4-SpE chimera remains glycosylated in an in vitro transcription system. (A) Schematic diagram of isolation of DNA-bound GST-Gal4-SpE. (B) In vitro transcription on immobilized DNA templates. Equal amounts of empty vector (Vector), Gal4-binding site-deleted template [pAd(G−)], or pG5Ad(G−) were coupled to Dynabeads mixed with vGST-Gal4-SpE in reactions. A reaction with dissolved pG5Ad(G−) template (Free) served as positive control. (C) The binding of vGST-Gal4-SpE to immobilized DNA templates was analyzed by immunoblot with anti-GST antibody. (D) Mass spectra of tryptic digests of vGST-Gal4-SpE either bound to DNA or in supernatant.
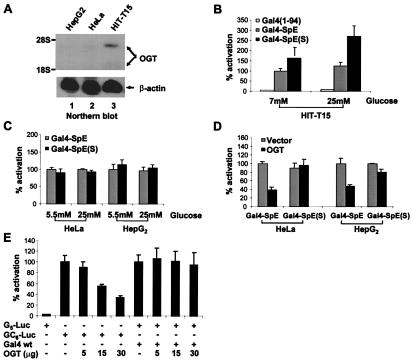
In a previous study, the transcriptional activity of Gal4-SpE could not be distinguished from Gal4-SpE(S) in intact HeLa cells (12). Unlike the persistent glycosylation of the wild-type protein in vitro, its glycosylation in vivo could be sufficiently transient to allow transcriptional activation because of the dynamic and reversible nature of O-GlcNAc modification (1). To shift the cycle of modification to favor the glycosylation of Gal4-SpE, we expressed the chimera in HIT-T15, a cell line derived from pancreatic β cells. Insulin-secreting β cells have been shown to express high levels of OGT (3, 22, 26). Indeed, the HIT-T15 cells exhibited a much higher content of OGT mRNA than HeLa or HepG2 cells (Fig. 4A). These cell lines were cotransfected with the vectors expressing Gal4-SpE or Gal4-SpE(S) and a reporter (pG5-Luc) containing tandem Gal4-binding sites upstream of the firefly luciferase reporter gene. In HIT-T15 cells, Gal4-SpE(S) stimulated transcription significantly more than Gal4-SpE, and this difference was accentuated when the cells were exposed to 25 mM glucose (Fig. 4B). In contrast, HeLa and HepG2 displayed no difference in transcriptional activation by Gal4-SpE and its glycosylation-site mutant, even under high glucose conditions (Fig. 4C). However, when an OGT expression vector was introduced into HeLa and HepG2 cells, the transcriptional activity of Gal4-SpE(S) was 2-fold greater than that of Gal4-SpE (Fig. 4D). These results suggest that the kinetics that control the level of SpE glycosylation can be altered both by the availability of UDP-GlcNAc derived from the flux of glucose into the hexosamine biosynthetic pathway (27, 28) and by the level of activity of OGT, the enzyme that catalyzes the modification. The finding that OGT overexpression in HeLa and HepG2 cells significantly reduced transcriptional activity of the wild-type Gal4-SpE but not of the mutant (Fig. 4D) suggests that increased Gal4-SpE glycosylation contributes to the loss of its activity.
Figure 4.
Transcriptional regulation in vivo by O-GlcNAc. (A) Northern blot analysis of endogenous OGT expression in distinct cell lines. (B and C) Distinct cell lines were cotransfected with 20 μg of G5-Luc reporter and 10 μg of expression vectors for Gal4(1–94) alone, the wild type, and mutant Gal4-SpE, respectively, and then treated with glucose as indicated. (D) HeLa or HepG2 cells were cotransfected with 20 μg of G5-Luc reporter and 10 μg of the wild type or mutant Gal4-SpE expression vector, together with 10 μg of the empty vector or OGT expression vector. The cells were grown in normal glucose (5.5 mM) medium. (E) Overexpression of OGT represses native Sp1-activated transcription. HepG2 cells were cotransfected with 20 μg of GC6-Luc reporter or 20 μg of G5-Luc reporter plus 10 μg of the expression vector for wild-type Gal4 (Gal4 wt), together with increasing amounts of OGT expression vector. Cells were transfected with equal amounts of DNA by inclusion of the parental expression vector.
To determine whether OGT affected the transcriptional activity of native Sp1, the enzyme was overexpressed in HepG2 cells, and Sp1 transcriptional activity was monitored by means of a luciferase reporter (pGC6-Luc) whose expression was driven by tandem GC boxes in the promoter. Overexpression of OGT markedly inhibited endogenous Sp1-activated transcription in a dose-dependent fashion, suggesting that elevated glycosylation of native Sp1 results in a decrease in its activity (Fig. 4E). It is also possible that OGT repressed Sp1-induced transcription by glycosylating other proteins involved in transcription, such as the carboxyl-terminal domain of RNA polymerase II (29). However, Gal4-stimulated transcription on a similar template was unaffected by the OGT overexpression, suggesting that most of the observed transcriptional repression by OGT was a result of altered transcriptional activity of Sp1.
Discussion
Most transcription factors appear to be O-GlcNAcylated by OGT in the transcriptional activation domains (11), and Sp1 is no exception (10, 12), suggesting that OGT plays a critical role in the control of protein—protein interactions involved in transcriptional activation. Using an in vitro transcription system, O-GlcNAcylation of the SpE peptide markedly inhibited its ability to activate transcription. SpE(S), with a mutation at the dominant glycosylation site (serine 484), was glycosylated at an alternative site when expressed in the vaccinia virus system in the presence of STZ. This finding indicates the flexibility of OGT in recognizing this domain of Sp1 as a substrate. Furthermore, this alternative placement of the O-GlcNAc modification has the same inhibitory effect on transcriptional activation and, presumably, the protein interactions that underlie this transcriptional activation. The flexibility of OGT in the placement of the modification suggests that this enzyme recognizes conformational features of numerous transcription factors rather than a specific sequence motif, which implies a ubiquitous role of OGT in regulating transcription.
In contrast to the inhibitory effect of O-GlcNAc on SpE-driven transcriptional activation seen in the in vitro transcription system, it was more difficult to observe this effect in vivo. Previously, we observed no difference in the activity of Gal4-SpE and its serine 484 mutant, Gal4-SpE(S), as transcriptional activators in HeLa cells (12). We had postulated that the SpE(S) mutant might behave similarly to the wild-type SpE in vivo either because the mutant can be glycosylated at an alternative site or the modification on the wild-type peptide occurred cyclically as a result of the sequential actions of OGT and O-GlcNAcase. Indeed, we know now that the SpE(S) mutant can be glycosylated at an alternative site. Furthermore, O-GlcNAcylated Gal4-SpE added to the in vitro transcription assay maintained its modification state in the nuclear extract, preventing its conversion to the unmodified and more transcriptionally active state. The absence of an O-GlcNAcase in vitro allowed a more clear distinction of transcriptional capabilities of the modified vs. unmodified peptides than might be observed in vivo, where both OGT and O-GlcNAcase activities are present. To accentuate the effect of O-GlcNAcylation on the SpE peptide in vivo, we tested the transcriptional activities of the peptide and the SpE(S) mutant in cells that express more OGT. With more OGT, we reasoned that SpE would tend to remain in the modified state, differentiating it from the less easily modified SpE(S) mutant. Using a cell line derived from pancreatic β cells that naturally express OGT at high levels (3, 22, 26), we could distinguish the activities of SpE from SpE(S). Even in HeLa cells and HepG2 cells in which the activities could not be distinguished normally, cotransfection of an OGT expression plasmid did result in differential behavior of the peptides in transcriptional activation. Furthermore, overexpression of OGT in HepG2 cells also was capable of suppressing native Sp1-driven transcription. Taken together, these results indicate that O-GlcNAcylation of the Sp1 transcriptional activation domain(s) represses the ability of this transcription factor to activate transcription, presumably as a consequence of the inhibition of the protein interactions that underlie transcriptional activation.
Sp1 and other transcription factors also can be phosphorylated. For Sp1, there is some evidence that a reciprocal relationship exists between phosphorylation and O-GlcNAcylation (9, 13). Because phosphorylation also plays an important role in transcriptional activation for many transcription factors, changes in the O-GlcNAc state of intact Sp1 may correlate with changes in transcriptional activation only because the phosphorylation state of the intact protein is changed in a coordinated and reciprocal manner. Although this possibility must be considered for the full-length Sp1, the SpE model peptide used in this study is not subject to phosphorylation. Thus, it can be concluded with some certainty that the O-GlcNAcylation of the second activation domain of Sp1 represses the transcriptional activation capability of this domain.
The glycosylation status of intracellular proteins including Sp1 can be modulated by the availability of extracellular glucose (9, 13, 22, 30). Meanwhile, because various protein kinases appear to regulate the key steps in the hexosamine biosynthetic pathway (31, 32), the signaling cascades targeting these protein kinases might modulate Sp1 glycosylation as well. Along with the studies on the proteosomal regulation of intracellular Sp1 abundance (23, 30), these studies support the model that the glycosylation of Sp1 serves as a sensor of physiological stimuli, such as nutrient availability, that allows the coupling of the metabolic state to the transcription of a broad variety of genes.
Recent studies reveal that glucose flux regulates the transcription of the genes encoding transforming growth factor α (TGF-α), TGF-β, leptin, and plasminogen activator inhibitor-1 via the hexosamine biosynthetic pathway (5, 6, 8, 9, 33). Given that most RNA polymerase II transcription factors examined so far bear O-GlcNAc modification, O-GlcNAc may play differential roles in modifying activities of these transcription factors. Glucose-induced changes in levels of transcription of a gene would reflect combinatorial effects of O-GlcNAc on multiple transcription factors that bind to the promoter. Even for Sp1, the role of the hexosamine pathway may be complex. Although we have shown that direct O-GlcNAcylation of a domain of this protein represses transcriptional activation, we also have shown that the stability of Sp1 is indirectly regulated by the O-GlcNAc state of a proteosomal component. Hence, the net effect of the hexosamine pathway on Sp1-mediated transcription may result from a complex relationship between the direct and indirect effects of O-GlcNAc on this and other transcription factors.
Another plausible model for functions of Sp1 O-GlcNAcylation is that the cyclical addition and removal of O-GlcNAc from Sp1 might provide temporal and spatial control of Sp1 homo- and heteromultimerization. Cotranslational addition of O-GlcNAc to Sp1 might prevent inappropriate protein interactions in the cytoplasm, whereas the removal of O-GlcNAc from Sp1 might permit the assembly of the transcription preinitiation complex on cognate DNA. In concert with this idea, it has been observed that the density of O-GlcNAcylated proteins along the length of Drosophila polytene chromosomes was markedly reduced in transcriptionally active “puff” regions (34), implying an inverse relationship between transcription and O-GlcNAcylation and, therefore, a vital role of O-GlcNAcase in the formation of transcriptionally competent complexes.
Acknowledgments
We thank G. W. Hart for the cDNA encoding rat OGT, J. Kadonaga for the cDNA encoding Sp1, and G. Fuller for critical comments on the manuscript. These studies were supported by a grant to J.E.K. from the National Institutes of Health.
Abbreviations
- O-GlcNAc
O-linked GlcNAc
- OGT
O-GlcNAc transferase
- O-GlcNAcase
O-GlcNAc-selective N-acetyl-β-d-glucosaminidase
- STZ
streptozotocin
- TAFII110
TATA-binding protein-associated factor II 110
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hart G W. Annu Rev Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- 2.Shafi R, Iyer S P, Ellies L G, O'Donnell N, Marek K W, Chui D, Hart G W, Marth J D. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. . (First Published May 9, 2000; 10.1073/pnas.100471497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu K, Paterson A J, Chin E, Kudlow J E. Proc Natl Acad Sci USA. 2000;97:2820–2825. doi: 10.1073/pnas.97.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yki-Jarvinen H, Virkamaki A, Daniels M C, McClain D, Gottschalk W K. Metabolism. 1998;47:449–455. doi: 10.1016/s0026-0495(98)90058-0. [DOI] [PubMed] [Google Scholar]
- 5.Sayeski P P, Kudlow J E. J Biol Chem. 1996;271:15237–15243. doi: 10.1074/jbc.271.25.15237. [DOI] [PubMed] [Google Scholar]
- 6.Roos M D, Han I O, Paterson A J, Kudlow J E. Am J Physiol. 1996;270:C803–C811. doi: 10.1152/ajpcell.1996.270.3.C803. [DOI] [PubMed] [Google Scholar]
- 7.McClain D A, Paterson A J, Roos M D, Wei X, Kudlow J E. Proc Natl Acad Sci USA. 1992;89:8150–8154. doi: 10.1073/pnas.89.17.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Liu R, Hawkins M, Barzilai N, Rossetti L. Nature (London) 1998;393:684–688. doi: 10.1038/31474. [DOI] [PubMed] [Google Scholar]
- 9.Du X L, Edelstein D, Rossetti L, Fantus I G, Goldberg H, Ziyadeh F, Wu J, Brownlee M. Proc Natl Acad Sci USA. 2000;97:12222–12226. doi: 10.1073/pnas.97.22.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson S P, Tjian R. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 11.Comer F I, Hart G W. Biochim Biophys Acta. 1999;1473:161–171. doi: 10.1016/s0304-4165(99)00176-2. [DOI] [PubMed] [Google Scholar]
- 12.Roos M D, Su K, Baker J R, Kudlow J E. Mol Cell Biol. 1997;17:6472–6480. doi: 10.1128/mcb.17.11.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haltiwanger R S, Grove K, Philipsberg G A. J Biol Chem. 1998;273:3611–3617. doi: 10.1074/jbc.273.6.3611. [DOI] [PubMed] [Google Scholar]
- 14.Pugh B F, Tjian R. Genes Dev. 1991;5:1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- 15.Kadonaga J T, Courey A J, Ladika J, Tjian R. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 16.Pascal E, Tjian R. Genes Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- 17.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 18.Gill G, Pascal E, Tseng Z H, Tjian R. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Courey A J, Holtzman D A, Jackson S P, Tjian R. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 20.Ferreri K, Gill G, Montminy M. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cress W D, Triezenberg S J. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 22.Roos M D, Xie W, Su K, Clark J A, Yang X, Chin E, Paterson A J, Kudlow J E. Proc Assoc Am Physicians. 1998;110:422–432. [PubMed] [Google Scholar]
- 23.Su K, Roos M D, Yang X, Han I, Paterson A J, Kudlow J E. J Biol Chem. 1999;274:15194–15202. doi: 10.1074/jbc.274.21.15194. [DOI] [PubMed] [Google Scholar]
- 24.Holt G D, Hart G W. J Biol Chem. 1986;261:8049–8057. [PubMed] [Google Scholar]
- 25.Dong D L, Hart G W. J Biol Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 26.Hanover J A, Lai Z, Lee G, Lubas W A, Sato S M. Arch Biochem Biophys. 1999;362:38–45. doi: 10.1006/abbi.1998.1016. [DOI] [PubMed] [Google Scholar]
- 27.Hawkins M, Barzilai N, Liu R, Hu M, Chen W, Rossetti L. J Clin Invest. 1997;99:2173–2182. doi: 10.1172/JCI119390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawkins M, Angelov I, Liu R, Barzilai N, Rossetti L. J Biol Chem. 1997;272:4889–4895. doi: 10.1074/jbc.272.8.4889. [DOI] [PubMed] [Google Scholar]
- 29.Kelly W G, Dahmus M E, Hart G W. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- 30.Han I, Kudlow J E. Mol Cell Biol. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreppel L K, Blomberg M A, Hart G W. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 32.Chang Q, Su K, Baker J R, Yang X, Paterson A J, Kudlow J E. J Biol Chem. 2000;275:21981–21987. doi: 10.1074/jbc.M001049200. [DOI] [PubMed] [Google Scholar]
- 33.Kolm-Litty V, Sauer U, Nerlich A, Lehmann R, Schleicher E D. J Clin Invest. 1998;101:160–169. doi: 10.1172/JCI119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly W G, Hart G W. Cell. 1989;57:243–251. doi: 10.1016/0092-8674(89)90962-8. [DOI] [PubMed] [Google Scholar]