As we near a complete understanding of plant transcriptomes and their corresponding proteomes, a remaining frontier in plant biology will be the in-depth definition of the posttranslational modifications that help generate the diverse array of appropriately functioning proteins from more finite genomic information. It is now obvious that plant proteins are subject to a wide array of possible modifications that regulate their structure, activity, interactions, location, and/or half-life. These alterations are often genetically predetermined, transient, and highly dynamic, thus providing near unlimited layers of control across the life span of individual proteins. The continually expanding list of over 200 possibilities include the covalent addition of methyl, acetyl, phosphate, and glycosyl moieties, fatty acids, vitamin cofactors, nucleosides, and even other proteins.
The first polypeptide modifier to be discovered was ubiquitin (Ub), a highly conserved, 76-amino acid protein ubiquitously present in eukaryotes (Hershko and Ciechanover, 1998; Smalle and Vierstra, 2004; Vierstra, 2009). Via an intricate enzymatic cascade, Ub becomes attached to a multitude of targets through an isopeptide bond between its C-terminal Gly residue and one or more accessible Lys residues in its targets. This addition can then be reversed by a family of deubiquitylating proteases (DUBs) that uniquely recognize and cleave the bond linking the two moieties, thus generating a reaction cycle akin to those involving protein kinases and phosphatases.
Elucidation of the Ub enzymatic paradigm was followed by the discovery that it represents just one member from a constellation of peptide tags that become reversibly attached to other intracellular constituents, including lipids and prenyl groups, in some cases using parallel chemistries indicative of a common ancestor (Downes and Vierstra, 2005; Kerscher et al., 2006; Burroughs et al., 2012). In plants, these Ub-like modifiers (UBLs) currently include RUB (for related to ubiquitin, or Nedd8 in yeast [Saccharomyces cerevisiae] and animals), SUMO (for small ubiquitin-like modifier), ATG8 (for autophagy8) and ATG12, MUB (for membrane-anchored ubiquitin-fold protein), UFM1 (for ubiquitin-fold modifier1), URM1 (for ubiquitin-related modifier1), HUB1 (for homology to ubiquitin1), and a diverse assortment of proteins that harbor structurally related folds fused translationally to other domains. The varied processes managed by these modifiers are extraordinary, ranging from cellular housekeeping, nutrient recycling, and sulfur chemistry to selective protein turnover, transcriptional regulation, chromatin remodeling, and RNA metabolism (Miura et al., 2007a; Hochstrasser, 2009; Vierstra, 2009; Li and Vierstra, 2012). In this review, I provide a glimpse into the importance of these modifiers in plant biology, an emerging appreciation for how these modifiers intersect to expand their influence, and how they might have evolved from prokaryotic progenitors. Specific examples supporting these themes are also well illustrated by the collection of papers and other reviews included in this Focus issue on “Ubiquitin in Plant Biology.”
Ub AND THE UBL SUPERFAMILY
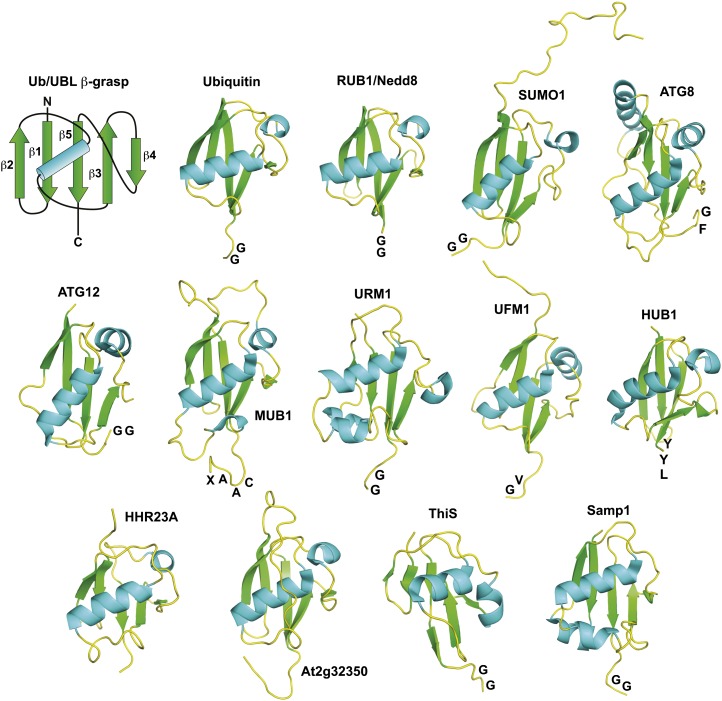
Ub and UBLs are characterized by a core β-grasp fold of approximately 70 amino acids that is phyletically widespread and of ancient origin (Burroughs et al., 2012). The fold contains secondary structure elements arranged in a ββαβββ order and is so named because the resulting five-stranded β-sheet appears to grasp the α-helix cradled diagonally (Fig. 1). Often, a short α-helix intervenes between the β4 and β5 strands. This compact arrangement generates a highly stable tertiary structure that folds rapidly and is often resistant to environmental perturbation such as heat. Sometimes elaborating this fold are additional α-helices, loops, and N-terminal extensions that likely endow unique binding surfaces to individual members. Whereas the Ub/UBLs often share little sequence identity, remarkable conservation is seen within each family across eukaryotes (e.g. Ub is 96%–97% identical among the plant, yeast, and animal kingdoms), strongly suggesting that most surfaces of each β-grasp fold participate in essential interactions during the reaction cycle of each modifier. With the exception of HUB1 and MUB, a short C-terminal sequence protrudes from the β-grasp core that ends upon proteolytic maturation with a Gly residue essential to each Ub/UBL reaction scheme (Fig. 1).
Figure 1.
Three-dimensional ribbon diagrams of the Ub/UBL β-grasp folds from eukaryotes and prokaryotes. Shown are the processed forms with the exposed C-terminal amino acid indicated. The exceptions are HHR23A and At2g32350, which are synthesized as domain fusions. At top left is a schematic of the Ub/UBL β-grasp showing the arrangement of the α-helix and β-strand secondary structures. The Protein Database accession numbers and references for the structures are as follows: yeast ATG8, 2KWC (Kumeta et al., 2010); Arabidopsis ATG12, 1WZ3 (Suzuki et al., 2005); yeast HUB1, 1M94 (Ramelot et al., 2003); Arabidopsis MUB1, 1SE9 (Downes et al., 2006); Arabidopsis RUB1, 1BT0 (Rao-Naik et al., 1998); H. volcanii Samp1, 3PO0 (Jeong et al., 2011); human SUMO1, 1AR5 (Bayer et al., 1998); E. coli ThiS, 1F0Z (Wang et al., 2001); Avena sativa Ub, 1UBQ (Vijay-Kumar et al., 1987); UBL domain from human HHR23A, 1P98 (Mueller and Feigon, 2003); human UFM1, 1WXS (Sasakawa et al., 2006); yeast URM1, 2AX5 (Xu et al., 2006); and the UBL domain from Arabidopsis At2g32350, 2KAN.
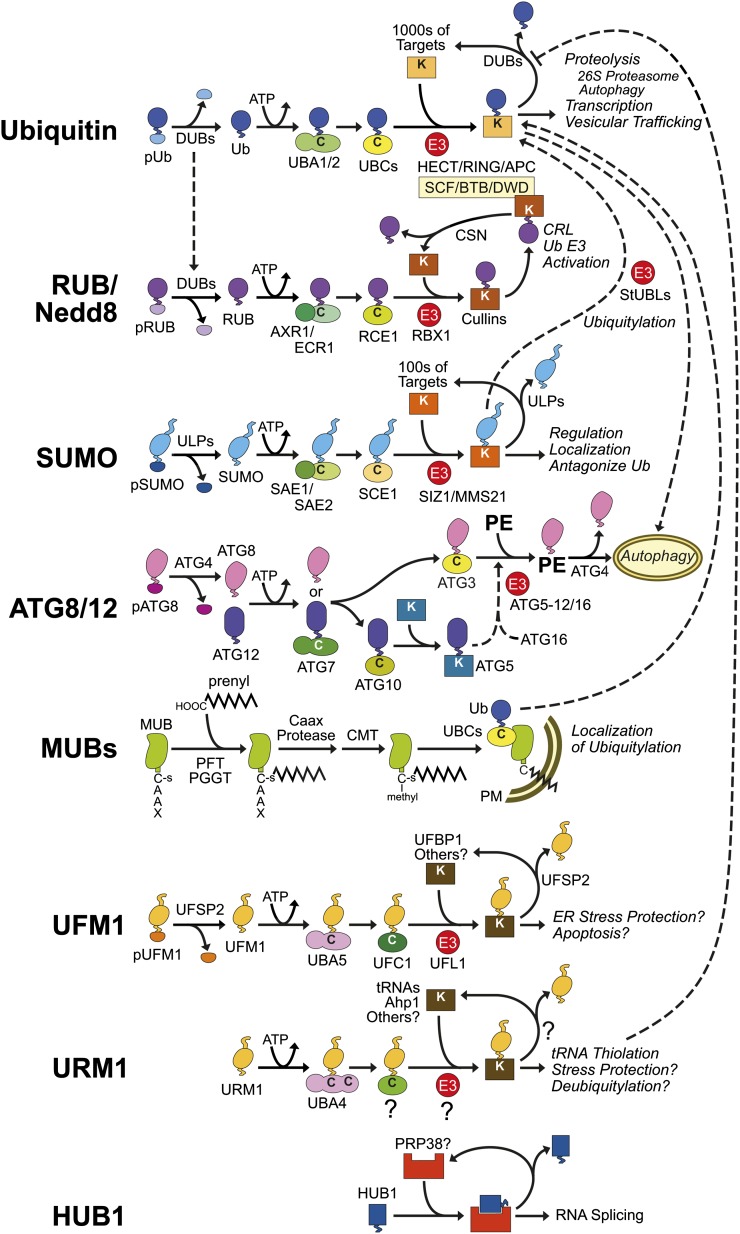
Ub and most UBLs enter an E1→E2→E3 reaction cascade that involves a signature high-energy thioester intermediate (Fig. 2). The cascade begins with activation of the Ub/UBL C-terminal Gly by an E1 that consumes ATP to first form an adenyl adduct followed by transfer of the Ub/UBL moiety onto an acceptor Cys sulfhydryl in the E1 (Smalle and Vierstra, 2004; Hochstrasser, 2009). This thioester product is then transferred to a unique Cys in an E2 via transthiolation before attachment to the target through an isopeptide bond. Although not universally required, the E3 has the crucial role of identifying appropriate targets and then spatially arranging Ub/UBL transfer. Most often, the ε-amine group in accessible Lys residues becomes conjugated to the C-terminal Gly, but at least for Ub, instances have been reported where the free N-terminal amino group or internal Cys, Ser, and Thr residues are acceptors (Iwai and Tokunaga, 2009; Shimizu et al., 2010; Okumoto et al., 2011). Like Ub, conjugation of most UBLs is transient and can be reversed by unique proteases that release the isopeptide-linked UBL moiety (Kerscher et al., 2006). These “deconjugases” are highly specific for the corresponding tag and fail to recognize or poorly recognize even closely related members (e.g. Ub and RUB), and in some cases they are highly selective for the modified target as well (Fig. 2).
Figure 2.
Schematic diagrams of the enzymology, targets, and function(s) of individual Ub/UBL pathways and known interactions among them (dashed lines). Where available, the protein names for the various components are indicated with the Arabidopsis nomenclature used if known. Question marks indicate components that might exist but have not been identified.
Interestingly, the appearance of Ub/UBL-specific deconjugases likely influenced by facultative evolution the unique arrangement of the corresponding UBQ/UBL genes, which often express precursors with C-terminal extensions of random sequence and, in the case of Ub and RUB, as head-to-tail polymers with varying numbers of monomers (Callis et al., 1990, 1995; Doelling et al., 2002; Kurepa et al., 2003). These novel genes presumably arose by mutation or unequal crossover but were genetically benign to Ub/UBL function given that the cognate deconjugases could easily correct the defect proteolytically following translation of the aberrant transcripts.
Ub AND ITS CENTRAL ROLE IN PROTEIN TURNOVER
Besides its founding status, both its depth of influence and striking genomic diversity easily place Ub at the top of the Ub/UBL pantheon. This β-grasp protein (Fig. 1) is synthesized by a complex assemblage of UBQ genes that express the full-length sequence fused either with other proteins or with varying numbers of Ub monomers concatenated head-to-tail, with the last monomer capped at its C terminus by extra amino acids. Following translation, the fusions are rapidly processed by DUBs to release functional Ub monomers with an exposed C-terminal Gly. In Arabidopsis (Arabidopsis thaliana), for example, 15 UBQ genes exist, with one expressing six Ub units linked in frame (Callis et al., 1995). Its E1→E2→E3 cascade is generated by a hierarchical organization of activities that ultimately reflects the myriad likely targets (Vierstra, 2009; Hua and Vierstra, 2011). Whereas Arabidopsis encodes for only two E1s and 37 E2s, it can theoretically express over 1,500 different E3s (Kraft et al., 2005; Stone et al., 2005; Hua and Vierstra, 2011). Remarkably, the vast number of predicted E3s in Arabidopsis is surpassed only by the number of transcription factors, thus illustrating the importance of ubiquitylation to plant biology.
A range of outcomes is possible for the conjugation cascade, including monoubiquitylation, the attachment of multiple Ubs to different Lys residues, or the assembly of poly-Ub chains in which any of the seven Ub Lys residues provides the site to link additional Ubs (Saracco et al., 2009; Komander and Rape, 2012). The architecture of the poly-Ub chains imbues additional information related to the fate of the modified protein. As examples, Lys-48 and Lys-11-linked chains commit proteins to degradation, while Lys-63 linked chains direct nonproteolytic outcomes related to DNA repair and endocytosis (Komander and Rape, 2012). The bound Ubs are then recognized by over 16 structurally distinct Ub recognition motifs, depending on the process (Harper and Schulman, 2006; Fatimababy et al., 2010). Numerous DUBs also exist to reverse ubiquitylation, with as many as 70 identified thus far in Arabidopsis (Yan et al., 2000; Vierstra, 2009). Whereas some DUBs help in general Ub recycling and poly-Ub disassembly (Doelling et al., 2001; Book et al., 2010), others uniquely act on specific ubiquitylated targets.
Proteomic studies in turn have identified at least 1,000 Arabidopsis proteins modified with Ub (Maor et al., 2007; Saracco et al., 2009; D.Y. Kim and R.D. Vierstra, unpublished data), strongly suggesting that many of the approximately 1,500 potential E3s are paired with specific targets. The lists of substrates include numerous nuclear proteins involved in chromatin structure, transcription, and RNA processing, enzymes that control rate-limiting steps in metabolism, ribosomal proteins and factors that effect translation, and transporters at the plasma membrane. Alignments of Ub attachment sites mapped by mass spectrometry from human cell cultures and Arabidopsis failed to identify even weak consensus sequences, except for an overrepresentation of acidic residues (Kim et al., 2011, D.Y. Kim and R.D. Vierstra, unpublished data), strongly suggesting that the information that directs Ub ligation is outside the primary sequence context of the affected Lys(s).
Based on both genetic and biochemical studies, E3s represent the key factors that determine substrate specificity. To date, three main E3 types are known in plants based on their mechanisms of action and subunit organization: HECT (for homology to E6-AP C terminus), RING (for really interesting new gene)/U-box, and the CRL (for Cullin-RING ligases) E3s, with the CRLs further divided into four subtypes (Downes et al., 2003; Mudgil et al., 2004; Stone et al., 2005; Lee et al., 2008; Lima et al., 2010; Hua and Vierstra, 2011). The assembly, activity, and specificity of each CRL is defined by the combinatorial assembly of adaptors that recruit appropriate substrates and the RBX1 protein that tethers the activated Ub-E2 intermediate onto one of several Cullin scaffolds. This assembly is dynamically controlled by a modification cycle requiring another β-grasp protein, RUB (see below). Substrate recognition by the adaptors is accomplished by a diverse array of protein-protein-binding domains that often look and act like “catcher’s mitts,” including familiar domains such as Leu-rich, Kelch, tetratricopeptide, and armadillo repeats, and MATH, ankryin, NPH3, Jumonji C, and Tubby domains (Gagne et al., 2002; Figueroa et al., 2005; Gingerich et al., 2005; Stone et al., 2005; Hua et al., 2011).
In line with the vast number of targets, the consequences of Ub addition are highly diverse. The most common is to commit ubiquitylated proteins to breakdown by the 26S proteasome, a 2.5-MD proteolytic machine structurally conserved among eukaryotes (Shibahara et al., 2002; Finley, 2009; Book et al., 2010). It is assembled from two particles, a 28-subunit 20S core protease (CP) and an 18-or-more-subunit 19S regulatory particle that caps one or both ends of the CP. The CP is a hollow cylinder that houses the protease active sites internally; this chamber is accessed by a gated axial channel that restricts entry to only those proteins that are deliberately unfolded and threaded inside (Finley, 2009). The regulatory particle has a set of Ub-binding receptors (RPN1, RPN10, and RPN13) that recognize appropriate substrates, particularly those modified with Lys-48 and Lys-11 poly-Ub chains, several associated DUBs that rescue the Ub moieties before target degradation, and a hexameric ring of AAA-ATPase activities that opens the CP gate and unfolds and imports substrates. A number of accessory proteins present at substoichiometric levels are also essential to the assembly of the 26S proteasome, recruitment of targets, and regulation of its proteolytic activity (Finley, 2009; Book et al., 2010).
Through the ubiquitin/26S proteasome system (UPS), Ub controls many facets of plant growth, development, and cytoplasmic and nuclear housekeeping. Examples include signaling by most, if not all, of the major plant hormones, the cell cycle, circadian rhythms, photomorphogenesis, floral and leaf homeosis, self-incompatibility, defense against pathogens, and survival during environmental stress (Dreher and Callis, 2007; Vierstra, 2009; Hua and Vierstra, 2011). Notably, Ub E3s have been shown to serve as receptors for the hormones auxin (Tan et al., 2007) and jasmonic acid (Sheard et al., 2011) as well as to provide numerous control points in the pathways required for the synthesis and/or perception of ethylene, abscisic acid, salicylic acid, strigolactone, and GA (Dreher and Callis, 2007; Santner and Estelle, 2010; Hua and Vierstra, 2011). The continuing arms race between plant pathogens and their hosts also exploits the UPS. While the host synthesizes E3s to ward off infection by ubiquitylating and removing pathogenic factors injected inside, the pathogen retaliates by injecting proteins and small molecule inhibitors that disregulate ubiquitylation and the UPS to favor pathogen invasion (Vierstra, 2009).
Additional roles for Ub outside of the 26S proteasome are also prominent and include the regulation of various DNA functions and trafficking proteins to their appropriate destinations. Studies with yeast and mammalian cells revealed that reversible ubiquitylation of core and linker histones is part of the code that controls nucleosome assembly and thus the accessibility of genes to transcriptional regulators (Weake and Workman, 2008), while the modification of various DNA repair factors such as proliferating cell nuclear antigen appears critical for fixing DNA breaks induced by genotoxic stress (Ulrich and Walden, 2010). H2B ubiquitylation/deubiquitylation is driven in Arabidopsis by the RING E3 BRE1 (or HUB1) and the UBP26 DUB, with null mutants displaying severe problems in the cell cycle, ploidy, transcription, and small interfering RNA-mediated gene silencing (Fleury et al., 2007; Liu et al., 2007; Sridhar et al., 2007). Moreover, the action of at least some transcription factors may require cycles of ubiquitylation to promote each round of transcriptional initiation (Geng et al., 2012). A number of membrane-bound receptors and transporters are also strongly ubiquitylated upon ligand engagement (Mukhopadhyay and Riezman, 2007). This modification involves either monoubiquitylation or polyubiquitylation via Lys-63-linked chains and appears to control signaling by cycling the receptors to internal compartments and/or directing their breakdown by the vacuole/lysosome. In this regard, the Arabidopsis DUB AMSH3 may be particularly important for ESCRT-mediated protein sorting (Katsiarimpa et al., 2011). As will be described below, ubiquitylation of supramolecular complexes and organelles too large for the proteasome may be widely used by plants to direct autophagic turnover (Li and Vierstra, 2012).
REGULATION OF CRL Ub E3S BY RUB
RUB (or Nedd8 in yeast and animals) was first discovered as part of a novel gene (UBQ7 in Arabidopsis) that expresses Ub appended in frame to a closely related polypeptide (55% amino acid sequence identity) that is released upon translation (Callis et al., 1995). X-ray crystallography studies of a free RUB revealed a β-grasp fold that is nearly identical to that of Ub, including a C-terminal extension ending in a di-Gly (Fig. 1). Subsequent genetic analyses of Arabidopsis RUBs (designated RUB1–RUB3) uncovered a distinct but chemically similar conjugation pathway with the surprising finding that its main, if not exclusive, targets are the Cullin subunits from CRL E3s, thus intimately connecting rubylation to ubiquitylation (del Pozo and Estelle, 1999; Hakenjos et al., 2011; Fig. 2). In fact, it is now clear that RUB is part of two dueling cycles that dynamically navigate CRL complexes through assembly, conjugation, and disassembly steps as they ubiquitylate substrates (Hotton and Callis, 2008; Hua and Vierstra, 2011).
One cycle employs CAND1 (for Cullin-associated Nedd8-dissociated1) as a clamp to maintain a pool of disassembled CRL complexes by binding reversibly to the Cullin-RBX1 subcomplex in a way that inhibits the association of Cullin-RBX1 with substrate adaptors in the absence of cognate targets. Possibly in the presence of a substrate-occupied adaptor, the Cullin scaffold becomes rubylated at a particular Lys by an E1→E2→E3 reaction cascade involving the heterodimeric E1 encoded by the AXR1/AXR1-LIKE and ECR1 genes and the RCE1 E2. The RBX1 subunit in association with the Cullin is thought to provide the E3 activity that directs RUB transfer. The conjugated RUB occludes CAND1 binding to the Cullin, which in turn allows the CRL to enter a second cycle, where the substrate-bound adaptor binds to Cullin-RBX1 followed by substrate ubiquitylation. After release of the appropriately ubiquitylated protein, the eight-subunit COP9/signalosome (CSN) enters to remove the RUB moiety using the deconjugase activity provided by the CSN5 subunit (Lyapina et al., 2001; Zhang et al., 2008). Finally, the CRL is disassembled and CAND1 rebinds to Cullin-RBX1.
Paradoxically, the opposing CAND1/RUB cycles are both essential for proper CRL activity, likely by maintaining a revolving pool of uncommitted Cullin-RBX1 scaffolds despite the presence of hundreds of waiting substrate adaptors (Hua and Vierstra, 2011). Accordingly, Arabidopsis mutants missing any components in the RUB scheme are lethal while weaker alleles have pleiotropic effects on plant growth and development and compromised UPS substrate turnover (del Pozo et al., 2002; Schwechheimer et al., 2002; Feng et al., 2004; Zhang et al., 2008).
Recent proteomic studies ectopically expressing tagged versions of RUB/Nedd8 in Arabidopsis and mammals reported other potential targets of rubylation besides Cullins, including numerous ribosomal subunits (Jones et al., 2008; Xirodimas et al., 2008; Hakenjos et al., 2011). Whether these represent authentic conjugates remains to be confirmed, given the observations that human Nedd8 will inappropriately enter the ubiquitylation system when in excess or when Ub is depleted (Kim et al., 2011).
SUMO AND NUCLEAR ACTIVITIES ASSOCIATED WITH PLANT STRESS DEFENSE
SUMO was first discovered in animals from studies on the nuclear pore protein RanGAP, which upon sequencing was found to have two N termini but only one C terminus, indicative of one protein attached to another (Matunis et al., 1996). The appended protein was named SUMO for its weak sequence homology to Ub (12% identity) and was later confirmed as a UBL by its β-grasp fold and analogous conjugation chemistry. However, SUMO differs from other UBLs by a long N-terminal extension that appears unstructured in solution (Fig. 1). Plants and animals typically express several SUMO isoforms (four in Arabidopsis) with evidence for subfunctionalization (Kurepa et al., 2003; van den Burg et al., 2010). The sumoylation pathway in Arabidopsis includes a heterodimeric E1 encoded by SAE1a/b and SAE2 genes, a single E2 SCE1, at least two E3s encoded by the SIZ1 and HYP2/MMS21 loci, and a family of desumoylating proteases (Fig. 2; Kurepa et al., 2003; Colby et al., 2006; Miura et al., 2007a). These desumoylating proteases not only disassemble SUMO conjugates, they are needed to generate mature SUMO by trimming extra amino acids beyond the C-terminal di-Gly. Whereas most targets bear a single SUMO moiety, some can be modified with multiple SUMOs or poly-SUMO chains via iterative cycles of conjugation analogous to ubiquitylation (Miller et al., 2010; Wilkinson and Henley, 2010). Based on alignments of known SUMO attachment sites, sumoylation appears to employ several loose consensus motifs; one prevalent in plants, yeast, and mammals encompasses a ΨKXE motif (where Ψ is a large hydrophobic residue and K is the Lys where SUMO is linked), and two others identified thus far in mammals contain extended motifs rich in phosphorylated or negatively charged residues (Miller et al., 2010; Wilkinson and Henley, 2010).
The functions of sumoylation are diverse and mostly nonproteolytic and include controls on localization, interaction, and activity of the modified protein (Miura et al., 2007a; Wilkinson and Henley, 2010). One unique situation has also been reported in mammalian cells, where SUMO addition protects the IκBα protein from Ub addition by blocking accessible Lys residues (Desterro et al., 1998). Some of these effects are mediated by a collection of binding partners bearing SUMO-interacting motifs.
Particularly important to plants is the role of SUMO in stress defense. The first connection came from the discovery that sumoylation is strongly up-regulated by various types of stress, including heat shock, freezing, drought, ethanol, oxidants, amino acid analogs, and pathogen exposure (Kurepa et al., 2003; Saracco et al., 2007; Conti et al., 2008). In fact, a dramatic rise in SUMO conjugates can be detected within minutes after subjecting Arabidopsis seedlings to a mild heat shock (37°C), making it one of the fastest stress responses observed, and is so strong that it consumes almost all available free SUMO. This stress-induced sumoylation is reversible, such that a few hours after the stress most conjugates are disassembled and the free SUMO pool is regenerated, and has a “memory” that prevents a second burst if too close in time to the first (Kurepa et al., 2003; Saracco et al., 2009; Miller et al., 2012).
The second connection of SUMO to stress came from phenotypic analysis of Arabidopsis pathway mutants. Whereas null mutants for the two main SUMOs (SUMO1 and -2), the E1 subunit SAE2, and the E2 SCE1 are embryo lethal (Saracco et al., 2007), mutants missing the SIZ1 E3 display pleiotropic stress phenotypes, including a hypersensitivity to heat, cold, drought, salt, and low phosphate and nitrogen, a hyposensitivity to pathogens, and altered signaling by the stress hormones abscisic acid and salicylic acid (Miura et al., 2005, 2007b, 2009; Lee et al., 2007; Conti et al., 2008; van den Burg et al., 2010; Park et al., 2011).
At present, it is unclear how SUMO helps plants survive adverse environments. Clues have come from that fact that most sumoylated proteins are nuclear (Saracco et al., 2007) and recent proteomic and interactome analyses that uncovered a deep catalog of SUMO targets and SUMO-interacting proteins (Elrouby and Coupland, 2010; Miller et al., 2010). Included in the list of approximately 350 conjugates are a “who’s who” of key transcriptome regulators and chromatin modifiers, including histones H1 and H2B, histone acetyl- and SET-domain-containing histone-methyl transferases, SAGA complex subunits, proteins involved in RNA-directed DNA methylation, SWI/SNF chromatin remodelers, DNA repair components, TOPLESS-type corepressors, numerous transcription factors required for developmental- and/or stress-induced gene expression, and nuclear pore proteins. Collectively, these targets imply that SUMO is crucial to chromatin architecture, transcription, cytoplasm/nuclear partitioning, and RNA retention, especially during stress (Miller et al., 2010; Meier, 2012). Subsequent quantitative proteomic studies on heat, oxidative, and ethanol stress discovered that proteins involved in RNA biology are some of the most robustly sumoylated targets during stress (as much as 15-fold within 30 min of heat shock), suggesting that RNA-related processes are particularly affected (Miller et al., 2012). From studies of individual SUMO targets outside of plants, it is noteworthy that only a small proportion of a substrate needs to be sumoylated to achieve maximal effect (Miura et al., 2007a; Wilkinson and Henley, 2010). This enigma implies that sumoylation does not activate/inactivate proteins per se but may drive cycles critical for their functions (e.g. assembly/disassembly of transcription complexes or nuclear/cytoplasm shuttling).
Surprisingly, proteomic studies with yeast, mammalian cell cultures, and Arabidopsis also discovered that a subset of sumoylated proteins becomes ubiquitylated during stress, with evidence that Ub can be directly linked to the SUMO moieties (Uzunova et al., 2007; Miller et al., 2010, 2012; Tatham et al., 2011). This dual modification implies a cross talk between the Ub and SUMO systems along with the added possibility that the bound SUMOs act as secondary degrons for directing UPS-mediated turnover. In yeast and metazoans, this Ub addition is driven by a family of SUMO-targeted Ub ligases containing both a SUMO-interacting motif and a RING motif (Sun et al., 2007; Uzunova et al., 2007). Possible SUMO-targeted Ub ligase orthologs are apparent in various plant genomes, including at least three in Arabidopsis.
ROLES OF ATG8 AND ATG12 IN DIRECTING AUTOPHAGIC RECYCLING
Autophagy, or “self-eating,” represents a major recycling route in eukaryotes whereby cytoplasmic constituents and organelles are sequestered in small double membrane-bound vesicles, which are then deposited into the vacuole (or lysosome in animals) for eventual breakdown by resident hydrolases (Bassham, 2009; Li and Vierstra, 2012). Even though autophagy was described in morphological terms decades ago, only recently have the underpinning mechanisms become apparent. Particularly informative were genetic screens of yeast and Pichia pastoris designed to discover loci required for autophagic transport (Klionsky, 2007). In addition to identifying various kinase signaling components required to direct autophagy during nutrient starvation and factors that promote hemifusion of the resulting vesicles with the tonoplast, these screens unexpectedly discovered a pair of UBLs named ATG8 and ATG12 whose conjugation is essential for autophagy (Ohsumi, 2001). Both ATG8 and ATG12 are faintly similar to Ub (8% and 5% amino acid sequence identity for the Arabidopsis versions [Doelling et al., 2002]) but contain obvious β-grasp folds that are decorated with additional helices, loops, and N-terminal extensions (Fig. 1). ATG12 is translated in its mature form with a protruding Gly and thus does not need processing by a deconjugase. ATG8, on the other hand, is typically synthesized with a variable C-terminal extension that must be removed by the ATG4 protease to expose its Gly.
Both ATG8 and ATG12 are activated by the same E1 (ATG7), but the thioester intermediates are transferred to separate E2s, ATG3 and ATG10, respectively (Fig. 2). ATG12 is then conjugated through an isopeptide bond to a single protein target, ATG5, without the assistance of an E3. In a unique twist, ATG8 is attached instead to the amine group of the lipid phosphatidylethanolamine (PE) and becomes membrane bound (Ohsumi, 2001; Klionsky, 2007). The E3 driving PE conjugation is composed of the ATG12-ATG5 conjugate in association with the dimeric ATG16 protein, thus generating an unusual scheme in which one UBL conjugate directs modification by another UBL (Fujioka et al., 2008; Chung et al., 2010). Toward the end of autophagic vesicle assembly, the ATG12-ATG5 conjugate dissociates from the membrane, while the ATG8 moiety lining the outer surface is delipidated by ATG4 and released. The ATG8-PE adduct bound to the inner surface enters the vacuole and is eventually consumed.
Why the ATG autophagic system culminates in generating the ATG8-PE adduct remained a puzzle until recently. A key breakthrough was the realization that ATG8, upon coating autophagic membranes, provides a docking platform to recruit other factors needed to build the engulfing vesicle and encapsulate cargo (Noda et al., 2010; Johansen and Lamark, 2011). Humans alone express almost 70 different ATG8-interacting proteins (Behrends et al., 2010). These factors are loosely defined by a signature ATG8-interacting motif (AIM) with a consensus core W/Y/F-XX-L/I/V sequence that tightly binds a hydrophobic cleft in ATG8 (Noda et al., 2008). Importantly, a number of AIM proteins have been identified that bind specific protein cargo and organelles, thus revising our appreciation of autophagy from one committed to bulk degradation to one that selectively recycles. Two AIM proteins of particular interest in mammals are SQSTM1 (for sequestosome1) and NBR1 (for neighbor of BRAC1-1), which have the capacity to bind ubiquitylated proteins through ubiquitin-associated (UBA) domains (Johansen and Lamark, 2011). This connection appears to provide an alternative recycling route for ubiquitylated proteins too large for the 26S proteasome, such as protein aggregates, large macromolecular complexes, and even organelles and invading pathogens following ubiquitylation of their surface proteins.
Descriptions of ATG components in Arabidopsis revealed that a remarkably similar autophagic system exists in plants (Thompson and Vierstra, 2005; Bassham, 2009; Li and Vierstra, 2012), including the presence of an NBR1 ortholog (but not SQSTM1) that connects the autophagic and ubiquitylation systems (Svenning et al., 2011; Zientara-Rytter et al., 2011). Genetic analyses showed that although ATG-mediated autophagy is not essential to plants, it is important for natural senescence, survival during nutrient starvation, and pathogen defense (Doelling et al., 2002; Hanaoka et al., 2002; Liu et al., 2005; Thompson et al., 2005; Hofius et al., 2009; Lenz et al., 2011). ATG-mediated autophagy also has been connected to chloroplast and mitochondria turnover (Wada et al., 2009; F. Li and R.D. Vierstra, unpublished data), the removal of oxidized or aggregated proteins (Toyooka et al., 2006; Xiong et al., 2007), the clearing of cytotoxic heme metabolites (Vanhee et al., 2011), and possibly the elimination of defective ribosomes and proteasomes (Hillwig et al., 2011; R.M. Marshal and R.D. Vierstra, unpublished data).
The main organizational difference is the fact that many plant ATG components are encoded by small gene families as opposed to single genes in yeast. For example, Arabidopsis expresses a pair of ATG12 proteins and nine ATG8 isoforms that have subfunctionalized expression patterns (Doelling et al., 2002; Hanaoka et al., 2002). Moreover, two ATG8 isoforms lack C-terminal extensions and thus act without ATG4 intervention. The ATG12-ATG5 conjugate appears to be constitutively synthesized at high levels (Thompson et al., 2005; Phillips et al., 2008). Conversely, the levels of the ATG8-PE adduct are more dynamic, being present at low levels in nonstressed plants and then accumulating to high levels in senescing leaves and in plants deprived of nitrogen or fixed carbon (Chung et al., 2009, 2010; Suttangkakul et al., 2011). Following starvation, numerous ATG8-decorated autophagic vesicles can be seen in the vacuole, implying that cytoplasmic recycling by this route is substantial (Yoshimoto et al., 2004; Thompson et al., 2005).
MUBS
MUBs were added to the UBL collection based on structural studies that discovered an obvious β-grasp fold in an Arabidopsis member (Downes et al., 2006). However, MUBs uniquely differed from the other UBLs by long N- and C-terminal loops and by ending not in a protruding Gly but in an invariant CAAX motif (Fig. 1). CAAX motifs are essential signatures for prenylation, a membrane-anchoring modification in which the Cys becomes conjugated with 15- or 20-carbon-long farnesyl or geranylgeranyl moieties by two distinct families of heterodimeric prenyl transferases, PFT and PGGT, respectively. The prenylated protein is proteolytically trimmed by a CAAX protease to expose the Cys, and its carboxyl group is subsequently methylated by a prenyl Cys methyltransferase (Crowell and Huizinga, 2009; Fig. 2). Sequence database searches subsequently found MUBs in other plant species, animals, and filamentous fungi but not in any unicellular fungi or prokaryotes (Downes et al., 2006). In vitro assays, inhibitor studies, and biochemical analyses of prenyl transferase mutants confirmed that most, if not all, MUBs are substrates for prenylation and in some cases also become palmitoylated. At least in Arabidopsis, the MUBs then become specifically anchored to the plasma membrane (Downes et al., 2006).
The function(s) of MUBs are not yet clear, as the presence of gene families has complicated genetic analyses. Of likely relevance are the observations that Arabidopsis MUBs selectively bind a subfamily of Ub E2s that then dock these E2s to the plasma membrane in planta (Dowil et al., 2011). Consequently, MUBs like RUB, SUMO, and ATG8/12 (Fig. 2) might intersect with the Ub system, in this case by recruiting specific components of the Ub conjugation machinery to the plasma membrane, presumably to promote the ubiquitylation of similarly localized targets.
URM1
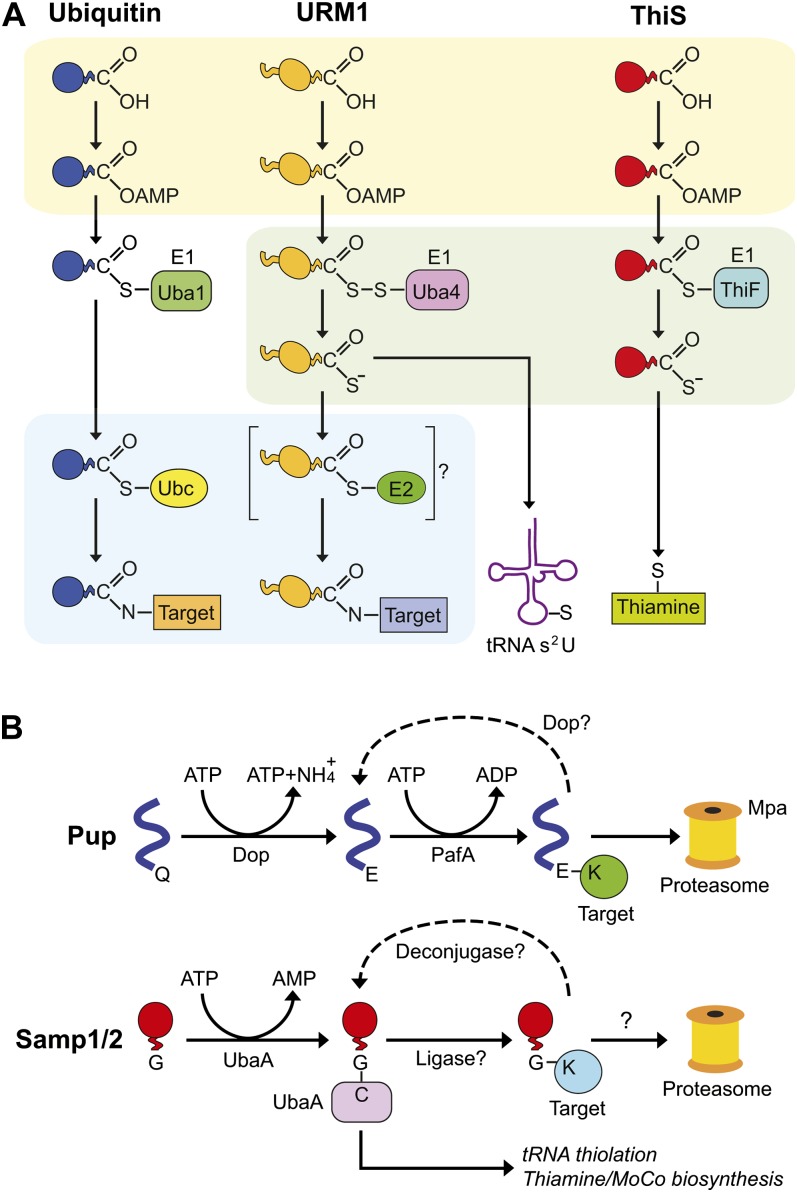
The emerging picture of URM1 is that it represents an evolutionary bridge between the ancient roles of UBLs in prokaryotic sulfur chemistry (see below) and the presumably more advanced roles of Ub and UBLs as protein modifiers (Fig. 3A). Unlike several others in the Ub/UBL family, URM1 is synthesized in its mature form with an exposed di-Gly sequence protruding from its β-grasp fold (Fig. 1). Studies with yeast and mammalian URM1 showed that it is activated by a novel E1 (UBA4) that comprises a fusion between the canonical E1 that adenylates the C terminus of Ub/UBLs and a rhodanese domain that directs sulfur transfer (Furukawa et al., 2000; Leidel et al., 2009; Van der Veen et al., 2011). By a two-Cys reaction cascade much like those employed by the Escherichia coli ThiS and MoaD sulfur transferases used for thiamine and molybdenum cofactor (MoCo) synthesis, respectively, URM1 is first adenylated, then forms an acyl disulfide intermediate with UBA4, before final release to generate an activated URM1 bearing a C-terminal Gly thiocarboxylate (Figs. 2 and 3A). This URM1 thiocarboxylate then participates in two sets of modifications. One likely more ancestral reaction donates the sulfur moiety to the wobble uridine in several transfer RNAs (tRNAs; Leidel et al., 2009). The second involves attachment of the thiocarboxylated URM1 to protein targets through an isopeptide linkage with free Lys residues concomitant with the release of the sulfur moiety (Goehring et al., 2003; Van der Veen et al., 2011). Whether E2, E3, or deurmylating activities also participate in the URM1 pathway is not yet known.
Figure 3.
Evolution of the eukaryotic Ub/UBL modification pathways from prokaryotic reaction schemes. A, The reaction mechanisms for E. coli ThiS involved in sulfur transfer chemistry, yeast UFM1 involved in tRNA thiolation and protein modification, and Ub involved in protein modification. B, Schematics for the prokaryotic conjugation pathways involving H. volcanii Samp1/2 and M. tuberculosis Pup. A was adapted from Petroski et al. (2011).
tRNA thiolation by URM1 is proposed to stabilize codon-anticodon interactions on ribosomes to improve translation efficiency (Leidel et al., 2009). Why proteins would be urmylated is not yet clear. Proteomic studies have identified only a handful of targets, including in yeast the peroxiredoxin Ahp1 and, in human cell lines, other components of the tRNA thiolation pathway, the cellular apoptosis susceptibility factor involved in nuclear transport, and several DUBs, thus providing another link between Ub and UBLs (Goehring et al., 2003; Van der Veen et al., 2011). Intriguingly, protein urmylation dramatically rises upon exposing yeast and mammalian cells to oxidative stress, and urmylation mutants are more sensitive to nutrient starvation, oxidants, and heat shock (Furukawa et al., 2000; Goehring et al., 2003; Leidel et al., 2009; Van der Veen et al., 2011).
At present, little is known about urmylation in plants, and no protein targets have been reported. Arabidopsis contains two active genes encoding URM1, with 37% to 39% identity to their yeast counterpart. Nakai et al. (2012) recently showed that one Arabidopsis isoform but the not the other participates in cytosolic tRNA thiolation and can functionally replace yeast URM1. In a search for Arabidopsis mutants resistant to sirtinol, a small molecular activator of auxin-inducible genes and a promoter of auxin-related developmental phenotypes, an ortholog of the UBA4 E1 (called SIR1) was unexpectedly discovered (Zhao et al., 2003). Subsequent sirtinol screens identified additional loci involved in sulfur transfer and MoCo synthesis, suggesting that the sirtinol resistance of UBA4/SIR1 loss-of-function plants is caused by compromised MoCo assembly (Dai et al., 2005); however, additional effects on protein urmylation cannot be ruled out.
UFM1
UFM1 closely follows the Ub enzymatic paradigm, but knowledge of its corresponding targets and associated functions in any eukaryote is lacking. Even with only 14% amino acid sequence identity to Ub, UFM1 adopts a remarkably similar β-grasp fold (Fig. 1), and like several other UBLs, it is synthesized as a precursor (seven residues longer for Arabidopsis UFM1) that requires processing by the UFSP2 protease to expose the functioning C-terminal Gly (Komatsu et al., 2004). Ufmylation as deduced in mammalian cells involves an E1 (UBA5), a single E2 (UFC1), and only one known E3 (UFL1), which will conjugate UFM1 to targets both in vivo and in vitro (Fig. 2; Komatsu et al., 2004; Tatsumi et al., 2010). Although driving the same biochemical reactions, both UFC1 and UFL1 have little sequence in common with the E2s and E3s from other Ub/UBL conjugation systems, implying that they work differently. Biochemical studies with mammalian cells suggest a limited number of UFM1 substrates; currently, the only bona fide target is the endoplasmic reticulum-resident protein UFBP1, which is modified at one or more Lys residues via isopeptide linkages (Tatsumi et al., 2010; Lemaire et al., 2011).
Comparable ufmylation pathways, including the UFBP1 target, can be readily found encoded in the genomes of other metazoans and in plants such as Arabidopsis but not in yeast or Schizosaccharomyces pombe, suggesting a unique role for this UBL in multicellular organisms. Uba5−/− mice are embryonic lethal, possibly caused by defective erythroid development (Tatsumi et al., 2011), and cultured human cells silenced for Ufm1 and Ufl1 show accelerated apoptosis (Lemaire et al., 2011), possibly due to heightened endoplasmic reticulum stress. Little is currently known about the phenotypic functions of the UFM1 pathway in plants.
HUB1
Despite being nearly identical to Ub structurally (Fig. 1), the HUB1 proteins are the most unconventional members of the UBL family. They invariably terminate in a di-Tyr sequence followed by a nonconserved amino acid, all of which remains linked to the active polypeptide (Dittmar et al., 2002). Rather then becoming covalently attached to other proteins, studies using yeast and mammalian cells show that HUB1 likely acts noncovalently, and accordingly no E1s, E2s, E3s, or processing enzymes have been identified for HUB1 in any eukaryote (Fig. 2). Genetic analyses of yeast HUB1 revealed roles in polarized growth, cell cycle progression, and the mitochondrial unfolded protein response, which might be collectively manifested by defective mRNA splicing (Dittmar et al., 2002; Mishra et al., 2011). In particular, HUB1 appears essential for cleaving noncanonical 5′ splice sites through interactions with the spliceosomal tri-small ribonucleoprotein particle, which in Arabidopsis likely involves binding to the Prp38 subunit (Mishra et al., 2011). Whereas Δhub1 mutants in yeast are viable, consistent with its strong avoidance of alternative splicing, S. pombe Δhub1 mutants are lethal (Wilkinson et al., 2004; Mishra et al., 2011). Arabidopsis contains two loci that encode HUB1 proteins with 95% amino acid sequence identity; one (At5g42300) shows strong expression, whereas the other (At3g45180) is expressed poorly, if at all, and may be a pseudogene (A. Lomax and R.D. Vierstra, unpublished data). Whether HUB1 is essential to Arabidopsis or other plants is not yet known.
UBL DOMAIN FUSIONS
In line with the versatile roles of β-grasp domains in promoting protein-protein interactions or encouraging protein folding, numerous proteins have been found that contain a Ub/UBL domain translationally linked to other domains, thus skirting the need for subsequent covalent attachment. In the Arabidopsis genome alone, over 50 loci are predicted to encode proteins with sequence homology to Ub, and where examined, these domains assume an obvious β-grasp fold (e.g. At2g23250 [Fig. 1]). Two essential ribosomal proteins are expressed only as Ub fusions (Arabidopsis UBQ1/2 and UBQ3/4 loci) but then are rapidly processed cotranslationally to release the ribosomal proteins and Ub in free, functional forms (Callis et al., 1990). In yeast, this cosynthesis dramatically elevates expression, presumably by exploiting the efficient folding of Ub to encourage folding of the more recalcitrant ribosomal subunit sequences downstream (Finley et al., 1989).
Examples of stable UBL fusions in Arabidopsis include the UBL-UBA family (RAD23 [HHR23A in humans], DSK2, DDI1, NUB1, and UBL1 [Farmer et al., 2010]), the Ub HECT E3 UPL5, the proteasome-associated deubiquitylating enzyme UBP6, and the RNA splicing factor 3A, as well as a number of uncharacterized proteins (e.g. At2g23250). To prevent inadvertent release by deconjugases, these UBL domains lack the C-terminal Gly. Members of the UBL-UBA family are uniquely organized in having an N-terminal UBL domain followed by one or more UBA domains that recognize Ub (Farmer et al., 2010; Fatimababy et al., 2010). By simultaneously binding ubiquitylated proteins via the UBA domain and the proteasomal receptors RPN1, RPN10, or RPN13 through the UBL domain, these proteins chaperone Ub targets to the 26S complex for turnover (Fu et al., 2010).
EVOLUTION OF THE Ub/UBL SUPERFAMILY
A long-standing question has been the evolutionary origins of these complex Ub/UBL systems. A major breakthrough was the discovery that the prokaryotic pathways directing the acquisition of inorganic sulfur during thiamine and MoCo biosynthesis use the structurally related β-grasp proteins, ThiS and MoaD (Lake et al., 2001; Wang et al., 2001), which also share common adenylate/thioester intermediates generated by E1-related enzymes, ThiF and MoaE, respectively (Figs. 1 and 3A). It is easy to imagine that the URM1 pathway evolved directly from these sulfur transferase reactions and thus represents the ancestral bridge between sulfur chemistry and protein conjugation (Fig. 3A).
Remarkably, as in eukaryotes, it also appears that prokaryotes further elaborated upon the ThiS/MoaD reaction scheme to generate their own protein-tagging systems, some of which might incorporate E1→E2→E3 conjugation cascades as well as deconjugases to reverse attachment (Hochstrasser, 2009; Nunoura et al., 2011; Burroughs et al., 2012). Where fully understood, conjugation also directs turnover of the modified protein. One recently discovered in Archaea species such as Haloferax volcanii revolves around two β-grasp proteins, Samp1 (for small archaeal modifier protein1) and Samp2 (Fig. 1), and their corresponding E1 UbaA (Humbard et al., 2010; Miranda et al., 2011). Samp1 and Samp2 have retained their participation in thiamine/MoCo and tRNA thiolation much like ThiS/MoaD and URM1, respectively (Fig. 3B). And like URM1, they also become isopeptide linked via their di-Gly C terminus to a collection of protein substrates by a streamlined pathway absent of E2 or E3 activities. Sampylation affects numerous targets in H. volcanii, including self-modification, to form poly-Samp chains much like eukaryotic Ub and SUMO (Humbard et al., 2010). The consequences of Samp1/2 addition are not yet known; that the levels of Samp1/2 conjugates rise during nitrogen starvation and are possibly turned over by a stripped-down archaeal version of the proteasome imply a role in amino acid recycling during nutrient deprivation (Humbard et al., 2010).
An intriguing variation on the theme is Pup (for prokaryotic Ub-like protein) found in a number of actinobacteria and nitrospira, including Mycobacterium tuberculosis (Pearce et al., 2008; Wang et al., 2010). Pup is structurally unrelated to β-grasp proteins and is in fact intrinsically disordered. However, like Ub/UBLs, M. tuberculosis Pup becomes conjugated to proteins and even directs their degradation by a primitive eubacterial proteasome consisting of just the CP capped by a ring of the AAA-ATPase protein Mpa, which serves as the Pup receptor (Wang et al., 2010). Surprisingly, the conjugation chemistry is markedly distinct from those used by Ub/UBLs, suggestive of convergent evolution (Fig. 3B). The C-terminal Gln residue of Pup is first deamidated by the Dop deamidase, and then Pup is isopeptide linked via the resulting Glu γ-carboxyl group to accessible target Lys residues using a Gln synthetase-like PafA ligase (Striebel et al., 2009). Interestingly, Dop also has depupylating activity that can reverse Pup addition (Burns et al., 2010). The extensive catalog of Pup substrates from M. tuberculosis and Mycobacterium smegmatis imply significant proteolytic roles for this modifier in metabolism, respiration, and adaptation (Pearce et al., 2008; Poulsen et al., 2010).
CONCLUSION
Certainly, the expanding collection of Ub/UBL proteins and their plethora of targets place these polypeptide modifiers at the center of many aspects of plant biology. In fact, based on the sheer number of associated components and phenotypic impact, Ub and the UPS are rivaled only by transcription factors and protein kinase cascades as the dominant regulators of plant growth, development, and survival. It is also intriguing that while these Ub/UBLs have substantially diversified with respect to the biological processes they control and their underpinning enzymology, several points of cross talk have evolved that connect some Ub/UBLs to each other, particularly between Ub and RUB, Ub and SUMO, Ub and MUBs, ATG8 and ATG12, and Ub and autophagy mediated by ATG8/12. Given the facts that (1) animals have UBLs (ISG15 and FAT10 [Hochstrasser, 2009]) not yet found in plants, (2) more UBLs will likely emerge in prokaryotes, and (3) our limited structural knowledge of most plant proteins, it is highly possible that more plant UBLs will be forthcoming. Furthermore, using bacterial Pup for illustration, it is also possible that new polypeptide modifiers will emerge that are independent of the hallmark β-grasp fold. Consequently, we should expect that the known functional space encompassed by these polypeptide modifiers in the plant realm will continue to increase.
Acknowledgments
I thank past and present members of the Vierstra laboratory for helpful discussions and for generating the data described herein from our laboratory. Special thanks goes to Aaron Lomax for unpublished information on various UBLs. I apologize to those whose work was not cited due to space constraints.
Glossary
- Ub
ubiquitin
- DUBs
deubiquitylating proteases
- UBL
ubiquitin-like modifier
- CP
core protease
- UPS
ubiquitin/26S proteasome system
- PE
phosphatidylethanolamine
- AIM
ATG8-interacting motif
- UBA
ubiquitin-associated
- MoCo
molybdenum cofactor
- tRNA
transfer RNA
References
- Bassham DC. (2009) Function and regulation of macroautophagy in plants. Biochim Biophys Acta 1793: 1397–1403 [DOI] [PubMed] [Google Scholar]
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280: 275–286 [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. (2010) Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Book AJ, Gladman NP, Lee SS, Scalf M, Smith LM, Vierstra RD. (2010) Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem 285: 25554–25569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Cerda-Maira FA, Wang T, Li H, Bishai WR, Darwin KH. (2010) “Depupylation” of prokaryotic ubiquitin-like protein from mycobacterial proteasome substrates. Mol Cell 39: 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L. (2012) The natural history of ubiquitin and ubiquitin-related domains. Front Biosci 17: 1433–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Carpenter T, Sun CW, Vierstra RD. (1995) Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 139: 921–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Raasch JA, Vierstra RD. (1990) Ubiquitin extension proteins of Arabidopsis thaliana: structure, localization, and expression of their promoters in transgenic tobacco. J Biol Chem 265: 12486–12493 [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD. (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung T, Suttangkakul A, Vierstra RD. (2009) The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby T, Matthäi A, Boeckelmann A, Stuible HP. (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142: 318–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A. (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN, Huizinga DH. (2009) Protein isoprenylation: the fat of the matter. Trends Plant Sci 14: 163–170 [DOI] [PubMed] [Google Scholar]
- Dai X, Hayashi K, Nozaki H, Cheng Y, Zhao Y. (2005) Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc Natl Acad Sci USA 102: 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M. (1999) The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA 96: 15342–15347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. (1998) SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 2: 233–239 [DOI] [PubMed] [Google Scholar]
- Dittmar GA, Wilkinson CR, Jedrzejewski PT, Finley D. (2002) Role of a ubiquitin-like modification in polarized morphogenesis. Science 295: 2442–2446 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD. (2001) The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Dowil RT, Lu X, Saracco SA, Vierstra RD, Downes BP. (2011) Arabidopsis membrane-anchored ubiquitin-fold (MUB) proteins localize a specific subset of ubiquitin-conjugating (E2) enzymes to the plasma membrane. J Biol Chem 286: 14913–14921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes B, Vierstra RD. (2005) Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem Soc Trans 33: 393–399 [DOI] [PubMed] [Google Scholar]
- Downes BP, Saracco SA, Lee SS, Crowell DN, Vierstra RD. (2006) MUBs, a family of ubiquitin-fold proteins that are plasma membrane-anchored by prenylation. J Biol Chem 281: 27145–27157 [DOI] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD. (2003) The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35: 729–742 [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Coupland G. (2010) Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA 107: 17415–17420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer LM, Book AJ, Lee KH, Lin YL, Fu H, Vierstra RD. (2010) The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22: 124–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatimababy AS, Lin YL, Usharani R, Radjacommare R, Wang HT, Tsai HL, Lee Y, Fu H. (2010) Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J 277: 796–816 [DOI] [PubMed] [Google Scholar]
- Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP, Deng XW. (2004) Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16: 1870–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, Feng S, Bostick M, Callis J, Hellmann H, et al. (2005) Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17: 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A. (1989) The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–401 [DOI] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, et al. (2007) The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19: 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Lin YL, Fatimababy AS. (2010) Proteasomal recognition of ubiquitylated substrates. Trends Plant Sci 15: 375–386 [DOI] [PubMed] [Google Scholar]
- Fujioka Y, Noda NN, Fujii K, Yoshimoto K, Ohsumi Y, Inagaki F. (2008) In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J Biol Chem 283: 1921–1928 [DOI] [PubMed] [Google Scholar]
- Furukawa K, Mizushima N, Noda T, Ohsumi Y. (2000) A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem 275: 7462–7465 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F, Wenzel S, Tansey WP. (2012) Ubiquitin and proteasomes in transcription. Annu Rev Biochem 81: 177–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich DJ, Gagne JM, Salter DW, Hellmann H, Estelle M, Ma LG, Vierstra RD. (2005) Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J Biol Chem 280: 18810–18821 [DOI] [PubMed] [Google Scholar]
- Goehring AS, Rivers DM, Sprague GF., Jr (2003) Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell 2: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenjos JP, Richter R, Dohmann EM, Katsiarimpa A, Isono E, Schwechheimer C. (2011) MLN4924 is an efficient inhibitor of NEDD8 conjugation in plants. Plant Physiol 156: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Schulman BA. (2006) Structural complexity in ubiquitin recognition. Cell 124: 1133–1136 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, Macintosh GC. (2011) RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci USA 108: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458: 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M. (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Hotton SK, Callis J. (2008) Regulation of cullin RING ligases. Annu Rev Plant Biol 59: 467–489 [DOI] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD. (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Hua Z, Zou C, Shiu SH, Vierstra RD. (2011) Phylogenetic comparison of F-box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 6: e16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA. (2010) Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F. (2009) Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep 10: 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YJ, Jeong BC, Song HK. (2011) Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii. Biochem Biophys Res Commun 405: 112–117 [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T. (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, Huang L. (2008) A targeted proteomic analysis of the ubiquitin-like modifier Nedd8 and associated proteins. J Proteome Res 7: 1274–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A, Anzenberger F, Schlager N, Neubert S, Hauser MT, Schwechheimer C, Isono E. (2011) The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell 23: 3026–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937 [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T, Tanaka K. (2004) A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J 23: 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumeta H, Watanabe M, Nakatogawa H, Yamaguchi M, Ogura K, Adachi W, Fujioka Y, Noda NN, Ohsumi Y, Inagaki F. (2010) The NMR structure of the autophagy-related protein Atg8. J Biomol NMR 47: 237–241 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis: accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872 [DOI] [PubMed] [Google Scholar]
- Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. (2001) Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature 414: 325–329 [DOI] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW. (2008) Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. (2009) Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458: 228–232 [DOI] [PubMed] [Google Scholar]
- Lemaire K, Moura RF, Granvik M, Igoillo-Esteve M, Hohmeier HE, Hendrickx N, Newgard CB, Waelkens E, Cnop M, Schuit F. (2011) Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE 6: e18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M, Bassham DC, Vierstra RD, Parker JE, Bautor J, et al. (2011) Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J 66: 818–830 [DOI] [PubMed] [Google Scholar]
- Li F, Vierstra RD. (2012) Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci (in press) [DOI] [PubMed] [Google Scholar]
- Lima MdF, Eloy NB, Pegoraro C, Sagit R, Rojas C, Bretz T, Vargas L, Elofsson A, de Oliveira AC, Hemerly AS, et al. (2010) Genomic evolution and complexity of the anaphase-promoting complex (APC) in land plants. BMC Plant Biol 10: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Koornneef M, Soppe WJ. (2007) The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. (2001) Promotion of Nedd8-Cul1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Maor R, Jones A, Nühse TS, Studholme DJ, Peck SC, Shirasu K. (2007) Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol Cell Proteomics 6: 601–610 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I. (2012) mRNA export and sumoylation: lessons from plants. Biochim Biophys Acta 1819: 531–537 [DOI] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA 107: 16512–16517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Scalf M, Smith LM, Vierstra RD. (2012) Quantitative proteomics reveal factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol Cell Proteomics (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda HV, Nembhard N, Su D, Hepowit N, Krause DJ, Pritz JR, Phillips C, Söll D, Maupin-Furlow JA. (2011) E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in Archaea. Proc Natl Acad Sci USA 108: 4417–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Ammon T, Popowicz GM, Krajewski M, Nagel RJ, Ares M, Jr, Holak TA, Jentsch S. (2011) Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing. Nature 474: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM. (2007a) Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10: 495–502 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. (2007b) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TD, Feigon J. (2003) Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J 22: 4634–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Nakai Y, Harada A, Hashiguchi Y, Nakai M, Hayashi H. (2012) Arabidopsis molybdopterin biosynthesis protein Cnx5 collaborates with the ubiquitin-like protein Urm11 in the thio-modification of tRNA. J Biol Chem (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F. (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, et al. (2011) Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39: 3204–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Misono S, Miyata N, Matsumoto Y, Mukai S, Fujiki Y. (2011) Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic 12: 1067–1083 [DOI] [PubMed] [Google Scholar]
- Park BS, Song JT, Seo HS. (2011) Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun 2: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322: 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Salvesen GS, Wolf DA. (2011) Urm1 couples sulfur transfer to ubiquitin-like protein function in oxidative stress. Proc Natl Acad Sci USA 108: 1749–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD. (2008) The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C, Akhter Y, Jeon AH, Schmitt-Ulms G, Meyer HE, Stefanski A, Stühler K, Wilmanns M, Song YH. (2010) Proteome-wide identification of mycobacterial pupylation targets. Mol Syst Biol 6: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Cort JR, Yee AA, Semesi A, Edwards AM, Arrowsmith CH, Kennedy MA. (2003) Solution structure of the yeast ubiquitin-like modifier protein Hub1. J Struct Funct Genomics 4: 25–30 [DOI] [PubMed] [Google Scholar]
- Rao-Naik C, delaCruz W, Laplaza JM, Tan S, Callis J, Fisher AJ. (1998) The Rub family of ubiquitin-like proteins: crystal structure of Arabidopsis Rub1 and expression of multiple Rubs in Arabidopsis. J Biol Chem 273: 34976–34982 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2010) The ubiquitin-proteasome system regulates plant hormone signaling. Plant J 61: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Hansson M, Scalf M, Walker JM, Smith LM, Vierstra RD. (2009) Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD. (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa H, Sakata E, Yamaguchi Y, Komatsu M, Tatsumi K, Kominami E, Tanaka K, Kato K. (2006) Solution structure and dynamics of Ufm1, a ubiquitin-fold modifier 1. Biochem Biophys Res Commun 343: 21–26 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Deng XW. (2002) Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Missan G, Hinds TR, Kobayashi Y, Hsu F-F, Sharon M, Browse J, et al. (2011) Jasmonate perception by inositol phosphate-potentiated COI1-JAZ coreceptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara T, Kawasaki H, Hirano H. (2002) Identification of the 19S regulatory particle subunits from the rice 26S proteasome. Eur J Biochem 269: 1474–1483 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Okuda-Shimizu Y, Hendershot LM. (2010) Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell 40: 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK. (2007) Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. (2009) Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol 16: 647–651 [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26: 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD. (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F. (2005) The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy 1: 119–126 [DOI] [PubMed] [Google Scholar]
- Svenning S, Lamark T, Krause K, Johansen T. (2011) Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic receptor NBR1 and p62/SQSTM1. Autophagy 7: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Matic I, Mann M, Hay RT. (2011) Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal 4: rs4. [DOI] [PubMed] [Google Scholar]
- Tatsumi K, Sou YS, Tada N, Nakamura E, Iemura S, Natsume T, Kang SH, Chung CH, Kasahara M, Kominami E, et al. (2010) A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem 285: 5417–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Yamamoto-Mukai H, Shimizu R, Waguri S, Sou YS, Sakamoto A, Taya C, Shitara H, Hara T, Chung CH, et al. (2011) The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat Commun 2: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD. (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Toyooka K, Moriyasu Y, Goto Y, Takeuchi M, Fukuda H, Matsuoka K. (2006) Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2: 96–106 [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Walden H. (2010) Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol 11: 479–489 [DOI] [PubMed] [Google Scholar]
- Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282: 34167–34175 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Kini RK, Schuurink RC, Takken FLW. (2010) Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 22: 1998–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh HL, Jentsch S. (2011) Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci USA 108: 1763–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee C, Zapotoczny G, Masquelier D, Ghislain M, Batoko H. (2011) The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23: 785–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S, Bugg CE, Wilkinson KD, Vierstra RD, Hatfield PM, Cook WJ. (1987) Comparison of the three-dimensional structures of human, yeast, and oat ubiquitin. J Biol Chem 262: 6396–6399 [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Xi J, Begley TP, Nicholson LK. (2001) Solution structure of ThiS and implications for the evolutionary roots of ubiquitin. Nat Struct Biol 8: 47–51 [DOI] [PubMed] [Google Scholar]
- Wang T, Darwin KH, Li H. (2010) Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat Struct Mol Biol 17: 1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. (2008) Histone ubiquitination: triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Dittmar GA, Ohi MD, Uetz P, Jones N, Finley D. (2004) Ubiquitin-like protein Hub1 is required for pre-mRNA splicing and localization of an essential splicing factor in fission yeast. Curr Biol 14: 2283–2288 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. (2010) Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. (2008) Ribosomal proteins are targets for the Nedd8 pathway. EMBO Rep 9: 280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang J, Wang L, Zhou J, Huang H, Wu J, Zhong Y, Shi Y. (2006) Solution structure of Urm1 and its implications for the origin of protein modifiers. Proc Natl Acad Sci USA 103: 11625–11630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD. (2000) The ubiquitin-specific protease family from Arabidopsis: AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124: 1828–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ito H, Quint M, Huang H, Noël LD, Gray WM. (2008) Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc Natl Acad Sci USA 105: 8470–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J. (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110 [DOI] [PubMed] [Google Scholar]
- Zientara-Rytter K, Lukomska J, Moniuszko G, Gwozdecki R, Surowiecki P, Lewandowska M, Liszewska F, Wawrzyńska A, Sirko A. (2011) Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]