Abstract
Explaining how the small molecule auxin triggers diverse yet specific responses is a long-standing challenge in plant biology. An essential step in auxin response is the degradation of Auxin/Indole-3-Acetic Acid (Aux/IAA, referred to hereafter as IAA) repressor proteins through interaction with auxin receptors. To systematically characterize diversity in degradation behaviors among IAA|receptor pairs, we engineered auxin-induced degradation of plant IAA proteins in yeast (Saccharomyces cerevisiae). We found that IAA degradation dynamics vary widely, depending on which receptor is present, and are not encoded solely by the degron-containing domain II. To facilitate this and future studies, we identified a mathematical model able to quantitatively describe IAA degradation behavior in a single parameter. Together, our results demonstrate the remarkable tunability conferred by specific configurations of the auxin response pathway.
Auxin directs almost every aspect of plant biology, yet how specificity is generated from auxin signaling components remains largely unresolved. A range of auxin-associated phenotypes, including profound disruptions in development and severely compromised responses to environmental signals, are caused by single amino acid substitutions that stabilize transcriptional corepressor proteins called the Auxin/Indole-3-Acetic Acids (Aux/IAAs, referred to hereafter as IAAs; Chapman and Estelle, 2009). The diversity of these phenotypes and the size of the IAA family suggest that IAAs may provide specificity in auxin responses (Lokerse and Weijers, 2009). Functional studies support this idea, as stabilized IAAs provoke different phenotypes even when expressed from the same promoter (Weijers et al., 2005; Muto et al., 2007).
Auxin activates gene expression by enhancing IAA turnover through interaction with auxin receptors, a family of F-box proteins called TRANSPORT INHIBITOR RESISTANT1 (TIR1)/AUXIN SIGNALING F-BOX PROTEINS (AFBs; Dharmasiri et al., 2005a; Kepinski and Leyser, 2005), referred to here collectively as AFBs. Variation in the affinities of IAA|AFB pairs has recently been observed (Calderón Villalobos et al., 2012). How such differences relate to degradation kinetics is still unclear. Labor-intensive seedling studies on a small number of IAA proteins, in combination with analysis of stabilized IAA mutants, uncovered the importance of a conserved region, termed domain II, in determining protein stability. The degron-containing IAA domain II is both necessary and sufficient for interaction with TIR1 and the resulting auxin-induced degradation (Ramos et al., 2001; Dharmasiri et al., 2005a; Kepinski and Leyser, 2005; Tan et al., 2007). In addition, IAA-reporter fusions with diverse domain II sequences show a range of degradation rates when overexpressed in Arabidopsis (Arabidopsis thaliana) seedlings (Dreher et al., 2006). However, the ubiquity of the auxin pathway in plants and the difficulty in reconstituting the complete degradation machinery in vitro have hindered further characterization of the molecular determinants of IAA degradation rates.
As a complement to existing systems and to systematically characterize the potential tunability of different IAA|AFB pairs, we engineered the auxin-induced degradation of IAA proteins in the yeast Saccharomyces cerevisiae. Our synthetic system has several advantages: precise control of auxin input levels, the ability to study IAA|AFB pairs in isolation, and the absence of the many other plant pathways known to impact auxin signaling (Stewart and Nemhauser, 2010). Our system allowed a comprehensive survey of IAA protein turnover while recapitulating behaviors observed in plants. We discovered that the particular AFB receptor used greatly impacted the rate of degradation and that sequences outside of the degron-containing domain II accelerated or decelerated IAA degradation in an IAA-specific manner. Moreover, we identified a mathematical model that provides a single parameter to quantitatively describe degradation behavior. The synthetic toolkit described here will facilitate rapid testing of hypotheses about the ubiquitylation of IAA proteins and suggests a means to characterize other hormone-induced protein degradation pathways.
RESULTS
A Synthetic Yeast System Recapitulates Auxin-Induced IAA Degradation Dynamics
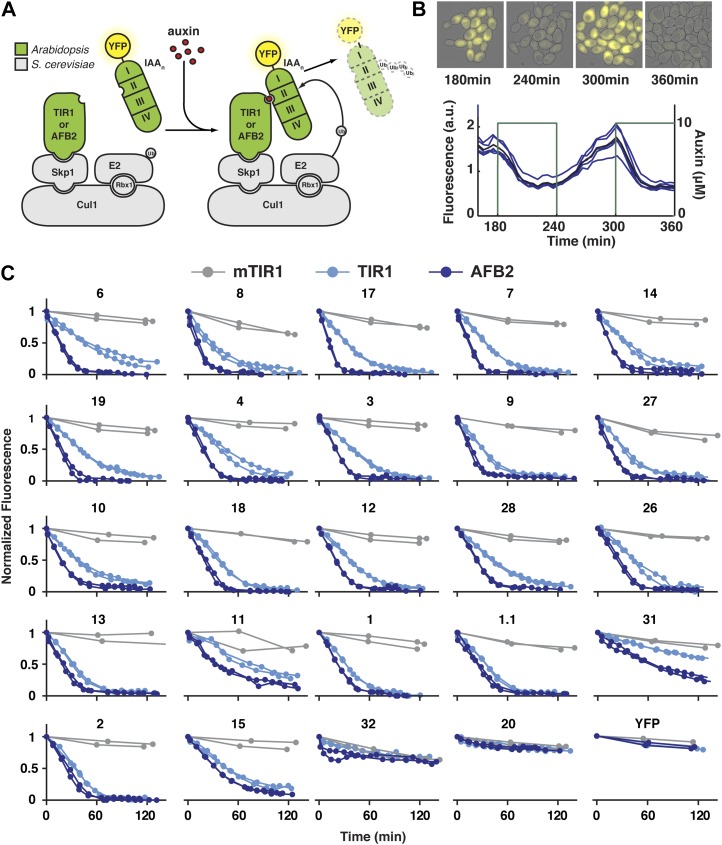
Our engineered auxin response system consisted of pairwise matings of yeast expressing either yellow fluorescent protein (YFP)-IAA fusion proteins or AFBs (Fig. 1A). In the presence of a functional AFB, YFP fluorescence (a proxy for IAA protein levels) could be both up- and down-regulated by modulating the levels of indole-3-acetic acid, hereafter referred to as auxin (Fig. 1B; Supplemental Fig. S1). The timing and extent of degradation were comparable to experimental systems relying on a much higher concentration of a synthetic auxin (Nishimura et al., 2009). Flow cytometry provided high-resolution IAA degradation profiles for each IAA|AFB pair with improved time-resolution measurements at the single cell level (Fig. 1C). In contrast to the “basal degradation” rates observed in plants (Dreher et al., 2006), YFP-IAA proteins were essentially stable in yeast in the absence of auxin or a functional AFB (Supplemental Figs. S2 and S3). This may reflect the difficulty of completely clearing auxin from plant cells or the presence of additional components in plants that are absent from our synthetic system. Following the major auxin-induced degradation events, a more gradual decline in YFP can be observed. As this behavior was also observed in strains expressing mTIR1, we believe that this decrease in fluorescence is caused by physiological changes associated with increasing culture density and not auxin-induced degradation of YFP-IAAs.
Figure 1.
IAA degradation is highly variable. A, Plant auxin receptors (TIR1 or AFB2) and YFP-tagged IAA repressors were integrated into the yeast ubiquitin pathway, shown in gray. B, Yeast cells were imaged while exposed to a square wave of auxin. Auxin leads to a rapid decrease in YFP (fluorescence of individual microcolonies in blue, average value in black), which can be recovered with auxin removal. C, A range of IAA|receptor degradation rates were obtained using time-lapse flow cytometry. Degradation curves were normalized to starting fluorescence. IAAs are listed in order of the relative difference in degradation in the presence of TIR1 versus AFB2. Strains expressing the F-box-deficient mTIR1 show no auxin-dependent degradation. [See online article for color version of this figure.]
The fine time resolution of our measurements resulted in complex degradation profiles that included an initial delay in degradation prior to an exponential decay of YFP levels (Fig. 1, B and C; Supplemental Fig. S9). Standard half-life calculations, therefore, were insufficient to describe the dynamics of degradation in our system. In order to quantitatively characterize the degradation behavior of every IAA|AFB pair, we identified a second-order nonlinear model that captures the dynamic auxin response in both time-course and dose-response experimental data (Fig. 2A; Supplemental File S1; Supplemental Figs. S9–S11; Supplemental Tables S3 and S7). This model accounts for the complex degradation behavior we observed and the nonlinear relationship between auxin concentration and steady-state YFP intensity (Supplemental Fig. S1). Among the candidate models tested, this model had the least complexity while still fitting the data with low residual error (Supplemental File S1). Auxin response is represented by YFP-IAA fluorescence intensity output y and a hypothesized internal state x, dependent on the auxin input concentration u. The hypothesized internal state x is not directly measured in our experiments and does not necessarily equate with specific active species, although one interpretation is that x is a complex formed between auxin and an AFB. Similarly, parameters k1, k2, k3, k4, and k5 are not intended to have direct association with physical values in the system. One possibility is that these rates correlate with synthesis (k1 for the internal state and k3 for the IAA), degradation of the internal state (k2), basal degradation/dilution of the IAA (k4), and AFB-induced degradation (k5). With this interpretation and applying the principles of global curve fitting, we reduced the total number of parameters needed to fit the entire data set (Supplemental File S1).
Figure 2.
Degradation dynamics can be described using few parameters. A, Our model is described by two ordinary differential equations. Degradation curves for AFB2 strains expressing IAA1 or yeast codon-optimized IAA1.1 are shown. B, k5 is largely independent of expression levels. IAA1 and IAA1.1 degradation curves overlap after normalization, although there is an approximately 2-fold difference in k3 values. C, IAA|AFB2 pairs have increased degradation rates (k5), a different rank order when compared with IAA/TIR1 pairs, and an increased dynamic range between the slowest and fastest pairs. Parameters were estimated for two independent replicates. All error bars represent 1 sd. Additional parameters are listed in Supplemental Tables S4 and S5. a.u., Arbitrary units. [See online article for color version of this figure.]
Modeling distilled the differences observed in degradation between the IAA|AFB pairs into a single parameter, k5. Importantly, the k5 value for each IAA|AFB pair is consistent with the qualitative behaviors present in the experimental data (Figs. 1C and 2C). For example, the faster degrading IAAs had the largest k5 values, while more stable IAAs had the lowest k5 values. Of all the parameters, k5 best captures IAA|AFB degradation behavior and is hereafter called the degradation rate.
IAA Proteins Exhibit a Range of Degradation Rates
In our system, auxin-induced degradation differs across IAA|AFB pairs (Figs. 1C and 2C). This is consistent with previous work in Arabidopsis seedlings, where half-lives of overexpressed IAA-LUC fusions were calculated by blocking new protein production with cycloheximide and treating exogenously with the synthetic auxin 2,4-dichlorophenoxyacetic acid (Dreher et al., 2006; Supplemental Table S1). In these assays, a strong match to the consensus domain II sequence was correlated with a short protein half-life in the presence of 2,4-dichlorophenoxyacetic acid. For example, IAA17 and IAA28 have strong matches to the consensus domain II and half-lives of 5 and 15 min, respectively. In contrast, IAA31, with a diverged domain II, has a half-life of approximately 4 h. IAA20 lacks a recognizable domain II sequence and is highly stable. We observed similar patterns of degradation in yeast (Figs. 1C and 2C). In yeast expressing either TIR1 or AFB2, IAAs with consensus-matching domain II sequences degraded rapidly, IAA31 was slow to degrade, and IAA20 showed no degradation. Of the IAAs with consensus domain II sequences, most IAA|TIR1 pairs had rates similar to the fast-degrading IAA17|TIR1 (Fig. 2C). We observed a few IAAs outside of this general trend, including the slow-degrading IAA11.
Despite being expressed from the same promoter and singly integrated in the same genomic location, IAAs displayed different basal fluorescence levels in yeast (Supplemental Fig. S3). Our model predicts that rates of IAA expression and degradation are independent of each other (Supplemental File S1). To test this prediction, we synthesized a variant allele of IAA1 (IAA1.1) with yeast-optimized codons. Basal expression of IAA1.1 was twice that of IAA1 (Fig. 2A), with a similar fold change in the estimated k3 values (Fig. 2B). In contrast, normalized degradation curves and k5 values overlapped (Fig. 2, B and C). This result validates our model and demonstrates that IAA degradation rates are indeed robust to fluctuations in IAA expression levels. Challenges in plant assays, including random location of insertions and multiple cell types contributing to variation in transgene expression, make this type of quantitative analysis quite difficult and highlight the benefits of using yeast as an additional resource for dynamic analysis of auxin responses.
IAA Degradation Rates Are Receptor Specific
For the majority of IAAs tested, AFB2 promoted faster degradation than TIR1 (Figs. 1C and 2C). This resulted in IAA|AFB2 pairs having degradation rates up to three times higher than IAA|TIR1 pairs. IAA|AFB2 pairs also had a wider range of degradation rates between all IAAs. Excluding the IAAs with divergent domain IIs, the fastest IAA|TIR1 pair (IAA8) had a degradation rate 3.3-fold higher than the slowest, IAA11. In contrast, the fastest IAA|AFB2 pair (IAA17) had a degradation rate 5.5-fold higher than IAA11. Strikingly, the rank order of degradation rates was not maintained between strains expressing different receptors. IAA6 showed one of the slowest rates of degradation with TIR1, yet it had one of the fastest degradation rates for IAA|AFB2 pairs. A subset of IAAs showed little difference in degradation between auxin receptors, leading to some of the widest discrepancies in relative rank order. For example, there was little or no change in IAA1 degradation when the receptor was switched from TIR1 to AFB2. This resulted in IAA1 being one of the fastest degrading IAAs in combination with TIR1 yet among the slowest when expressed with AFB2. While it is possible that AFB2 functions more efficiently than TIR1 in yeast, the identification of a subset of IAAs that show no change in degradation between the two receptors suggests that these two proteins are intrinsically different in their IAA interactions.
While TIR1 and AFB2 conferred rapid auxin-induced degradation of IAAs, AFB1 and AFB3 had little effect on IAA degradation rates (Supplemental Fig. S4). Genetic analysis suggests that TIR1 and AFB2 are the major auxin receptors in Arabidopsis, but it is still unclear the degree to which each TIR1/AFB protein contributes to specific auxin responses (Dharmasiri et al., 2005b; Parry et al., 2009). Mutations in TIR1 or AFB2 lead to stronger overall auxin-related phenotypes than mutations in AFB1 or AFB3, although the loss of AFB1 or AFB3 can enhance mutations in other AFB family members (Dharmasiri et al., 2005b; Parry et al., 2009). We reasoned that AFBs might differ in their ability to interact with IAAs. This hypothesis is consistent with our findings as well as with in vitro pull-down assays showing that AFB1 and AFB3 have lower levels of interaction with IAAs than TIR1 and AFB2 (Parry et al., 2009). In addition, IAA-reporter fusions are strongly stabilized in afb2 mutants, while loss of AFB1 or AFB3 alone has little effect on turnover rates (Dharmasiri et al., 2005b). An additional factor may be that our heterologous degradation assay is less sensitive than other assays to weak or transient IAA-AFB interactions. Indeed, in vitro pull-down assays and yeast two-hybrid screens have shown low levels of interaction between some IAA|AFB pairs even in the absence of auxin (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005; Parry et al., 2009; Calderón Villalobos et al., 2012), while we did not see any change in IAA stability without auxin addition (Supplemental Figs. S2 and S3). Moreover, auxin can increase interactions between AFB1-DNA-binding domain fusion proteins and IAA-activation domain fusion proteins when both constructs are highly expressed in yeast (Calderón Villalobos et al., 2012), suggesting that these weaker interactions may contribute to auxin responses in plants. Additional work in plants will be needed to discriminate among these different possibilities.
Receptor expression levels did not influence IAA degradation in our yeast assays. Degradation rates were not correlated with receptor abundance (Supplemental Fig. S5), nor could they be increased by adding a second copy of the same receptor to the genome (Supplemental Fig. S6). In addition, when TIR1 and AFB2 were coexpressed, the degradation rate of IAA6 closely matched that of AFB2 alone (Supplemental Fig. S6). Genetic studies indicate that TIR1 is the primary auxin receptor, and AFB2 is not able to substitute for TIR1 even when expressed from the TIR1 promoter (Parry et al., 2009). This suggests that degradation rate differences are not the sole distinguishing characteristic of receptors and that further functional studies of the dynamics of IAA degradation in receptor mutant backgrounds could be fruitful.
Residues Outside of Domain II Differentially Affect Degradation Rates
Residues outside of domain II have been found to contribute to IAA|AFB auxin-binding affinity in vitro (Calderón Villalobos et al., 2012) and high basal IAA degradation rates in seedlings (Dreher et al., 2006). We engineered truncations in IAAs with disparate degradation rates (Fig. 3A; Supplemental Fig. S7) to directly test the role of nondomain II residues in auxin-induced degradation. The N-terminal half of the protein (T1) or a smaller region restricted to domain II (T2) was fused to an SV40 nuclear localization signal (NLS; Supplemental Table S2). Degradation rates of truncated proteins were compared with full-length constructs fused to the same NLS (Fig. 3, B and C). We found that sequences outside of domain II could accelerate or decelerate degradation rates in an IAA-specific manner. Relative rank order of full-length IAA degradation rates was not conserved in the truncations. IAA28.T2 was the fastest degrading of the T2 truncations, yet IAA28 showed much slower degradation rates than IAA1 or IAA6. Moreover, parallel truncations in different IAAs did not have the same effect on degradation rates. IAA6.T1 was slower than full-length IAA6, but IAA28.T1 was faster than full-length IAA28.
Figure 3.
Residues outside of domain II contribute to auxin-induced degradation rates. A, Schematic of IAA truncations. B and C, Degradation dynamics of full-length proteins are not maintained in truncations. B, Degradation rates of truncations expressed with TIR1 or AFB2, normalized to the starting fluorescence for each strain. C, Parameters k3 and k5 were determined using parameters k1, k2, and k4 from previous model fitting. Additional parameters are listed in Supplemental Table S6.
The recently reported DII-VENUS auxin sensor is similar to IAA28.T2 but shifted 15 amino acids toward the IAA28 N terminus (Vernoux et al., 2011; Brunoud et al., 2012). To test whether this small difference in sequence had any effect on degradation rates, we engineered the identical IAA28 truncation into our system (IAA28.T2V; Fig. 3A; Supplemental Table S2). IAA28.T2V degraded far slower than all other constructs tested (Fig. 3B). This effect is opposite to what we observed with IAA28.T1 and IAA28.T2, both of which had increased rates of degradation compared with the full-length protein. The markedly slower degradation rate we observed for IAA28.T2V could explain why it was the brightest reporter tested (Brunoud et al., 2012). In fact, our analysis predicted that IAA8 and IAA9, the other IAA truncations tested in that study, would degrade faster than IAA28 (Fig. 2; Supplemental Table S1). This is consistent with the much dimmer fluorescence observed for the DII reporters made with these IAA proteins (Brunoud et al., 2012). To directly test whether our yeast assays could predict relative degradation rates in plants, we generated transgenic seedlings expressing a modified DII-VENUS reporter where we replaced the IAA28.T2V sequence with the IAA28.T2 sequence. Consistent with the higher rate of turnover of the IAA28.T2 fusion protein in yeast, we observed significantly lower levels of fluorescence of the IAA28.T2 reporter in transgenic plants (Supplemental Fig. S8).
DISCUSSION
The size and diversity of the IAA and AFB protein families suggest that auxin specificity can be conferred by specific configurations of IAA and AFB family members (Lokerse and Weijers, 2009). In this study, we present a new method for investigating the range of diversity encoded by these large families. By porting plant proteins into yeast, we could directly test the variability in degradation rates between specific IAA|AFB pairs. We were able to reproduce auxin-induced degradation and generate high-resolution real-time data. Our yeast platform was able to recapitulate behaviors previously observed in studies of IAA turnover and allowed for an extensive survey of IAA degradation behavior.
Assessing degradation with each receptor individually, we showed that IAA degradation is highly influenced by which receptor is present and that these receptor effects are IAA specific (Figs. 1C and 2C). Our data provide evidence of receptor choice on regulating the turnover of IAAs and show that each member of the IAA|AFB pair plays a role in determining auxin sensitivity. The high sequence similarity between TIR1 and AFB2, in combination with their shared substrates, should provide a platform to dissect how F-box proteins influence the rate of ubiquitylation, a factor known to vary among other F-box proteins (Pierce et al., 2009). The lack of detectable IAA degradation in yeast expressing AFB1 or AFB3, despite their ability to bind auxin, may have important implications for calibrating auxin responses. This implies that different combinations of receptors may produce varied response thresholds, which may each trigger a specific auxin-regulated process (Reinhardt et al., 2003; Del Bianco and Kepinski, 2011).
Surprisingly, IAA degradation rates were not strongly correlated with the few recently reported in vitro dissociation constants (Calderón Villalobos et al., 2012; Supplemental Table S1). This lack of correlation could simply be the result of the artificial nature of both systems: dissociation constants are a measure of complex formation and are determined independent of a complete ubiquitin complex, and our heterologous system has a mixture of yeast and plant components (Fig. 1A). However, a testable alternative hypothesis is that the interaction strength between TIR1 and an IAA is not a direct reflection of how quickly the IAA is degraded. Conserved sequences outside of the interaction domain have recently been shown to impact the rate of degradation of a number of substrates of the anaphase-promoting complex (Williamson et al., 2011). While similar sequences have not been identified in IAA proteins, the fact that truncations have such varied degradation rates clearly shows that additional residues play a crucial role in modulating interaction with the ubiquitin machinery (Fig. 3; Dreher et al., 2006). Identification of IAA degradation rate determinants could be accelerated by combining information from studies in yeast, in vitro, and in plants.
By utilizing a small, data-driven model, we were able to quantitatively characterize the complex degradation behavior of each IAA|AFB pair in response to auxin (Fig. 2). Mathematical modeling allowed us to distinguish IAA and AFB contributions to degradation and thereby demonstrate how auxin perception can be tuned. We chose a small, empirical model because large mechanistic models (Bridge et al., 2012) require more parameters than could be identified from the low-dimensional output available in our experiments. Small models can nevertheless be quite useful. For example, a new negative feedback loop was discovered in yeast osmoadaptation using a small-model approach (Mettetal et al., 2008). Similarly, our simple model showed that unknown molecular interactions beyond complex affinity are required to describe IAA degradation dynamics. Moreover, small models such as ours can provide a simple description of the input-output properties of a module and facilitate the rational design of new systems in synthetic biology.
In this study, we have demonstrated the utility of porting a pathway to an orthogonal organism to characterize its function. As a single-celled eukaryote with conserved cellular machinery, yeast provides a seminatural context that facilitates the study of complex signaling pathways. The rapid generation time, control of insertion site and number, and high-throughput methods for quantitative analysis, combined with the absence of other known confounding factors like auxin transport, auxin metabolism, and the coexpression of AFB and IAA family members, make studies in yeast a strong complement to plant studies. Given the obvious artificiality of our system, it is quite promising that the rank order of auxin-induced degradation rates parallels the limited number of half-lives observed in plants (Supplemental Table S1). The fact that the IAA28.T2 construct behaved as predicted when expressed in seedlings (Fig. 3; Supplemental Fig. S8) also points to overall conservation in degradation determinants between plant cells and engineered yeast. A full analysis of the similarities and differences between the systems will require more plant studies and likely better tools for measuring dynamic behaviors in plants. Our heterologous system provides a new method to investigate auxin signaling as well as suggests a means to study the many other plant pathways that rely on ubiquitin-mediated protein degradation.
MATERIALS AND METHODS
Yeast Methods
Yeast transformations were performed using a standard lithium acetate protocol (Gietz and Woods, 2002) into MATa W303-1A or MATα W814-29B, a gift from the Gottschling laboratory. Yeast Peptone Dextrose (YPD) and Synthetic Complete (SC) medium supplemented with 80 mg mL−1 adenine were made according to standard protocols. All strains used in this study are listed in Supplemental Table S9.
Strain Construction
IAAs, TIR1, and AFB were amplified from Arabidopsis (Arabidopsis thaliana) complementary DNA (Columbia ecotype) using the primers listed in Supplemental Table S8. A partial attB1 site and Kozak sequence (AAA) were added to the 5′ end of each forward primer (5′-AAAAAGCAGGCTTCAAA-3′), and a partial attB2 site was added to the 5′ end of each reverse primer (5′-AGAAAGCTGGGTG-3′). The remaining attB1 and attB2 sequences were added with a second PCR using generic forward and reverse adapter primers (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′). Products were subcloned into a Gateway pDONR221 plasmid using a standard Gateway BP reaction (BP Clonase II; Life Technologies). Each complementary DNA was fully sequenced and then cloned into destination vectors with a standard Gateway LR reaction (LR Clonase II; Life Technologies). IAAs were cloned into pGP4GY-ccdB (Trp selection), and auxin receptors were cloned into pGP5G-ccdB (Leu selection; K.A. Havens, N. Bolten, J.L. Nemhauser, and E. Klavins, unpublished data). Approximately 300 ng of each plasmid was digested with PmeI and transformed: pGP4GY-IAA into W303-1A, pGP5G-AFB into W814-29B. Integrations were confirmed by PCR. Strains to be mated were coinoculated at low density into YPD medium, grown overnight at 30°C, and struck out to single colonies on SC-His-Trp to select for diploids.
IAA truncations were fused to an N-terminal YFP and C-terminal SV40 NLS repeat using Gly-Ala linkers (GAGAGAGAGAGP and GAGA, respectively; Nishimura et al., 2009). The IAA17.T1 construct was synthesized with partial EYFP and the complete NLS sequence (www.genewiz.com) and then cloned into the pGP4GY-ccdB vector backbone via Gibson assembly (Gibson et al., 2009). The cloning scheme is outlined in Supplemental Figure S7. Primers are listed in Supplemental Table S8. Gateway acceptor sites were removed by this process. Further truncation constructs were amplified from full-length IAA sequences and cloned in place of IAA17 using Gibson assembly (Gibson et al., 2009).
The DII-VENUS plasmid was a gift from Teva Vernoux. The IAA28.T2-VENUS plasmid was constructed by replacing the DII region of DII-VENUS with the IAA28.T2 degron region using Gibson assembly (Gibson et al., 2009).
Flow Cytometry
YFP intensity measurements were taken with a BD Accuri C6 flow cytometer with a CSampler plate adapter using excitation wavelengths of 488 and 640 nm and an emission detection filter at 533 nm (FL1 channel). A total of 10,000 events above a 400,000 FSC-H threshold (to exclude debris) were measured for each sample at a flow rate of 66 μL min−1 and core size of 22 μm using the Accuri C6 CFlow Sampler software. Cytometry data were exported as FCS 3.0 files and processed using the flowCore R software package and custom R scripts to obtain the mean FL1-A value at each time point. The script applies two polygon gates on the data to isolate single yeast cells. One gate separates the total yeast population from debris on the SSC-A and FSC-A channels. The second gate isolates single cells from cell aggregates (doublet discrimination) via their higher FSC-H (peak height) to FSC-A (peak area) ratio. Scripts are available upon request.
Degradation Assays
Cells were prepared by transferring a freshly grown colony from YPD plates into SC. The cell density (in events μL−1) was estimated using cytometry data gated for yeast by a custom R script. Each culture was then diluted to 0.5 events μL−1 in 15 mL of SC. This dilution was split into duplicate 4-mL aliquots with the exception of controls. For IAA17 without a YFP tag and YFP without an IAA, only a single 4-mL aliquot was prepared. YFP-IAA17 was split into three aliquots to serve as an internal replicate control within each experiment. Aliquots were incubated for 16 h at 30°C with shaking. At 16 h, duplicate aliquots of each strain were mixed and split again into two tubes. Cultures were in log phase at the beginning of each experiment (density measured in the cytometer at approximately 500 events μL−1) and remained in log phase for the duration of each experiment (Supplemental Fig. S2).
Measurements were taken at two time points prior to the addition of any treatment. For each strain, one replicate was mock treated (95% [v/v] ethanol) and one replicate was treated with 10 μm indole-3-acetic acid (the minimal concentration of auxin needed to promote complete IAA degradation during log-phase growth of the yeast; Supplemental Figs. S1 and S2). As soon as possible after the addition of auxin, fluorescence for the 0-min time point was recorded. Subsequent measurements were acquired at 10-min intervals for the first 2 h after auxin addition and every 30 min thereafter until the fluorescence level in most strains had plateaued (approximately 3.5 h). Controls were measured every hour for the duration of the experiment.
Modeling and Quantitative Analysis
Modeling methods and quantitative analysis are described in Supplemental File S1.
Microscopy
Yeast cells grown overnight in SC at 30°C were diluted 1:100 in SC, incubated for 4 to 5 h, and then diluted 1:20 before loading onto a Y04D plate (CellASIC). Using the CellASIC-ONIX microfluidic system and associated software, cells were pulsed with a square wave of 10 μm auxin in SC medium over a period of 2 h. An inverted Nikon Eclipse Ti microscope with a 60×, numerical aperture 1.4 oil objective was used to image the yeast cells at 5-min intervals using a YFP-HYQ 535 bandpass filter (Nikon; excitation at 515 nm, detection from 520 to 550 nm) and a CoolSNAP HQ2 14 bit camera. Image processing was done with custom MATLAB scripts, available upon request. Briefly, a segmentation algorithm was applied to bright-field images to produce a binary mask for each microcolony. This binary mask was then applied to the YFP image to calculate the average YFP intensity value within the colony. Background fluorescence level was estimated using the average fluorescence of a 100- × 100-pixel square away from the yeast colony and subtracted from total fluorescence values.
Generation and Analysis of Transgenic Plants
Columbia ecotype plants were transformed using the floral dip method (Clough and Bent, 1998). T1 plants were selected on 0.5× LS agar plates containing 30 μg mL−1 hygromycin B. Plates were stratified for 2 d, exposed to light for 6 h, and then grown in the dark for 3 d following a modification of the method of Harrison et al. (2006). Resistant seedlings were transferred to plates containing no antibiotics and allowed to recover for an additional 3 d. DII-VENUS seeds provided generously by Teva Vernoux were grown in identical conditions to allow a direct comparison of the IAA28.T2 and IAA28.T2V constructs in plants.
Plants were imaged using a Leica DMI 3000B microscope fitted with a Leica long-working 20× HCX PL FLUORTAR objective and illuminated with a Lumencor SOLA light source. Images were captured using Leica LAS AF version 2.6.0 software and a Leica DFC 345FX camera. Seven independent IAA28.T2 transformants were examined and compared with 10 DII-VENUS seedlings. Fiji software was used to quantify fluorescence in a region of interest centered on each image.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Addition of 10 μm auxin is sufficient for maximal degradation of IAAs.
Supplemental Figure S2. Fluorescence levels decrease drastically as yeast cells enter the stationary phase.
Supplemental Figure S3. AFBs do not have differential effects on basal degradation of IAAs.
Supplemental Figure S4. AFB1 and AFB3 do not promote degradation of IAA2.
Supplemental Figure S5. TIR1 is expressed at a similar level to AFB2.
Supplemental Figure S6. AFB2 expression is not rate limiting.
Supplemental Figure S7. Cloning scheme for domain II truncation constructs.
Supplemental Figure S8. Fluorescence accumulation differs between IAA28 truncations in plants.
Supplemental Figure S9. Sample time-course IAA degradation data and model fits of IAA14|TIR1.
Supplemental Figure S10. Sample dose-response data and model predicted dose response of IAA17|AFB2.
Supplemental Figure S11. Parameter variations study of the preferred model.
Supplemental Table S1. Comparison of degradation rates, half-lives, and affinities from yeast, in vitro, and plant studies.
Supplemental Table S2. Table of amino acids included in each IAA truncation.
Supplemental Table S3. The residuals and the number of distinct parameters for all candidate models.
Supplemental Table S4. Estimated parameters for IAA|TIR1 pairs using the preferred model interpretation.
Supplemental Table S5. Estimated parameters for IAA|AFB2 pairs using the preferred model interpretation.
Supplemental Table S6. Estimated parameters for degron comparison study using the preferred model interpretation.
Supplemental Table S7. Average, minimum, and maximum values of the estimated parameters.
Supplemental Table S8. Oligonucleotides used in this study
Supplemental Table S9. Yeast strains used in this study.
Supplemental File S1. Quantitative Analysis.
Supplementary Material
Acknowledgments
We thank Mark Estelle, Ning Zheng, and members of the Nemhauser and Klavins groups for helpful discussions and Alec Nielson, Selma Alkafeef, and Brandi House for technical assistance.
Glossary
- IAA
Auxin/Indole-3-Acetic Acid
- YFP
yellow fluorescent protein
- NLS
nuclear localization signal
- SC
synthetic medium supplemented with 80 mg mL−1 adenine
References
- Bridge LJ, Mirams GR, Kieffer ML, King JR, Kepinski S. (2012) Distinguishing possible mechanisms for auxin-mediated developmental control in Arabidopsis: models with two Aux/IAA and ARF proteins, and two target gene-sets. Math Biosci 235: 32–44 [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Del Bianco M, Kepinski S. (2011) Context, specificity, and self-organization in auxin response. Cold Spring Harb Perspect Biol 3: a001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, III, Smith HO. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A. (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Lokerse AS, Weijers D. (2009) Auxin enters the matrix: assembly of response machineries for specific outputs. Curr Opin Plant Biol 12: 520–526 [DOI] [PubMed] [Google Scholar]
- Mettetal JT, Muzzey D, Gómez-Uribe C, van Oudenaarden A. (2008) The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319: 482–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT. (2007) Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol 144: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922 [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. (2009) Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462: 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce E-R, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Stewart JL, Nemhauser JL. (2010) Do trees grow on money? Auxin as the currency of the cellular economy. Cold Spring Harb Perspect Biol 2: a001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G. (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Banerjee S, Zhu X, Philipp I, Iavarone AT, Rape M. (2011) Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Mol Cell 42: 744–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.