Abstract
Mutations in human (Homo sapiens) ETHYLMALONIC ENCEPHALOPATHY PROTEIN1 (ETHE1) result in the complex metabolic disease ethylmalonic encephalopathy, which is characterized in part by brain lesions, lactic acidemia, excretion of ethylmalonic acid, and ultimately death. ETHE1-like genes are found in a wide range of organisms; however, the biochemical and physiological role(s) of ETHE1 have not been examined outside the context of ethylmalonic encephalopathy. In this study we characterized Arabidopsis (Arabidopsis thaliana) ETHE1 and determined the effect of an ETHE1 loss-of-function mutation to investigate the role(s) of ETHE1 in plants. Arabidopsis ETHE1 is localized in the mitochondrion and exhibits sulfur dioxygenase activity. Seeds homozygous for a DNA insertion in ETHE1 exhibit alterations in endosperm development that are accompanied by a delay in embryo development followed by embryo arrest by early heart stage. Strong ETHE1 labeling was observed in the peripheral and chalazal endosperm of wild-type seeds prior to cellularization. Therefore, ETHE1 appears to play an essential role in regulating sulfide levels in seeds.
Mutations in the human (Homo sapiens) ETHYLMALONIC ENCEPHALOPATHY PROTEIN1 (ETHE1) gene result in the severe infantile metabolic disease ethylmalonic encephalopathy (EE; Tiranti et al., 2004). Common symptoms of EE patients include chronic diarrhea, a delay in neural development, symmetric brain lesions, lactic acidemia, and acrocyanosis of the extremities. Biochemical alterations associated with EE include elevated levels of C4 and C5 plasma acylcarnitines, and elevated urinary excretion of high levels of ethylmalonic acid and thiosulfate (Tiranti et al., 2004). The disease is ultimately lethal within the first decade of life (Burlina et al., 1991, 1994).
ETHE1 proteins are found in a wide range of organisms, including bacteria, plants, and animals. They belong to the β-lactamase fold family of proteins and show considerable similarity to glyoxalase II, which is involved in the detoxification of 2-oxoaldehydes and plays a role in abiotic stress response and survival in plants (Singla-Pareek et al., 2003, 2006; Yadav et al., 2005a, 2005b). A recent analysis showed that sulfur dioxygenase activity was absent from EE patients and Ethe1−/− mice and that increased levels of sulfur dioxygenase activity are present in HeLa and Escherichia coli cells overexpressing human ETHE1, suggesting that ETHE1 is a mitochondrial sulfur dioxygenase involved in the catabolism of sulfide (Tiranti et al., 2009).
Sulfide occupies a central position in sulfur assimilation in plants (Rennenburg, 1989). Sulfur typically enters a plant as sulfate, which is reduced via a multistep process to sulfide in the chloroplast. Sulfide is then incorporated into Cys and further metabolized to produce thioethers, sulfoxides, and methylsulfonium compounds (for review, see Leustek et al., 2000; Hell and Leustek, 2005). Cys degradation generates hydrogen sulfide, a potent inhibitor of aerobic respiration, which can act as a substrate for ATP production and also has important physiological functions as a signaling molecule. For example, hydrogen sulfide can help improve drought resistance and regulate flower senescence (Jin et al., 2011; Zhang et al., 2011). However, hydrogen sulfide is also highly toxic to cells and therefore its levels are tightly regulated (Bouillaud and Blachier, 2011). Hydrogen sulfide can react spontaneously with gluthathione disulfide to produce S-sulfanylgutathione, which is then converted to sulfite by sulfur dioxygenase. Most of the information available on sulfur dioxygenases has come from studies on the sulfur-oxidizing bacteria (Sugio et al., 1989; Rohwerder and Sand, 2003, 2007). However, a mitochondrial pathway that catalyzes the oxidation of sulfide to thiosulfate and appears to involve a putative ETHE1 enzyme has also been characterized in rat liver (Hildebrandt and Grieshaber, 2008). In contrast, nothing is known about sulfur dioxygenases in plants. Therefore, to gain insights into the role of ETHE1 in plant growth and development, we analyzed the putative Arabidopsis (Arabidopsis thaliana) ETHE1 gene and the effect of an ETHE1 knockout mutation. Similar to the situation in humans, Arabidopsis ETHE1 is localized in the mitochondrion and exhibits sulfur dioxygenase activity. We further found that ETHE1 is essential for endosperm and embryo development. Seeds lacking ETHE1 exhibit endosperm defects beginning approximately 24 h after fertilization accompanied by a delay in embryo development and ultimately embryo arrest by early heart stage.
RESULTS
ETHE1 Encodes a Mitochondrial Protein with Sulfur Dioxygenase Activity
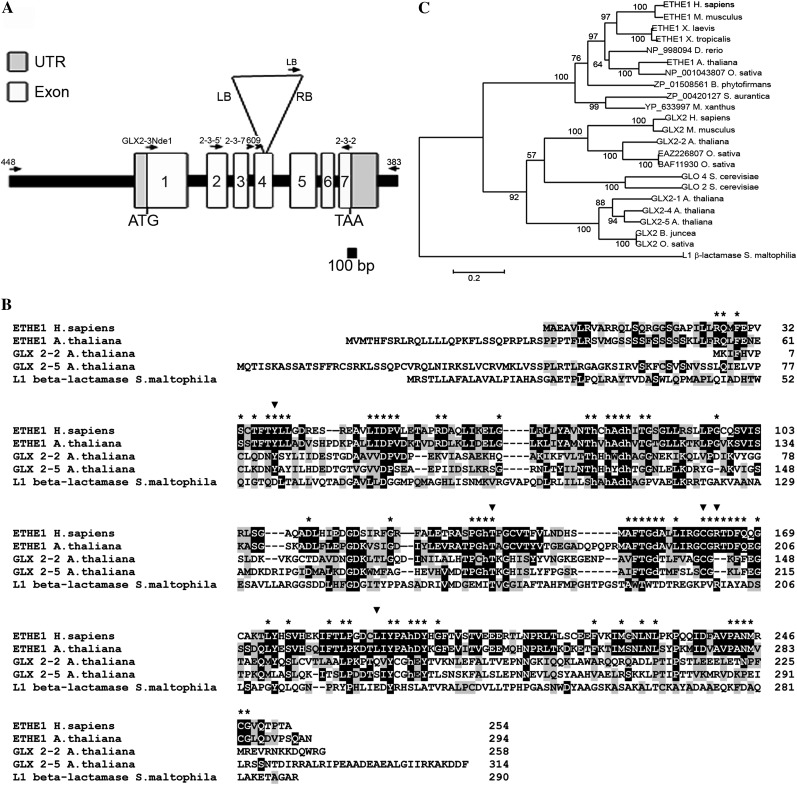
The Arabidopsis gene At1g53580 (Fig. 1A) had previously been predicted to encode a glyoxalase II isozyme (GLX2-3; Maiti et al., 1997). However, after further analysis we show here that it represents Arabidopsis ETHE1. A complementary DNA (cDNA) for At1g53580 (NM_202289) has the potential to encode a 294 amino acid protein that belongs to the β-lactamase family of proteins. The predicted protein showed only 13% identity with the cytoplasmic glyoxalase II isozyme from Arabidopsis (GLX2-2), but 54% identity with human ETHE1, indicating that At1g53580 is likely Arabidopsis ETHE1 (Fig. 1B).
Figure 1.
Molecular characterization of ETHE1. A, Map of the ETHE1 locus. The positions and directions of primers used in this study are shown as arrows. The T-DNA insert is shown as an inverted triangle. B, Alignment of ETHE1 with select β-lactamase fold proteins. Identical and similar residues are highlighted in black and gray, respectively. Residues labeled with an asterisk are conserved in ETHE1-like enzymes. Residues labeled with an inverted triangle are mutated in EE patients. Lowercase residues are metal-binding ligands. C, Phylogenic analysis of ETHE1. The tree was derived from the alignment of multiple β-lactamase fold proteins with Clustal W followed by a neighbor-joining analysis conducted with MEGA version 3.1. Bootstrap values are shown at branch points.
A neighbor-joining tree was generated based on a Clustal W alignment of select metallo-β-lactamase proteins, including ETHE1-like and GLX2 proteins from several organisms. Analysis of the phylogenic tree, rooted in the common ancestor β-lactamase, demonstrates that β-lactamases diverged through ancient duplication events into separate ETHE1 and GLX2 lineages (Fig. 1C). As expected, Arabidopsis ETHE1 grouped together with human ETHE1 and predicted ETHE1 proteins from mouse (Mus musculus), frog (Xenopus laevis), fish (Danio rerio), rice (Oryza sativa), and bacteria. Likewise, Arabidopsis GLX2 enzymes grouped with GLX2 proteins from human, mouse, rice, mustard (Brassica juncea), and yeast (Saccharomyces cerevisiae). In addition to conserved metal-binding residues found in the metallo-β-lactamases, ETHE1 proteins share a number of conserved residues, including R163, C161, Y38, L185, and T136 (Fig. 1B) that have been associated with EE in humans, suggesting that they are required for catalytic activity (Tiranti et al., 2006). Likewise, ETHE1 enzymes lack conserved GLX2 substrate-binding ligands.
To further demonstrate that At1g53580 is Arabidopsis ETHE1, we examined the subcellular localization and enzymatic activity of the protein. Transgenic Arabidopsis cell cultures that express a FLAG-tagged version of ETHE1 were generated and used in subcellular fractionation studies. Western blots containing total protein and protein isolated from purified mitochondrial and cytoplasmic fractions, were probed with either anti-FLAG antibody or an antibody to mitochondrial cytochrome c oxidase (COX) as a control. ETHE1 and COX were both found in the total protein and mitochondrial fractions, but not the cytosolic fraction (Fig. 2A). Therefore, similar to human ETHE1, the Arabidopsis protein is localized to the mitochondrion.
Figure 2.
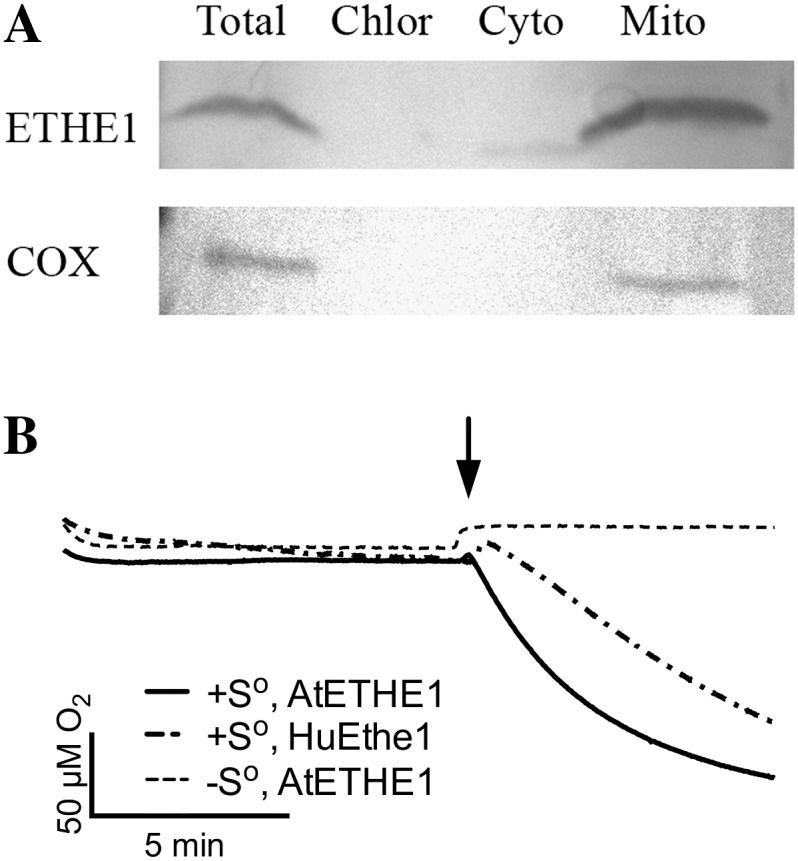
Arabidopsis ETHE1 is localized to the mitochondrion and exhibits sulfur dioxygenase activity. A, The subcellular localization of ETHE1-FLAG proteins in transgenic Arabidopsis cell cultures was determined through western-blot analysis using COX as a mitochondrial fraction control. B, Oxygraphic traces of oxygen consumption of purified human and Arabidopsis ETHE1. Arrow denotes the time of sulfur or buffer (control) addition.
We next investigated whether the Arabidopsis protein, like human ETHE1, exhibits sulfur dioxygenase activity. Arabidopsis ETHE1 was overexpressed in E. coli, purified, and assayed for glyoxalase II and sulfur dioxygenase activity. Recombinant human ETHE1 was used as a positive control. As predicted, Arabidopsis ETHE1 showed no activity with S-lactoylglutathione, the standard glyoxalase II substrate. In contrast, it did exhibit sulfur-dependent dioxygenase activity at a level similar to that of human ETHE1 (Fig. 2B). Specifically, Arabidopsis ETHE1 catalyzed the glutathione-persulfide-dependent consumption of oxygen at a rate of 7.95 ± 0.711 μmol O2 min−1 mg−1 compared with 4.93 ± 0.989 μmol O2 min−1 mg−1 for human ETHE1. Therefore, At1g53580 encodes the Arabidopsis ortholog of human ETHE1.
ETHE1 Is Essential for Early Seed Development
An Arabidopsis line containing a transferred DNA (T-DNA) insertion in ETHE1 was isolated and characterized to gain insights into the role of ETHE1. PCR screening of a population of T-DNA insertion mutants resulted in the identification of a line that contains a T-DNA insertion in exon four of ETHE1 (Fig. 1A). Segregation analyses of progeny of ETHE1/ethe1 plants failed to identify plants homozygous for the T-DNA insertion; 312 (32%) were homozygous for the wild-type locus and 673 (68%) were heterozygous for the ETHE1 T-DNA insertion. The 1:2:0 segregation ratio [c2 = 1.33 < c20.05(1) = 3.841] suggested that ETHE1 is an essential gene and that the mutation may result in embryo lethality.
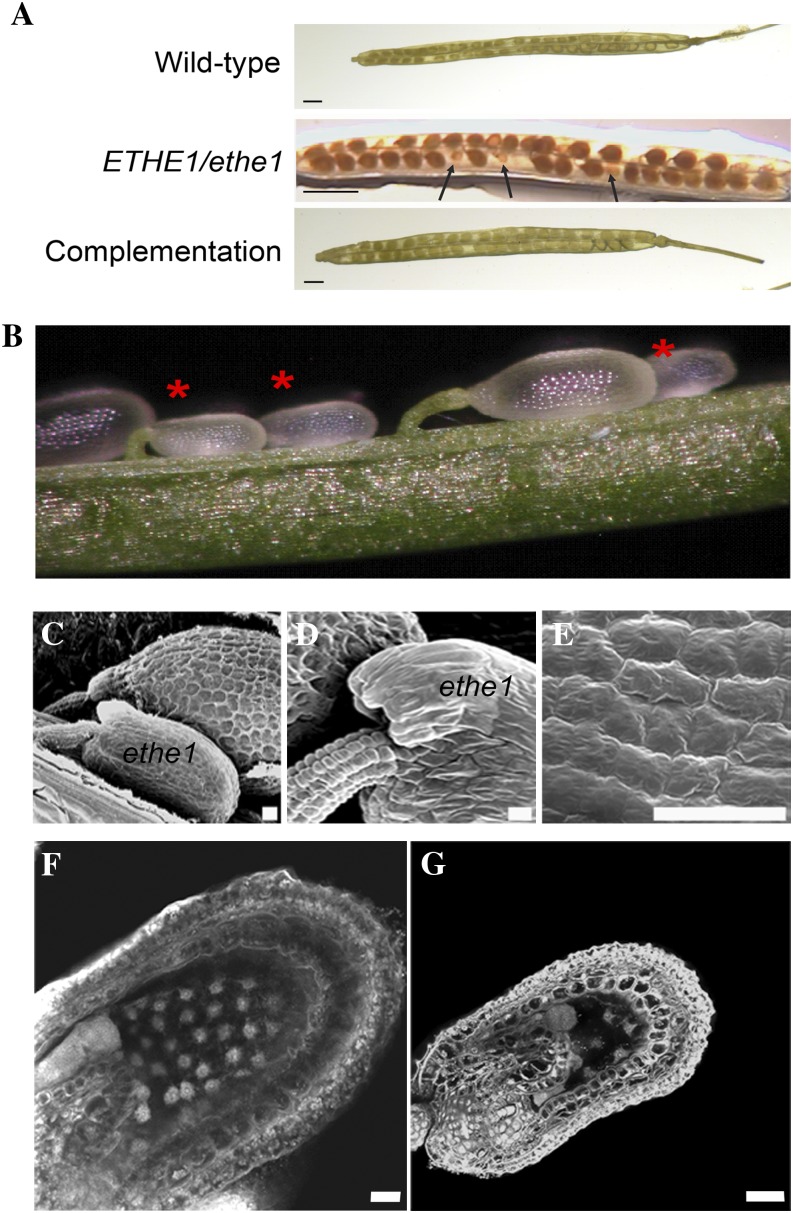
Approximately 150 mature siliques from >50 self-pollinated ETHE1/ethe1 plants were examined and found to contain 24.2% (805/3,353) aborted seeds [c2 = 1.81 < c20.05(1) = 3.841], which appeared as shrunken, shriveled masses (Fig. 3A). In contrast, approximately 4% (27/638) of the seeds in wild-type siliques appeared aborted. Likewise, approximately 25% of the seeds in immature siliques (7–8 mm in length and longer) of ETHE1/ethe1 plants were smaller in size than their wild-type counterparts (Fig. 3B). Scanning electron microscopy (SEM) revealed no differences between seeds in the siliques of ETHE1/ethe1 plants at very early stages in seed development. However, size differences soon became apparent with the outer integument cells in the smaller ethe1 seeds appearing smoother and less well defined than those of wild-type and heterozygous seeds in the same silique (Fig. 3, C and D). The appearance of the integument on these seeds is similar to that of wild-type seeds during early stages of development (Fig. 3E), suggesting that seed coat development is delayed or arrests relatively early in the development of ethe1 seeds.
Figure 3.
Inactivation of ETHE1 disrupts seed development. A, Mature siliques of wild-type, ETHE1/ethe1, and ethe1 complementation plants observed by light microscopy. Aborted seeds are indicated by arrows. Size bar = 1.5 mm. B, ETHE1/ethe1 developing siliques observed by light microscopy. Ethe1−/− seeds are indicated with an asterisk. C to E, SEM of developing seeds in ETHE1/ethe1 siliques. C and D, The outer integument in post-globular-staged wild-type and ethe1 seeds. Higher magnification image of the outer integument of an earlier staged wild-type seed is shown in section E. F and G, Confocal microscopy of Wt (F) and ethe1 (G) seeds from the same silique at approximately octant stage. Size bar = 10 μm. [See online article for color version of this figure.]
Embryo and endosperm development was investigated in siliques of ETHE1/ethe1 plants using laser-scanning confocal microscopy to more specifically determine the nature of the defect. Although differences in seed size were already apparent in approximately 25% (129 of 560) of the seeds, which we predict to be ethe1 seeds, no obvious alterations were observed in early embryo development (Fig. 3, F and G). The smaller ethe1 seeds did, however, contain fewer endosperm nuclei than wild-type seeds. These differences are most pronounced in the peripheral endosperm (PEN) region, which accounts for the majority of the size of the seed.
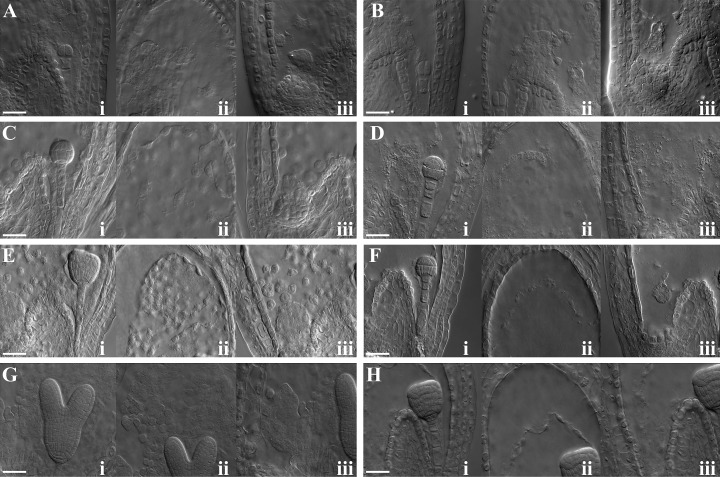
Whole-mount clearing was used to further investigate embryo and endosperm development in siliques of ETHE1/ethe1 plants. In wild-type Arabidopsis plants, a single-celled zygote and a primary endosperm nucleus are normally observed along with degenerated and persistent synergid cells after fertilization of the egg and central cell. Endosperm development begins with the division of the fertilized central cell before the first division of the zygote. Several synchronized syncytial nuclear divisions, accompanied by cell growth, result in a large multinucleate cell that is divided into three distinct domains: the micropylar endosperm (MCE; Fig. 4Ai) surrounding the embryo, the PEN (Fig. 4Aii) in the central chamber, and the chalazal endosperm (CZE; Fig. 4Aiii) opposite to the developing embryo (Brown et al., 1999; Olsen, 2004). After the endosperm nucleus has undergone three to four rounds of nuclear division the zygote becomes elongated and divides asymmetrically, generating a smaller apical cell and a larger basal cell. Subsequent divisions produce an embryo composed of the embryo proper and the suspensor, which is composed of an enlarged basal cell and a file of six to eight additional cells. The embryo proper undergoes successive rounds of division forming the two-cell, quadrant (Fig. 4Ai), octant, dermatogen (Fig. 4Di), and globular (Fig. 4Ci) embryo. By the time the embryo has reached early globular stage, the endosperm, which consists of roughly 100 to 200 free nuclei, begins to cellularize, beginning first with the MCE and progressing through the PEN and CZE (Brown et al., 1999; Nguyen et al., 2000). Continued development of the embryo results in the early heart (Fig. 4Ei), heart, torpedo (Fig. 4Gi), and mature embryo, respectively. Cellularization of the endosperm is complete by the time the embryo reaches late heart stage. The endosperm is then gradually depleted as the embryo continues to develop.
No obvious alterations were observed early in embryo and endosperm development, including at the zygote and early quadrant stages of development (Fig. 4, A and B). However, clear differences in the endosperm and subtle differences in the embryo were apparent, beginning at approximately octant/dermatogen stage (Fig. 4, C and D). Wild-type seeds at the eight-cell octant stage contain approximately 100 free endosperm nuclei in the PEN. In contrast, few endosperm nuclei were observed in the PEN of ethe1 seeds. In addition, few chalazal nodules were observed, and in some instances the CZE appeared to degenerate (Fig. 4Diii). At approximately this time, embryo development also appeared to slow in ethe1 seeds. Specifically, embryos in ethe1 seeds had only progressed to the dermatogen stage when the rest of the seeds in the silique contained embryos at the globular stage (Fig. 4, Ci and Di). By the time wild-type embryos had reached the triangular stage the endosperm contained approximately 300 cellularized nuclei in the PEN and the CZE was well developed (Fig. 4E, i–iii). In contrast, ethe1 embryos in the same silique had only progressed to the globular stage with 20 to 30 nuclei observed in the PEN and no development of the CZE (Fig. 4F, i–iii). As seed development progressed, ethe1 embryo development continued to lag that of the wild type, and development of the endosperm arrested. When wild-type embryos were at the torpedo stage, ethe1 embryos had only reached the triangular stage (Fig. 4, Gi and Hi). At this point 40 to 50 endosperm nuclei could be found in the PEN with few, if any, nuclei present in the CZE of ethe1 seeds (Fig. 4H, ii and iii). Cellularization of endosperm never occurred in ethe1 seeds. Instead, premature degeneration of the PEN and CZE was observed in ethe1 seeds, with embryo development eventually arresting at approximately early heart stage.
Figure 4.
Embryo and endosperm development in ethe1 siliques. Embryo and endosperm images from wild-type and ethe1 seeds analyzed by whole-mount clearing are shown. Images of wild-type (A, C, E, and G) and corresponding ethe1 (B, D, F, and H) embryo sacs from the same siliques. Adjacent images of embryos (i), PEN (ii), and CZE (iii) were taken from the same embryo sac. A, Wild-type four-cell embryo (i), PEN (ii), and CZE (iii). B, ethe1 four-cell embryo (i), PEN (ii), and CZE (iii). No abnormalities were observed at this stage. C, Wild-type globular embryo (i), PEN (ii), and CZE (iii). D, ethe1 dermatogen embryo (i), PEN with only few endosperm nuclei visible (ii), and CZE (iii). E, Wild-type early heart embryo (i), PEN that is undergoing cellularization (ii), CZE (iii). F, ethe1 early globular embryo (i), PEN (ii), and CZE (iii). Very few nuclear cytoplasmic domains (NCDs) are visible in the PEN and CZE. G, Wild-type torpedo stage embryo (i), PEN that has completed the cellularization process (ii), and CZE (iii). H, ethe1 triangular embryo (i), PEN (ii), and CZE (iii). NCDs in the PEN and CZE are in the process of degradation. Scale bars = 10 μm.
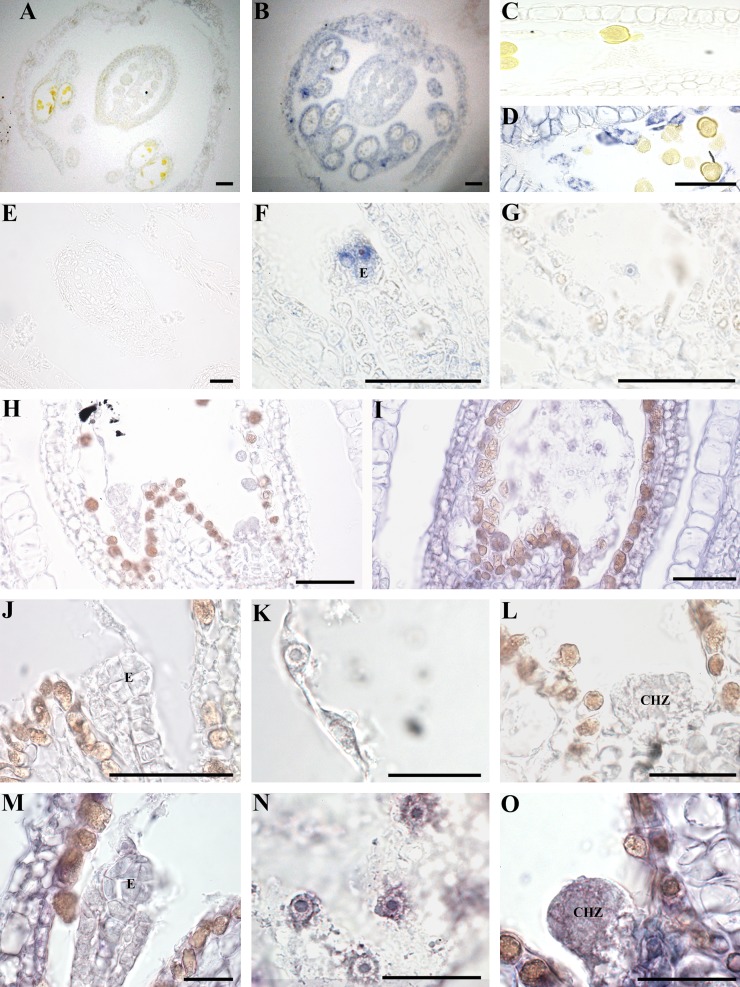
We next investigated the distribution of ETHE1 during wild-type flower and seed development to help understand the observed seed defects. Immunolocalization studies on buds, flowers, and developing siliques revealed ETHE1 signals above background levels in all tissues examined (Fig. 5). ETHE1 is present in flower buds with the strongest ETHE1 signals observed in the tapetal cells, the nutrient cells of the anther (Fig. 5, A–D). In early stages of seed growth, including from the zygote to globular embryo stages, ETHE1 signal is present in the developing embryo and throughout the endosperm (Fig. 5, E–I). Closer examination revealed that early in development the strongest ETHE1 signals are observed in the embryo proper and the PEN of young, zygote-stage seeds (Fig. 5, F and G). As development progresses to the globular stage, the ETHE1 signal becomes more pronounced and is the strongest in the PEN and CZE tissues (Fig. 5, M–O). At this stage in development, the MCE has begun to cellularize, while the PEN and CZE are still rapidly dividing.
Figure 5.
Immunolocalization of ETHE1 in wild-type buds. Sections of wild-type buds (A and B), anthers (C and D), and seeds (E–O) were prepared and treated with preimmune serum (A, C, E, H, J–l) or ETHE1 antibody (B, D, F, G, I, M–O). ETHE1 cross-reactive material is purple, nonspecific signals from the detection system are brown. ETHE1 is present in the tapetal cells of the anther (B and D), zygote (F), and PEN nuclei (G). ETHE1 is also present in globular-staged seeds (I), in particular, in the globular embryo (M), the PEN nuclei (N), and the chalazal cyst (O). The embryo and chalazal region are labeled with e and chz, respectively. Size bar = 50 μm.
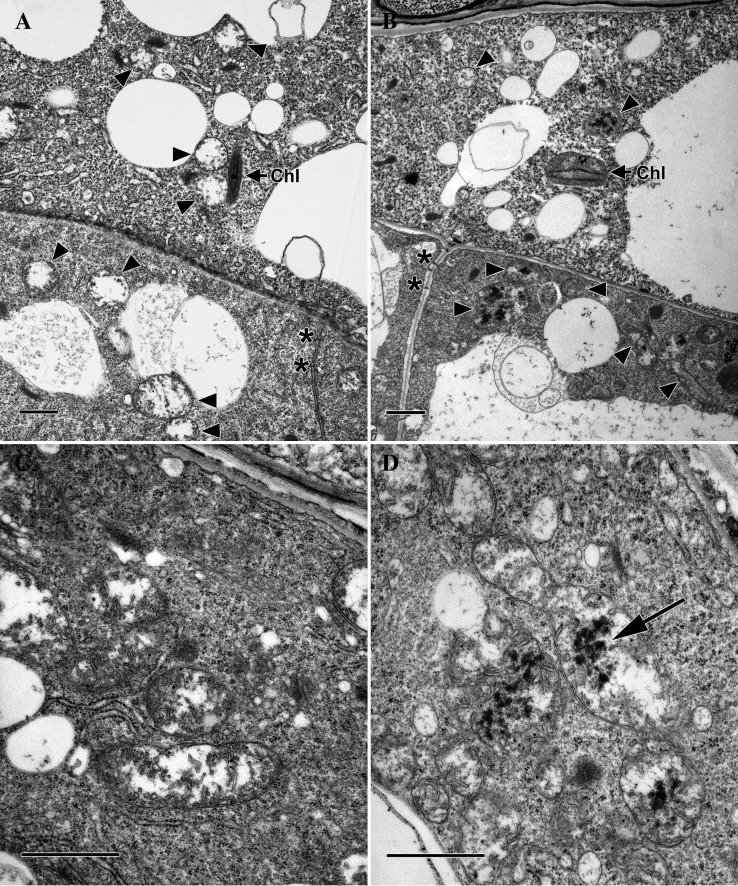
Transmission electron microscopy (TEM) was then performed to further define the differences in the endosperm and embryos of ethe1 and wild-type seeds. Light microscopy indicated that embryo arrest occurs by approximately early heart stage in ethe1 seeds. The ultrastructure of both endosperm and embryo cells in ethe1 ovules at this stage was consistently poor, even though the ultrastructure of the surrounding cells of the ovule wall was consistently good (not shown). This is consistent with cellular degeneration of the endosperm and embryo at the time of arrest. In contrast, the ultrastructure of embryo and endosperm cells in ethe1 ovules at earlier stages of development was relatively normal. For example, although the size of ovules containing wild-type and ethe1 globular-staged embryos was quite different, the ultrastructure of the embryo and suspensor cells was similar (Fig. 6, A and B), with the exception of the appearance of mitochondria. Adjacent cells of the suspensor are connected by plasmodesmata, and the suspensor cells and endosperm contain numerous large and small vacuoles. Small plastids with a few thylakoid membranes and plastoglobuli are present in both the suspensor and endosperm, visible in the cytoplasm of the endosperm shown in Figure 6, A and B. While mitochondria in the suspensor and endosperm of the wild type have electron-lucent interiors and relatively few lamellar cristae that are restricted to the periphery of the organelles (Fig. 6, A and C), mitochondria in ethe1 suspensors and endosperm often have electron-dense inclusions in the mitochondrial matrix (Fig. 6, B and D). The nuclear cytoplasmic domains of the PEN are also smaller in ethe1 seeds than in the wild type. Therefore, with the exception of the size of these nuclear cytoplasmic domains, mitochondrial alterations are the first alterations observed at the ultrastructural level.
Figure 6.
TEMs of wild-type and ethe1 seeds at late globular stage. Micrographs of endosperm (oriented to bottom; A and B) and adjacent basal cells of the suspensor (C and D) in the wild type (A and C) and ethe1 (B and D). Plasmodesmata (asterisk) are present between cells of the suspensor. Small chloroplasts are visible in the endosperm (Chl) and mitochondria (arrowheads) and large and small vacuoles are present in both endosperm and suspensor cells. Wild-type mitochondria have electron lucent interiors with cristae located at the periphery. The interior of most ethe1 mitochondria contain electron-dense inclusions. Size bars = 1 mm.
The tight genetic linkage between the ETHE1 T-DNA insertion and the observed seed defects indicated that ETHE1 is an essential gene that is required for early embryo and endosperm development. A complementation study was performed to verify this conclusion. A 3.4-kb genomic DNA fragment containing ETHE1 was transformed into a segregating population of ETHE1/ethe1 plants. Three independent lines that were homozygous for the ethe1 T-DNA mutation and contained the ETHE1 complementation construct were identified (Fig. 3A). To further verify that complete complementation had occurred, individual siliques from ethe1 plants that contained the complementation construct were analyzed for the presence of abnormal/aborted seeds. A total of 59 (3.7%) out of 1,341 seeds examined in siliques from the complementation lines were aborted, which is similar to the number of aborted seeds typically observed in wild-type siliques. These results confirm that ETHE1 is required for endosperm and, ultimately, normal embryo development.
DISCUSSION
In this report we show that the protein encoded by At1g53580 encodes the Arabidopsis ortholog of human ETHE1, and that Arabidopsis ETHE1 is an essential protein required for endosperm and embryo development. The first defects associated with ethe1 seeds are observed in the endosperm. Wild-type seeds at the eight-cell octant stage contain approximately 50 to 75 free endosperm nuclei in the PEN, and chalazal nodules have formed (Ingouff et al., 2005). At this time wild-type seeds have begun to enlarge. In contrast, ethe1 seeds are significantly smaller with relatively few nuclei in the PEN; chalazal nodules were typically also absent (Figs. 3G and 4D). Early endosperm development is coupled to cell elongation and seed coat development (Sundaresan, 2005; Ingouff et al., 2006) and several studies have shown that the endosperm plays a direct role in early seed size (Garcia et al., 2003; Weijers et al., 2003; Wang et al., 2010). For example, the haiku2 mutant shows defects in endosperm development, integument elongation, and seed size (Garcia et al., 2003, 2005; Luo et al., 2005). However, the endosperm of ethe1 seeds arrests at a much earlier stage of development than in haiku2 seeds, which exhibit precocious endosperm cellularization, reduced proliferation of the endosperm, and a reduction of embryo growth at the torpedo stage (Garcia et al., 2003). Interestingly, haiku2 seeds while smaller than the wild type are still viable. The delayed/restricted development of the integument and reduced size of ethe1 seeds is consistent with the theory that early endosperm development plays an important role in seed size and integument development.
A delay in ethe1 embryo development becomes apparent by the time wild-type embryos reach the globular stage. At this point ethe1 seeds contain embryos that had progressed to the dermatogen stage (Fig. 4, Ci and Di). As seed development continues the alterations in ethe1 seeds become more apparent. By the time wild-type embryos reach the triangular stage the endosperm contained approximately 300 cellularized nuclei in the PEN and the CZE was well developed (Fig. 4E, i–iii). In contrast, ethe1 embryos in the same silique had only progressed to the early globular stage with 20 to 30 nuclei observed in the PEN and no development of the CZE (Fig. 4F, i–iii). As seed development progressed further, ethe1 embryo development continued to slow and development of the endosperm arrested. When wild-type embryos were at the torpedo stage, ethe1 embryos had only reached the triangular stage (Fig. 4, Gi and Hi) with 40 to 50 endosperm nuclei in the PEN with few, if any nuclei present in the CZE (Fig. 4H, ii and iii). Cellularization of endosperm never occurred in ethe1 seeds; rather, degeneration of the PEN and CZE was observed in most ethe1 seeds by the time embryo development arrested at the triangular/early heart stage.
Several mutants have been isolated showing defects in late endosperm development, including the absence of endosperm cellularization. The majority of these mutants such as pilz, titan, knolle, and hinkel are in genes required for nuclear or cellular division and affect both the embryo and endosperm (Lukowitz et al., 1996; Liu and Meinke, 1998; Mayer et al., 1999; Strompen et al., 2002). In several other mutants, including spatzle, fis1, and fis2, the endosperm fails to cellularize, but no apparent cytokinesis defects are observed in the developing embryo (Luo et al., 1999, 2000; Ohad et al., 1999; Sørensen et al., 2002). Interestingly, even though endosperm cellularization is blocked in these mutants, the resulting seed is comparable in size to the wild type and is able to develop into a fully functional plant, suggesting that late endosperm development is not required for embryo viability (Sørensen et al., 2002). Therefore, the absence of endosperm cellularization that we observe is likely not the cause of the embryo arrest in ethe1 seeds.
With the exception of the developmental delay, major alterations were not observed in ethe1 embryos (Fig. 4, B, D, F, and H). The observation that the earliest and most dramatic effects are on the endosperm indicates that ETHE1 is critical for early endosperm development and suggests that ETHE1 may play a lesser/later role in embryo development. Consistent with this theory is our observation that while ETHE1 is present in both the embryo and endosperm during early seed development (Fig. 6, G and H), the strongest ETHE1 signals are observed in the PEN and the chalazal cyst (Fig. 6, M–O).
The endosperm plays a number of roles in embryo development, including serving as a physical support for the growing embryo, protecting the embryo from physical and osmotic stress, and serving a critical role in nutrient flux and storage (Nguyen et al., 2000; Garcia et al., 2003). Metabolic and transcriptional profiling along with classical biochemical studies all provide strong support for the role of the endosperm acting as a sink for the storage, processing, and delivery of nutrients to the embryo (for review, see Berger et al., 2006; Ingram, 2010). Therefore, the slow growth of the embryo in ethe1 seeds is likely due in part to the early alterations we observe in the endosperm. ETHE1 may also play an important role in the suspensor and/or embryo proper. Mitochondrial alterations in the suspensor and endosperm are the first ultrastructural abnormalities that we observe. The suspensor and in particular the basal cell of the suspensor is metabolically very active. During mid to late globular stages giant mitochondria, which are hypothesized to form when the nutritional needs of the developing embryo outstrip the ability of the endosperm to provide nutrition, are observed in the basal cells in Arabidopsis (Mansfield and Briarty, 1991). Therefore, the slow growth and ultimate arrest of ethe1 embryos could result from a reduced flow of nutrients from the endosperm and suspensor that results from mitochondrial dysfunction in these cells.
Humans with EE and ethe1−/− mice excrete large amounts of thiosulfate and human ETHE1 exhibits sulfur dioxygenase activity (Tiranti et al., 2006, 2009). Micromolar concentrations of sulfide inhibit mitochondrial COX and short-chain acyl-CoA dehydrogenase (Tiranti et al., 2009; Bouillaud and Blachier, 2011), and long-term exposure to sulfide can result in degradation of COX and mitochondrial dysfunction (Di Meo et al., 2011). We have shown here that Arabidopsis ETHE1 also exhibits sulfur dioxygenase activity. Electron-dense inclusions are observed in the matrix of mitochondria in the endosperm and suspensor cells of ethe1 seeds when examined by TEM (Fig. 6, B and D). Sulfide readily forms precipitates with metal ions, raising the possibility that the precipitates we observed in ethe1 mitochondria during the TEM analysis arise from high sulfide concentrations. Attempts to identify the nature of the mitochondrial inclusions using energy-dispersive x-ray spectrometry were unsuccessful. Similar mitochondrial inclusions were observed in mouse mammary epithelial cell lines treated with selenium (Medina et al., 1983). The nature of these mitochondrial inclusions was not determined, but proposed to be protein:selenium complexes since selenium reacts readily with mercapto groups of organic compounds. High concentrations of selenium can inhibit a number of enzymes, in particular mitochondrial succinic dehydrogenase (Klug et al., 1953). Therefore, the endosperm defects and ultimately the embryo arrest that we observe are likely the direct result of alterations in mitochondrial function.
However, further experiments are necessary to test this hypothesis and determine more specifically the cause of endosperm and embryo arrest in ethe1 plants. For example, the observed defects could arise directly from the inhibition of mitochondrial respiration. However, the alternative oxidase is resistant to inhibition by sulfide and therefore should maintain respiration but with the loss of energy (Bahr and Bonner, 1973). At this time it is not clear if the alternative oxidase is active in the endosperm and developing embryo. It is also possible that inhibition of the mitochondrial respiratory chain enhances superoxide formation, causing oxidative stress in the cells (Bouillaud and Blachier, 2011). This could inhibit endosperm division and ultimately embryo growth. Likewise, little to nothing is known about the sulfur assimilatory pathway in Arabidopsis seeds. Therefore, it is also possible that inactivation of ETHE1 disrupts an aspect of secondary sulfur metabolism, which in turn could affect the levels of brassinosteroid and jasmonate hormones and/or other sulfonated compounds necessary for embryo growth (Klein and Papenbrock, 2004; Halkier and Gershenzon, 2006; Amano et al., 2007). EE patients display a wide range of metabolic alterations, many of which are not easily explained by the sulfur dioxygenase activity of ETHE1 (Burlina et al., 1991, 1994; Tiranti et al., 2004, 2006, 2009). We expect that this will also be the situation in plants and that more detailed studies on ETHE1 and sulfur metabolism during seed development will be required before the specific basis of the seed defect in ethe1 plants is fully understood.
MATERIALS AND METHODS
Plant Material
The ETHE1 loss-of-function mutant was obtained from the DuPont p2800 T-DNA-tagged seed pool. Seeds were vernalized at 4°C for 48 h prior to growth on commercial potting soil at 23°C with a 16/8-h light/dark cycle. Wild-type Wassilewskija was used as the control in development and immunolocalization studies.
Arabidopsis (Arabidopsis thaliana; ecotype Landsberg erecta) cell suspensions were grown in 50 mL liquid medium (1× Murashige and Skoog basal salts, 1× Gamborg’s B5 vitamins, 3% [w/v] Suc, 0.59 g/L MES, 0.5 mg/L 1-naphthaleneacetic acid, and 0.05 mg/L benzylaminopurine, pH 5.7) at 25°C with gentle agitation (130 rpm) in 16/8-h light/dark cycles. A 6-mL aliquot was transferred to 50 mL fresh medium each week.
Phylogenetic Analysis of β-Lactamase Proteins
A phylogenetic tree was derived from multiple alignments of β-lactamase fold-containing proteins using Clustal W version 1.82. A neighbor-joining analysis was conducted with MEGA version 3.1 using the Poisson correction amino acid substitution model and the complete deletion gaps option (Kumar et al., 2004). Bootstrap values from 500 replicates were calculated and are indicated at branch points on the neighbor-joining tree.
Molecular Analysis of ETHE1
PCR was performed on pooled genomic DNA isolated from a population of Arabidopsis T-DNA insertion lines. T1 seeds from pool p2800, which was found to contain the ETHE1 T-DNA line, were obtained from the Arabidopsis Biological Resource Center. Genotyping was performed by PCR using a combination of T-DNA insertion and wild-type ETHE1 primer pairs. Amplification products were verified through DNA sequencing.
Complementation studies were conducted using pCAMBIA-1390 containing a 3,400-bp ETHE1 genomic DNA fragment. The construct was transformed into GV3101/PMP90 A. tumefaciens cells and transferred into a segregating population of heterozygous ETHE1/ethe1 plants. Transformants were selected on hygromycin (25 mg/L) and confirmed by PCR screening. Genotyping was performed by PCR using primers specific to the T-DNA insertion and the wild-type ETHE1 locus.
The Arabidopsis ETHE1 cDNA was cloned into pFAST (Ge et al., 2005) to generate a C-terminal FAST-tag fusion for expression in plant cells. The construct was sequenced and transformed into GV3101/PMP90 A. tumefaciens cells, which were used to transform Arabidopsis suspension cells (Holdorf, 2008). Transformants were selected on solid medium containing kanamycin (50 mg/L) and cefotaxime (200 mg/L) and confirmed by PCR screening. Positive calli were reintroduced into liquid medium containing kanamycin (50 mg/L) and cefotaxime (200 mg/L).
Biochemical Studies
The Arabidopsis ETHE1 cDNA was cloned into pET15b, sequenced, and introduced into Escherichia coli BL21-RIL cells for protein overexpression. The human ETHE1 overexpression clone was used as a control (Tiranti et al., 2009). ETHE1 plasmid-containing cells were grown, induced, and ETHE1 purified as described previously (Holdorf et al., 2008). Protein purity was assessed by SDS-PAGE gels and quantified at Abs280 using molar extinction coefficients of 10,240 m−1 cm−1 and 19,275 m−1 cm−1 for Arabidopsis and human ETHE1, respectively. Purified ETHE1 (280 mg) was resuspended in 1 mL phosphate-buffered saline and mixed with either Freund’s complete or incomplete adjuvant and used to raise polyclonal antibody in rabbits using standard procedures.
Sulfur-dependent dioxygenase activity was measured on purified ETHE1 using the assay of Hildebrandt and Grieshaber (2008) on an Oxygraph system (Hansatech Instruments). The reaction mixture (2 mL) contained 0.1 m potassium phosphate buffer (pH 7.4), 1 mm reduced glutathione, and 12 μg/mL purified ETHE1. The assay was started by addition of 30 μL of acetonic sulfur or acetone as a control. Acetonic sulfur was a saturated solution containing approximately 17 mm sulfur. The rate of O2 consumption was determined and reported as μmol min−1 mg−1. The data reflect the average rate of at least three different protein preparations for Arabidopsis ETHE1 (n = 14 ± sulfur) and a single preparation for human ETHE1 (n = 8 + sulfur and n = 5 − sulfur). The enzymatic activity of ETHE1 with S-D-lactoylglutathione was measured as previously described (Marasinghe et al., 2005).
ETHE1-FAST cell cultures were fractionated using differential centrifugation and analyzed by western blotting to determine the subcellular location of ETHE1. Harvested cells were lysed in isolation buffer (0.35 m sorbitol, 25 mm MOPS, 0.1% bovine serum albumin, 2 mm EDTA, 0.1% 2-mercaptoethanol, 1% polyvinylpyrrolidone-40) and centrifuged at 2,500g for 5 min. Mitochondria were pelleted and washed three times in wash buffer (0.4 m Suc, 2 mm EDTA, 10 mm MOPS, 0.1% bovine serum albumin) using alternating low- and high-speed spins. The mitochondrial pellet was lysed in 50 mm TRIS pH 8.0, 0.5% Triton X-100, and centrifuged for 30 min at 12,000g. Protein was quantitated using a bicinchoninic acid assay, and 10 mg of total, cytoplasmic, and mitochondrial protein was separated by SDS-PAGE gel for western detection using anti-FLAG or antimitochondrial COX antibody.
Microscopy
Mature siliques from ETHE1/ethe1 and wild-type plants (20 each) were treated with a 4% (w/v) sodium hydroxide solution for 16 h at room temperature and the numbers of aborted and healthy seeds were determined under a dissecting microscope.
Endosperm development was analyzed in siliques from individual ethe1 heterozygous plants using laser-scanning confocal microscopy essentially as described (Braselton et al., 1996). Whole-mount clearing was also used to analyze embryo and endosperm development. Siliques from ethe1 heterozygous plants were dissected and cleared in Herr’s solution containing lactic acid:chloral hydrate:phenol:clove oil:xylene (2:2:2:2:1, w/w). Embryo and endosperm development was studied microscopically with a Nikon microscope equipped with differential interference contrast optics.
SEM was performed on staged flowers/siliques fixed overnight at room temperature in 2.5% (v/v) glutaraldehyde, 0.1 m HEPES buffer (pH 7.2), 0.02% (v/v) Triton X-100, rinsed in 0.1 m HEPES buffer, and postfixed in 1% (w/v) aqueous OsO4 overnight. Individual gynoecia were dissected in buffer and ovules placed into a critical point drier specimen basket submerged in buffer. Samples were dehydrated through a graded ethanol series (10% increments, 1 h each) ending with three changes of 100% ethanol, and critical point dried with a Balzers CPD 020 using CO2 as the transitional fluid. Dried specimens were mounted, sputter coated with gold, and examined with a Hitachi S-570 SEM operating at 15 kV. Digital images were captured using an Oxford Link ISIS microanalysis system.
Immunolocalization studies were conducted on flowers and siliques (0.4–0.8 cm). Blocks were sectioned at 12 microns on a microtome and adhered to poly-l-Lys coated slides (Hong et al., 1996). Immunochemistry was performed essentially as described (Shiba et al., 2001) using anti-ETHE1 antibody (1:1,000) and alkaline-phosphatase goat-anti-rabbit secondary antibody (1:1,000). Samples were observed and analyzed as described above.
For TEM, siliques from ETHE1/ethe1 and wild-type plants were opened, individual ovules were removed with a fine needle and transferred to separate cell strainer baskets immersed in 2.5% (v/v) glutaraldehyde in 0.1 m HEPES buffer (pH 7.2) and 0.02% (v/v) Triton X-100. Ovules were fixed overnight at room temperature and rinsed in three changes of 0.1 m HEPES buffer (pH 7.2). The rinsed ovules were then submerged in 1% (w/v) aqueous OsO4 and postfixed overnight at room temperature. After rinsing with distilled, deionized water individual ovules were moved to a pad of solidified 4% agar and encased in additional warm but still molten 4% agar. The agar solidified was cut into cubes each containing a single ovule. The cubes were dehydrated in acetone and infiltrated in Spurr’s resin modified by the addition of Quetol 651 according to Table 3 of Ellis, 2006, with the exception that 0.05 g 2(dimethylamino)ethanol was used in place of the 0.20 mL N,N-dimethylbenzylamine. Individual ovules were oriented for longitudinal sectioning in flat embedding molds and polymerized. Silver sections were cut with a Reichert Ultracut E ultramicrotome, stained with uranyl acetate and Reynold’s lead citrate, examined, and imaged with a Hitachi H-600 TEM operating at 75 kV.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers human (Homo sapiens) ETHE1 (NP_055112), mouse (Mus musculus) ETHE1 (NP_075643), frog (Xenopus laevis) ETHE1 (NP_001079404), Xenopus tropicalis ETHE1 (NP_001005706), fish (Danio rerio) ETHE1 (NP_998094), Arabidopsis ETHE1 (NP_974018), rice (Oryza sativa) Os01g066200 (NP_001043807), Burkholderia phytofirmans (ZP_01508561), Stigmatella aurantiaca ETHE1 (ZP_01459266), Myxococcus xanthus (YP_633997), Arabidopsis GLX2-1 (NP_973679), Arabidopsis GLX2-2 (NP_187696), Arabidopsis GLX2-4 (NP_849599), Arabidopsis GLX2-5 (NP_850166), human GLX2 (CAA62483), mouse GLX2 (NP_077246), rice GLX2 (AAL14249), Brassica juncea GLX2 (AA026580), yeast GLO2 (CAA71335), yeast (Saccharomyces cerevisiae) GLO4 (CAA99230), rice Os03g0332400 (BAF11930), rice OsJ_010290 (EAZ26807), and Stenotrophomonas maltophilia L1 β-lactamase (CAB63489).
Acknowledgments
Thanks go to Valeria Tiranti for the human ETHE1 cDNA clone, Sue Gibson for the pooled DNAs, and the Arabidopsis Biological Resource Center for the seed stocks. We would also like to thank Rich Moore, Lara Strittmatter, and Ling Jiang for helpful discussions, Joey Van Rossum for performing the SEM analysis, and Richard Edelman and Matthew Duley for support with the microscopes.
Glossary
- EE
ethylmalonic encephalopathy
- COX
cytochrome c oxidase
- SEM
scanning electron microscopy
- PEN
peripheral endosperm
- MCE
micropylar endosperm
- CZE
chalazal endosperm
- TEM
transmission electron microscopy
- cDNA
complementary DNA
- T-DNA
transferred DNA
References
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. (2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr JT, Bonner WD., Jr (1973) Cyanide-insensitive respiration. II. Control of the alternate pathway. J Biol Chem 248: 3446–3450 [PubMed] [Google Scholar]
- Berger F, Grini PE, Schnittger A. (2006) Endosperm: an integrator of seed growth and development. Curr Opin Plant Biol 9: 664–670 [DOI] [PubMed] [Google Scholar]
- Bouillaud F, Blachier F. (2011) Mitochondria and sulfide: a very old story of poisoning, feeding, and signaling? Antioxid Redox Signal 15: 379–391 [DOI] [PubMed] [Google Scholar]
- Braselton JP, Wilkinson MJ, Clulow SA. (1996) Feulgen staining of intact plant tissues for confocal microscopy. Biotech Histochem 71: 84–87 [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen OA. (1999) Development of endosperm in Arabidopsis thaliana. Sex Plant Reprod 12: 32–42 [Google Scholar]
- Burlina AB, Dionisi-Vici C, Bennett MJ, Gibson KM, Servidei S, Bertini E, Hale DE, Schmidt-Sommerfeld E, Sabetta G, Zacchello F, et al. (1994) A new syndrome with ethylmalonic aciduria and normal fatty acid oxidation in fibroblasts. J Pediatr 124: 79–86 [DOI] [PubMed] [Google Scholar]
- Burlina AB, Zacchello F, Dionisi-Vici C, Bertini E, Sabetta G, Bennet MJ, Hale DE, Schmidt-Sommerfeld E, Rinaldo P. (1991) New clinical phenotype of branched-chain acyl-CoA oxidation defect. Lancet 338: 1522–1523 [DOI] [PubMed] [Google Scholar]
- Di Meo I, Fagiolari G, Prelle A, Viscomi C, Zeviani M, Tiranti V. (2011) Chronic exposure to sulfide causes accelerated degradation of cytochrome c oxidase in ethylmalonic encephalopathy. Antioxid Redox Signal 15: 353–362 [DOI] [PubMed] [Google Scholar]
- Ellis E. (2006) Solutions to the problem of substitution of ERL 4221 for vinyl cyclohexene dioxide in Spurr low viscosity embedding formulations. Microscopy Today 14: 32–33 [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F. (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Saingery V, Chambrier P, Mayer U, Jürgens G, Berger F. (2003) Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol 131: 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XC, Dietrich C, Matsuno M, Li GJ, Berg H, Xia YJ. (2005) An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep 6: 282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Hell R, Leustek T. (2005) Sulfur metabolism in plants and algae—a case study for an integrative scientific approach. Photosynth Res 86: 297–298 [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM, Grieshaber MK. (2008) Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J 275: 3352–3361 [DOI] [PubMed] [Google Scholar]
- Holdorf MM. (2008) Characterization of Arabidopsis ETHE1, a gene associated with ethylmalonic encephalopathy. PhD thesis. Miami University, Oxford, OH
- Holdorf MM, Bennett B, Crowder MW, Makaroff CA. (2008) Spectroscopic studies on Arabidopsis ETHE1, a glyoxalase II-like protein. J Inorg Biochem 102: 1825–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Takano M, Liu CM, Gasch A, Chye ML, Chua NH. (1996) Expression of three members of the calcium-dependent protein kinase gene family in Arabidopsis thaliana. Plant Mol Biol 30: 1259–1275 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Haseloff J, Berger F. (2005) Polycomb group genes control developmental timing of endosperm. Plant J 42: 663–674 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Jullien PE, Berger F. (2006) The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell 18: 3491–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram GC. (2010) Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247: 195–214 [DOI] [PubMed] [Google Scholar]
- Jin ZP, Shen JJ, Qiao ZJ, Yang GD, Wang R, Pei YX. (2011) Hydrogen sulfide improves drought resistance in Arabidopsis thaliana. Biochem Biophys Res Commun 414: 481–486 [DOI] [PubMed] [Google Scholar]
- Klein M, Papenbrock J. (2004) The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J Exp Bot 55: 1809–1820 [DOI] [PubMed] [Google Scholar]
- Klug HL, Moxon AL, Petersen DF, Painter EP. (1953) Inhibition of rat liver succinic dehydrogenase by selenium compounds. J Pharmacol Exp Ther 108: 437–441 [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP. (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Liu CM, Meinke DW. (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16: 21–31 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84: 61–71 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti MK, Krishnasamy S, Owen HA, Makaroff CA. (1997) Molecular characterization of glyoxalase II from Arabidopsis thaliana. Plant Mol Biol 35: 471–481 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. (1991) Early embryogenesis in Arabidopsis thaliana. 2. The developing embryo. Can J Bot 69: 461–476 [Google Scholar]
- Marasinghe GPK, Sander IM, Bennett B, Periyannan G, Yang KW, Makaroff CA, Crowder MW. (2005) Structural studies on a mitochondrial glyoxalase II. J Biol Chem 280: 40668–40675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Herzog U, Berger F, Inzé D, Jürgens G. (1999) Mutations in the pilz group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur J Cell Biol 78: 100–108 [DOI] [PubMed] [Google Scholar]
- Medina D, Miller F, Oborn CJ, Asch BB. (1983) Mitochondrial inclusions in selenium-treated mouse mammary epithelial cell lines. Cancer Res 43: 2100–2105 [PubMed] [Google Scholar]
- Nguyen H, Brown RC, Lemmon BE. (2000) The specialized chalazal endosperm in Arabidopsis thaliana and Lepidium virginicum (Brassicaceae). Protoplasma 212: 99–110 [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. (1999) Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell (Suppl) 16: S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennenburg H. (1989) Synthesis and emission of hydrogen sulfide by higher plants. In ES Saltzman, WJ Cooper, eds, Biogenic Sulfur in the Environment. American Chemical Society, New Orleans, pp 44–57
- Rohwerder T, Sand W. (2003) The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149: 1699–1710 [DOI] [PubMed] [Google Scholar]
- Rohwerder T, Sand W. (2007) Oxidation of inorganic sulfur compounds in acidophilic prokaryotes. Eng Life Sci 7: 301–309 [Google Scholar]
- Shiba H, Takayama S, Iwano M, Shimosato H, Funato M, Nakagawa T, Che FS, Suzuki G, Watanabe M, Hinata K, et al. (2001) A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-incompatibility of Brassica species. Plant Physiol 125: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla-Pareek SL, Reddy MK, Sopory SK. (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci USA 100: 14672–14677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla-Pareek SL, Yadav SK, Pareek A, Reddy MK, Sopory SK. (2006) Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol 140: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MB, Mayer U, Lukowitz W, Robert H, Chambrier P, Jürgens G, Somerville C, Lepiniec L, Berger F. (2002) Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129: 5567–5576 [DOI] [PubMed] [Google Scholar]
- Strompen G, El Kasmi F, Richter S, Lukowitz W, Assaad FF, Jürgens G, Mayer U. (2002) The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol 12: 153–158 [DOI] [PubMed] [Google Scholar]
- Sugio T, Katagiri T, Inagaki K, Tano T. (1989) Actual substrate for elemental sulfur oxidation by sulfur-ferric ion oxidoreductase purified from Thiobacillus ferrooxidans. Biochim Biophys Acta 973: 250–256 [Google Scholar]
- Sundaresan V. (2005) Control of seed size in plants. Proc Natl Acad Sci USA 102: 17887–17888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V, Briem E, Lamantea E, Mineri R, Papaleo E, De Gioia L, Forlani F, Rinaldo P, Dickson P, Abu-Libdeh B, et al. (2006) ETHE1 mutations are specific to ethylmalonic encephalopathy. J Med Genet 43: 340–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V, D’Adamo P, Briem E, Ferrari G, Mineri R, Lamantea E, Mandel H, Balestri P, Garcia-Silva MT, Vollmer B, et al. (2004) Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein. Am J Hum Genet 74: 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V, Viscomi C, Hildebrandt T, Di Meo I, Mineri R, Tiveron C, Levitt MD, Prelle A, Fagiolari G, Rimoldi M, et al. (2009) Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat Med 15: 200–205 [DOI] [PubMed] [Google Scholar]
- Wang AH, Garcia D, Zhang HY, Feng K, Chaudhury A, Berger F, Peacock WJ, Dennis ES, Luo M. (2010) The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J 63: 670–679 [DOI] [PubMed] [Google Scholar]
- Weijers D, van Hamburg JP, van Rijn E, Hooykaas PJJ, Offringa R. (2003) Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol 133: 1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SK, Singla-Pareek SL, Ray M, Reddy MK, Sopory SK. (2005a) Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem Biophys Res Commun 337: 61–67 [DOI] [PubMed] [Google Scholar]
- Yadav SK, Singla-Pareek SL, Reddy MK, Sopory SK. (2005b) Transgenic tobacco plants overexpressing glyoxalase enzymes resist an increase in methylglyoxal and maintain higher reduced glutathione levels under salinity stress. FEBS Lett 579: 6265–6271 [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu SL, Zhang ZJ, Hu LY, Jiang CX, Wei ZJ, Liu JA, Wang HL, Jiang ST. (2011) Hydrogen sulfide acts as a regulator of flower senescence in plants. Postharvest Biol Technol 60: 251–257 [Google Scholar]