Abstract
Plant carotenoids play essential roles in photosynthesis, photoprotection, and as precursors to apocarotenoids. The plastid-localized carotenoid biosynthetic pathway is mediated by well-defined nucleus-encoded enzymes. However, there is a major gap in understanding the nature of protein interactions and pathway complexes needed to mediate carotenogenesis. In this study, we focused on carotene ring hydroxylation, which is performed by two structurally distinct classes of enzymes, the P450 CYP97A and CYP97C hydroxylases and the nonheme diiron HYD enzymes. The CYP97A and HYD enzymes both function in the hydroxylation of β-rings in carotenes, but we show that they are not functionally interchangeable. The formation of lutein, which involves hydroxylation of both β- and ε-rings, was shown to require the coexpression of CYP97A and CYP97C enzymes. These enzymes were also demonstrated to interact in vivo and in vitro, as determined using bimolecular fluorescence complementation and a pull-down assay, respectively. We discuss the role of specific hydroxylase enzyme interactions in promoting pathway flux and preventing the formation of pathway dead ends. These findings will facilitate efforts to manipulate carotenoid content and composition for improving plant adaptation to climate change and/or for enhancing nutritionally important carotenoids in food crops.
Carotenoids are a large class of isoprenoid pigments synthesized by all photosynthetic organisms as well as some bacteria, fungi, and aphids (Cuttriss et al., 2011). In plants, carotenoids serve essential roles in photosynthesis and photoprotection (Jahns and Holzwarth, 2012) and are precursors to apocarotenoids that function in stress and developmental responses (Walter et al., 2010). Plant-derived carotenoids also provide nutritional benefits to humans (von Lintig, 2010; Wurtzel et al., 2012).
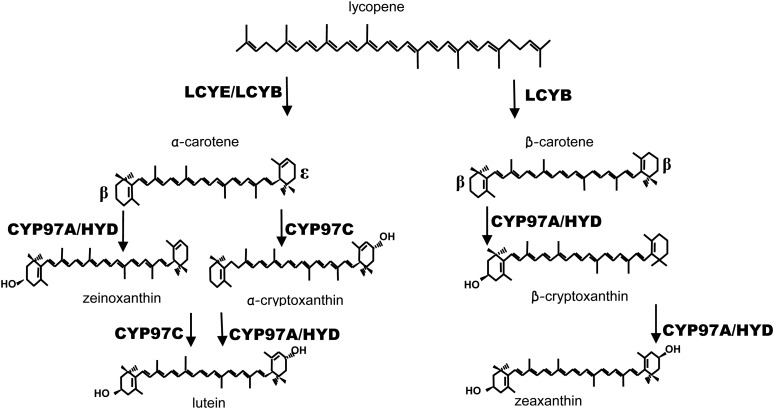
The plastid-localized carotenoid biosynthetic pathway is mediated by well-defined nucleus-encoded enzymes. The product of the first committed biosynthetic step, phytoene, is enzymatically converted into all-trans-lycopene, the major branch point precursor for downstream carotenoids (Fig. 1). The linear lycopene is enzymatically converted to carotenes by the formation of an ε-ring or β-ring at each end of lycopene. The rings differ only in the position of a double bond. Hydroxylation of the carotene rings is mediated by ring-specific hydroxylases and leads to xanthophylls such as lutein and zeaxanthin.
Figure 1.
Conversion of carotenoids to xanthophylls. Lycopene cyclases (LCYE and LCYB) convert lycopene to α-carotene and β-carotene. Formation of the xanthophylls zeaxanthin and lutein is mediated by two separate stereospecific β- and ε-ring hydroxylases (CYP97 and diiron HYD).
Although the individual biosynthetic enzymes are known, there is a gap in the fundamental understanding of complexes and protein interactions involved in mediating carotenogenesis. The pathway likely functions as a multienzyme complex(es) to facilitate metabolite channeling, as predicted by the absence of pathway intermediates and the presence of complexes containing carotenoid biosynthetic enzymes (Maudinas et al., 1977; Camara et al., 1982; Kreuz et al., 1982; Al-Babili et al., 1996; Bonk et al., 1997; Lopez et al., 2008). In this study, we examined an intriguing portion of the pathway, where hydroxylation of rings in carotenes is catalyzed by two structurally distinct enzymes, P450 heme (CYP97) and nonheme diiron (HYD) enzymes (Sun et al., 1996; Tian and DellaPenna, 2001, 2004; Kim and DellaPenna, 2006; Quinlan et al., 2007). Hydroxylation of the two β-ionone rings in β-carotene leads to a formation of zeaxanthin, while hydroxylation of the one β-ring and one ε-ring in α-carotene leads to lutein. Hydroxylation of the β-rings in the carotenes is potentially mediated by either the P450-type CYP97A or diiron HYD β-ring hydroxylase enzymes. Hydroxylation of the ε-ring of α-carotene is performed by another P450 enzyme, CYP97C. Theoretically, a single β-ring hydroxylase should suffice for hydroxylation of the β-ring in both α-carotene and β-carotene. It is unknown why two different β-ring hydroxylases have been maintained throughout evolution; it is possible that their respective activities are not entirely interchangeable. We hypothesized that hydroxylation of each of the carotene rings does not happen independently but that a synergistic interaction occurs between partner enzymes (CYP97A and CYP97C) to facilitate the carotene hydroxylation of α-carotene. To provide support for this hypothesis, we investigated whether carotene hydroxylase enzyme coexpression influenced the biosynthesis of enzyme products. We also determined which enzyme partners showed evidence of physical interaction.
RESULTS
Functional Complementation in Escherichia coli to Test for CYP97 and HYD Substrate Specificities
A widely used functional complementation method employed in our previous studies demonstrated activity of the P450 and HYD carotene ring hydroxylases from grasses (Quinlan et al., 2007; Vallabhaneni et al., 2009). Compared with β-ring hydroxylation by HYD, CYP97A4 and CYP97C2 were less effective in hydroxylating carotene rings in E. coli accumulating β-carotene or ε-ε-carotene, their respective β-ring or ε-ring substrates. We considered two reasons for the low activity of the CYPs. The first possibility was that the enzymes were not presented with their optimal substrate, α-carotene, which contains both β- and ε-rings. The second possibility was that perhaps the CYP97 enzymes did not function optimally as individual enzymes but required coexpression and interaction, which would allow for efficient hydroxylation of a mixed-ring compound, such as α-carotene. Biochemical phenotypes of plant knockouts support the hypothesis that CYP97 enzymes act sequentially (first CYP97A and then CYP97C) to hydroxylate α-carotene (Kim and DellaPenna, 2006).
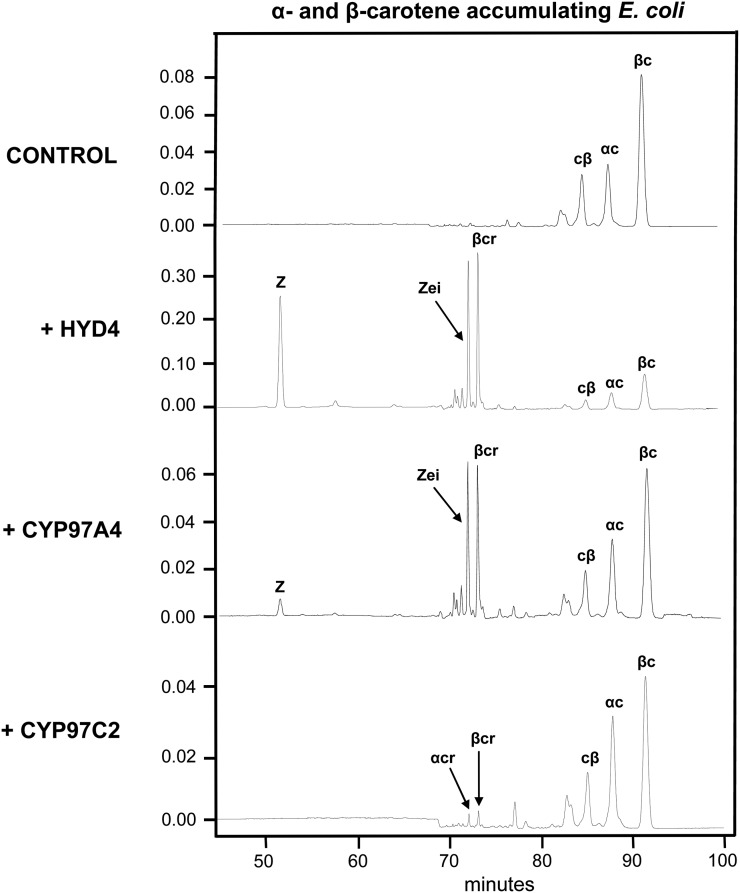
We first tested the effectiveness of α-carotene as a substrate, which can only be produced by engineering bacteria to synthesize both α-carotene and β-carotene. We expressed rice (Oryza sativa) CYP97A4 and CYP97C2 (Quinlan et al., 2007) and maize (Zea mays) HYD4 (Vallabhaneni et al., 2009) in E. coli that accumulated both α-carotene (β-ε-rings) and β-carotene (β-β-rings). Carotenoid products were analyzed by HPLC and/or liquid chromatography-mass spectrometry (LC-MS). In cells accumulating both α- and β-carotene, the expectation was that hydroxylation of both β-rings in β-carotene by the β-ring hydroxylases (CYP97A and HYD) would lead to formation of the monohydroxylated intermediate, β-cryptoxanthin, as well as the end product, zeaxanthin. This was the case for HYD4; cells expressing this enzyme accumulated approximately 30% zeaxanthin. By contrast, cells expressing CYP97A4 mainly accumulated the monohydroxylated intermediate β-cryptoxanthin (17% total carotenoids), while only 3% zeaxanthin was generated (Fig. 2; Table I). Similar results were observed when cells were engineered to accumulate β-carotene only (Table II). It was also expected that these β-ring hydroxylases would hydroxylate α-carotene to form zeinoxanthin, and indeed, this product was detected in cells expressing either CYP97A or HYD4, although the HYD4 enzyme was twice as active as CYP97A. In addition, we expected that cells transformed with the ε-ring hydroxylase CYP97C2 would accumulate the monohydroxylated product α-cryptoxanthin. However, this compound was barely detected (approximately 0.7% total carotenoids). These results show that HYD4 was most effective in producing a dihydroxylated carotene, in this case zeaxanthin, which was produced from β-carotene. The above results only partially confirmed the hypothesis that P450 carotene hydroxylases (CYP97A and CYP97C) prefer α-carotene over β-carotene as a substrate. CYP97A appeared to function as a monohydroxylase for either β-carotene or α-carotene, but CYP97C was marginally functional, regardless of the substrate. These experiments also showed that CYP97C could not efficiently hydroxylate carotene β-rings. Such a finding was inconsistent with the proposal that CYP97C could hydroxylate both rings of α-carotene to explain the formation of lutein in mutants lacking other known β-ring hydroxylases (Kim et al., 2009). Therefore, we next tested our second hypothesis, that CYP97A and CYP97C must be coexpressed and physically interact to convert α-carotene to lutein.
Figure 2.
Functional complementation tests of individually expressed CYP97 and HYD enzymes in cells accumulating α- and β-carotene. E. coli cells accumulating both α-carotene and β-carotene were transformed with test plasmids encoding HYD4, CYP97A4, or CYP97C2. Isolated pigments were separated by reverse-phase HPLC, with spectra extracted at 450 nm. Control, Empty pCOLADuet; Z, zeaxanthin; Zei, zeinoxanthin; αcr, α-cryptoxanthin; βcr, β-cryptoxanthin; cβ, 13-cis-β-carotene; αc; α-carotene; βc, β-carotene.
Table I. Major products in α- and β-carotene-accumulating E. coli with individually expressed hydroxylases.
Values are expressed as a percentage of total carotenoids. Each value is the mean of three replicates ± sd. ND, Not detectable.
| Hydroxylase | Zeaxanthin | α-Cryptoxanthin | Zeinoxanthin | β-Cryptoxanthin |
|---|---|---|---|---|
| CYP97A4 | 3.38 ± 0.27 | ND | 13.63 ± 2.97 | 16.76 ± 2.14 |
| CYP97C2 | ND | 0.71 ± 0.21 | ND | 1.14 ± 0.30 |
| HYD4 | 30.74 ± 1.85 | ND | 23.03 ± 2.72 | 24.03 ± 0.36 |
| Empty vector control | ND | ND | ND | ND |
Table II. Major products in β-carotene-accumulating E. coli with individually expressed hydroxylases.
Values are expressed as a percentage of total carotenoids. Each value is the mean of three replicates ± sd. ND, Not detectable.
| Hydroxylase | Zeaxanthin | β-Cryptoxanthin |
|---|---|---|
| CYP97A4 | 11.08 ± 1.21 | 26.19 ± 0.53 |
| CYP97C2 | ND | 0.78 ± 0.12 |
| HYD4 | 29.34 ± 3.86 | 24.14 ± 1.92 |
| Empty vector control | ND | ND |
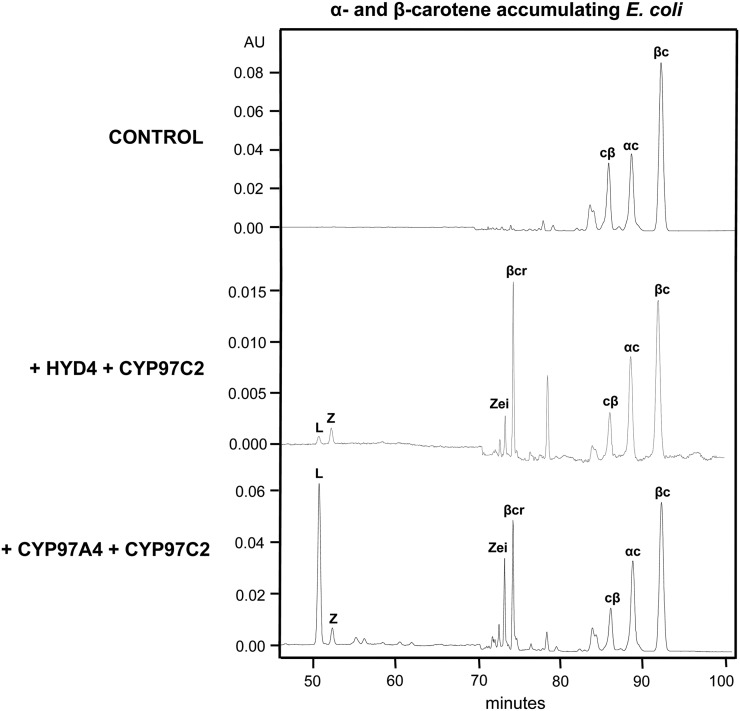
When P450 hydroxylases were coexpressed in the presence of α-carotene and β-carotene, their combined activity was dramatically increased, as evidenced by the formation of lutein (29% of total carotenoids), representing hydroxylation of the ε-ring in α-carotene by CYP97C2 and the β-ring by CYP97A4 (Fig. 3; Table III). This level of dihydroxylated product was comparable to that found for zeaxanthin formation by HYD4 (Fig. 2; Table I). In contrast, coexpression of the β-ring hydroxylase HYD4 with the ε-ring hydroxylase CYP97C2 did not lead to significant levels of hydroxylated carotenes. Perhaps there was a synergistic interaction occurring between P450 enzymes that did not occur between HYD4 and CYP97C2, since creating a monohydroxylated substrate by HYD4 was insufficient for CYP97C2 to efficiently hydroxylate the remaining ε-ring. Our results showed that the CYP97 enzymes must be coexpressed in order for α-carotene to be fully hydroxylated to form lutein and that the nonheme diiron β-ring hydroxylase (HYD) was not functionally equivalent to the P450 β-ring hydroxylase CYP97A.
Figure 3.
Functional complementation tests of CYP97/HYD enzyme combinations in cells accumulating α- and β-carotene. E. coli cells accumulating both α-carotene and β-carotene were transformed with the combinations of test plasmids HYD4 + CYP97C2 and CYP97A4 + CYP97C2. Extracted pigments were separated by reverse-phase HPLC, with spectra extracted at 450 nm. Control, Empty pCOLADuet; L, lutein; Z, zeaxanthin; Zei, zeinoxanthin; βcr, β-cryptoxanthin; cβ, 13-cis-β-carotene; αc; α-carotene; βc, β-carotene.
Table III. Major products in α- and β-carotene-accumulating E. coli with coexpressed hydroxylases.
Values are expressed as a percentage of total carotenoids. Each value is the mean of three replicates ± sd. ND, Not detectable.
| Hydroxylase | Lutein | Zeaxanthin | α-Cryptoxanthin | Zeinoxanthin | β-Cryptoxanthin |
|---|---|---|---|---|---|
| CYP97A4 + CYP97C2 | 28.99 ± 2.90 | 2.98 ± 0.44 | ND | 7.86 ± 1.28 | 13.32 ± 1.90 |
| HYD4 + CYP97C2 | 1.58 ± 0.14 | 3.16 ± 0.13 | ND | 3.49 ± 0.47 | 17.93 ± 1.57 |
| Empty vector control | ND | ND | ND | ND | ND |
The requirement for coexpression suggested that the CYP97 enzymes might interact with each other to produce the dihydroxylated carotenes. We predicted that the interacting enzymes should have similar patterns of plastid localization. Moreover, we expected to detect physical interactions in planta between CYP97A and CYP97C but not between CYP97C and HYD enzymes. To test these predictions, we carried out the following localization experiments.
Plastid Localization of Carotene Hydroxylases Based on Chloroplast Import Studies
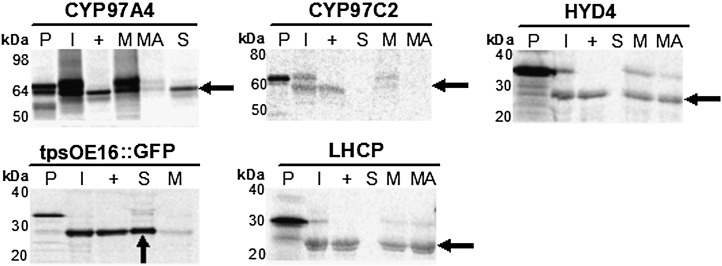
Recent proteomic methods utilizing liquid chromatography-tandem mass spectrometry showed CYP97A and CYP97C localized to the Arabidopsis (Arabidopsis thaliana) chloroplast envelope (Joyard et al., 2009; Ferro et al., 2010). However, no data were available for the location of HYD enzymes. Using the online prediction server TMHMM (Krogh et al., 2001), HYD4 was predicted to have four transmembrane helices, which would be expected to confer an integral membrane localization. The CYP97 structures were not predicted to have transmembrane helices. To test whether the hydroxylases were integrally or peripherally associated with membranes, we conducted in vitro chloroplast import assays (Fig. 4). Radioactively labeled protein precursors were imported into isolated chloroplasts, and then chloroplasts were fractionated into membrane and soluble fractions. CYP97A4 and CYP97C2 proteins were found in the membrane fraction and dissociated from it upon alkaline treatment, indicating that these proteins were peripherally associated. In addition, a significant amount of the CYP97A4 protein was found in the soluble fraction, which also suggested that the peripheral association of the protein is quite weak, allowing the protein to dissociate into a soluble fraction during the fractionation procedure. In contrast, HYD4, found in the membrane fraction as well, proved to be an integral protein, as evidenced by its resistance to alkaline treatment. These data are consistent with our structural predictions and earlier studies of a citrus HYD (Inoue et al., 2006).
Figure 4.
Chloroplast import assays of CYP97 and diiron HYD proteins. Isolated pea chloroplasts were used for in vitro import of [35S]Met-radiolabeled protein precursors. Chloroplasts harboring imported proteins were then reisolated and subjected to thermolysin treatment to distinguish between proteins that were peripherally bound to the outer chloroplast envelope and those that had been imported and thus processed to remove the transit peptide. The mature proteins were recovered as protease-resistant forms (arrows), confirming the import of these proteins into chloroplasts. Chloroplasts containing imported proteins were hypotonically lysed and fractionated into soluble and membrane fractions. The pellet fractions were then treated with an alkaline buffer to wash away peripherally associated membrane proteins. The purity of fractions was controlled by import and fractionation analysis of a chloroplast lumen protein, tpsOE16::GFP (Marques et al., 2003), and the integral thylakoid membrane-bound protein, LHCP (Tan et al., 2001). SDS-PAGE analysis of chloroplasts and their fractions indicated that CYP97A4 and CYP97C2 were synthesized as precursors of about 69 and 62 kD and then processed to 64 and 59 kD, respectively. HYD4 was synthesized as a precursor of roughly 34 kD and processed to 27 kD. P, Translation products; I, imported protein; +, thermolysin treatment; S, soluble proteins; M, membrane proteins; MA, alkaline-treated membrane fraction.
Testing Plastid-Localized Interactions of Partner Hydroxylases
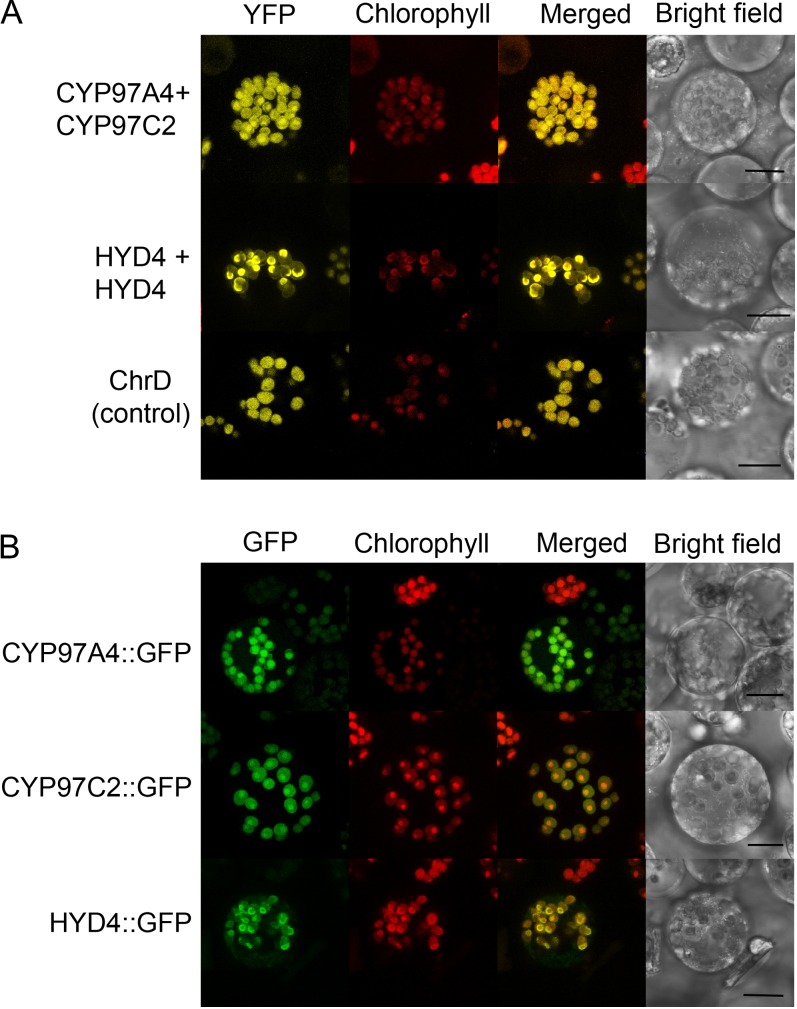
The functional complementation experiments in E. coli suggested that a synergistic interaction between CYP97A and CYP97C facilitates lutein formation from α-carotene. Enzyme interactions were tested in planta using the approach of bimolecular fluorescence complementation (BiFC; Citovsky et al., 2006) by transient expression in isolated maize protoplasts. Protoplasts maintain their tissue specificity and reflect in vivo conditions (Faraco et al., 2011; Denecke et al., 2012) and therefore are valuable for examining enzyme interactions. In addition, transient expression is an advantageous approach for monitoring the localization of low-abundance carotenoid biosynthetic enzymes that evade detection in proteomic studies. In BiFC, putative interacting proteins are fused to nonfluorescent N-terminal and C-terminal halves of the yellow fluorescent protein (YFP); interacting proteins bring together the nonfluorescent fragments, thereby restoring the yellow fluorescence. Constructs encoding fusion proteins were created for CYP97A4, CYP97C2, or HYD4, such that each was fused at their C termini to N- or C-terminal halves of YFP. The resulting constructs were transiently coexpressed in maize protoplasts and examined using confocal microscopy (Fig. 5A). CYP97A4 and CYP97C2 were found to interact with each other, as shown by restored YFP fluorescence. The interaction of CYP97A4 and CYP97C2 was additionally confirmed by an in vitro pull-down assay (Fig. 6). We also detected a HYD4 + HYD4 interaction, which suggested that HYD4 formed a homodimer. We did not detect homodimers for CYP97A4 or CYP97C2 or heterodimers for CYP97A4 + HYD4 or CYP97C2 + HYD4 (Supplemental Fig. S2). Interaction results for all tested protein combinations are summarized in Table IV. For comparison, we also individually expressed the enzymes as GFP fusions to confirm plastid localization in our protoplast system (Fig. 5B). A similar fluorescence pattern indicates that the interaction does not change protein localization as seen for the individually expressed proteins.
Figure 5.
Interactions and localization of carotene hydroxylases. A, BiFC detection of protein-protein interactions in maize protoplasts. CYP97A4 + CYP97C2 and HYD4 + HYD4 are interacting with each other, as seen by restored YFP fluorescence. Fusions of N-terminal and C-terminal halves of YFP with ChrD protein from cucumber (Cucumis sativus), which is known to form homodimer complexes in plastids (Libal-Weksler et al., 1997), were used as positive controls. B, Transient expression of GFP-fused proteins in maize protoplasts. CYP97 proteins are localized throughout etioplasts and concentrated at the spot of red chlorophyll autofluorescence of prolamellar bodies, as would be expected for proteins with stromal/weak peripheral membrane association. HYD4 is strictly colocalized with prolamellar bodies, consistent with integral thylakoid membrane binding. Chlorophyll, Chlorophyll autofluorescence. Bars = 10 µm.
Figure 6.
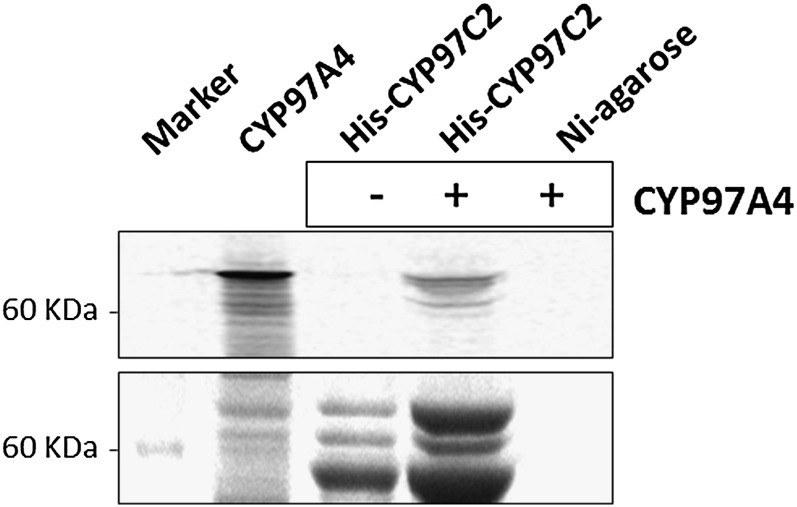
Pull-down assay. Interaction of CYP97A4 and CYP97C2 was shown in vitro by pull-down assay. CYP97C2 was expressed and purified from E. coli cells carrying pET23-CYP97C2, and CYP97A4 (carried by pTnT-A4) was translated in vitro using [35S]Met (see “Materials and Methods”). CYP97C2 was bound to Ni-NTA in the column and used as bait for CYP97A4. Radioactively labeled CYP97A4 interacted with CYP97C2, and interacting proteins eluted from the column together. Control loading of CYP97A4 to pure Ni-NTA did not show any nonspecific binding. Top, autoradiograph of a SDS-PAGE gel, showing CYP97A4 from the in vitro translation reaction and CYP97A4 in the eluate from the Ni-NTA + CYP97C2 column; bottom, Coomassie blue staining of the same gel.
Table IV. Interaction of proteins in BiFC experiments.
| Hydroxylase | CYP97A4 | CYP97C2 | HYD4 |
|---|---|---|---|
| CYP97A4 | − | + | − |
| CYP97C2 | + | − | − |
| HYD4 | − | − | + |
DISCUSSION
Interacting Proteins Exhibit Synergistic Effects on Carotene Dihydroxylation
Using a bacterial assay system, we showed that dihydroxylation of α-carotene to lutein requires the coexpression of CYP97A and CYP97C, enzymes that interact in planta and interact in vitro in pull-down assays. In contrast, lutein does not form in the case of enzymes that do not exhibit interaction, such as HYD and CYP97C. We hypothesize that a synergistic interaction between CYP97A and CYP97C is required to drive lutein biosynthesis for the purpose of channeling pathway substrates, stabilizing the enzyme-substrate complex and/or promoting interaction with other enzymes or components. We also found that the most efficient dihydroxylation of β-carotene to zeaxanthin was achieved by HYD, an enzyme that could form homodimers. CYP97A, which could not form homodimers, was less efficient in the dihydroxylation of β-carotene to zeaxanthin. Although further research is needed to understand the connection between interaction and the efficiency of dihydroxylation in planta, we hypothesize that the ability to form a protein complex improves the efficiency of hydroxylation of dual-ringed carotene substrates.
Do CYP97A and CYP97C Work Simultaneously or Sequentially?
The hypothesis based on plant mutants is that in carotenoid biosynthesis, CYP97A functions first to produce the monohydroxylated carotene, zeinoxanthin, which is transferred by some unknown mechanism as the substrate for CYP97C (Kim and DellaPenna, 2006; Kim et al., 2009). In those studies, CYP97C mutants accumulated substantially higher levels of the monohydroxylated carotene, compared with CYP97A mutants, although the levels were not what would be expected on the basis of wild-type levels of lutein formed in leaf tissue. In CYP97A mutants, the monohydroxylated α-cryptoxanthin barely accumulated, indicating that CYP97C was unable to hydroxylate the ε-ring of α-carotene. Using the bacterial assay system, we found that CYP97A produced approximately 20-fold more monohydroxylated carotene (zeinoxanthin) as compared with levels of monohydroxylated carotene (α-cryptoxanthin) catalyzed by CYP97C, when either enzyme was expressed alone in bacterial cells producing α- and β-ring-containing carotenes. These results show that in bacteria, CYP97A can accept the α-carotene substrate to produce the monohydroxylated product, while CYP97C is limited. Of course, there are many reasons why CYP97C might show poor activity in E. coli (e.g. nonoptimized codon usage, missing cofactors). However, the fact that CYP97C can function in bacteria when coexpressed with CYP97A suggests that CYP97A is the missing factor that must be present in order for CYP97C to function and, together with CYP97A, to produce lutein. The observed interaction between these two enzymes may reflect a stabilizing complex required in the case of CYP97A activity and a multienzyme structure needed to channel the zeinoxanthin substrate in the case of CYP97C. We propose that protein-protein interaction between CYP97A and CYP97C facilitates the recruitment of CYP97C2 to access the zeinoxanthin substrate. Such interaction might also serve in a regulatory role to control pathway flux. It would be intriguing to learn whether these enzymes exist only as a heterodimer or if other proteins, including other carotenoid enzymes, also participate in the formation of a metabolon in vivo.
Substrate Specificity?
From genetic and functional complementation studies (Tian et al., 2004; Kim and DellaPenna, 2006; Quinlan et al., 2007), it is generally accepted that CYP97C is an ε-ring hydroxylase and CYP97A is a β-ring hydroxylase. Mutant phenotypes suggested that CYP97A hydroxylates the β-rings of α-carotene to form zeinoxanthin, which is the preferred substrate for ε-ring hydroxylation by CYP97C (Kim and DellaPenna, 2006). However, biochemical phenotypes of mutants carrying only a subset of carotene ring hydroxylases suggest broader substrate specificity for these enzymes. Arabidopsis mutant plants with only CYP97C (and not CYP97A or the two nonheme carotene hydroxylases) produced significant levels of lutein, approximately 75% of the wild type, and plants with only CYP97A contained lutein at about 5% of the wild type level (Kim and DellaPenna, 2006; Kim et al., 2009). The explanation given for the significant level of lutein, which requires both ring-specific hydroxylases although only one is present, is that the remaining CYP has additional activity toward the other ring type. CYP97C, in particular, was thought to have significant β-ring hydroxylase activity, given that the triple mutants still produced 75% of wild-type levels of lutein (Kim et al., 2009). Broad substrate specificity for CYPs was also suggested by the results of CYP97A overexpression (Kim et al., 2010). However, in our earlier E. coli functional complementation studies (Quinlan et al., 2007), we found no evidence that the ε-ring hydroxylase CYP97C could hydroxylate β-rings, and in this study, β-ring hydroxylation by CYP97C was minimal. One explanation (Kim et al., 2009) for the disparity between the apparent function of CYP97C in planta and in E. coli was that the engineered E. coli contained only the individual β-ring or ε-ring substrates but not the mixed-ring substrates found in plants. Therefore, if CYP97C did indeed hydroxylate both β- and ε- rings, as postulated, we should have obtained lutein accumulation in bacteria that produce both β- and ε-ring substrates as evidence of such postulated broad substrate specificity. However, even when simultaneously presented with both the β- and ε-rings, neither CYP97C nor CYP97A, when expressed alone, produced detectable levels of lutein and in general barely produced a dihydroxylated product. Therefore, we found no evidence for broad substrate specificity to explain lutein formation by a single CYP enzyme, as suggested by the plant studies.
The question remains, why is lutein still formed in mutants containing only CYP97C? Recent studies (Kim et al., 2010) suggest that another poorly studied paralog, CYP97B, which is evolutionarily related to CYP97A and CYP97C, might exhibit a carotene β-ring hydroxylase activity. If so, then an explanation for the production of lutein in the Arabidopsis triple mutants is the functional CYP97B that is present in the mutant genetic background. If CYP97B is indeed a β-ring hydroxylase, it might function together with CYP97C to form lutein. Similarly, if CYP97B has some minor ε-ring hydroxylase activity, CYP97B could form lutein in partnership with CYP97A. In fact, in triple Arabidopsis mutants, levels of lutein were lower when the “only” ring hydroxylase was CYP97A as compared with when the “only” ring hydroxylase was CYP97C. Therefore, one could speculate that CYP97B is more efficient in hydroxylating β-rings as compared with ε-rings. Furthermore, we might expect CYP97B to form functional enzyme partnerships, as we observed for CYP97A and CYP97C. That is, expression of CYP97B alone would be predicted to be insufficient to mediate carotene dihydroxylation (e.g. to produce xanthophylls such as lutein). In support of this hypothesis, quadruple mutants (CYP97A, CYP97C, and the two nonheme carotene hydroxylases) of Arabidopsis were albino and showed 10% of wild-type levels of carotenes, but xanthophylls were completely blocked (Kim et al., 2009). Absence of a requisite partner enzyme could explain why xanthophylls are not produced in the quadruple mutant even in the presence of CYP97B. Further enzyme analysis of CYP97B and studies to test for synergistic interaction with the other known carotene hydroxylases are warranted.
Enzyme Interchangeability and Pathway Dead Ends
Since both HYD and CYP97A have the ability to produce zeinoxanthin by hydroxylation of the β-ring of α-carotene, CYP97A and HYD were expected to be functionally interchangeable. However, the results of enzyme coexpression did not support this. Combined expression of HYD and CYP97C was not productive in lutein formation in bacteria, showing that HYD could not substitute for CYP97A, which is consistent with a similar conclusion based on the biochemical phenotypes of plant mutants (Kim and DellaPenna, 2006). If interaction between CYP97C and CYP97A is needed to stabilize carotene ring hydroxylation by CYP97C (e.g. by providing zeinoxanthin substrate), then the inability to form a complex could account for the inability of HYD and CYP97C to function together. It is possible that a noninteraction has nonbiological explanations. However, the absence of interaction was consistent with the lack of synergy seen in bacterial coexpression and may be further explained by the fact that the enzymes exist in different membrane settings. Therefore, if and when HYD catalyzes the formation of zeinoxanthin from α-carotene in planta, this zeinoxanthin will be a pathway dead end in terms of further conversion to lutein.
CONCLUSION
Based on our results, we conclude that in the branched pathway in plants, the primary route to the formation of lutein from α-carotene is mediated by interacting CYP97 enzymes, whereas the primary route for zeaxanthin formation from β-carotene is mediated by HYD enzymes with some contribution from CYP97A. Our studies also support the widely held notion that carotenoid biosynthesis involves protein complexes to maximize pathway flux. Such an interaction between proteins is a useful regulatory mechanism that allows plants to direct the pathway toward various metabolites, as required in certain tissues or conditions. Our studies showing an interaction of these proteins, to our knowledge for the first time, lay the foundation for further investigations into the roles and topologies of the putative carotenoid metabolons. Further understanding of protein-protein interactions in the pathway will provide insight for more efficient engineering of carotenoid composition to avoid dead-end products and improve plant stress responses or nutritional content.
MATERIALS AND METHODS
Cloning of CYP97A4, CYP97C2, and HYD4
Amplification of open reading frames for cloning was performed by Platinum PCR Supermix High Fidelity Master Mix (Invitrogen) according to the manufacturer’s instructions. PCR conditions were as follows: one cycle of 95°C for 3 min; 35 to 40 cycles of 95°C for 45s, 58°C for 45s, and 72°C for 2 to 2.5 min; and one cycle of 72°C for 10 min. For primers, see Supplemental Table S1.
pCOLADuet and pCDFDuet Constructs Used for Expression in Escherichia coli
For cloning into pCOLADuet-1 vector (Novagen), full copies of complementary DNA (cDNA) of CYP97A4 and CYP97C2 were amplified from rice (Oryza sativa) cDNA (Quinlan et al., 2007). pCOLADuet-1-CYP97A4 was renamed pRT-A4. CYP97C2 was amplified from pCOLADuet-1-CYP97C2 using primers 2370 and 2371, cloned into NdeI and Acc65I sites of pCDFDuet-1 vector (Novagen), and renamed pRQ-C2. HYD4 was amplified from pTHYD4 (Vallabhaneni et al., 2009) using primers 1932 and 1933 and cloned into pCOLADuet-1. pCOLADuet-1-HYD4 was renamed pRQ-H4.
pTnT Constructs Used for in Vitro Transcription/Translation
A full-length cDNA of CYP97A4 was amplified from the pRT-A4 vector via PCR using primers 2175 and 2176. CYP97C2 was amplified from rice cDNA using primers 2140 and 2168. HYD4 was amplified from pRQ-H4 with primers 2165 and 2166. CYP97A4, CYP97C2, and HYD4 were cloned into the XhoI and XbaI sites of the pTnT vector (Promega) and named pTnT-A4, pTnT-C2, and pTnT-H4, respectively.
pUC35S-GUS-Nos Constructs Used for in Planta Localization
A full-length cDNA of CYP97A4 without a stop codon was amplified from the pRT-A4 vector with primers 2634 and 2635. CYP97C2 was amplified from pGEMT-C2 (pGEMT-Easy vector [Promega] harboring a full-length cDNA of CYP97C2) using primers 2879 and 2880. HYD4 was amplified from the pRQ-H4 using primers 2640 and 2641. CYP97A4, CYP97C2, and HYD4 were cloned in frame with GFP into the XbaI and BamHI sites of the pUC35S-sGFP-Nos vector (based on pUC35S-GUS-Nos and pBIG121 vectors; Okada et al., 2000) and named A4-GFP, C2-GFP, and H4-GFP, respectively.
pSAT Constructs Used for BiFC Experiments
For cloning into pSAT-2236 [pSAT4(A)-nEYFP-N1], a full-length cDNA without the stop codon of the CYP97A4 open reading frame was amplified from pRT-A4 using primers 2455 and 2426 (Citovsky et al., 2006). CYP97C2 was amplified from pGEMT-C2 using primers 3025 and 3026. HYD4 was amplified from pRQ-H4 using primers 2469 and 2470. CYP97A4, CYP97C2, and HYD4 were cloned into the XhoI and EcoRI sites of pSAT-2236 and named A4_2236, C2_ 2236, and H4_2236, respectively.
For cloning into pSAT-1476 (pSAT6-cEYFP-N1), a full copy of cDNA without the stop codon of CYP97A4 was amplified from pRT-A4 using primers 3023 and 3024. CYP97C2 was amplified from pGEMT-C2 using primers 2459 and 2460. HYD4 was amplified from pRQ-H4 using primers 2848 and 2849. CYP97A4 was cloned into XhoI and EcoRI sites of pSAT-1476 and named A4_1476. CYP97C2 was cloned into NcoI and EcoRI sites of the pSAT-1476 and named C2_1476, and HYD4 was cloned into BspHI and EcoRI sites of pSAT-1476 and named H4_1476.
pET23b+ Construct Used for Expression and Purification
CYP97C2 was PCR amplified from pTnT-C2 with primers 3209 and 3210 and cloned into pET23b+ (Promega) to give a translational fusion with a His tag. The resulting plasmid was named pET23-CYP97C2.
Functional Analysis of Hydroxylases in E. coli
For testing of substrate specificity for individual enzymes, pRT-A4, pRQ-C2, or pRQ-H4 was transformed into E. coli BL21 (DE3) cells (Novagen) harboring either of the following plasmids: (1) pAC-BETA-At (Cunningham et al., 2007) only, containing Erwinia uredovora (Pantoea ananas) crt EIB and Arabidopsis (Arabidopsis thaliana) lcyb, which confer β-carotene accumulation; or (2) pAC-BETA-At + plasmid y2 (encoding Arabidopsis lcyE; Cunningham et al., 1996), which together confer the accumulation of α- and β-carotene.
For testing of substrate specificity for enzyme combinations, the pRT-A4 + pRQ-C2 and pRQ-C2 + pRQ-H4 constructs were cotransformed into E. coli BL21 (DE3) cells (Novagen) harboring both pAC-ΒETA-At + plasmid y2. For negative controls, α- and β-carotene-accumulating cells were transformed with empty vectors.
For carotenoid analyses, overnight cultures in Luria-Bertani medium were diluted 50-fold into 50 mL of fresh medium, grown in the dark at 250 rpm at 37°C until an optical density of 0.6, induced with 10 mm isopropylthio-β-galactoside, and further cultured for a total of 3 d. Negative controls never generated any hydroxylated products.
Extraction of Carotenoids from E. coli cells, HPLC, and LC-MS Analysis
Fifty-milliliter cultures containing antibiotics for selection of plasmids (ampicillin, 50 μg mL−1; chloramphenicol, 34 μg mL−1; kanamycin, 30 μg mL−1; streptomycin, 30 μg mL−1) were centrifuged at 3,000g for 10 min. Bacterial cell pellets were extracted in 5 mL of methanol using a sonicator (Vibra Cell) and pelleted down at 3,000g for 10 min. Supernatants were transferred to 100-mL Pyrex flasks and evaporated under nitrogen gas, then dissolved in 300 µL of methanol and frozen at −80°C for 30 min, and pelleted down at 4°C. Supernatants were transferred to HPLC vials (Waters).
HPLC separation was carried out using a Waters system equipped with a 2695 Alliance separation module, a 996 photodiode array detector, a column heater, a fraction collector II, Empower software (Millipore), and a Develosil C30 RP-Aqueous (5 μm, 250 × 4.6 mm) column with a Nucleosil C18 (5 μm, 4 × 3.0 mm) guard column (Phenomenex), with mobile phase consisting of mixtures of acetonitrile:methanol:water (84:2:14, v/v/v [A]) and methanol:ethyl acetate (68:32, v/v [B]), with a gradient to obtain 100% B at 60 min (flow rate, 0.6 mL min−1), 100% B at 71 min with flow rate changing to 1.2 mL min−1, followed by 100% A (flow rate, 1.2 mL min−1) at 110 min. Peaks were identified on the basis of retention times/spectra matching those of authentic standards (Indofine) and standards purified from bacteria expressing genes encoding carotenoid biosynthetic enzymes (Cunningham et al., 1996, 2007). Integrated peak areas for extracted metabolites were calculated, and carotenoids were quantified as a percentage of total carotenoids. All data were collected at 450 nm.
LC-MS was performed on a Waters 2695 HPLC apparatus equipped with a 2998 PDA detector coupled to a Waters LCT Premiere XE TOF-MS system using electrospray ionization in positive ion mode. Separation was performed as described above.
Accumulated carotenoids and standards for lutein, zeaxanthin, β-cryptoxanthin, and α- and β-carotene were analyzed via HPLC; α-cryptoxanthin, β-cryptoxanthin, and zeinoxanthin were identified/confirmed by LC-MS. Cryptoxanthin isomers were identified as described (Kim and DellaPenna, 2006; Supplemental Fig. S1).
Chloroplast Isolation and in Vitro Import
Chloroplasts used in import assays were isolated from 10- to 14-d-old pea (Pisum sativum) plants as described (Bruce et al., 1994). Approximately 25 g of leaves was homogenized at 4°C with a blender in 75 mL of cold grinding buffer (50 mm HEPES, pH 8, 0.33 m sorbitol, 1 mm MgCl2, 1 mm MnCl2, 2 mm Na2EDTA, pH 8, 0.1% bovine serum albumin, and 0.1% Na-ascorbate) by three to five bursts of 1 s each. All further operations were performed on ice using cold buffers. The homogenate was filtered through two layers of cheesecloth and one layer of nylon mesh (60 µm), and the filtrate was centrifuged at 2,000g for 2 min. Pellets were carefully resuspended in 1 mL of grinding buffer, overlaid on top of 36 mL of Percoll gradient (prepared by centrifugation of 50% Percoll [Sigma] in grinding buffer, 40,000g for 30 min at 4°C), and centrifuged at 12,000g for 11 min at 4°C. Intact chloroplasts in the lower band were gently collected with a pipette, washed with 3 volumes of import buffer (50 mm HEPES, pH 8, and 0.33 m sorbitol), pelleted at 2,000g for 2 min at 4°C, resuspended in import buffer to yield a chloroplast concentration of 0.5 mg mL−1, and kept on ice until use.
Plasmid constructs pTnT-A4, pTnT-C2, and pTnT-H4 were used as templates for in vitro transcription/translation performed with the TnT Coupled Reticulocyte Lysate System (Promega) in the presence of [35S]Met, according to the manufacturer’s instructions. Reaction mixtures were prepared containing purified chloroplasts (0.5 mg mL−1), 1× import buffer, 4 mm Met, 4 mm ATP, 4 mm MgCl2, 10 mm KAc, 10 mm NaHCO3, and 10 µL of reticulocyte lysate translation product in a total volume of 150 µL. Reaction mixtures were incubated for 25 min at 25°C in light. Import reactions were stopped by adding 500 µL of import buffer and centrifuged at 800g for 2 min at 4°C to obtain a pellet of intact chloroplasts. Pellets were resuspended in 200 µL of import buffer supplemented by 1 mm CaCl2, and each reaction mixture was divided into two equal aliquots. Thermolysin was added to one aliquot to a concentration of 125 ng μL−1 and incubated for 30 min at 4°C. The reaction was terminated by the addition of EDTA to a concentration of 10 mm. The other aliquot was used as a control of the import reaction. For fractionation, chloroplasts were washed twice with import buffer and then diluted with HL buffer (10 mm HEPES-KOH and 10 mm MgCl2, pH 8); the total mixture was frozen in liquid nitrogen, thawed three times, and then centrifuged at 16,000g for 20 min. Alkaline treatment of membrane fractions was performed with 200 mm Na2CO3, pH > 10, for 10 min on ice, and pellets containing treated membranes were separated from the supernatant by centrifugation at 16,000g for 20 min. All fractions including soluble, membrane, and purified membrane pellets were analyzed by SDS-PAGE. Radiolabeled protein bands were visualized using a Storm Phosphorimager (Amersham Biosciences).
Isolation and Transformation of Maize Protoplasts
Isolation and transformation of maize (Zea mays) protoplasts was performed using protocols from Sheen (1991) and van Bokhoven et al. (1993) as a guide, with modifications. Maize var B73 plants were grown in the dark at 26°C for 12 d (12 h of day, 12 h of night). Middle parts of second leaves of 20 plants were cut into razor-thin sections and transferred to 50 mL of calcium/mannitol solution (10 mm CaCl2, 0.6 m mannitol, and 20 mm MES, pH 5.7) to which was added 1% cellulase (Trichoderma viride), 0.3% Rhizopus sp. pectinase (Sigma), 5 mm β-mercaptoethanol (Sigma), and 0.1% bovine serum albumin (Sigma). Vacuum was applied for 5 min, followed by shaking at 60 rpm at room temperature in the dark for 3 h. The supernatant was filtered by 60-µm nylon mesh and collected in a 50-mL Falcon centrifuge tube. Protoplasts were pelleted at 60g for 5 min at room temperature, washed three times with 25 mL of calcium/mannitol solution, divided into aliquots of 150 µL of 106 cells, and 10 µg of ice-cold plasmid DNA was added to each reaction. Protoplasts were then continuously mixed with 500 µL of polyethylene glycol solution [40% PEG 6000, 0.5 m mannitol, and 0.1 m Ca(NO3)2] for 10 s followed by the addition of 4.5 mL of mannitol/MES solution (15 mm MgCl2, 0.1% MES, pH 5.5, and 0.5 m mannitol) and incubated at room temperature for 25 min. The suspension was then centrifuged at 60g for 5 min at room temperature, and the supernatant was discarded. Sediment was washed with calcium/mannitol solution and pelleted at 60g for 5 min at room temperature. The supernatant was discarded, and protoplasts were resuspended in 1 mL of calcium/mannitol solution. Protoplasts were transferred to a 24-well plate and incubated overnight at 25°C under dim light. The transformational efficiency for protoplasts was 80% to 90%.
Confocal Microscopy
Transient expression of GFP or YFP fusion proteins was visualized using a DM16000B inverted confocal microscope with the TCS SP5 system (Leica Microsystems). An oil- or water-immersion objective (63×) was used in all cases. A 488-nm argon laser was used to excite the fluorescence of GFP and chlorophyll. Chlorophyll autofluorescence was detected between 664 and 696 nm. GFP fluorescence was detected between 500 and 539 nm and always confirmed by recording the emission spectrum by wavelength scanning (λ scan) between 500 and 600 nm with a 3-nm detection window. A 514-nm line of the argon laser was used to excite the fluorescence of YFP, and the emission spectrum was detected and confirmed by λ scan between 524 and 575 nm. LAS AF software (Leica Microsystems) was used for image acquisition. Images were obtained by combining several confocal Z-planes.
In Vitro Pull-Down Assay
pET23-CYP97C2, encoding a His-tagged CYP97C2 fusion protein, was expressed in the C43 (DE3) strain of E. coli (Lucigen). Cells were grown in 2× yeast extract and tryptone (2× YT) medium containing 50 mg mL−1 ampicillin until an optical density of 0.6, and protein expression was induced by adding 1 mm isopropylthio-β-galactoside, followed by incubation at 28°C overnight. Pelleted cells were resuspended in 25 mL of resuspension buffer (50 mm HEPES, pH 7.8, 300 mm NaCl, 20 mm imidazole, pH 7.8, and 1 mm MgCl2) containing 0.5 mm Tris(2-carboxyethyl) phosphine hydrochloride, 4 units of DNaseI (Invitrogen), and 0.12% 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride. Lysed cells were then sonicated and diluted with 10 mL of resuspension buffer containing 1.5% n-dodecyl β-d-maltoside, 0.5 mm Tris(2-carboxyethyl) phosphine hydrochloride, and 0.12% 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride. Samples were rotated at 4°C for 1 h. Insoluble material was removed by centrifugation at 5,000g for 20 min at 4°C. Lysate, containing CYP97C2, was mixed with 600 µL of equilibrated nickel-nitrilotriacetic acid agarose (Ni-NTA) slurry (Qiagen) and incubated overnight at 4°C. Two hundred microliters of the Ni-NTA slurry containing immobilized CYP97C2 was used for the pull-down assay.
Fifteen microliters of 35S-labeled CYP97A4 protein precursor translated in vitro from pTnT-A4 was added to 400 µL of diluting buffer (25 mm HEPES, pH 7.8, 150 mm NaCl, and 40 mm imidazole). CYP97A4 solution was mixed with the CYP97C2 agarose slurry and incubated for 2 h at 4°C. The same amount of CYP97A4 was mixed with 200 µL of pure Ni-NTA slurry as a negative control. After incubation, slurries were loaded onto 1-mL polypropylene columns (Qiagen) and washed three times with wash buffer (25 mm HEPES, pH 7.8, 25 mm NaCl, and 40 mm imidazole). Interacting proteins were eluted with the same buffer but containing 200 mm imidazole and 5% glycerol. Samples were analyzed by SDS-PAGE, and the gel was stained with Coomassie blue for total protein and dried before phosphorimaging.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: for rice, CYP97A4 (AK068163) and CYP97C2 (AK065689); for maize, HYD4 (BG320875/AY844956).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Extracted ion chromatogram of mass spectrometry traces corresponding to the major quasimolecular ions or fragmented ions for carotenoid extracts from E. coli accumulating α- and β-carotene.
Supplemental Figure S2. BiFC assays demonstrating no protein interaction for the indicated CYP97/HYD combinations tested using protoplasts isolated from etiolated maize leaf tissue.
Supplemental Table S1. Primers used in the study.
Supplementary Material
Acknowledgments
We thank Dr. Francis Cunningham for the Erwinia expression plasmids, Dr. Vitaly Citovsky for the pSAT vectors, and Drs. Akira Kawamura, Gema Flores, and Keyvan Dastmalchi for technical advice on LC-MS analysis.
Glossary
- LC-MS
liquid chromatography-mass spectrometry
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- cDNA
complementary DNA
References
- Al-Babili S, von Lintig J, Haubruck H, Beyer P. (1996) A novel, soluble form of phytoene desaturase from Narcissus pseudonarcissus chromoplasts is Hsp70-complexed and competent for flavinylation, membrane association and enzymatic activation. Plant J 9: 601–612 [DOI] [PubMed] [Google Scholar]
- Bonk M, Hoffmann B, Von Lintig J, Schledz M, Al-Babili S, Hobeika E, Kleinig H, Beyer P. (1997) Chloroplast import of four carotenoid biosynthetic enzymes in vitro reveals differential fates prior to membrane binding and oligomeric assembly. Eur J Biochem 247: 942–950 [DOI] [PubMed] [Google Scholar]
- Bruce BD, Perry S, Froehlich J, Keegstra K. (1994) In vitro import of protein into chloroplasts. In SB Gelvin, RB Schilperoort, eds, Plant Molecular Biology Manual, Vol J1. Kluwer Academic Publishers, Boston, pp 1–15
- Camara B, Bardat F, Monéger R. (1982) Sites of biosynthesis of carotenoids in Capsicum chromoplasts. Eur J Biochem 127: 255–258 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee L-Y, Vyas S, Glick E, Chen M-H, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362: 1120–1131 [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Jr, Lee H, Gantt E. (2007) Carotenoid biosynthesis in the primitive red alga Cyanidioschyzon merolae. Eukaryot Cell 6: 533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FX, Jr, Pogson B, Sun Z, McDonald KA, DellaPenna D, Gantt E. (1996) Functional analysis of the β and ε lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8: 1613–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttriss AJ, Cazzonelli CI, Wurtzel ET, Pogson BJ. (2011) Carotenoids. In F Rébeillé, R Douce, eds, Biosynthesis of Vitamins in Plants (Advances in Botanical Research, Part A), Vol 58. Elsevier, Amsterdam, The Netherlands, pp 1–36.
- Denecke J, Aniento F, Frigerio L, Hawes C, Hwang I, Mathur J, Neuhaus JM, Robinson DG. (2012) Secretory pathway research: the more experimental systems the better. Plant Cell 24: 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco M, Di Sansebastiano GP, Spelt K, Koes RE, Quattrocchio FM. (2011) One protoplast is not the other! Plant Physiol 156: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Furbee KJ, Uratsu S, Kato M, Dandekar AM, Ikoma Y. (2006) Catalytic activities and chloroplast import of carotenogenic enzymes from citrus. Physiol Plant 127: 561–570 [Google Scholar]
- Jahns P, Holzwarth AR. (2012) The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim Biophys Acta 1817: 182–193 [DOI] [PubMed] [Google Scholar]
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N. (2009) Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol Plant 2: 1154–1180 [DOI] [PubMed] [Google Scholar]
- Kim J, DellaPenna D. (2006) Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc Natl Acad Sci USA 103: 3474–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Smith JJ, Tian L, Dellapenna D. (2009) The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol 50: 463–479 [DOI] [PubMed] [Google Scholar]
- Kim J-E, Cheng KM, Craft NE, Hamberger B, Douglas CJ. (2010) Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a β-carotene ketolase provides insight into in vivo functions. Phytochemistry 71: 168–178 [DOI] [PubMed] [Google Scholar]
- Kreuz K, Beyer P, Kleinig H. (1982) The site of carotenogenic enzymes in chromoplasts from Narcissus pseudonarcissus L. Planta 154: 66–69 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Libal-Weksler Y, Vishnevetsky M, Ovadis M, Vainstein A. (1997) Isolation and regulation of accumulation of a minor chromoplast-specific protein from cucumber corollas. Plant Physiol 113: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AB, Yang Y, Thannhauser TW, Li L. (2008) Phytoene desaturase is present in a large protein complex in the plastid membrane. Physiol Plant 133: 190–198 [DOI] [PubMed] [Google Scholar]
- Marques JP, Dudeck I, Klösgen RB. (2003) Targeting of EGFP chimeras within chloroplasts. Mol Genet Genomics 269: 381–387 [DOI] [PubMed] [Google Scholar]
- Maudinas B, Bucholtz ML, Papastephanou C, Katiyar SS, Briedis AV, Porter JW. (1977) The partial purification and properties of a phytoene synthesizing enzyme system. Arch Biochem Biophys 180: 354–362 [DOI] [PubMed] [Google Scholar]
- Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y. (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol 122: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan RF, Jaradat TT, Wurtzel ET. (2007) Escherichia coli as a platform for functional expression of plant P450 carotene hydroxylases. Arch Biochem Biophys 458: 146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (1991) Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell 3: 225–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Gantt E, Cunningham FX., Jr (1996) Cloning and functional analysis of the beta-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 271: 24349–24352 [DOI] [PubMed] [Google Scholar]
- Tan BC, Cline K, McCarty DR. (2001) Localization and targeting of the VP14 epoxy-carotenoid dioxygenase to chloroplast membranes. Plant J 27: 373–382 [DOI] [PubMed] [Google Scholar]
- Tian L, DellaPenna D. (2001) Characterization of a second carotenoid beta-hydroxylase gene from Arabidopsis and its relationship to the LUT1 locus. Plant Mol Biol 47: 379–388 [DOI] [PubMed] [Google Scholar]
- Tian L, DellaPenna D. (2004) Progress in understanding the origin and functions of carotenoid hydroxylases in plants. Arch Biochem Biophys 430: 22–29 [DOI] [PubMed] [Google Scholar]
- Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D. (2004) The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid ε-ring hydroxylation activity. Proc Natl Acad Sci USA 101: 402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni R, Gallagher CE, Licciardello N, Cuttriss AJ, Quinlan RF, Wurtzel ET. (2009) Metabolite sorting of a germplasm collection reveals the hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiol 151: 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H, Verver J, Wellink J, van Kammen A. (1993) Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J Gen Virol 74: 2233–2241 [DOI] [PubMed] [Google Scholar]
- von Lintig J. (2010) Colors with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annu Rev Nutr 30: 35–56 [DOI] [PubMed] [Google Scholar]
- Walter MH, Floss DS, Strack D. (2010) Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232: 1–17 [DOI] [PubMed] [Google Scholar]
- Wurtzel ET, Cuttriss A, Vallabhaneni R. (2012) Maize provitamin a carotenoids, current resources, and future metabolic engineering challenges. Front Plant Sci 3: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.