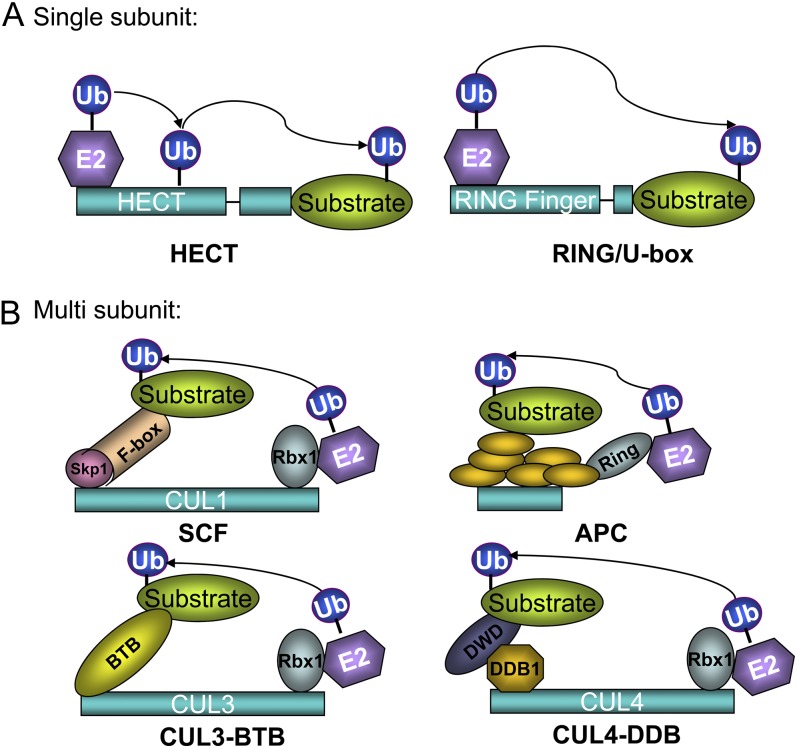
The ubiquitin-proteasome system (UPS) is involved in the selective degradation of proteins in the cells of eukaryotic organisms and consists of three main enzymes: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3). In a typical ubiquitination reaction, E1, E2, and E3 enzymes participate in the transfer of the ubiquitin monomers to the ε-amino group of a Lys residue in the target protein. In the first step of the reaction, E1 catalyzes the activation of ubiquitin via ATP, creating a high-energy thiol intermediate. After activation, ubiquitin is transferred to the E2 and then to the E3 ligase for the final step in the process, which involves the creation of an isopeptide bond between the C-terminal Gly of ubiquitin and a Lys residue in the substrate protein (for review, see Vierstra, 2009; Ye and Rape, 2009). The UPS in plants has a hierarchical structure for the three enzymes: there are few E1s, about 50 E2s, but over 1,000 E3s. For example, the rice (Oryza sativa) genome encodes six E1 genes, 49 E2 and E2-like genes, and over 1,300 E3 genes (Du et al., 2009). The abundance of the E3 proteins in the UPS system allows plants to target substrates for many biological processes, because each E3 ubiquitin ligase acts for the ubiquitination of only one or a few target proteins. Based on the subunit component and action modes, E3 ligases can be divided into two major types: the single-subunit type (Homology to E6 carboxyl terminus [HECT] and RING/U-box) and the multisubunit type (Skp1 [for S-phase kinase-associated protein1]-CUL1 [for Cullin1]-F-box [SCF], Anaphase promoting complex [APC], CUL3-BTB [for Bric-a-Brac, tramtrack broad complex], CUL4-DDB [for damage-specific DNA binding protein] complex, and others; Vierstra, 2009; Fig. 1).
Figure 1.
Two types of E3 ligases, their interaction components, and their modes of action.
Depending on the number (single or multiple) of ubiquitins attached to the substrates and on the manner in which multiple ubiquitins are attached to the substrates, the ubiquitin-mediated protein modifications can be classified as monoubiquitination, multiubiquitination, and polyubiquitination (for review, see Hochstrasser, 2006; Mukhopadhyay and Riezman, 2007; Vierstra, 2009). Monoubiquitination and multiubiquitination usually alter substrate protein localizations, affect protein-protein interactions, and modulate protein activities. The polyubiquitination can differ in structure and function depending on how the seven Lys residues in ubiquitin (Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, and Lys-63) are connected to each other and to the substrates. For example, Lys-48-mediated and Lys-11-mediated polyubiquitination target substrates for degradation by the 26S proteasome, while Lys-63-mediated polyubiquitination usually mediates DNA repair, membrane trafficking, and chromatin remodeling (for review, see Ye and Rape, 2009; Liu and Chen, 2011). The function of polyubiquitination mediated by the other four Lys residues is not clear.
The UPS functions in nearly all aspects of plant life, including the cell cycle, embryogenesis, photomorphogenesis/flowering, hormone signaling, and abiotic and biotic stress responses (for review, see Stone and Callis, 2007; Vierstra, 2009; Santner and Estelle, 2010). In this paper, we focus on the function of the rice U-box protein SPOTTED LEAF11 (SPL11) and its Arabidopsis (Arabidopsis thaliana) ortholog protein PLANT U-BOX13 (PUB13) in programmed cell death (PCD), defense, and flowering (Table I).
Table I. Summary of the rice and Arabidopsis genes discussed in this paper.
| Gene | Full Name | Species | Protein Encoding | Biological Function |
|---|---|---|---|---|
| SPL11 | Spotted leaf11 | Rice | U-box domain and ARM repeat domain protein | Negative regulator of PCD and defense, and positive regulator of flowering in LD conditions |
| SPIN1 | SPL11-interacting protein1 | Rice | K homology domain-containing RNA-binding protein | Suppressor of flowering in both SD and LD conditions |
| SPIN6 | SPL11-interacting protein6 | Rice | Small GTPase-activating protein | Component involved in SPL11-mediated PCD and defense |
| RBS1 | RNA-binding and SPIN1-interacting1 | Rice | RNA-binding protein | Component involved in SPL11/SPIN1-mediated flowering |
| OsRAC1 | Rice homolog of mammalian Rac-GTPase RAC1 | Rice | Small GTPase protein | Integrator of PTI and ETI |
| OsCERK1 | Chitin elicitor receptor kinase1 | Rice | LysM RLK | Positive regulator of defense by perceiving chitin elicitors |
| Pit | Pyricularia grisea resistance locus t | Rice | CC-nucleotide-binding site-LRR protein | Positive regulator by recognizing its cognate M. oryzae Avr effector AvrPit-triggered immune signaling |
| Hd1 | Heading date1 | Rice | Zinc finger protein and ortholog of Arabidopsis CO | Positive regulator of flowering in SD conditions but a negative regulator in LD conditions |
| Hd3a | Heading date3a | Rice | Similar to phosphatidylethanolamine-binding protein and ortholog of Arabidopsis FT | Positive regulator of flowering in both SD and LD conditions |
| MAPKs | Mitogen-activated protein kinases | Rice and Arabidopsis | Ser/Thr protein kinases | Components for signal transduction by phosphorylation |
| NOXs | NADPH oxidases | Rice and Arabidopsis | Membrane-bound enzyme complex | Regulator of ROS generation |
| PUB13 | Plant U-box13 | Arabidopsis | U-box and ARM repeat domain protein and ortholog of rice SPL11 | Negative regulator of plant PCD, defense, and flowering in LD conditions |
| PAD4 | Phytoalexin deficient4 | Arabidopsis | Triacylglycerol lipase-like protein | Positive regulator of SA-dependent defense signaling |
| SID2 | SA induction deficient2 | Arabidopsis | Isochorismate synthase | Positive regulator of SA-dependent defense signaling and type I SA gene required for SA synthesis |
| WIN3 | HOPW1-1-interacting3 | Arabidopsis | Member of the firefly luciferase family | Positive regulator of SA-dependent defense signaling and type II SA gene regulating SA level |
| FLS2 | Flagellin-sensing2 | Arabidopsis | LRR-RLK | Positive regulator of plant defense by perceiving PAMPs (e.g. flagellin) |
| BAK1 | Brassinosteroid-insensitive1-associated receptor kinase1 | Arabidopsis | LRR-RLK | Component involved in hormone BRI1-mediated growth and development and plant PTI immune signaling |
| FLC | Flowering locus C | Arabidopsis | MADS box transcription factor | Suppressor of the flowering signal from the vernalization and autonomous pathways |
| FT | Flowering locus T | Arabidopsis | Similar to phosphatidylethanolamine-binding protein | Positive regulator of flowering in both SD and LD conditions |
| SOC1 | Suppressor of overexpression of constans1 | Arabidopsis | MADS box transcription factor | Integrator of multiple flowering signals from photoperiod, temperature, hormone, and age-related signals |
| HFR1 | Long hypocotyl far-red light1 | Arabidopsis | Atypical basic helix-loop-helix protein | Component involved in plant photomorphogenesis |
THE FUNCTION OF PCD IN PLANT DEFENSE
During the course of coevolution, plants have developed two layers of innate immune systems that rely on the pattern-recognition receptors (PRRs) and resistance (R) proteins to defend against pathogen attack (for review, see Jones and Dangl, 2006; Dodds and Rathjen, 2010). The first line of defense depends on the activation of PRRs by the perception of pathogen-associated molecular patterns (PAMPs). The PAMP-triggered immunity (PTI) causes the accumulation of reactive oxygen species (ROS) and the deposition of phenolic compounds. To suppress PTI, pathogens have evolved effector proteins that can be secreted into the cytoplasm of host cells. In response, plants employ R proteins, such as nucleotide-binding site leucine-rich repeat (LRR) proteins (or NLRs), to monitor the entry of these effector proteins directly or indirectly, resulting in the second layer of defense, which is called effector-triggered immunity (ETI). ETI is more robust and effective than PTI and is often associated with a hypersensitive response (HR), which is characterized by the rapid death of cells in the local region surrounding an infection. Several recent studies, however, have challenged the distinction between PTI and ETI and have provided evidence for a continuum between the two types of immunities (Lee et al., 2009; Thomma et al., 2011).
Lesion-mimic (LM) mutants displaying spontaneous HR-like cell death under normal growth conditions represent a unique kind of PCD in plants. Many LM mutants have been identified and characterized in maize (Zea mays), Arabidopsis, rice, barley (Hordeum vulgare), and tobacco (Nicotiana tabacum). One of the common characteristics of numerous LM mutants is that they exhibit enhanced resistance to biotrophic or hemibiotrophic pathogens. Cloning and molecular characterization have revealed that the mutated genes encode various proteins involved in defense-related signaling pathways, including ion channels, ROS generation, sphingolipid metabolism, porphyrin/phenolics/chorophyll biosynthesis and metabolism, ubiquitination, and other signaling transduction (for review, see Love et al., 2008; Moeder and Yoshioka, 2008).
An intimate relationship between LM mutation and ETI was indicated by the genetic analysis of the Arabidopsis LM mutant lesion simulating disease1 (lsd1). This analysis revealed that the phenotypes of the mutant were nullified by mutation in EHANCED DISEASE SUSCEPTIBILITY1 and PHYTOALEXIN DEFICIENT4 (PAD4), two important genes required for the Toll-IL-1 receptor (TIR)-type NLR R gene-mediated resistance and also for basal resistance (Rustérucci et al., 2001; Wiermer et al., 2005). Both genes are also essential for cell death regulation in Arabidopsis under intense light and for the ROS- and salicylic acid (SA)-dependent defense signal amplification loop (Glazebrook, 2005; Mühlenbock et al., 2008). Recently, a suppressor screening of the lsd1 mutant revealed that PHOENIX21 is a positive regulator of lsd1 runaway cell death and is a member of the ACTIVATED DISEASE RESISTANCE1 family of the coiled coil (CC)-type NLR proteins (Bonardi et al., 2011). Thus, these results suggested that the HR and cell death in some LM mutants share common signaling pathways.
THE FUNCTION OF UPS IN PCD AND DEFENSE RESPONSES IN PLANTS
The role of UPS in regulating apoptosis in animals has been well established (for review, see Broemer and Meier, 2009). Recently, the role of UPS in plant PCD and defense responses has become clearer based on many studies with dicot plants. One of the first studies reported that Arabidopsis SGT1, a homolog of yeast SUPPRESSOR OF G2 ALLELE OF SKP1 (SGT1), interacts with SUPPRESSOR OF KINETOCHORE PROTEIN1 (SKP1) and CULLIN1 (CUL1), subunits of the Skp1-Cullin-F-box (SCF) E3 ligase, and is required for defense signaling mediated by multiple NLR-type R genes (Kitagawa et al., 1999; Austin et al., 2002; Azevedo et al., 2002; Tör et al., 2002; Moon et al., 2004). In tobacco, a set of E3 ligase genes induced by the fungal avirulence protein Avr9 are required for the HR of Cf9-mediated resistance (González-Lamothe et al., 2006; Yang et al., 2006; van den Burg et al., 2008). In Arabidopsis, two RING finger E3 ligases, RPM1-INTERACTING PROTEIN2 (RIN2) and RIN3, have also been implicated in the HR of both CC-type NLR R proteins RPM1- and RPS2-mediated resistance (Kawasaki et al., 2005), whereas a PUB E3 ligase, PUB17, is required for CC-type NLR R protein RPM1- and TIR-type NLR R protein RPS4-mediated resistance (Yang et al., 2006), indicating the critical role of UPS in ETI. Similarly, a homologous triplet of PUBs (PUB22, PUB23, and PUB24) and PUB12/13 in Arabidopsis have been demonstrated to negatively regulate PTI (Trujillo et al., 2008; Li et al., 2012a), suggesting the involvement of the UPS in plant PTI signaling.
The role of UPS in the regulation of PCD and defense of the monocot rice is also becoming clearer. Our laboratory’s identification and characterization of SPL11 in rice as a U-box protein with E3 ligase activity provided, to our knowledge, the first direct evidence that ubiquitination controls resistance and PCD in rice (Zeng et al., 2004). In addition, a RING finger E3 ligase, BLAST AND BTH-INDUCED1, was found to positively regulate resistance against Magnaporthe oryzae by modifying the rice cell wall (Li et al., 2011). Another interesting protein is the RING finger E3 ligase XA21-BINDING PROTEIN3, which is required for the accumulation of the rice bacterial blight R protein XA21 and XA21-mediated defense signaling against Xanthomonas oryzae pv oryzae (Wang et al., 2006). Surprisingly, XA21 was identified to be a PRR that binds a PAMP-like type I-secreted sulfated peptide, AxYS22, derived from the Ax21 protein (Lee et al., 2009). Further characterization of these PCD- and defense response-related UPS components will shed light on the molecular mechanisms underlying HR cell death through the UPS in rice.
THE FUNCTION OF UPS IN FLOWERING TIME REGULATION
Flowering is a well-defined developmental process that is controlled by environmental cues and intrinsic biological rhythms (for review, see Amasino, 2010). Extensive investigations in Arabidopsis have identified four major pathways that perceive and process different signals. The autonomous and GA pathways perceive and transduce internal signals to promote flowering. The external stimuli of variations in daylength and temperature mediate signal transduction in the photoperiodic and vernalization pathways, respectively. Ultimately, signaling in all pathways converges at the floral pathway integrators, a group of genes that are turned on or off in a manner that is consistent with the decision to flower. Among these integrators, the best characterized are FLOWERING LOCUS T (FT; Kardailsky et al., 1999) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1; Borner et al., 2000; Lee et al., 2000; Moon et al., 2003). Both FT and SOC1 are activated by the photoperiodic protein CONSTANS (CO) and are repressed by FLOWERING LOCUS C (FLC), a negative regulator of the autonomous and vernalization pathways (for review, see Lee and Lee, 2010).
In recent years, the UPS has been found to contribute to the regulation of flowering time mainly by regulating the accumulation and stability of CO, GIGANTEA (GI), and FLC. The evening stability and morning instability of CO are affected by the blue light receptor CRYPTOCHROME2 and the light receptor PHYTOCHROME B, respectively (Valverde et al., 2004; Liu et al., 2008). Furthermore, the stability of the CO protein is controlled by the 26S proteasome, because the degradation of CO in the morning and in the dark is inhibited by proteasome inhibitors (Valverde et al., 2004). Intriguingly, the photomorphogenesis-related RING finger protein CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) was identified as the E3 ligase responsible for the degradation of CO in the dark (Liu et al., 2008), while another RING finger E3 ligase, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1, was recently determined to interact synergistically with COP1 and to negatively regulate CO abundance, especially during the day (Lazaro et al., 2012). The stability of GI, a circadian-associated protein and promoter of CO, is also mediated by COP1, which acts with the clock-associated protein EARLY FLOWERING3 (ELF3) to regulate circadian function and photoperiodic flowering by degrading GI (Yu et al., 2008). GI stability is also mediated by COP1, which acts with the clock-associated protein ELF3 to regulate circadian function and photoperiodic flowering by degrading GI (Yu et al., 2008). In addition, two E2 ubiquitin-conjugating enzymes (UBC), AtUBC1 and AtUBC2, contribute to flowering time regulation by working with the E3 ligases HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2 to monoubiquitinate the histone protein H2B; monoubiquitination of H2B is required for the transcriptional activation of FLC expression in Arabidopsis (Cao et al., 2008; Xu et al., 2009). Taken together, these studies have revealed a pivotal role for the UPS in flowering time regulation.
CONVERGENCE OF DEFENSE AND FLOWERING IN ARABIDOPSIS
As discussed above, defense and flowering are two distinct signaling pathways in plants. Interestingly, emerging evidence suggests that the two pathways are connected through the SA pathway. SA is an essential hormone for plant disease resistance (for review, see Vlot et al., 2009) and plant growth and development (Martínez et al., 2004; Wada et al., 2010; Rivas-San Vicente and Plasencia, 2011). SIZ1, which encodes a SUMO E3 ligase in Arabidopsis, suppresses SA- and PAD4-mediated signaling to regulate innate immunity (Lee et al., 2007a, 2007b). The siz1 mutant flowers early under short-day (SD) conditions with elevated SA levels. Further study indicated that SIZ1 regulates flowering by operating a SA-dependent floral promotion pathway and repressing FLD activity through sumoylation to promote FLC expression (Jin and Hasegawa, 2008; Jin et al., 2008). Recently, HOPW1-1-INTERACTING3 (WIN3) was also found to regulate both plant innate immunity and flowering time in Arabidopsis (Wang et al., 2011). Among its multiple functions, WIN3 confers broad-spectrum disease resistance to biotrophic and necrotrophic pathogens, modulates cell death in the SA signaling mutant accelerated cell death6-1, and contributes to flg22-induced PTI (Wang et al., 2011). Additionally, WIN3 negatively modulates flowering under long-day (LD) conditions via the regulation of FLC and FT (Wang et al., 2011). These data indicated that WIN3 might be one of the connecting points for plant defense and flowering signaling pathways.
THE FUNCTIONS OF SPL11 AND PUB13 IN PCD AND DEFENSE
The rice LM mutant spl11, which was identified from an ethyl methanesulfonate-mutagenized population of IR68, is genetically controlled by a recessive gene (Singh et al., 1995; Yin et al., 2000). Disease resistance evaluations indicated that spl11 confers enhanced non-race-specific resistance to both X. oryzae pv oryzae and M. oryzae. When the LMs develop, the resistance is correlated with a constitutive activation of defense-related genes, including pathogenesis-related (PR) genes (PR1, PBZ1, chintinaseIII), oxalate oxidase genes involved in the production of ROS (HvOxOa, HvOxOLP), and genes encoding peroxidases (POX8.1, POX22.3; Yin et al., 2000). Subsequent comparison of the global transcripts between the wild-type and spl11 plants in three types of leaf tissues with different lesion-development phenotypes revealed that the spl11 mutation causes significant changes in the rice transcriptome (Zeng et al., 2006). Over 300 genes are highly induced in the fully expanded leaves of spl11, and nearly half are classified as oxidative stress/cell death- or defense-related genes. These results suggested a strong correlation between the non-race-specific resistance and the constitutive activation of genome-wide defense signaling in the spl11 mutant. In addition, Kojo et al. (2006) showed that the ROS accumulation induced by elicitors is significantly suppressed by the NADPH oxidase (NOX) inhibitor imidazole in spl11, suggesting that SPL11 is important for the regulation of NOX-mediated ROS generation.
The Spl11 gene was mapped on chromosome 12 (Zeng et al., 2002) and subsequently cloned by a map-based strategy (Zeng et al., 2004). Protein sequence analysis revealed that Spl11 encodes a U-box and armadillo (ARM) repeat domain E3 ligase protein, which is a member of the PUB-ARM family (Zeng et al., 2004, 2008). The protein is localized in both the nucleus and cytoplasm. An E3 ligase activity assay indicated that the intact U-box domain is essential for SPL11 E3 ligase activity (Zeng et al., 2004).
To search for other components in the rice SPL11-mediated signaling pathway, we used the ARM domain in SPL11 as the bait in a yeast two-hybrid screen and identified eight SPL11-interacting proteins (SPINs; Vega-Sánchez et al., 2008). Among them, SPIN6 is a putative small GTPase-activating protein (GAP) and interacts with SPL11 in vivo (J.L. Liu and G.-L. Wang, unpublished data). GAP hydrolyzes the active GTP-bound small G protein to inactive the GDP-bound state, which leads to the activation of the cycle of small GTPase-mediated GTP-GDP exchange and downstream signaling (Tcherkezian and Lamarche-Vane, 2007). In rice, a small GTPase protein named OsRAC1 has been implicated in NOX-mediated ROS generation signaling (Kawano et al., 2010). Interestingly, the spl11 mutant is defective in the regulation of the activity of NADPH oxidases (Kojo et al., 2006). We also found that OsRac1 expression is induced in the Spin6 RNA interference and knockout mutant plants and that Spin6 expression is down-regulated in the spl11 mutant (J.L. Liu and G.-L. Wang, unpublished data). These results suggested that SPL11/SPIN6 may regulate NOX-mediated ROS accumulation through OsRAC1. It will be interesting to see whether there is a direct interaction between the SPL11/SPIN6 complex and the OsRAC1-associated defensome and/or between the GAP protein SPIN6 and the small GTPase OsRAC1 in regulating ROS generation and immunity signaling. In addition, six suppressors of spl11-mediated lesion development have been identified. These mutants show reduced or no lesion development in the spl11 background (G. Shirsekar and G.-L. Wang, unpublished data). Three have been molecularly mapped and phenotypically characterized. The reduction or elimination of lesions in spl11 is closely related to the disease resistance level and defense gene expression. The SA level is significantly lower in the spl11 suppressors than in the spl11 plants, indicating that SA might play a role in the regulation of cell death and defense in rice. Cloning of these genes will provide new insight into the SPL11-mediated defense signaling pathway.
Among the 61 putative PUB genes in Arabidopsis (Zeng et al., 2008), PUB13 is the closest ortholog of the rice U-box ligase gene Spl11. PUB13 has 73% identity in amino acids with SPL11 and also contains a U-box/ARM structure with E3 ligase activity. We recently characterized the PUB13 gene and found that disruption of the gene by T-DNA insertion results in spontaneous cell death and accumulation of hydrogen peroxide (H2O2) and SA. Consistent with the phenotypes of spl11, the pub13 mutant plants showed elevated resistance to biotrophic or hemibiotrophic pathogens but increased susceptibility to necrotrophic pathogens (Li et al., 2012a). We also found that cell death is suppressed in the pub13sid2-2 double mutant compared with the clear cell-death phenotype in pub13 under LD and high-humidity conditions. When another SA-deficient mutant, pad4, is crossed with pub13, the cell death in the pub13pad4 double mutant is also suppressed under both conditions. These results demonstrated that the increase in cell death and H2O2 accumulation in pub13 depends on the SA signal. In addition, the LM cell-death phenotype of pub13 is partially rescued to the wild-type level after the overexpression of Spl11 under LD and high-humidity conditions, indicating that PUB13 and Spl11 are functionally conserved in regulating cell death and defense.
Remarkably, Lu et al. (2011) recently reported that the PRR FLAGELLIN-SENSING2 (FLS2), a LRR receptor-like kinase (RLK), is a target of the E3 ligases PUB12 and PUB13 via the UPS. When induced by the bacterial PAMP elicitor flagellin, FLS2 is polyubiquitinated and degraded by PUB12/13. Moreover, the FLS2-PUB12/13 association is required for a PUB12/13 phosphorylation targeted by the LRR-RLK BRASSINOSTEROID-INSENSITIVE1 (BRI1)-ASSOCIATED RECEPTOR KINASE1 (BAK1), an important component in the plant hormone brassinosteroid receptor BRI1-mediated signaling. These results revealed a significant role for E3 ubiquitin ligase PUB12/13-mediated protein degradation in plant PTI signaling.
Environmental factors such as light, temperature, and humidity affect the lesion development of LM mutants (Roden and Ingle, 2009; Mosher et al., 2010; Alcázar and Parker, 2011). In rice spl11 mutant plants, LM formation is greater under SD conditions than under LD conditions (Fig. 2A). Conversely, cell-death phenotypes are enhanced in pub13 under LD conditions (Fig. 2, C and D), suggesting that lesion development in both spl11 and pub13 depends on light period or the circadian clock. Additionally, pub13 exhibits increases in cell death, H2O2 accumulation, and resistance to biotrophic and hemibiotrophic pathogens after pretreatment with high humidity (Li et al., 2012a). Similarly, LM formation is accelerated when the spl11 mutant is kept in a growth chamber with high humidity and under strong sunlight in a greenhouse in summer. These results suggested that both SPL11 and PUB13 are involved in responses to multiple environmental cues.
Figure 2.
Phenotypes of spl11, the Spin1 overexpression line, and pub13. A, Cell-death phenotypes of spl11 under SD (10 h of light and 14 h of dark) and LD (14 h of light and 10 h of dark) conditions in growth chambers. B, Flowering phenotype of japonica rice ‘Nipponbare’ Nipponbare and Spin1 overexpression (OX) plants under natural LD conditions (13 h of light and 11 h of dark). C and D, Cell death in pub13 under LD conditions (16 h of light and 8 h of dark) in a growth room. Col-0 (ecotype Columbia) is the wild-type control. Trypan blue staining was used to detect the cell death of Col-0 and pub13 in D. E, Flowering phenotype of pub13 under LD conditions in a growth room. Both Col-0 and pub13 were 32 d old.
SPL11 REGULATES FLOWERING TIME VIA SPIN1
In addition to its function in cell death and defense regulation, SPL11 is also involved in regulating flowering time (Vega-Sánchez et al., 2008). The spl11 mutant shows a delayed-flowering phenotype only under LD conditions. In contrast, the spl11 suppressors partially recover the delayed flowering under the same conditions, suggesting that SPL11 is likely involved in flowering time regulation. Direct evidence for SPL11 involvement in flowering came from the analysis of the Spin1 gene, which encodes a nuclear, RNA/DNA-binding protein that is a member of the STAR (for signal transduction and activation of RNA) family. SPL11 interacts with SPIN1 in yeast and in planta and monoubiquitinates but does not degrade SPIN1 (Vega-Sánchez et al., 2008). SPL11 negatively regulates Spin1 expression during the light phase under both SD and LD conditions (Vega-Sánchez et al., 2008). Overexpression of Spin1 causes late flowering under both SD and LD conditions in the growth chamber and field (Fig. 2B). SPIN1 represses flowering time via transcriptional perturbation of Heading date3a (Hd3a), which is daylength independent (Vega-Sánchez et al., 2008). These results demonstrated that SPL11 controls flowering time through the monoubiquitination of the flowering suppressor SPIN1 and that ubiquitination and RNA metabolism are linked (via the interaction between SPL11 and SPIN1) in the regulation of flowering time.
To identify the target(s) of SPIN1 in the regulation of flowering time, we performed a yeast two-hybrid screen using SPIN1 as the bait and found another RNA-binding protein, named RBS1 (for RNA-binding and SPIN1-interacting1; Y. Cai and G.-L. Wang, unpublished data). Preliminary analysis of the overexpression and RNA interference lines of Rbs1 revealed that it also positively regulates flowering time (Y. Cai and G.-L. Wang, unpublished data). Based on this result and those mentioned above, we speculate that SPL11 might enter into the nucleus to form a protein complex with the nuclear proteins SPIN1 and RBS1 and that the modification of these two proteins by SPL11 can alter the flowering signaling pathway in rice.
PUB13 NEGATIVELY REGULATES FLOWERING TIME IN A PHOTOPERIOD- AND SA-DEPENDENT MANNER
We recently found that PUB13 is also involved in the regulation of flowering time in Arabidopsis (Li et al., 2012a). Unlike the rice mutant spl11, which has a delayed-flowering phenotype under LD conditions (Vega-Sánchez et al., 2008), the Arabidopsis mutant pub13 displays an early-flowering phenotype under LD or middle-day conditions (Li et al., 2012a). The opposite functions of PUB13 and SPL11 in flowering time control are probably caused by differences in the photoperiodism of Arabidopsis (a LD plant) and rice (a SD plant) or by the divergence in molecular mechanisms controlling flowering in these two types of photoperiodic plants. For instance, GI is a promoter of flowering time in Arabidopsis, while its ortholog in rice, OsGI, is a suppressor of flowering time under both SD and LD conditions (Fowler et al., 1999; Hayama et al., 2003). In addition, CO is a positive regulator of flowering time under SD or LD conditions in Arabidopsis, but its ortholog in rice, Hd1, functions as a negative regulator of flowering time under LD conditions but as a positive regulator under SD conditions (Yano et al., 2000). Intriguingly, overexpression of Spl11 in pub13 can complement the early-flowering phenotype of pub13 (Li et al., 2012a, 2012b), suggesting that the function of these two E3 ligases, PUB13 and SPL11, in flowering time control is conserved and that the E3 ligase activity of the two homologous proteins may be similar in dicot and monocot plants.
Plant hormones determine many biological processes, such as growth, development, and responses to environmental stresses (for review, see Depuydt and Hardtke, 2011). The hormone SA is not only pivotal in plant responses to biotic stress but is also involved in flowering (for review, see Martínez et al., 2004). SA probably controls flowering time through the photoperiod and vernalization flowering pathways that are independent of FLC, CO, and FCA. The SA level is elevated in pub13, but once the increased level of SA in pub13 is reduced by knockout of the two important components (SID2 and PAD4) in the SA pathway, the early-flowering phenotype in pub13 is restored to the wild-type level (Li et al., 2012a). These results suggested that PUB13 regulates flowering time mainly through an SA-dependent pathway. The function of SA in flowering control in pub13 is consistent with previous reports that SA accelerates the transition from the vegetative to the reproductive phase by suppressing the floral repressor genes and activating the positive regulator of flowering (Martínez et al., 2004).
PUB13 INTERACTS WITH HFR1, A POSITIVE REGULATOR OF PHOTOMORPHOGENESIS
It is clear that SPL11 regulates flowering time through SPIN1 in rice (Vega-Sánchez et al., 2008). To identify a SPIN1-like ortholog in Arabidopsis, we performed a yeast two-hybrid screen using the PUB13 mutant protein PUB13V273R, an E3 activity-compromised mutant, as the bait because we did not find any SPIN1-like interactor when the full-length PUB13 was used (Li et al., 2012b). One of the interacting proteins identified in the screen is LONG HYPOCOTYL IN FAR-RED LIGHT1 (HFR1), which encodes a basic helix-loop-helix-type transcription factor (Duek et al., 2004). The interaction between PUB13 and HFR1 was confirmed by a glutathione S-transferase pull-down assay (Li et al., 2012b). HFR1 promotes photomorphogenesis and is ubiquitinated and degraded by the E3 ligase COP1 in darkness, a protein that is involved in photomorphogenesis, plant growth, flower shape, and flowering time (Jang et al., 2008). How PUB13 regulates HFR1 to control flowering time and whether the regulation is associated with the COP1 protein complex are being investigated.
WORKING MODELS FOR SPL11- AND PUB13-MEDIATED REGULATION OF DEFENSE AND FLOWERING AND FUTURE RESEARCH DIRECTIONS
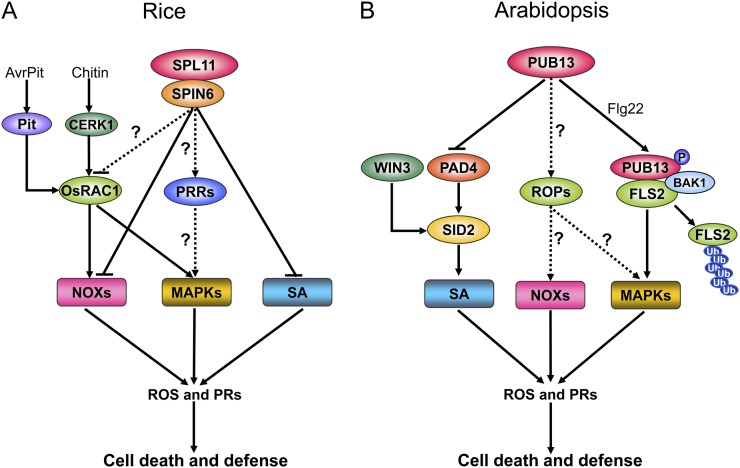
In the last 10 years, we have obtained considerable information on the function of SPL11 and PUB13 in the regulation of PCD, defense, and flowering in model monocot and dicot plants. Based on our results and those from other studies, we have proposed working models to illustrate the function of SPL11 and PUB13 and their interacting proteins in the regulation of defense (Fig. 3) and flowering (Fig. 4) in both species. Although the biological functions of SPL11 and PUB13 are similar, the components participating in SPL11- and PUB13-mediated signaling have become diverse during the course of speciation (Table I). In rice, SPL11, a negative regulator of cell death, physically associates with the putative GAP protein SPIN6 to suppress NOX-mediated ROS generation and PR gene activation and thereby to inhibit the autoactivation of defense responses (Fig. 3A). GAP regulates the activity of GTP-bound small GTPase proteins. The small G protein OsRAC1 is a signal integrator of OsCERK1-mediated PTI and Pit-mediated ETI signaling pathways (for review, see Kawano et al., 2010). Thus, we speculate that SPIN6 might biochemically inactive GTP-bound OsRAC1 to suppress the OsRAC1-mediated signaling and to prevent unnecessary cell death and defense activation when no pathogen is present. The SPL11 downstream defense signaling might be transduced to mitogen-activated protein kinase (MAPK) cascades and the NOX complex by OsRAC1. The SPL11 also negatively regulates SA accumulation, because mutation of Spl11 elevates the SA level in the spl11 mutant (Fig. 3A). In Arabidopsis, PUB13 negatively regulates the PAD4/SID2-dependent SA defense pathway and FLS2-mediated PTI signaling (Fig. 3B). SID2 is also regulated by WIN3, a type II regulator of the SA pathway (Wang et al., 2011). PUB13 interacts with BAK1 and is phosphorylated by BAK1. The PAMP elicitor flg22 stimulates PUB13 to associate with FLS2, an association that is required for the BAK1-mediated PUB13 phosphorylation. Polyubiquitination of FLS2 by PUB13 causes FLS2 degradation. After FLS2 is activated by flg22, the signal is transduced to the MAPK cascades to regulate ROS burst and PR gene expression and thereby to trigger cell death and defense responses. Because of the conserved function of SPL11 and PUB13 in defense, some FLS2-like PRRs might be targeted by SPL11 in rice or some OsRAC1-like proteins (called Rho-related GTPases from plants) might be targeted by PUB13 in Arabidopsis. These interesting questions remain to be answered in future work.
Figure 3.
Proposed working models of cell death and defense signaling mediated by rice SPL11 (A) and Arabidopsis PUB13 (B). See details in the text.
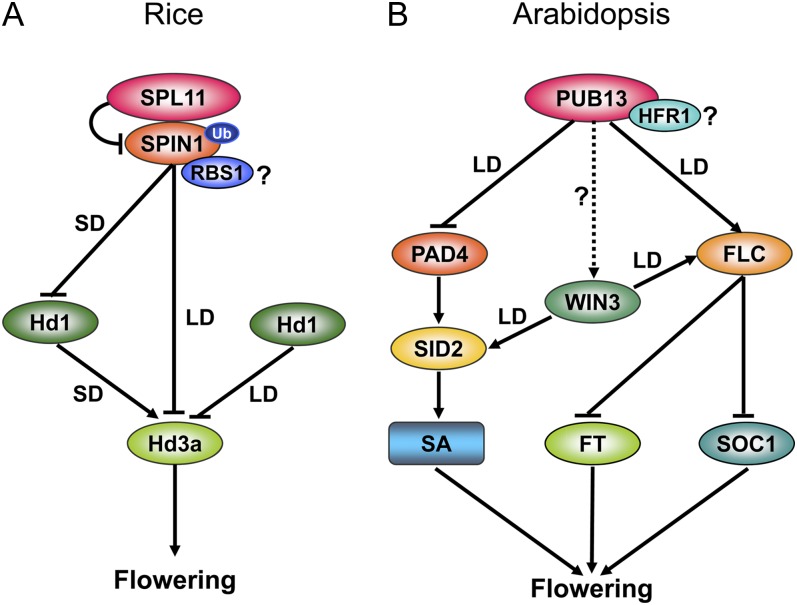
Figure 4.
Proposed working models of flowering time regulation meditated by rice SPL11 (A) and Arabidopsis PUB13 (B). See details in the text.
For flowering time regulation in rice, SPL11 monoubiquitinates SPIN1 and negatively regulates its activity (Fig. 4A). Under SD conditions, the suppression of SPIN1 by SPL11 promotes the H3a-dependent pathway by activating Hd1. Under LD conditions, SPIN1 and Hd1 may act additively to repress Hd3a, causing late flowering but high levels of SPL11 throughout the day in order to alleviate flowering repression. SPIN1 can also interact with RBS1 to regulate flowering time, but how the interaction affects flowering remains unknown. In Arabidopsis, however, PUB13 positively regulates the flowering suppressor FLC to delay flowering under LD conditions; FLC is a negative regulator of the flowering activator FT and SOC1 (Fig. 4B). PUB13 also physically associates with HFR1, a positive regulator of plant photomorphogenesis, but its role in PUB13-mediated flowering remains unclear. Furthermore, PUB13-mediated LD flowering also requires the PAD4/SID2-mediated SA pathway. Additionally, WIN3 positively regulates SID2-mediated SA signaling and the FLC-mediated flowering pathway under LD conditions (Wang et al., 2011). The relationship between PUB13 and WIN3 needs to be investigated.
Although much progress has been made in the dissection of the SPL11 and PUB13 pathways, several critical questions remain. How do the E3 ligases SPL11 and PUB13 regulate their substrates to modulate both defense and flowering? Is there an intimate connection between the SPL11/SPIN6 complex and the OsRAC1-mediated PTI and ETI defensome? Which genes are targeted by the SPL11/SPIN1-mediated RNA-processing complex in the regulation of flowering time? What is the relationship between SPL11/PUB13 and other hormone signaling pathways in plants? How does PUB13 regulate flowering through HRF1 and other COP1-associated proteins? Is PUB13 physically associated with resistance proteins, as has recently been demonstrated for FLS2 (Qi et al., 2011)? Answers to these questions will provide a better understanding of the dual functions of SPL11 and PUB13 in rice and Arabidopsis.
Acknowledgments
We thank Dr. Bruce Jaffee for his careful reading of the manuscript.
Glossary
- UPS
ubiquitin-proteasome system
- PCD
programmed cell death
- PRR
pattern-recognition receptor
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- ROS
reactive oxygen species
- ETI
effector-triggered immunity
- LRR
leucine-rich repeat
- HR
hypersensitive response
- LM
lesion-mimic
- SA
salicylic acid
- CC
coiled coil
- SD
short-day
- LD
long-day
- NOX
NADPH oxidase
- H2O2
hydrogen peroxide
- RLK
receptor-like kinase
- MAPK
mitogen-activated protein kinase
References
- Alcázar R, Parker JE. (2011) The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci 16: 666–675 [DOI] [PubMed] [Google Scholar]
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108: 16463–16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Broemer M, Meier P. (2009) Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol 19: 130–140 [DOI] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L. (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS. (2011) Hormone signalling crosstalk in plant growth regulation. Curr Biol 21: R365–R373 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Li L, Su Z. (2009) plantsUPS: a database of plants’ ubiquitin proteasome system. BMC Genomics 10: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C. (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JD. (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124: 27–34 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Hasegawa PM. (2008) Flowering time regulation by the SUMO E3 ligase SIZ1. Plant Signal Behav 3: 891–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, et al. (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through effects on FLC chromatin structure. Plant J 53: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Chen L, Shimamoto K. (2010) The function of Rac small GTPase and associated proteins in rice innate immunity. Rice 3: 112–121 [Google Scholar]
- Kawasaki T, Nam J, Boyes DC, Holt BF, III, Hubert DA, Wiig A, Dangl JL. (2005) A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. Plant J 44: 258–270 [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33 [DOI] [PubMed] [Google Scholar]
- Kojo K, Yaeno T, Kusumi K, Matsumura H, Fujisawa S, Terauchi R, Iba K. (2006) Regulatory mechanisms of ROI generation are affected by rice spl mutations. Plant Cell Physiol 47: 1035–1044 [DOI] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Piñeiro M, Jarillo JA. (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I. (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Lee J, Miura K, Bressan RA, Hasegawa PM, Yun DJ. (2007a) Regulation of plant innate immunity by SUMO E3 ligase. Plant Signal Behav 2: 253–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007b) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, Ronald PC. (2009) A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326: 850–853 [DOI] [PubMed] [Google Scholar]
- Li W, Ahn IP, Ning Y, Park CH, Zeng L, Whitehill JG, Lu H, Zhao Q, Ding B, Xie Q, et al. (2012a) The U-box/ARM E3 ligase PUB13 regulates cell death, defense and flowering time in Arabidopsis. Plant Physiol 159: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Dai L, Wang GL. (2012b) PUB13, a U-box/ARM E3 ligase, regulates plant defense, cell death, and flowering time. Plant Signal Behav 7: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhong S, Li G, Li Q, Mao B, Deng Y, Zhang H, Zeng L, Song F, He Z. (2011) Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res 21: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen ZJ. (2011) Expanding role of ubiquitination in NF-κB signaling. Cell Res 21: 6–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love AJ, Milner JJ, Sadanandom A. (2008) Timing is everything: regulatory overlap in plant cell death. Trends Plant Sci 13: 589–595 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L. (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C, Pons E, Prats G, León J. (2004) Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J 37: 209–217 [DOI] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K. (2008) Lesion mimic mutants: a classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav 3: 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I. (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, Urquhart W, Klessig DF, Kim SK, Nambara E, Yoshioka K. (2010) The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol 152: 1901–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S. (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Qi Y, Tsuda K, Glazebrook J, Katagiri F. (2011) Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol 12: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J. (2011) Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- Roden LC, Ingle RA. (2009) Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21: 2546–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Aviv DH, Holt BF, III, Dangl JL, Parker JE. (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2010) The ubiquitin-proteasome system regulates plant hormone signaling. Plant J 61: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Multani DS, Khush GS. (1995) A new spotted leaf mutant in rice. Rice Genet Newsl 12: 192–193 [Google Scholar]
- Stone SL, Callis J. (2007) Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol 10: 624–632 [DOI] [PubMed] [Google Scholar]
- Tcherkezian J, Lamarche-Vane N. (2007) Current knowledge of the large RhoGAP family of proteins. Biol Cell 99: 67–86 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH. (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tör M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Türk F, Can C, Dangl JL, Holub EB. (2002) Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Tsitsigiannis DI, Rowland O, Lo J, Rallapalli G, Maclean D, Takken FL, Jones JD. (2008) The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20: 697–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez ME, Zeng L, Chen S, Leung H, Wang G-L. (2008) SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell 20: 1456–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wada KC, Yamada M, Shiraya T, Takeno K. (2010) Salicylic acid and the flowering gene FLOWERING LOCUS T homolog are involved in poor-nutrition stress-induced flowering of Pharbitis nil. J Plant Physiol 167: 447–452 [DOI] [PubMed] [Google Scholar]
- Wang GF, Seabolt S, Hamdoun S, Ng G, Park J, Lu H. (2011) Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol 156: 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YS, Pi LY, Chen X, Chakrabarty PK, Jiang J, De Leon AL, Liu GZ, Li L, Benny U, Oard J, et al. (2006) Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18: 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Xu L, Ménard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH. (2009) The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J 57: 279–288 [DOI] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JD, Sadanandom A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Chen J, Zeng L, Goh M, Leung H, Khush GS, Wang G-L. (2000) Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact 13: 869–876 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Yin Z, Chen J, Leung H, Wang G-L. (2002) Fine genetic mapping and physical delimitation of the lesion mimic gene Spl11 to a 160-kb DNA segment of the rice genome. Mol Genet Genomics 268: 253–261 [DOI] [PubMed] [Google Scholar]
- Zeng LR, Park CH, Venu RC, Gough J, Wang G-L. (2008) Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant 1: 800–815 [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang G-L. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Vega-Sánchez ME, Zhu T, Wang G-L. (2006) Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res 16: 413–426 [DOI] [PubMed] [Google Scholar]