Abstract
Arabidopsis (Arabidopsis thaliana) mutants hypersensitive to far-red light were isolated under a light program of alternating red and far-red light pulses and were named eid (for empfindlicher im dunkelroten Licht). The dominant eid3 mutant carries a missense mutation in a conserved domain of PHYTOCHROME AND FLOWERING TIME1 (PFT1), an important component of the plant mediator coactivator complex, which links promoter-bound transcriptional regulators to RNA polymerase II complexes. Epistatic analyses were performed to obtain information about the coaction between the mutated PFT1eid3 and positively and negatively acting components of light signaling cascades. The data presented here provide clear evidence that the mutation mainly enhances light sensitivity downstream of phytochrome A (phyA) and modulates phyB function. Our results demonstrate that the Mediator component cooperates with CONSTITUTIVE PHOTORMORPHOGENIC1 in the regulation of light responses and that the hypersensitive phenotype strictly depends on the presence of the ELONGATED HYPOCOTYL5 transcription factor, an important positive regulator of light-dependent gene expression. Expression profile analyses revealed that PFT1eid3 alters the transcript accumulation of light-regulated genes even in darkness. Our data further indicate that PFT1 regulates the floral transition downstream of phyA. The PFT1 missense mutation seems to create a constitutively active transcription factor by mimicking an early step in light signaling.
Light is essential for the survival of plants in their natural environment, controlling the timing and the extent of many developmental transitions, including seed germination, seedling deetiolation, phototropism, shade-avoidance responses, circadian rhythms, and flowering time. To sense light quality, intensity, direction, and duration, higher plants have evolved several classes of photoreceptors. Among them, cryptochromes, phototropins and zeitlupe-like photoreceptors respond to UV-A and blue light, whereas UVR8 is the UV-B receptor (Sullivan and Deng, 2003; Chen et al., 2004; Franklin et al., 2005; Jenkins, 2009; Franklin and Quail, 2010; Rizzini et al., 2011). Phytochromes mainly function as receptors for red and far-red light (Bae and Choi, 2008; Franklin and Quail, 2010).
The phytochrome family in Arabidopsis (Arabidopsis thaliana) is composed of five members, phyA through phyE. Four members, phyB to phyE, are more stable in the light and predominantly regulate shade-avoidance responses (SAR) under low red/far-red light ratios, classical red/far-red reversible responses, and responses toward strong continuous red light. Among these light-stable, or type II, phytochromes, phyB exhibits the highest levels and dominates physiological responses in Arabidopsis. The light-labile, or type I, phytochrome is encoded by the PHYA gene of Arabidopsis. It accumulates to very high levels in darkness, enabling it to sense extremely low amounts of light, which trigger the so-called very-low-fluence responses (VLFR). Furthermore, phyA controls high-irradiance responses (HIR), which become maximally induced under strong, continuous far-red light (Chen et al., 2004; Bae and Choi, 2008; Franklin and Quail, 2010).
Genetic, biochemical, and molecular studies have identified a high number of DNA-binding proteins that function as negatively or positively acting regulators in light signaling (Jiao et al., 2007; Bae and Choi, 2008; Leivar and Quail, 2011). Among these factors, ELONGATED HYPOCOTYL5 (HY5), a basic Leu-zipper transcriptional regulator, acts at the beginning of the transcriptional cascades that regulate seedling photomorphogenesis downstream of many photoreceptors (Oyama et al., 1997; Ang et al., 1998; Saijo et al., 2003; Ulm et al., 2004; Lee et al., 2007; Lian et al., 2011; Liu et al., 2011; Rizzini et al., 2011; Zhang et al., 2011). In addition, phytochrome-interacting factors, members of the basic helix-loop-helix transcription factor superfamily, directly or indirectly interact with Pfr forms of phytochromes. Physiological studies with phytochrome-interacting protein mutants imply that these factors mainly function as negative regulators of phytochrome signaling (Castillon et al., 2007; Leivar et al., 2009; Leivar and Quail, 2011). Screening for mutants with impaired photomorphogenic development under far-red light led to the identification of several DNA-binding proteins that function as positive factors downstream of phyA, including the basic helix-loop-helix transcription factor LONG HYPOCOTYL IN FAR-RED1 (HFR1) and the R2R3-MYB transcription factor LONG AFTER FAR-RED1 (LAF1; Fairchild et al., 2000; Ballesteros et al., 2001; Duek and Fankhauser, 2003; Seo et al., 2003; Jang et al., 2007; Hornitschek et al., 2009). FAR-RED IMPAIRED RESPONSE1 (FAR1) and FAR-RED ELONGATED HYPOCOTYL3 (FAR3) belong to a class of transposon-derived transcription factors that participate in phyA signaling (Wang and Deng, 2002; Hudson et al., 2003; Lin et al., 2007; Saijo et al., 2008; Yang et al., 2009; Ouyang et al., 2011).
The repression of light responses is mainly achieved by ubiquitin-mediated proteolysis. CONSTITUTIVE PHOTORMORPHOGENIC1 (COP1) is an important component of E3 ubiquitin ligase complexes that include proteins of the SUPRESSOR OF PHYA-105 family (SPA1–SPA4), DAMAGED DNA-BINDING PROTEIN1 and -2, and CULLIN4 (Yi and Deng, 2005; Chen et al., 2010). COP1-containing E3 ubiquitin ligase complexes (ULC) function as general repressors of plant photomorphogenesis downstream of several photoreceptors by inducing proteolysis of positively acting factors involved in light signaling. Target proteins include transcription factors like HY5, LAF1, and HFR1 and photoreceptors such as phyA, phyB, and cryptochrome2 (cry2; Hardtke et al., 2000; Holm et al., 2002; Seo et al., 2003, 2004; Duek et al., 2004; Jang et al., 2005, 2010).
To identify additional components of the phyA signal transduction pathway, a specific irradiation program was established consisting of repetitive cycles of alternating 20-min-long red/far-red light pulses (Büche et al., 2000). The red light preirradiation decreases the level of light-labile phyA, which results in a loss of far-red light-dependent HIR. Using this screening program, several mutants were isolated that overcome red light-induced suppression of HIR. Because of their increased far-red light sensitivity, these mutants were called eid (for empfindlicher im dunkelroten Licht). The EID1 F-box protein is a component of an SCF E3 ubiquitin ligase complex that specifically functions as a negative regulator of phyA-dependent HIR (Büche et al., 2000; Dieterle et al., 2001; Zhou et al., 2002; Marrocco et al., 2006). The semidominant Eid4 phenotype is caused by the phyA-401 missense mutation in the PHYA gene (Dieterle et al., 2005). eid6 carries a missense mutation in COP1 that leads to an extremely enhanced light sensitivity but not to the constitutive photomorphogenic phenotype in darkness (Dieterle et al., 2003).
Here, we report the isolation and characterization of eid3, the only fully dominant mutant from the screening program. The hypersensitive mutant phenotype is caused by a missense mutation in a highly conserved domain of PHYTOCHROME AND FLOWERING TIME1 (PFT1), a component of a plant Mediator complex (Cerdán and Chory, 2003; Bäckström et al., 2007). The eukaryotic Mediator complex plays a central role in transcriptional initiation by linking DNA-binding proteins, a subset of transcription factors of the core promoter, and the RNA polymerase II complex (Björklund and Gustafsson, 2005; Conaway et al., 2005; Malik and Roeder, 2010; Kidd et al., 2011). Genetic and physiological analyses demonstrated that PFT1 regulates flowering, jasmonate signaling, and biotic and abiotic stress tolerance (Cerdán and Chory, 2003; Kidd et al., 2009; Elfving et al., 2011). Our data provide clear evidence that the Mediator component plays an important role in early steps of phytochrome-dependent gene expression and that the protein cooperates with COP1, HY5, and other known light signaling components to control the transition between skotomorphogenic and photomorphogenic seedling development. Furthermore, our results strongly indicate that PFT1 functions downstream of phyA to regulate flowering time and modulate phyB signaling.
RESULTS
Isolation of the eid3 Mutant
To identify specific mutants in phyA signaling in Arabidopsis, ethyl methylsulfonate-treated phyB-5 seedlings were treated with a multiple pulse program consisting of 20 min of strong red light followed by 20 min of strong far-red light for 3 d after germination induction (Büche et al., 2000). In phyB-5 seedlings, preirradiation with red light pulses reduces the amount of phyA, which in turn decreases the HIR mediated by subsequent treatments with far-red light. The isolated phyB-5 eid3 mutant was able to avoid this red light-dependent reduction of HIR (Fig. 1A). Compared with its phyB-5 background, the mutant exhibited reduced hypocotyl elongation, open cotyledons, and increased anthocyanin accumulation under screening conditions. Except for the opening of hypocotyl hooks, phyB-5 eid3 remained etiolated in darkness (Fig. 1B). Segregation analyses of backcrosses with phyB-5 and the corresponding Landsberg erecta (Ler) wild type revealed that the eid3 mutant behaved like a dominant monogenic locus under selective light conditions.
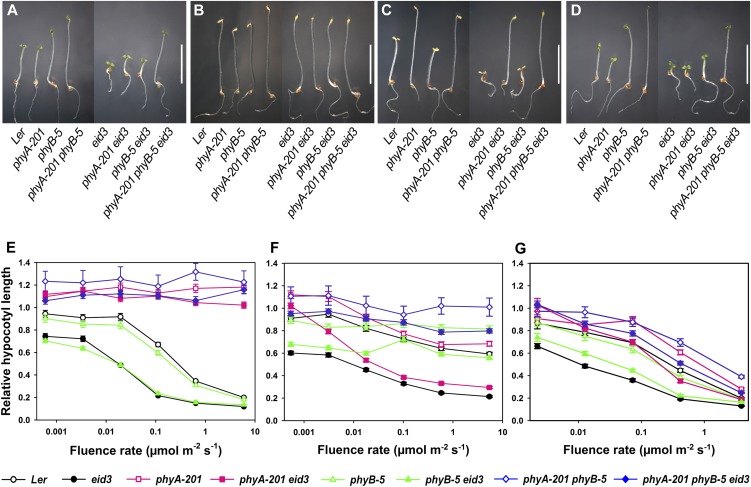
Figure 1.
Epistatic analyses with phyA and phyB loss-of-function mutants. A to D, Photographs of Ler wild-type, phyA-201, phyB-5, phyA-201 phyB-5, eid3, phyA-201 eid3, phyB-5 eid3, and phyA-201 phyB-5 eid3 seedlings grown under different light conditions for 4 d after induction of germination. Seedlings were kept under the screening program (cycles of 20 min of red light followed by 20 min of far-red light; A), in darkness (B), under weak far-red light (0.1 µmol m−2 s−1; C), and under weak red light (1 µmol m−2 s−1; D). Bars = 5 mm. E to G, Fluence rate response curves for the inhibition of hypocotyl elongation of Ler wild-type, phyA-201, phyB-5, phyA-201 phyB-5, eid3, phyA-201 eid3, phyB-5 eid3, and phyA-201 phyB-5 eid3 seedlings under continuous far-red light (E), continuous red light (F), or continuous blue light (G). Relative hypocotyl lengths were calculated in relation to the length of dark-grown seedlings for each line. Each data point represents the mean ± se of two independent experiments with at least 30 seedlings.
Epistasis between eid3 and phyA and phyB Loss-of-Function Mutants during Seedling Development
In order to test the influence of phyA and phyB on the expression of the Eid3 phenotype, an eid3 mutant in a PHYB wild-type background, a phyA-201 eid3 double mutant, and a phyA-201 phyB-5 eid3 triple mutant were created. The eid3 single and the phyB-5 eid3 double mutants exhibited an approximately 10-fold increase in light sensitivity compared with the wild type and phyB-5 under continuous far-red light (Fig. 1, C and E). In contrast, phyA-201 eid3 and phyA-201 phyB-5 eid3 seedlings did not respond to far-red light, similar to their phyA-201 and phyA-201 phyB-5 background lines.
Seedlings of the eid3 single mutant exhibited an extremely enhanced sensitivity to red light compared with the wild type (Fig. 1, D and F). Studies with phyA-201 eid3 and phyB-5 eid3 double mutants demonstrated that phyA is mainly responsible for the expression of the hypersensitive phenotype at photon fluence rates below approximately 0.01 µmol m−2 s−1, whereas the increased red light sensitivity can mainly be attributed to the presence of the phyB photoreceptor at higher photon fluence rates. The phyA-201 phyB-5 eid3 triple mutant exhibited a slightly increased red light response compared with its phyA-201 phyB-5 background.
Blue light is not only sensed by cryptochrome and phototropin photoreceptors but can also induce the formation of the active Pfr conformation of phytochromes. Among the phytochromes, phyA is the most potent blue light receptor and, correspondingly, its loss resulted in a clear reduction of blue light responses in phyA-201 eid3 compared with the eid3 mutant background (Fig. 1G). In contrast, the lack of phyB caused only a weak reduction in blue light responses in phyB-5 eid3 and phyA-201 phyB-5 eid3 with respect to background lines. Comparison of fluence rate response curves between phyA-201 phyB-5 and phyA-201 phyB-5 eid3 indicated that blue light sensitivity in the triple mutant background is only slightly increased compared with its phyA-201 phyB-5 background.
In contrast to the strong photomorphogenic phenotype during seedling development, the eid3 mutation caused only a mild alteration during vegetative growth. Rosettes of eid3, phyA-201 eid3, phyB-5 eid3, and phyA-201 phyB-5 eid3 plants exhibited a slightly decreased diameter and more rounded leaves compared with the wild type and the corresponding mutant backgrounds, indicative of a reduced SAR (Supplemental Fig. S1). The eid3 mutation also did not alter phyA degradation or subcellular localization (Supplemental Fig. S2).
Genetic Interaction with EID1 and COP1 Loss-of-Function Mutants and the phyA-401 Gain-of-Function Allele
In order to test the interaction between eid3 and negatively acting components of the light signaling cascade, the mutant was crossed with the strong eid1-1 loss-of-function allele and the cop1eid6 allele, which does not exhibit a constitutive photomorphogenic phenotype in the absence of light (Dieterle et al., 2001, 2003). Additionally, double mutants were generated with eid3 and the phyA-401 (eid4) gain-of-function allele. The extreme light sensitivity of eid3 single and double mutants hinders standard procedures used to compare light responses under continuous irradiation: it was nearly impossible to apply nonsaturating doses of light. To overcome this problem, a 3-d light program was used that consisted of hourly 5-min light pulse treatments interrupted by 55 min of darkness. Wavelengths that adjust the levels of the physiologically active form of phytochromes [Pfr/(Pfr + Pr)] were used and resulted in relative ratios of approximately 0.87 (659-nm filter), approximately 0.5 (692-nm filter), approximately 0.05 (719-nm filter), and below 0.001 (760-nm filter; Dieterle et al., 2005).
In accordance with the proposed role of EID1 as a specific regulator of phyA-dependent HIR (Dieterle et al., 2001; Zhou et al., 2002), a significant difference between wild-type and eid1-1 seedlings was only detectable under multiple 719-nm pulse treatments, which are known to be maximally effective at inducing HIR light responses (Fig. 2). Correspondingly, a strong synergistic effect was seen in the eid3 eid1-1 double mutant upon the application of multiple 719-nm pulses. Weaker synergistic effects also became detectable under pulses with 692- and 659-nm light. In addition, eid3 eid1-1 double mutants exhibited clear alteration in the morphology of rosette leaves of adult plants compared with background lines (Supplemental Fig. S3).
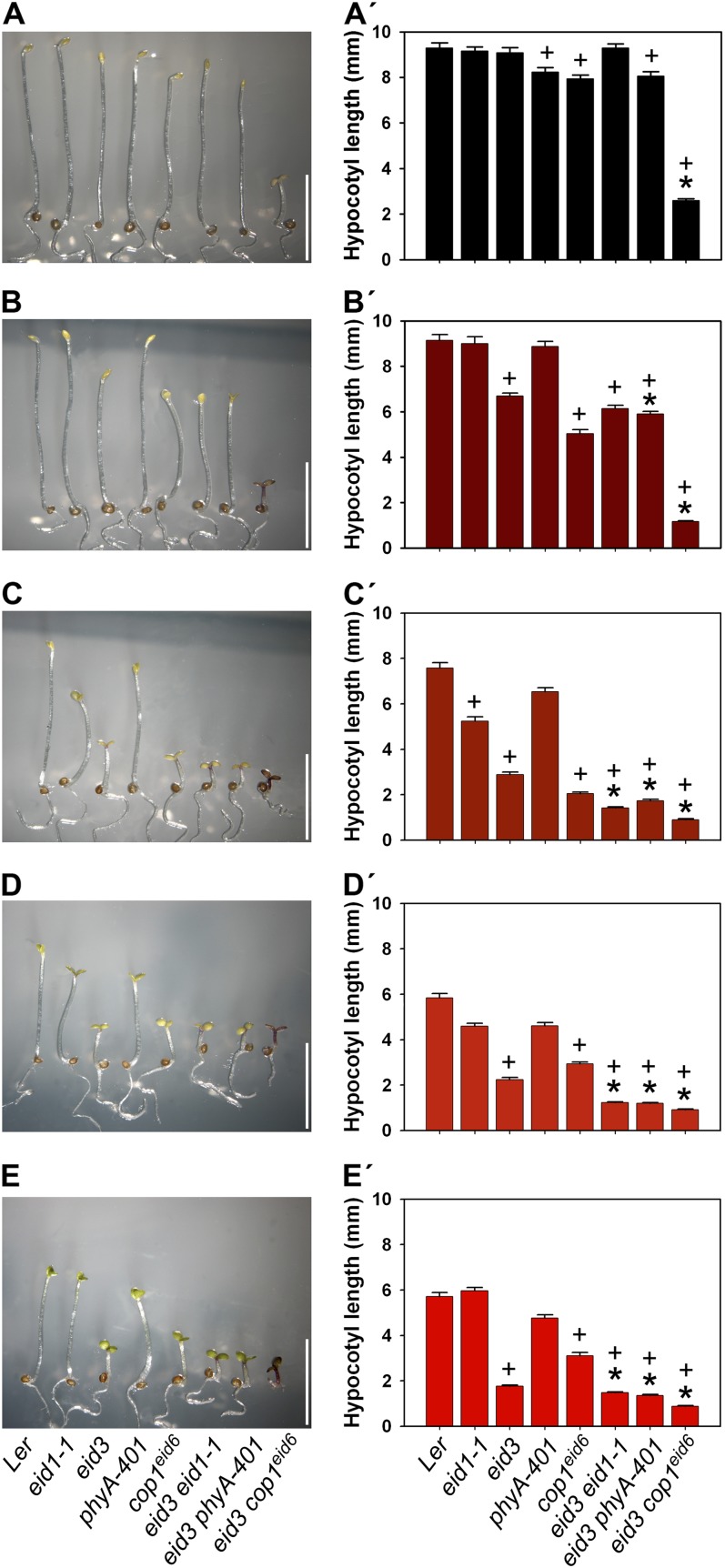
Figure 2.
The eid3 mutation cooperates with EID1 and COP1 to regulate photomorphogenic seedling development. Seedlings of Ler, eid1-1, eid3, phyA-401, cop1eid6, eid3 eid1-1, eid3 phyA-401, and eid3 cop1eid6 were grown for 3 d after induction of germination under repetitive pulse treatments consisting of 5 min of saturating light followed by 55 min of darkness. The light conditions used for the pulse treatment were darkness (A and A′), 760-nm light (B and B′), 719-nm light (C and C′), 692-nm light (D and D′), and 659-nm light (E and E′). A to E, Photographs of seedlings grown under the different multiple pulse treatment programs. Bars = 5 mm. A′ to E′, Absolute hypocotyl lengths of pulse-treated seedlings. Data represent means of two independent experiments ± se with 30 seedlings. +, significant difference from the Ler wild type; * significantly reduced hypocotyl lengths compared with the corresponding single mutant background lines (one-way ANOVA on ranks, P < 0.05).
The phyA-401 missense allele leads to an enhanced light sensitivity under continuous red and far-red light (Dieterle et al., 2005), but the mutant did not exhibit significant differences in light responses compared with the wild type under hourly light pulse treatments (Fig. 2). In contrast, eid3 phyA-401 double mutants showed an increased light response compared with their corresponding background lines and the wild type under all tested wavelengths. The eid3 phyA-401 double mutant did not show any further alteration in the shape of the rosette compared with the eid3 line (Supplemental Fig. S3).
The most severe synergistic effects were obtained with the eid3 cop1eid6 double mutant. Even though cop1eid6 and eid3 background lines remained almost completely etiolated in darkness, the double mutant exhibited a strong constitutive photomorphogenic phenotype (Fig. 2, A and A′). The eid3 cop1eid6 double mutant also showed an extremely enhanced light sensitivity compared with both background lines and the other eid3 double mutant lines. Seedlings were extremely short, had open and expanded cotyledons, and accumulated very high levels of anthocyanin under all applied pulse treatments (Fig. 2). The eid3 cop1eid6 mutant also showed a severe reduction in rosette size compared with the wild-type, eid3, and cop1eid6 background lines (Supplemental Fig. S3).
Epistatic Analyses with Positively Acting Factors Involved in Arabidopsis Light Signaling
In order to test for the interdependency between eid3 and the positive phyA-dependent light signaling effectors HFR1, FHY3, FAR1, and HY5 (Oyama et al., 1997; Fairchild et al., 2000; Wang and Deng, 2002; Hudson et al., 2003), double mutants were isolated to perform epistatic analyses with 4-d-old seedlings that were grown under continuous far-red light of variable photon fluence rates.
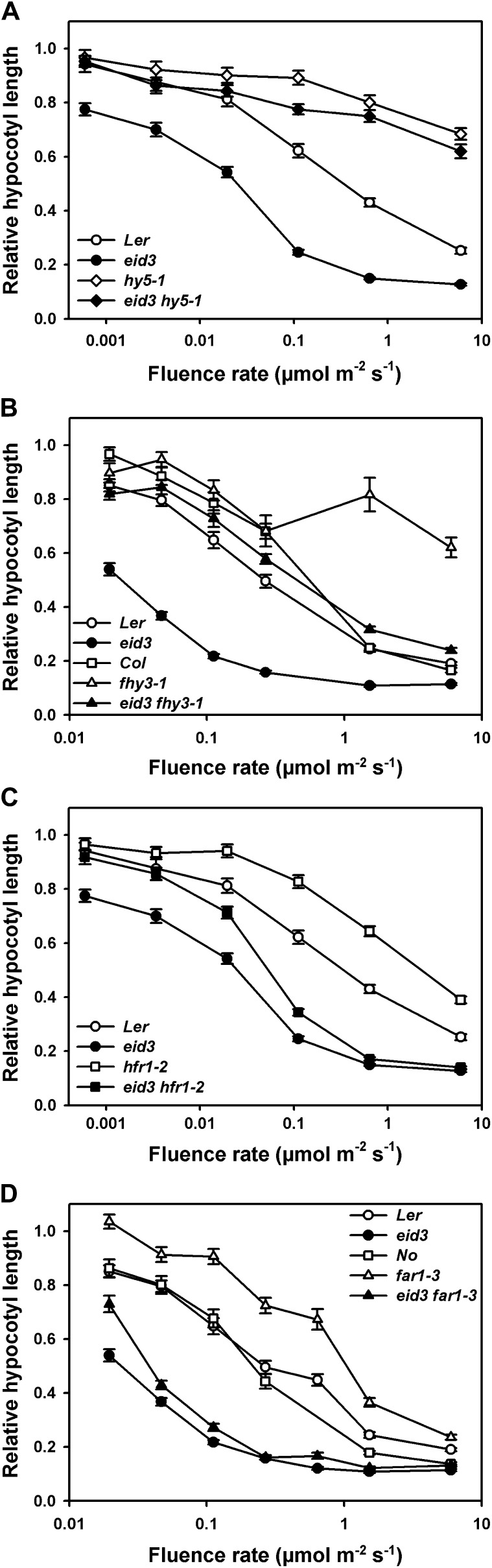
A full epistatic effect was only obtained with hy5 eid3 double mutants (Fig. 3A). The hy5 loss-of-function mutant reduced light responses to the level of the hy5 mutant background line under all applied far-red light intensities. Loss of FHY3 caused a clear reduction in light sensitivity at all applied photon fluence rates, but far-red light sensitivity was still increased compared with fhy3-1 under light intensities normally inducing HIR (Fig. 3B). The lack of HFR1 and FAR1 had only a weak influence on the expression of the Eid3 phenotype at low fluence rates of far-red light, whereas far-red light responses remained more or less unaltered at high light intensities (Fig. 3, C and D). This finding contrasts with the results obtained for the hfr1 and far1 single mutants, which showed strongest loss-of-function phenotypes at high photon fluence rates.
Figure 3.
Epistatic analyses with positively acting Arabidopsis light signaling factors. Fluence rate response curves for the inhibition of hypocotyl elongation are shown for seedlings grown under continuous far-red light for 3 d after germination induction. Relative hypocotyl lengths were calculated in relation to the lengths of dark-grown seedlings for each line. Each data point represents the mean of two independent experiments ± se with at least 30 seedlings. A, Ler wild type, eid3, hy5-1, and eid3 hy5-1 mutants. B, Ler wild type, Col wild type, eid3, fhy3-1, and eid3 fhy3-1 mutants. C, Ler wild type, eid3, hfr1-2, and eid3 hfr1-2 mutants. D, Ler wild type, Nossen (No) wild type, eid3, far1-3, and eid3 far1-3 mutants.
Isolation of the Mutated Gene
For mapping analyses, phyB-5 eid3 was crossed with a phyB-9 loss-of-function mutant in the Columbia ecotype (Col). Analyses of seedling phenotypes under the screening program revealed that all F1 seedlings exhibited a clear eid3 mutant phenotype, indicative of a dominant mutant. The F2 generation exhibited a segregation of seedling phenotypes under selective light conditions (Eid3:wild type = 88:39) that was consistent with the expected 3:1 segregation of a dominant mutation (P < 0.05, χ2 test). Because eid3 has a dominant genetic inheritance, the recessive phenotype of the wild-type allele was used to isolate homozygous plants. The observed linkage with markers on chromosome 1 indicated that eid3 might be a novel allele of PFT1 (Supplemental Fig. S4). Sequence analyses of amplified genomic fragments revealed that eid3 carries a point mutation in the PFT1 gene (Fig. 4A). The eid3 mutation results in the replacement of Thr-650 of PFT1 with a Met residue in a highly conserved motif in the herpesvirus protein16 (VP16)-like domain, which is thought to be involved in specific recognition of transcription factors bound to promoter elements (Fig. 4B). The base pair exchange enabled the design of a specific derived cleaved-amplified polymorphic sequence (dCAPS) marker for the mutated allele. Analyses with the eid3 dCAPS marker exhibited a perfect cosegregation with the hypersensitive Eid3 phenotype after examination of 15 homozygous phyB-5 eid3 plants. Taken together, these findings strongly indicate that the dominant eid3 mutation is caused by a missense mutation in PFT1.
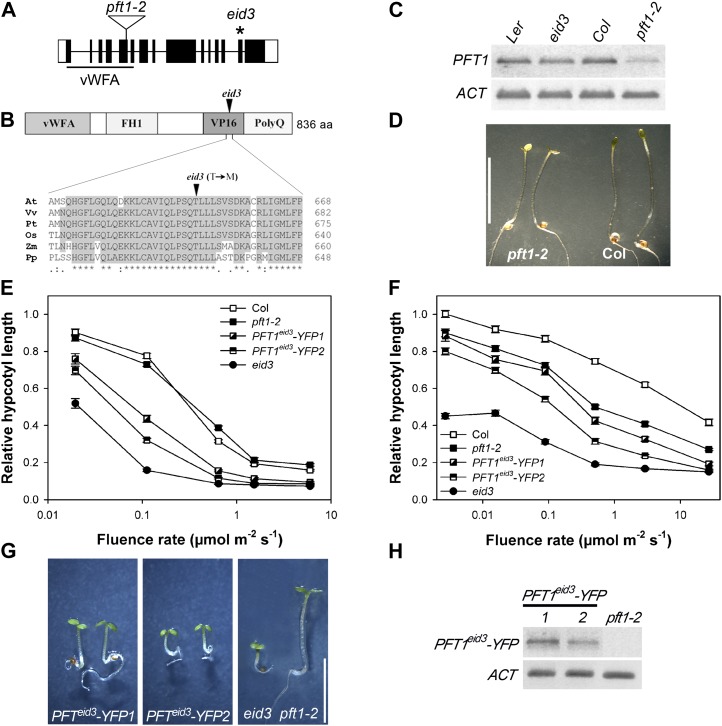
Figure 4.
Characterization of the pft1-2 mutant and reconstitution of the Eid3 phenotype. A, Schematic structure of the PFT1 gene. Black boxes represent exons, lines represent introns, and white boxes show untranslated regions of the mRNA. The positions of the van Willebrand factor type A domain (vWFA) and the T-DNA insertion in the pft1-2 mutant are indicated. The position of the eid3 missense mutation is indicated by the asterisk. B, Schematic structure of the PFT1 protein, and amino acid sequence alignment of the plant-specific, highly conserved region containing the eid3 mutation. FH1, Formin homology 1 domain; VP16, VP16-like interaction domain; PolyQ, Gln-rich domain; aa, amino acids; At, Arabidopsis; Vv, Vitis vinifera; Pt, Populus trichocarpa; Os, Oryza sativa; Zm, Zea mays; Pp, Physcomitrella patens. C, PFT1 transcript levels were monitored by RT-PCR using 4-d-old etiolated seedlings of the pft1-2 T-DNA line, eid3, and the corresponding wild types. ACT transcript levels are presented as a constitutive control. Gels were stained with SYBR-safe DNA gel stain and are shown inverted. The gel shows the representative result of three independent experiments. D, Photograph of pft1-2 mutant and Col wild-type seedlings grown under the eid screening program (cycles of 20 min of red light followed by 20 min of far-red light) for 4 d after germination induction. Bar = 5 mm. E and F, Fluence rate response curves for the inhibition of hypocotyl elongation of eid3, Col wild type, pft1-2, and pft1-2 mutant lines transformed with ProPFT1-PFT1eid3-YFP constructs. Seedlings were grown under continuous far-red light (E) or continuous red light (F). Relative hypocotyl lengths were calculated in relation to the lengths of dark-grown seedlings for each line. Each data point represents the mean ± se of two independent experiments with 30 seedlings. G, Photograph of pft1-2 mutant seedlings expressing PFT1eid3-YFP under the control of the PFT1 promoter and grown under the eid screening program (cycles of 20 min of red light followed by 20 min of far-red light) for 4 d. Seedlings of two independent transgenic lines are shown together with the eid3 mutant and the pft1-2 mutant background. Bar = 5 mm. H, Expression levels of the PFT1eid3-YFP transgene in seedlings of two independent transgenic lines and the corresponding pft1-2 mutant background line. Transcript levels were monitored as described for C. The gel shows representative results of three independent experiments. [See online article for color version of this figure.]
To further analyze the effect of PFT1 on light signaling, a transferred DNA (T-DNA) insertion line was isolated from the SALK collection (SALK_129555) that is identical to the published pft1-2 mutant (Kidd et al., 2009). The T-DNA was inserted into the fifth exon, which encodes for the van Willebrand factor type A domain (Fig. 4A). Reverse transcription (RT)-PCR analyses using primers downstream of the insertion site exhibited clear reduction in transcript accumulation in the pft1-2 T-DNA insertion line (Fig. 4C). With respect to transcript accumulation, pft1-2 resembles pft1-1, which accumulates reduced levels of a truncated transcript and carries a T-DNA insertion in the fourth intron (Cerdán and Chory, 2003).
To test whether the reduction of PFT1 gene expression leads to alterations in light sensitivity, the pft1-2 mutant was subjected to the multiple red/far-red pulse treatment used for the screening of eid mutants and to continuous irradiation with variable fluence rates of red and far-red light. The T-DNA line showed only a very weak hypersensitive response under the red/far-red pulse treatment compared with the Col wild-type control (Fig. 4D). No difference in light response was seen under continuous far-red light with pft1-2, which contrasts with the strong hypersensitive response of eid3 (Fig. 4E). The pft1-2 line exhibited an increase in light sensitivity under strong continuous red light (Fig. 4F), similar to published results for the pft1-1 allele (Cerdán and Chory, 2003).
To further prove whether the mutated form of PFT1 is responsible for the hypersensitive eid3 phenotype, genomic fragments spanning the promoter region and all exons and introns of PFT1 and PFT1eid3 were cloned in front of a Yellow Fluorescent Protein (YFP) cassette, and constructs were then introduced into the pft1-2 mutant. This approach was chosen because overexpression of PFT1 and PFT1eid3 causes the cosuppression of protein function in transgenic lines (Cerdán and Chory 2003; data not shown). Different homogenous lines were isolated that carried single T-DNA integrations. Analyses of light responses under the screening program demonstrated that the introduced ProPFT1-PFT1eid3-YFP constructs restore the hypersensitive Eid3 phenotype (Fig. 4G). The analyzed transgenic lines exhibited an increased sensitivity toward continuous red and far-red light compared with the Col wild type and the pft1-2 background (Fig. 4, E and F). The increase in light sensitivity was stronger in PFT1eid3-YFP2 compared with PFT1eid3-YFP1, even though transcript levels were lower in PFT1eid3-YFP2 (Fig. 4H). This finding, together with the increased red light sensitivity compared with the pft1-2 T-DNA line, argues against a dominant negative effect of PFTeid3 and further supports the notion that the eid3 missense mutation created a hyperactive transcriptional coregulator.
Temporal Expression Pattern of Light-Regulated Marker Genes during Seedling Deetiolation
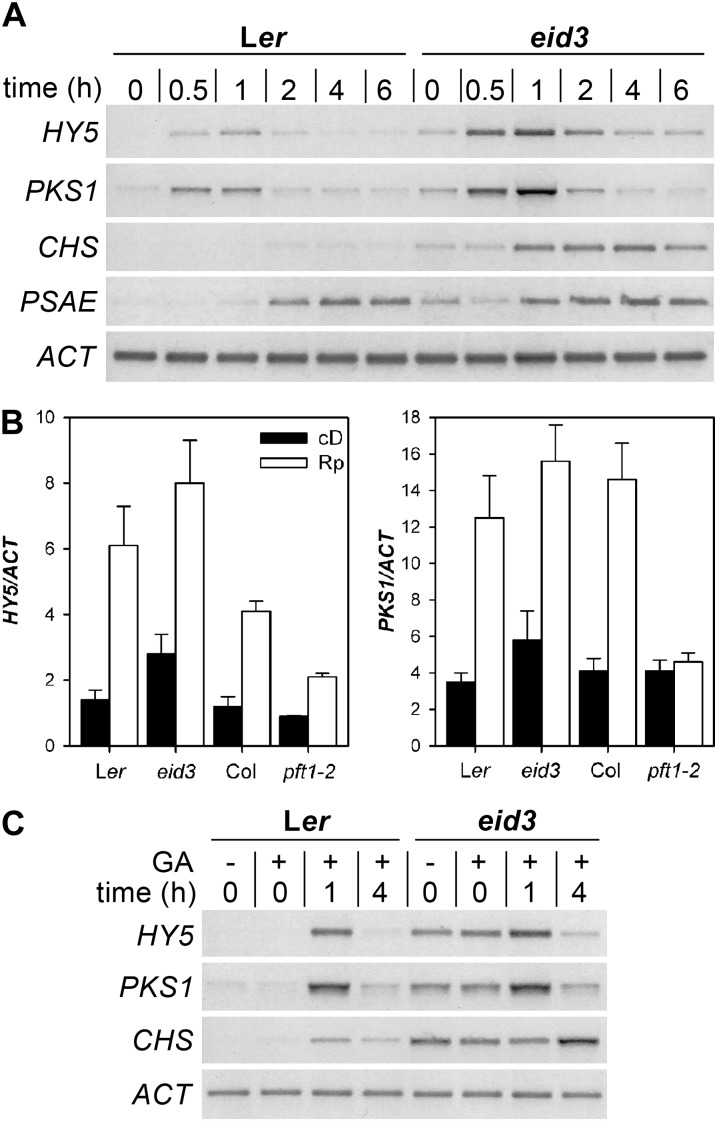
PFT1 was identified as a component of the Mediator transcriptional coregulator complex (Bäckström et al., 2007). In order to gain insight into the function of PFT1eid3 during light-regulated gene expression, temporal transcript accumulation patterns were analyzed by RT-PCR of different light-regulated marker genes and ACTIN1 (ACT).
HY5 and PHYTOCHROME KINASE-SUBSTRATE1 (PKS1) become rapidly increased upon light treatment and respond to even single pulses of red light (Peschke and Kretsch, 2011); they accumulated to very low levels in etiolated wild-type seedlings. In contrast, etiolated eid3 seedlings exhibited increased transcript accumulation for both genes. The red light pulse induced a rapid increase in HY5 and PKS1 transcript levels in the wild type, with maximum levels at 1 h (Fig. 5A). The eid3 mutant exhibited the same temporal accumulation pattern for both transcripts, but transcript levels were enhanced at all analyzed time points. Quantitative RT-PCR measurements verified the strong influence of eid3 on HY5 and PKS1 transcript accumulation in dark-grown and pulse-treated seedlings (Fig. 5B). The eid3 mutant exhibited increased transcript levels compared with the Ler wild type for both marker genes. In contrast, marker gene expression was reduced in pft1-2 compared with the corresponding Col wild type. Thus, eid3 behaves like a gain-of-function mutant, whereas pft1-2 shows characteristics of a loss-of-function allele.
Figure 5.
Expression pattern of light-regulated marker genes during seedling deetiolation. A, Expression of light-induced marker genes after a red light pulse. Ler wild-type and eid3 mutant seedlings were grown in darkness for 4 d after induction of germination. Expression of light-regulated genes was induced by a saturating 2-min red light pulse. The gel shows representative results of three independent experiments. HY5, PKS1, CHS, and PSAE transcript levels were monitored at the indicated time points after the red light pulse using RT-PCR. ACT transcript levels served as a constitutive control. Gels were stained with SYBR-safe DNA gel stain. Images are shown inverted. B, Quantification of transcript levels of light-induced HY5 and PKS1 marker genes in eid3, pft1-2, and the corresponding Ler and Col wild types. Seedlings were treated as described for A. Transcript levels were determined by quantitative real-time PCR analyses. Results of experiments were normalized according to the constitutively expressed ACT gene. Data represent means of three independent biological replicates ± se. Rp, 2-min red light pulse; cD, dark control. C, Light-independent expression of light-induced marker genes in the eid3 mutant and the Ler wild type. Germination was induced by either 2 h of continuous red light without GA (−GA) or without light treatment and application of 10 µm GA (+GA). Seedlings were grown in darkness for 4 d after germination induction. Expression of light-induced genes was initiated by a saturating 2-min red light pulse, and samples were harvested at 1 and 4 h or before light treatment (0 h). Transcript levels were monitored as described for A. The gel shows the representative result of two independent experiments.
Transcripts of SUBUNIT E OF PHOTOSYSTEM I (PSAE) and CHALCONE SYNTHASE (CHS) accumulate late compared with HY5 and PKS1 (Fig. 5A). PSAE gene expression is very light sensitive, whereas strong CHS expression is only obtained under prolonged irradiation with strong continuous blue and far-red light or upon UV-B pulse treatment (Peschke and Kretsch, 2011). In contrast to the wild type, eid3 seedlings accumulated enhanced transcript levels for PSAE, even in darkness. The red light pulse caused a weak preliminary accumulation of PSAE transcripts in eid3, but no major differences in signal intensities were detectable after 4 h, when maximum transcript levels were reached in wild-type and mutant seedlings (Fig. 5A). CHS transcripts remained below the detection level in etiolated wild-type seedlings and were faintly induced by red light pulse treatment. In striking contrast, eid3 seedlings exhibited enhanced CHS transcript levels in the dark and strong induction of CHS by a single red light pulse.
Constitutive Expression of Light-Regulated Marker Genes in Complete Darkness
All tested light-regulated marker genes exhibited weak constitutive expression in etiolated eid3 seedlings (Fig. 5, A and B), which had received 2 h of red light to induce homogeneous seed germination. Since eid3 exhibits an extremely enhanced light sensitivity, seeds were sown on 10 µm GA to enable germination and seedling development in complete darkness. The lack of red light pretreatment for germination induction did not alter the enhanced dark expression of light-regulated marker genes in eid3 (Fig. 5C). Furthermore, GA treatment did not alter HY5, PKS1, and CHS transcript accumulation in the wild type or eid3 upon the application of a red light pulse. HY5 transcript accumulation was also tested in both dark-grown phyA-201 phyB-5 eid3 seedlings and in GA-induced, dark-grown seedlings that had received pulses of extreme far-red light (red glass9 filter light) to photoconvert any remaining Pfr molecules back to the inactive Pr form. Neither the lack of the two dominant phytochromes nor the reduction of Pfr remaining from seed development resulted in a reduction of HY5 transcript levels in etiolated seedlings of eid3 mutant lines (Supplemental Fig. S5). Taken together, the data clearly indicate that the eid3 mutation of PFT1 enables the expression of light-regulated marker genes independent of any light input.
Transcript Accumulation Patterns in Dark-Grown eid3 Seedlings
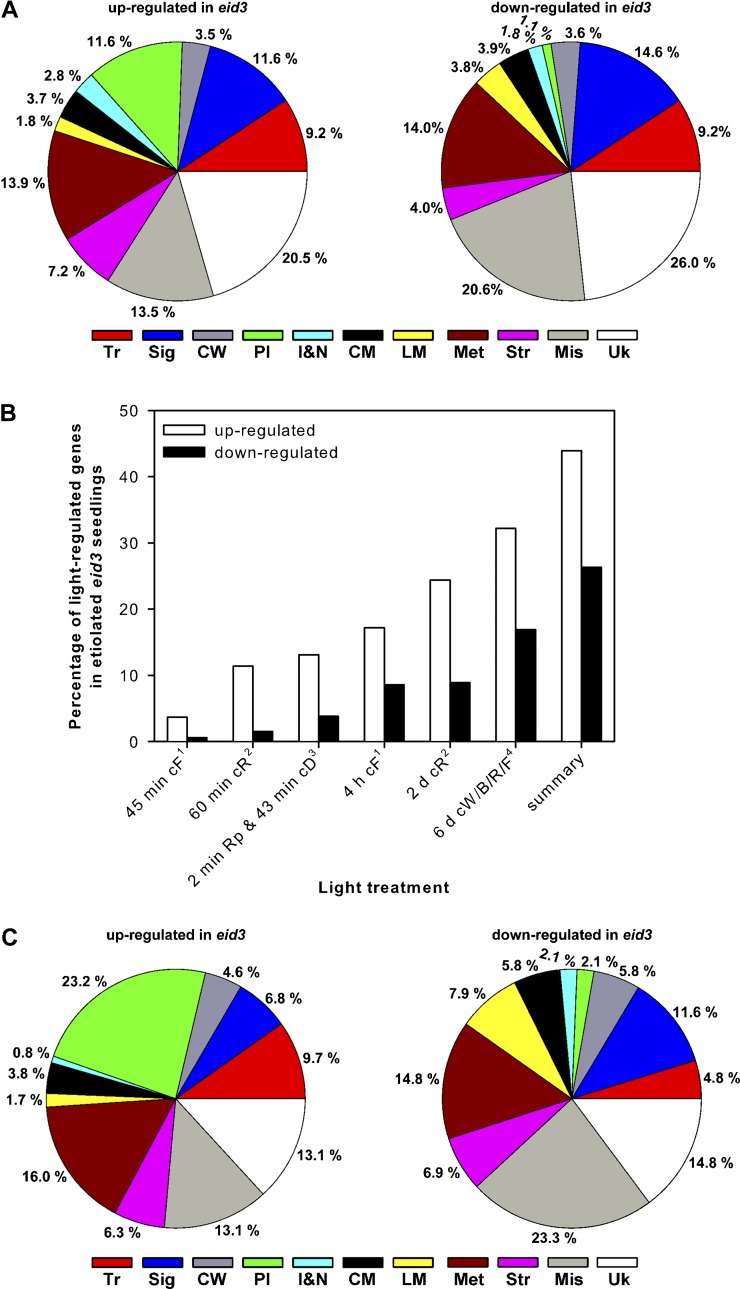
To obtain broader insight into the influence of eid3 on gene expression during skotomorphogenic development, transcript accumulation patterns were analyzed using Agilent 44K Arabidopsis gene expression microarrays, which include approximately 28,900 nuclear genes, 66 plastidic genes, and 67 mitochondrial genes. Three independent RNA samples were isolated from 4-d-old etiolated eid3 or wild-type seedlings and subjected to microarray analyses. Transcript levels were only regarded as differentially regulated between the wild type and eid3 if they exhibited both a 2-fold or greater change in signal intensities (up or down) and a statistically significant difference in expression values (t test, P < 0.05) adjusted for a false discovery rate of Q < 0.05 (Benjamini and Hochberg, 1995).
In etiolated eid3 plants, 542 genes exhibited significantly enhanced and 718 genes exhibited significantly reduced transcript levels compared with etiolated wild-type seedlings, indicating a dual role for PFT1 as a transcriptional activator and suppressor. The list of up-regulated genes includes PSAE and CHS marker genes, which exhibited enhanced transcript levels with RT-PCR analyses. Twelve percent of the up-regulated genes (66) encode for genes related to photosynthesis and chloroplast development (Fig. 6A; Supplemental Table S1). High numbers of the up-regulated genes belong to the group’s transcriptional regulators (50 of 542) and signaling components (62 of 542). Among these 112 genes, 34 are known to be related to light signaling (10), hormone function (13), leaf development (four), or regulation of meristematic activity and cell differentiation (six; Supplemental Table S2). Up-regulated genes involved in light signaling include ATTENUATED FAR-RED RESPONSE1, LAF1, and HY5 HOMOLOG, all of which function as important positive regulators of photomorphogenesis. Among the up-regulated genes related to hormone function, five genes encode for negative regulators of cytokinin that function by either inactivating the hormone (CYTOKININ OXIDASE5) or at the level of transcriptional regulation (ARABIDOPSIS RESPONSE REGULATOR7/15/16; BEL1-LIKE HOMEODOMAIN3). Up-regulated genes also included ALTERED MERISTEM1, which functions as a regulator of photomorphogenesis (cop2 mutant) and transition to flowering.
Figure 6.
Analyses of transcript accumulation patterns in dark-grown eid3 seedlings. A, Ler wild-type and eid3 seeds were grown in darkness for 4 d before extraction of RNA and determination of transcript accumulation patterns by microarray analyses. Pie charts show the distribution of more than 2-fold up- or down-regulated genes among functional categories. Distributions are expressed as percentages of 541 up-regulated and 718 down-regulated genes in etiolated eid3 compared with wild-type seedlings. B, Bar charts depict percentages of up- and down-regulated genes in etiolated eid3 seedlings that have been annotated as being light regulated in published data sets (Jiao et al., 2005; Leivar et al., 2009; Peschke and Kretsch, 2011) and upon red light pulse treatment of 4-d-old, etiolated Ler wild-type seedlings (this study). Ler wild-type seedlings received one saturating red light pulse before transfer back to darkness for 43 min. Transcript levels were compared with etiolated seedlings harvested at the same time. Genes exhibiting more than 2-fold up- or down-regulation in transcript levels were annotated as being light regulated: 1Peschke and Kretsch (2011); 2Leivar et al. (2009); 3this study; 4Jiao et al. (2005). cD, Continuous darkness; cF, continuous far-red light; cR, continuous red light; cW/B/R/F, continuous white/blue/red/far-red light; Rp, red light pulse. C, Pie charts show the distribution among functional categories of up- and down-regulated genes in etiolated eid3 seedlings that have been annotated as being light regulated in other data sets. Distributions are expressed as percentages of 337 up-regulated and 179 down-regulated genes. Tr, Transcription; Sig, signaling; CW, cell wall; Pl, plastids; I&M, ions and nutrition; CM, carbohydrate metabolism; LM, lipid metabolism; Met, metabolism miscellaneous; Str, stress; Mis, miscellaneous; Uk, unknown.
High numbers of the down-regulated genes encode for transcriptional regulators (63 of 718), signaling components (107 of 718), and factors related to cell wall metabolism (26 of 718; Fig. 6A; Supplemental Table S3). Many of the down-regulated genes encoding for signaling and transcription factors are related to cell cycle control (15), meristem organization (five), leaf development (four), and differentiation of epidermal cells (six; Supplemental Table S2). Down-regulated genes also included repressors of the floral transition, such as FLOWERING LOCUS C (FLC), AGAMOUS-LIKE42, and MADS AFFECTING FLOWERING1. These findings indicated that the eid3 mutation induces strong alterations in the expression of genes related to growth and development in the absence of light.
Furthermore, the eid3 mutant caused down-regulation of a high number of genes related to diverse metabolic processes (Fig. 6A; Supplemental Table S3). Among these genes, a high proportion encode for enzymes and proteins involved in the activation of storage compounds. Of the 28 genes involved in carbohydrate metabolism, 12 have been annotated as hydrolases. Of the 27 genes involved in lipid metabolism, 12 are lipases and 10 are lipid transfer/seed storage proteins. Furthermore, 17 genes encode for proteases (Supplemental Table S3).
Comparisons of Transcript Accumulation Patterns in Dark-Grown eid3 Seedlings with Data Sets of Light-Regulated Genes
To determine if differentially regulated transcripts in etiolated eid3 plants are known light-regulated genes, the above data set was compared with published microarray data of wild-type Arabidopsis seedlings subjected to various irradiation times under different light qualities (Jiao et al., 2005; Leivar et al., 2009; Peschke and Kretsch, 2011). Furthermore, transcript accumulation patterns were compared with data from 4-d-old, etiolated Ler wild-type seedlings that were treated with a saturating, 2-min red light pulse before transfer back to darkness for 43 min. Transcript levels of three independent replicates were compared with the corresponding dark controls using the above-described criteria (P < 0.05, Q < 0.05, 2-fold difference). In wild-type seedlings, transcript levels were significantly increased for 544 genes and significantly decreased for 214 genes by the red light pulse compared with dark controls (Supplemental Tables S4 and S5).
Transcript accumulation patterns from the three studies were most dissimilar at 45 min to 1 h after the onset of irradiation but showed increasing degrees of overlap at extended irradiation times (Fig. 6B; Supplemental Fig. S6; Supplemental Table S6). Analyses with the current data set showed that 44% (237 of 541) of up-regulated genes and 26% (189 of 718) of down-regulated genes in etiolated eid3 have been identified as being light regulated by other studies (Fig. 6B; Supplemental Tables S7 and S8). Of the overlapping genes, 23% of the up-regulated genes (55) encode for proteins related to photosynthesis and chloroplast development (Fig. 6C). Additionally, six enzymes involved in flavonoid biosynthesis and PORA, all known to be light regulated, were identified. The list of light-induced genes also includes most of the regulatory factors involved in light signaling that become up-regulated in etiolated eid3 seedlings (Supplemental Table S2). A high proportion of constitutively repressed, light-regulated genes encode for proteins involved in the mobilization of storage compounds, including seven glycosyl hydrolases, seven lipases, eight lipid transfer proteins, and seven proteases (Supplemental Table S3). These findings demonstrate that the eid3 mutant of PFT1 causes constitutive expression of an important subset of light-regulated genes and genes related to photomorphogenic development in the dark.
Since epistasis analyses showed that the hypersensitive Eid3 phenotype strictly depends on the presence of the transcription factor HY5, the list of differentially up-regulated genes in etiolated eid3 seedlings was compared with known HY5 target genes (Lee et al., 2007). In etiolated eid3, 24% (128 of 541) of the up-regulated genes are putative targets of HY5 (Supplemental Table S6). Interestingly, an even higher congruence (58%) was found among the 71 early light-induced genes that were significantly increased in dark-grown eid3 seedlings. These percentages clearly exceed the 14% ratio of putative HY5 target genes from the entire Arabidopsis genome.
Epistatic Regulation of the Floral Transition between eid3 and phyA and phyB Loss-of-Function Mutants
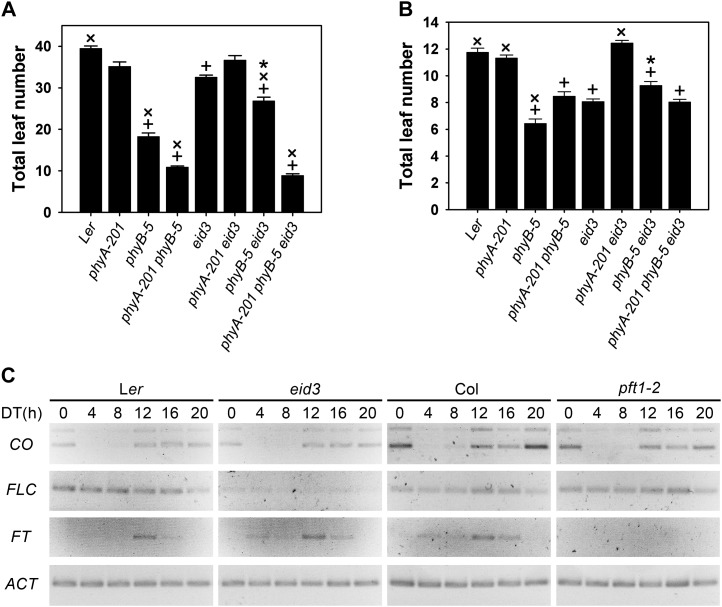
PFT1 was first described as a regulator of flowering time under high-density far-red light, which is mainly sensed by phyB (Cerdán and Chory, 2003). To test for the function of the eid3 allele of PFT1 in flowering induction downstream of phyA and phyB, the different single, double, and triple mutants were grown under short-day (8 h of light/16 h of dark) and long-day (16 h of light/8 h of dark) conditions. Numbers of leaves on the rosette and the main stem were counted to measure time until flowering.
Under both short- and long-day conditions, the phyA-201 loss-of-function mutant had no significant effect on flowering compared with the wild type, whereas the loss the phyB caused a strong reduction in leaf number (Fig. 7, A and B). The flowering time of the phyA-201 phyB-5 double mutant was slightly sooner in short days and delayed in long days compared with phyB-5. The eid3 mutant exhibited an early-flowering phenotype under both light regimes (Fig. 7, A and B). This finding differs from the results obtained with pft1-1 mutants in the Col ecotype, in which delays of flowering under long-day conditions and only a weak, if any, reduction in flowering time in short days occurred (Cerdán and Chory, 2003). The early-flowering phenotype of eid3 strictly depends on the presence of phyA, because the phyA-201 eid3 double mutant developed the same number of leaves at flowering as the corresponding phyA-201 mutant and the Ler wild type. Induction of flowering under both short and long days was significantly delayed in the phyB-5 eid3 double mutant compared with phyB-5, but double mutant plants still flowered earlier compared with the wild type. This result differs from epistatic analyses with pft1 T-DNA insertion lines, which show a complete suppression of early flowering induced by the loss of phyB under either short or long days (Cerdán and Chory, 2003; Wollenberg et al., 2008; Iñigo et al., 2012). Compared with eid3, leaf number was still significantly reduced in short days and remained unaltered in long days. The eid3-dependent delay of flowering in the phyB-5 background is masked in the phyA-201 phyB-5 eid3 triple mutant under long-day and, more severely, short-day conditions (Fig. 7, A and B). These data clearly indicate that the presence of phyA is necessary for the inhibitory effect of PFT1eid3on early flowering in a phyB mutant.
Figure 7.
The eid3 mutant affects the floral transition and the expression of flowering genes. A and B, Flowering time of eid3, eid3 phyA-201, eid3 phyB-5, eid3 phyA-201 phyB-5, and corresponding background lines under variable daylengths. Flowering time was determined in short days consisting of 8-h-light/16-h-dark cycles (A) and long days consisting of 16-h-light/8-h-dark cycles (B). Flowering time is presented as the sum of rosette and cauline leaves at the main stem. Data represent means of at least two independent experiments ± se with 20 or more plants. + Significant difference from the Ler wild type; ×, significant difference from eid3; * significant difference from the eid3 double and triple mutants to corresponding phyA, phyB, and phyA phyB mutant background lines (one-way ANOVA on ranks, P < 0.05). C, Diurnal transcript accumulation patterns of different flowering genes in Ler wild type, eid3, Col wild type, and pft1-2. Plants were grown under 12-h light/dark cycles for 2 weeks. On day 14, samples were taken every 4 h starting at the onset of light (DT 0 h). Transcript levels were determined by RT-PCR using identical cDNA samples. CO, FLC, and FT transcript levels were monitored at the indicated time points. ACT transcript levels served as a constitutive control. Gels were stained with SYBR-safe DNA gel stain. Images are shown inverted. The gel shows representative results of two independent experiments.
Expression Pattern of Flowering Genes
To analyze the influence of PFT1eid3 on the central regulatory components of the floral transition, transcript levels of CONSTANS (CO), FLC, and their downstream target FLOWERING LOCUS T (FT) together with a constitutive ACT control were followed by RT-PCR in eid3, pft1-2, and corresponding wild types. Plants were grown under 12-h light/dark cycles for 2 weeks. On day 14, samples were harvested every 4 h starting at the onset of light (daytime [DT] 0 h).
In accordance with published results (Cerdán and Chory, 2003; Wollenberg et al., 2008), CO transcript levels fell below detection levels at DT 4 h, started to accumulate at DT 8 h, and remained high during the dark phase (DT 12 h–DT 20 h; Fig. 7C). Compared with the corresponding wild-type controls, CO transcript levels were slightly reduced in pft1-2 and did not exhibit clear alterations in eid3. FLC transcripts did not show major diurnal fluctuations (Fig. 7C). The eid3 mutant exhibited a very strong reduction in FLC transcript levels compared with the Ler wild-type background, confirming results obtained with microarray analyses. In contrast to the strong effect observed with eid3, no clear difference in FLC expression was detected between pft1-2 and the corresponding Col wild type. FT exhibited a diurnal rhythm with maximum transcript levels at DT 12 h in wild-type plants (Fig. 7C), similar to published results (Cerdán and Chory, 2003; Wollenberg et al., 2008). Compared with the corresponding wild-type controls, FT transcript accumulation was enhanced in eid3 but remained below detection levels in pft1-2.
DISCUSSION
The study describes the characterization of the dominant eid3 mutant, which manifests as extremely enhanced light sensitivity during Arabidopsis seedling development. The phenotype is caused by the missense mutation T650M of the PFT1 subunit of the plant Mediator complex, as determined by mapping and sequencing analyses, cosegregation analyses of the eid3 dCAPS marker with the hypersensitive phenotype, and reconstitution of the Eid3 phenotype by transformation of pft1-2 T-DNA lines with the mutated version of the gene.
The missense mutation is localized to a domain of the PFT1 protein that shows weak similarity to the VP16 interaction domain of MEDIATOR SUB-UNIT25 (MED25) from higher eukaryotes (Bäckström et al., 2007). The VP16 interaction domain of MED25 enables interactions between the Mediator core complex and numerous transcriptional regulators downstream of different signaling cascades in mammalian systems in order to trigger RNA polymerase II-dependent gene expression (Malik and Roeder, 2010). Because of its weak similarity to the VP16 interaction domain, its conserved localization in front of the Q-rich domain, and its high conservation in the plant kingdom, the corresponding domain of PFT1 is hypothesized to perform a similar function in the transcriptional regulation of plant genes (Bäckström et al., 2007; Kidd et al., 2009). This hypothesis has been verified by several studies demonstrating that the corresponding domain interacts with transcription factors involved in the regulation of jasmonate signaling and pathogen responses (Elfving et al., 2011; Kidd et al., 2011; Ou et al., 2011).
The eid3 Mutant Is a Gain-of-Function Allele of PFT1 and Alters Gene Expression Patterns of Light-Regulated Genes in Darkness
The dominant genetic inheritance and the gain-of-function phenotype of the eid3 allele of PFT1 can most easily be explained by a constitutively active transcriptional coregulator that either escapes inactivation or does not depend on additional modifications to achieve full activity. This interpretation is consistent with the observed constitutive expression of light-regulated genes in darkness. When the transcript accumulation patterns in this study were compared with previously identified light-regulated genes (Jiao et al., 2005; Leivar et al., 2009; Peschke and Kretsch, 2011), 44% of up-regulated genes and 26% of down-regulated genes in dark-grown eid3 seedlings matched, in agreement with the observed weak COP-like phenotype for hook opening. The set of up-regulated transcripts includes genes related to chloroplast development and flavonoid accumulation, two processes known to be linked to photomorphogenic development. A high number of down-regulated genes in eid3 encode factors involved in transcriptional regulation, signaling, cell division, and mobilization of storage compounds. Genes related to the mobilization of storage compounds are expected to be suppressed upon the transition from skotomorphogenesis to photomorphogenesis, which is accompanied by a change from heterotrophic to photoautotrophic metabolism. These findings further confirm that the eid3 mutation creates a constitutively active transcriptional coregulator, which causes a mild constitutive photomorphogenic phenotype in the absence of light.
The hypersensitive light phenotype of eid3 is caused by the exchange of a Thr with a Met residue in a sequence of highly conserved amino acids that carries additional highly conserved Thr and Ser residues in the VP16-like domain of PFT1. It is worthwhile to speculate that the corresponding sequence can be modified by light-dependent kinase or phosphatase activities in wild-type PFT1, whereas the eid3 gain-of-function mutation either overcomes the necessity of such a modification for its activation or escapes an inactivation process.
PFT1eid3 Interferes with Positively and Negatively Acting Components Regulating Arabidopsis Light Signaling
Epistasis analyses demonstrated that a mutated PFT1 component of the Mediator complex cooperates with different DNA-binding proteins that function as positive effectors in Arabidopsis light signaling. Loss of FAR1, FHY3, HFR1, and HY5 strongly reduced light responses in eid3 under very low light intensities of far-red light normally sensed by phyA. These findings indicate that responses under very-low-fluence conditions need the presence of the complete set of these positively acting transcriptional regulators to enable expression of the Eid3 phenotype. In contrast, analyses with hfr1-1 eid3 and far1-1 eid3 double mutants demonstrate that loss of HFR1 and FAR1 does not alter expression of the hypersensitive Eid3 phenotype under strong continuous far-red light. Thus, loss of both transcription factors seems to be insignificant for PFT1eid3 function in the regulation of far-red HIR. A stronger but not fully epistatic loss-of-function phenotype was observed with eid3 fhy3-1 double mutant lines, indicating that FHY3 acts in concert with the mutated PFT1 factor regulating phyA-dependent HIR. The fhy3-1 loss-of-function allele might not be fully epistatic toward eid3, because the remaining FAR1 transcription factor might partially compensate for the loss of its homologous partner FHY3 (Wang and Deng, 2002; Wang et al., 2002; Hudson et al., 2003).
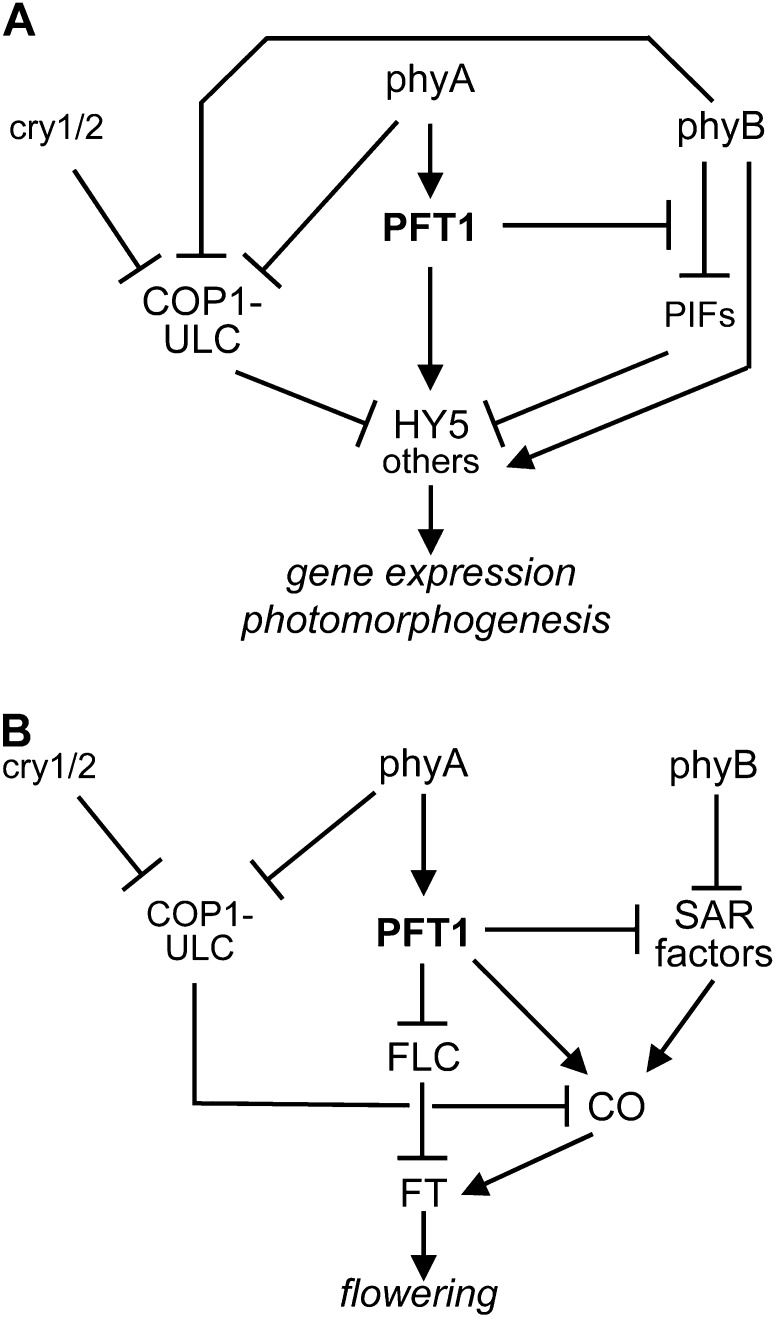
While neither the eid3 nor cop1eid6 background line shows a strong deetiolated phenotype in darkness, the combination of both alleles in double mutants enabled nearly full establishment of photomorphogenesis in the absence of light. The cop1eid6 allele carries an amino acid exchange in the zinc-finger domain of COP1 that is sufficient to suppress light responses in dark-grown seedlings but loses its functional integrity even upon the application of very low light fluences (Dieterle et al., 2003). Similar to weak light, PFT1eid3 is sufficient to surmount residual COP1-dependent ubiquitin ligase activity in eid3 cop1eid6 double mutants and, thus, seems to replace light stimulation. Because microarray data demonstrate that PFT1eid3 causes strong alterations in transcript accumulation patterns of light-regulated genes in etiolated seedlings, it is worthwhile to speculate that corresponding alterations in gene expression are responsible for the failure of COP1eid6 to repress photomorphogenesis. Conversely, intact COP1-ULCs seem to counteract PFT1eid3 function in darkness, most probably by targeting transcription factors and other regulatory proteins to degradation in the proteasome (Hardtke et al., 2000; Holm et al., 2002; Seo et al., 2003, 2004; Duek et al., 2004; Jang et al., 2005, 2010). According to this interpretation, PFT1eid3 would function upstream of COP1 and, thus, might be a good candidate for an immediate early target of light sensed by phytochromes and other photoreceptors (Fig. 8A).
Figure 8.
Models for the proposed function of PFT1 in light signaling during photomorphogenic seedling development and the regulation of flowering. A, Proposed function of PFT1 in light signaling downstream of different photoreceptors. PFT1 may function mainly by up-regulation of the HY5 basic Leu-zipper transcription factor gene, a central positive regulator of early light signaling. COP1-ULCs function as negative regulators that target positively acting factors like HY5 to degradation in the proteasome until COP1 inactivation by Pfr- and/or cry1/2-dependent mechanisms. COP1-ULC activity would act downstream of PFT1 by counteracting the protein accumulation of regulatory factors, for which transcription becomes up-regulated by the function of the respective subunit of a plant Mediator complex. PFT1 should also function as a negatively acting component in a phyA signaling cascade that inhibits phyB responses. PIFs, Phytochrome-interacting proteins. B, Proposed function of PFT1 in regulation of the floral transition. PFT1 is thought to mainly function downstream of phyA. As a component of the Mediator complex, PFT1 seems to increase the expression of the CO gene, a central regulator of the photoperiod pathway that is also involved in the acceleration of flowering under high far-red conditions sensed by phyB. CO protein levels are also regulated by phyA and cry1/2, both of which block COP1-dependent ubiquitin ligase activity in the light. Additionally, PFT1 might inhibit expression of the FLC gene, an important repressor of the FT florigen. The model further proposes that PFT1 inhibits phyB-dependent inhibition of SAR in a phyA-dependent manner.
The proposed model about coaction between PFT1eid3 and COP1eid6 also fits with the epistatic effect of the hy5 loss-of-function mutant on the pft1eid3 allele. HY5 takes a central role as a positive regulator at the base of transcriptional networks regulating early photomorphogenesis (Jiao et al., 2007; Lee et al., 2007; Zhang et al., 2011). COP1-ULCs function as major negative regulators of HY5 by targeting the protein to degradation in the proteasome in the absence of light (Ang et al., 1998; Hardtke et al., 2000; Osterlund et al., 2000; Saijo et al., 2003; Lian et al., 2011; Liu et al., 2011; Rizzini et al., 2011). In contrast, PFT1eid3 and, most probably, wild-type PFT1 act as positive regulators of HY5 gene expression. Because a high proportion of the light-dependent genes differentially regulated in etiolated eid3 seedlings belong to the set of identified HY5 target genes, it is worthwhile to speculate that the hypersensitive Eid3 phenotype is caused by the observed overexpression of the transcription factor in the eid3 mutant line in darkness and under weak light (Fig. 8A). Enhanced levels of HY5 and, as a consequence, HY5 target genes might overwhelm the residual function of COP1eid6 in the eid3 cop1eid6 double mutant. Alternatively, PFT1-containing Mediator complexes might directly interact with HY5-containing transcriptional initiation complexes formed at promoter elements of light-regulated genes. PFT1eid3 might then enable increased transcriptional activity by an altered recruitment of RNA polymerase II complexes to corresponding promoters but would lose its ability in the absence of HY5. However, we could not detect direct interactions between HY5 and PFT1 or PFT1eid3 with either the yeast two-hybrid system or in bimolecular fluorescence complementation assays in transfected mustard (Sinapis alba) seedlings (data not shown).
PFT1eid3 Functions Downstream of Phytochrome Photoreceptors
Data from double and triple mutant lines demonstrated that PFT1eid3 functions downstream of the phyA photoreceptor to regulate VLFR and far-red HIR. First, loss of phyA completely abolished the expression of the hypersensitive Eid3 seedling phenotype under weak and strong continuous far-red light, inducing VLFR and HIR. Second, a strong increase in light responses was observed upon hourly pulse treatments with extreme far-red light, which can only induce phyA-dependent VLFR (Yanovsky et al., 1997). Responses to hourly pulse treatments were enhanced in phyA-401 eid3 double mutants, indicating that both hypersensitive mutants cooperate synergistically to regulate VLFR. Third, eid3 eid1-1 double mutants exhibited a strong synergistic increase in light sensitivity. EID1 functions as a negative regulator that is specifically involved in the down-regulation of phyA-dependent HIR (Dieterle et al., 2001; Zhou et al., 2002). Finally, data on floral induction demonstrated that the effects of eid3 on floral transition strictly depend on the presence of an intact phyA photoreceptor.
The enhanced far-red light response of the dominant eid3 allele contrasts with the weak loss-of-function phenotype observed in pft1-1 (Cerdán and Chory, 2003) and the lack of strong far-red light sensitivity alterations in pft1-2 (this study). The pft1-1 and pft1-2 T-DNA insertion lines are regarded as loss-of-function alleles of PFT1. Nevertheless, because both T-DNA lines still exhibited an accumulation of PFT1 transcripts, it cannot fully be excluded that truncated versions of PFT1 or low levels of the full-length protein might be able to maintain some function necessary to enable residual light responses. Alternatively, it might be possible that light-induced activation of PFT1 is mainly involved in early light-dependent gene expression and becomes rapidly shut down in the wild type either by the reversion of protein modifications and/or by the downstream function of COP1-ULC. Thus, lack of PFT1 might be less severe under continuous light compared with the presence of a constitutively active PFT1eid3 transcriptional coregulator, which might escape inactivation to function as a dominant positive regulator of light responses during an extended time period.
Analyses of red light responses with pft1-2, eid3, eid3 phyA-201, eid3 phyB-5, and eid3 phyA-201 phyB-5 mutants revealed a very complex interaction pattern between PFT1, PFT1eid3, and phytochrome photoreceptors under this light condition. Fluence rate response curves demonstrated that loss of phyA causes a strong reduction in red light sensitivity at very low light intensities that induce VLFR, but it does not affect the expression of hypersensitive light responses under high red light intensities. This finding resembles results obtained with eid1 mutants (Büche et al., 2000). Loss of hypersensitivity in eid1 correlates with reduced steady-state levels of phyA due to its light-induced degradation being dependent on fluence rate. Because eid3 does not exhibit alterations in phyA stability, a similar inhibitory effect is expected under strong red light.
In contrast, loss of phyB did not alter responses toward weak red light but caused a strong reduction in the expression of the Eid3 phenotype at high photon fluence rates of red light-inducing, phyB-dependent HIR. Even though the loss of dominating phyA and phyB photoreceptors resulted in strong reductions in red light responses in eid3 phyA-201 phyB-5, the triple mutant still exhibited increased light sensitivity compared with its phyA-201 phyB-5 background. These findings indicate that PFT1eid3 acts not only as a positive regulator in light signaling downstream of phyB but also for other type II phytochromes. Assuming that eid3 is a gain-of-function allele of PFT1, the wild-type Mediator component would play a similar positive role in light signaling, although with reduced efficiency. This interpretation seems to contradict the results obtained with pft1-1 and pft1-2 T-DNA insertion lines that, although thought to represent loss-of-function alleles, also exhibited a hypersensitive light response toward high intensities of red light (Cerdán and Chory, 2003; this study). These data indicate that PFT1 functions as a negative regulator downstream of phyB. Nevertheless, it should be mentioned that introduction of ProPFT1-PFT1eid3-YFP constructs into pft1-2 clearly increases red light sensitivity above the level observed with the T-DNA insertion line. This finding argues in favor of the hypotheses that eid3 is a gain-of-function allele rather than a loss-of-function allele of PFT1 and that PFT1eid3 functions as a positive regulator of red light signaling downstream of phyB.
To explain these contradictory results, we propose a model that is based on three assumptions (Fig. 8A): first, PFT1 mainly functions as a positive factor in light signaling downstream of phyA; second, PFT1 mediates phyA-dependent inhibition of phyB-controlled light responses that have been described in several studies (Hennig et al., 1999, 2001; Cerdán and Chory, 2003); and third, phyB should be able to stimulate light-dependent gene expression on at least two separate pathways, one of which should be negatively regulated by PFT1 in a phyA-dependent manner, whereas the second should have a stimulatory effect on PFT1-dependent light signaling. The first pathway might be related to the phyB-dependent degradation of phytochrome-interacting factors (negative regulators of light signaling) or another positively acting pathway functioning on the level of transcriptional regulation. The second pathway might be mediated through the inhibition of COP1-ULC by phyB (Yi and Deng, 2005; Jang et al., 2010). According to this model, loss of PFT1 would release phyB responses from phyA-dependent suppression, which would result in the observed hypersensitive red light response. A similar release of phyB suppression would occur in phyA loss-of-function mutants or under light conditions that induce the degradation of phyA (strong continuous red light or red/far-red pulse treatment of the screening program). Reduction of phyA levels under strong red light would most probably also counteract any inhibition of phyB function in the presence of the hyperactive pft1eid3 allele. As a consequence, inhibition of COP1-ULC by phyB might come into play, which would release the downstream block suppressing hypersensitive light responses induced by PFT1eid3.
Loss of phyA caused a severe reduction in blue light sensitivity in eid3 phyA-201, indicating that the photoreceptor also plays a dominant role in the expression of hypersensitive blue light responses mediated by PFT1eid3. Because blue light sensitivity was still increased in eid3 phyA-201 and eid3 phyA-201 phyB-5 compared with corresponding background controls, the data indicate that the PFT1 protein is also involved in signaling downstream of blue light photoreceptors, most probably cry1 and cry2. Nevertheless, effects of blue light receptors are weak compared with the strong enhancement of light signaling downstream of phyA. Cryptochromes may function mainly by the suppression of COP1-ULC, which is shown to occur by direct interaction of activated photoreceptor molecules with the ubiquitin ligase complexes (Liu et al., 2011; Zuo et al., 2011).
PFT1eid3 Alters the Transition to Flowering
FT is a central floral activator in Arabidopsis that integrates signals downstream of the photoperiod, vernalization, and autonomous pathways and that directly regulates floral meristem identity genes upon its transport from leaves to the vegetative bud (Imaizumi and Kay, 2006; Turck et al., 2008; Amasino, 2010). FT transcript levels were up-regulated in eid3 and down-regulated in pft1-2 under diurnal conditions. This finding fits to the early-flowering phenotype observed with eid3 under short- and long-day conditions and the delay in flowering time observed with pft1 T-DNA lines at least under long days (Cerdán and Chory, 2003; Wollenberg et al., 2008; Kidd et al., 2009; Iñigo et al., 2012). These observations are consistent with interpretations that pft1eid3 is a hyperactive allele enhancing FT expression and floral transition, whereas the pft1-2 T-DNA line represents a loss-of-function allele.
Nevertheless, transcript accumulation patterns of CO and FLC exhibited a more complex interaction between different PFT1 alleles and diverse flowering pathways. Expression of CO, a key component of the photoperiod pathway (Imaizumi and Kay, 2006; Turck et al., 2008; Amasino, 2010), did not exhibit strong alterations in eid3 but is down-regulated in pft1-2, similar to results obtained with pft1-1 (Cerdán and Chory, 2003). CO is most important for the floral transition under extended light phases, which enable accumulation of the protein at dawn due to light-induced inhibition of COP1-ULC (Imaizumi and Kay, 2006; Turck et al., 2008; Amasino, 2010). This effect would explain why pft1 loss-of-function alleles mainly delay flowering under long-day conditions but show nearly no effect under short days (Cerdán and Chory, 2003). Even though eid3 does not seem to alter the photoperiod pathway via CO expression, a strong down-regulation of FLC transcripts was detected in the mutant. In contrast, pft1-2 did not exhibit a clear difference in FLC transcript accumulation. FLC is a negative regulator of FT expression, which is suppressed by the function of the vernalization and autonomous pathways to enable flowering (Michaels and Amasino, 1999; Sheldon et al., 2000, 2002; Sung and Amasino, 2005; Amasino, 2010). Analyses with lines expressing variable levels of FLC demonstrated that the transcription factor also modulates early-flowering responses under low red/far-red ratios through adjusting low levels of the active Pfr form of phyB and other type II phytochromes (Wollenberg et al., 2008; Adams et al., 2009). Accordingly, constitutive down-regulation of FLC in eid3 might result in the observed early-flowering phenotype under both short and long days. The proposed dual function of PFT1 in the photoperiod and FLC-dependent pathways is in accordance with findings that propose CO-dependent and -independent mechanisms of PFT1 operating downstream of phyB, phyD, and phyE (Iñigo et al., 2012).
Epistatic analyses of eid3 with phyA knockout mutants revealed that the early-flowering phenotype of the pft1eid3 allele strictly depends on the presence of the light-labile phytochrome. This phyA-dependent enhancement provides further evidence that PFT1eid3 is a constitutively active version of the transcriptional coregulator that functions downstream of phyA. In contrast to its positive effect on phyA-dependent flowering, data from eid3 phyB-5 and eid3 phyA-201 phyB-5 demonstrated that PFT1eid3 suppresses the early-flowering phenotype normally observed with phyB loss-of-function alleles in a phyA-dependent manner. Early flowering in phyB loss-of-function mutants is attributed to a constitutive SAR, because the mutants should completely eliminate active Pfr-B similar to high far-red light (Somers et al., 1991; Devlin et al., 1992; Reed et al., 1993; Franklin and Quail, 2010). Nevertheless, in contrast to the full epistatic effect of pft1 T-DNA lines on the early flowering of phyB mutants (Cerdán and Chory, 2003; Wollenberg et al., 2008; Kidd et al., 2009; Iñigo et al., 2012), double and triple mutants still flowered earlier compared with the wild type (in short and long days) and eid3 (short days). Epistasis of pft1 mutants also remains unaltered in phyA loss-of-function mutants (Cerdán and Chory, 2003). These findings indicate that the loss of PFT1 has a much stronger influence on flowering under high far-red light compared with the hypersensitive pft1eid3 allele and that these responses most probably follow different mechanisms. As mentioned above, the inhibition of early flowering in pft1 loss-of-function mutants can be attributed to the reduced expression of CO, which plays an important role not only in the photoperiodic pathway but also under high far-red light (Wollenberg et al., 2008). Additionally, loss of PFT1 might counteract the down-regulation of FLC at later developmental stages, which would suppress flowering under low red-far-red ratios (Wollenberg et al., 2008; Adams et al., 2009). In addition to its inhibitory effect on FLC expression, PFT1eid3 might have a direct stimulatory effect on CO expression but also an inhibitory function in the phyB-dependent blocking of SAR, similar to the model proposed for phyB-induced photomorphogenic seedling development (Fig. 8B). According to the model and in agreement with our results, inhibition of phyB function might predominate as long as sufficient levels of phyA remain in the system and will never result in a complete block of flowering induced by loss of the light-stable phytochrome.
In conclusion, our data demonstrate that the hypersensitive light response of eid3 is caused by a gain-of-function mutation in PFT1, an important component of the plant Mediator transcriptional coactivator complex. Epistatic analyses show that PFTeid3 functions downstream of phyA and phyB photoreceptors and cooperates with positively acting factors involved in phytochrome signaling in order to regulate light responses during seedling deetiolation and upon the floral transition. Our results show that the Mediator component cooperates with COP1 in the regulation of light responses and that the hypersensitive seedling phenotype strictly depends on the presence of HY5, an important positive regulator of light-dependent gene expression. RT-PCR and microarray analyses reveal that PFT1eid3 alters the expression of light-regulated genes in darkness, indicating that the missense mutation creates a constitutively active transcription factor by mimicking an early step in light signaling. These data provide evidence that the early-flowering phenotype of eid3 is caused by the down-regulation of FLC, a repressor of the floral transition. PFT1 seems to function in a phyA-dependent signaling cascade that modulates phyB function. Thus, regulatory modification of PFT1 is now a known point in the complex network controlling light-regulated gene expression.
MATERIALS AND METHODS
Plant Material, Mutagenesis, and Screening for eid Mutants
Plant propagation, mutagenesis of phyB-5, and screening for the eid3 mutant were done as described (Büche et al., 2000). Mutants were screened under alternating 20-min red/far-red light treatments (5.4 µmol m−2 s−1/5 µmol m−2 s−1) for 3 d after germination induction, which induced strong photomorphogenic development in eid but not in wild-type seedlings. For genetic crossing and physiological analyses, the following ecotypes and photomorphogenic mutants of Arabidopsis (Arabidopsis thaliana) were used: Ler wild type, Col wild type, Nossen wild type, eid1-1 (Büche et al., 2000; Dieterle et al., 2001), cop1eid6 (Dieterle et al., 2003), far1-3 (Hudson et al., 1999), fhy3-1 (Whitelam et al., 1993), hy5-1 (Oyama et al., 1997), pft1-2 (this study); phyA-201 (Nagatani et al., 1993), phyA-401 (Dieterle et al., 2005), phyB-5 and phyB-9 (Reed et al., 1993). To obtain double mutants, F2 generations of crosses were tested with cleaved-amplified polymorphic sequence and dCAPS markers (Supplemental Table S9).
Mapping and Isolation of the eid3 Mutant and the PFT1 T-DNA Line
The isolated phyB-5 eid3 line was crossed with phyB-9 in a Col background for mapping analyses using PCR-based simple sequence length polymorphisms and cleaved-amplified polymorphic sequence markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). Using oligonucleotides (Supplemental Table S9), the PFT1 gene was amplified as four overlapping fragments, which were subsequently sequenced. For detection of the eid3 mutation, a dCAPS marker (Neff et al., 1998) was developed using the eid3_BsaBI_F and eid3_BsaBI_R oligonucleotides (Supplemental Table S9). The PCR product was analyzed for the presence (eid3) or absence (wild type) of a BsaBI restriction site.
Cloning of Genomic Constructs and Plant Transformation
For the construction of ProPFT1:PFT1eid3:YFP chimeric genes, genomic PFT1 DNA was subcloned from a bacterial artificial chromosome F2J7 clone in four consecutive steps using the primers listed in Supplemental Table S9. First, PFT1-BamHI-F and PFT1-dT-EcoRI-R oligonucleotides were used to create a BamHI restriction site before the start codon and to replace the stop codon by introducing an EcoRI restriction site at the 3′ end of the last exon. The eid3 mutation was introduced into a genomic SalI-PmeI subclone by site-directed mutagenesis using muteid3-F and muteid3-R primers, and the obtained mutated fragment was integrated into the genomic clone. Second, the 1.5-kb PFT1 promoter fragment was amplified using PFT1Pro-SalI-F and PFT1Pro-BglII-R oligonucleotides to introduce SalI and BglII restriction sites. Third, all fragments were combined to obtain 6.6-kb genomic constructs in the pENTR3C vector. Finally, constructs were recombined into a modified pB7YWG2.0 plant transformation vector (Karimi et al., 2002), from which the P35S promoter has been deleted. Arabidopsis transformation was done as described (Clough and Bent, 1998). BASTA-resistant plants were grown to maturation, selfed, and F2 seeds were tested for a 3:1 segregation of the resistance gene and the presence of YFP fluorescence. F3 seeds of positive F2 lines were tested for homozygous genetic segregation of both marker genes.
Seedling Growth and Light Sources
Seeds were sown on four layers of Schleicher & Schüll 595 filter paper circles as described (Büche et al., 2000). The standard sowing procedure was followed by a 2-d cold treatment at 6°C in darkness, a 2-h red light induction of germination at 21°C, and 22 h of darkness before the onset of different light treatments for 3 d. For microarray analyses, seedlings were grown on one-half-strength Murashige and Skoog agar plates covered with one layer of filter paper. Standard red light (39 µmol m−2 s−1; λmax = 650 nm) and far-red light (20 µmol m−2 s−1; λmax = 730 nm) fields were used for microscopic studies (Kretsch, 2010). For all other light treatments, modified Leitz Prado 500-W universal projectors (Leitz) with Xenophot long-life lamps (Osram) were used. Red light was obtained by passing the light beam through KG65 filters (λmax = 650 nm; Balzers). Far-red light treatments were performed with 715-nm DAL interference filters (Schott). For hourly light pulse treatments, light was passed through narrow-banded DIL and DEPIL interference filters (Schott).
Determination of Hypocotyl Elongation, Anthocyanin Content, and Flowering Time
Measurements were done as described (Kretsch, 2010). Data represent means ± se of at least 40 seedlings analyzed in at least two independent experiments for determination of hypocotyl elongation and of five to 10 independent replicates for measurement of anthocyanin content. Flowering time is presented as the sum of rosette leaves and cauline leaves at the main stem. A minimum of 30 plants were analyzed in at least two independent experiments.
RNA Isolation and Determination of Transcript Levels
Sample preparation and RNA isolation were performed as described by Peschke and Kretsch (2011) using an RNeasy Plant Mini Kit including a DNase digestion step (Qiagen). SuperScript III reverse transcriptase (Invitrogen) was used with a dT20 oligomer for complementary DNA (cDNA) synthesis according to the manufacturer’s instructions. For quantitative analyses with SYBR Green (Invitrogen), cycle numbers were adapted to the amount of transcript present in the samples, and agarose gels were stained with dye. Quantitative RT-PCR was carried out as described (Peschke and Kretsch, 2011). TaqMan probes and primer pairs for each marker gene are listed in Supplemental Table S9. ACT served as a constitutive control.
Microarray Analyses
Four-day-old etiolated seedlings were either kept in darkness or irradiated for 2 min with red light (30 µmol m−2 s−1) to be harvested after a further 43 min in darkness. RNA was isolated as described above. Total RNA was quantified using a NanoDrop-1000 spectrometer (Peqlab), and quality was estimated on a 2100 Bioanalyzer (Agilent Technologies). RNA integrity number index was calculated for each sample using Agilent 2100 Expert software, and only RNA with an integrity index greater than 8.0 was further processed. Five hundred nanograms of total RNA was reverse transcribed into cDNA and labeled with Cy3 using the one-color Quick Amp Labeling Kit (Agilent) according to the supplier’s instructions. Hybridization on Arabidopsis V4 Gene Expression Microarrays 4 × 44K (Agilent) was performed with 1.5 µg of labeled cDNA per array at 65°C overnight. Arrays were scanned on Agilent Technologies Scanner G2505C at a resolution of 5 µm. Agilent Feature Extraction Software 10.5.1.1 was used to process and analyze array images. The raw data were analyzed using GeneSpring GX 10.0 (normalization shift to 75.0 percentile, baseline transformation, median of all samples). The normalized data were filtered to exclude probes flagged absent in all samples. The remaining probes were tested for statistical significance of expression using an unpaired t test with an asymptotic P value computation and a P value cutoff of 0.05. Multiple testing correction was performed according to Benjamini and Hochberg (1995) with a P value cutoff of 0.05. The fold change cutoff was 2 or greater.
Sequence data for the genes described in this article can be found in the GenBank/EMBL libraries under the following accession numbers: At1g25540 (PFT1), At1g65480 (FT), At2g02950 (PKS1), At2g37620 (ACT), At4g28750 (PSAE), At5g10140 (FLC), At5g11260 (HY5), At5g13930 (CHS), At5g15840 (CO).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Rosette leaf morphology of single, double, and triple mutants of eid3, phyA-201, and phyB-5.
Supplemental Figure S2. Degradation and subcellular localization of phyA in the eid3 mutant background.
Supplemental Figure S3. Rosette leaf morphology of single, double, and triple mutants of eid3, eid1-1, phyA-401, and cop1eid6.
Supplemental Figure S4. Mapping of the eid3 mutation.
Supplemental Figure S5. Expression of light-induced marker genes in etiolated eid3 seedlings.
Supplemental Figure S6. Venn diagrams depicting data from comparisons of transcript accumulation patterns of genes that were up- or down-regulated in etiolated eid3 seedlings compared with published data sets of light-regulated genes.
Supplemental Table S1. List of genes with significantly enhanced transcript levels in dark-grown eid3 seedlings compared with the Ler wild type.
Supplemental Table S2. Classification of differently regulated genes in dark-grown eid3 seedlings with known functions in transcriptional regulation and signaling.
Supplemental Table S3. List of genes with significantly reduced transcript levels in dark-grown eid3 seedlings compared with the Ler wild type.
Supplemental Table S4. List of genes with significantly enhanced transcript levels upon red light pulse treatment of dark-grown Ler wild-type seedlings.
Supplemental Table S5. List of genes with significantly reduced transcript levels upon red light pulse treatment of dark-grown Ler wild-type seedlings.
Supplemental Table S6. Comparisons with published data sets.
Supplemental Table S7. List of light-regulated genes with significantly enhanced transcript levels in dark-grown eid3 seedlings.
Supplemental Table S8. List of light-regulated genes with significantly reduced transcript levels in dark-grown eid3 seedlings.
Supplemental Table S9. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the Salk Institute and the Nottingham Arabidopsis Stock Centre for providing T-DNA insertion mutants of Arabidopsis, Genoscope for the BX816858 cDNA clone, Martina Krenz for helpful technical assistance, Thorsten Kurz for microarray analyses, and Anita K. Snyder for helpful comments on the manuscript.
Glossary
- SAR
shade-avoidance responses
- HIR
high-irradiance responses
- VLFR
very-low-fluence responses
- ULC
ubiquitin ligase complex
- Ler
Landsberg erecta
- dCAPS
derived cleaved-amplified polymorphic sequence
- RT
reverse transcription
- Col
Columbia ecotype
- DT
daytime
- cDNA
complementary DNA
- T-DNA
transferred DNA
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. KR2020/2-3), the Ministry of Science, Research, and the Arts, Baden-Württemberg, and the European Social Fund within the Schieben-Lange-Program (to C.K.), and the Freiburg Institute for Advanced Studies, School of Life Sciences (to T.K. and E.Z.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams S, Allen T, Whitelam GC. (2009) Interaction between the light quality and flowering time pathways in Arabidopsis. Plant J 60: 257–267 [DOI] [PubMed] [Google Scholar]
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G. (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH. (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Björklund S, Gustafsson CM. (2005) The yeast Mediator complex and its regulation. Trends Biochem Sci 30: 240–244 [DOI] [PubMed] [Google Scholar]
- Büche C, Poppe C, Schäfer E, Kretsch T. (2000) eid1: a new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell 12: 547–558 [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu Det al. (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. (2005) The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci 30: 250–255 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Rood SB, Somers DE, Quail PH, Whitelam GC. (1992) Photophysiology of the elongated internode (ein) mutant of Brassica rapa: ein mutant lacks a detectable phytochrome B-like polypeptide. Plant Physiol 100: 1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Bauer D, Büche C, Krenz M, Schäfer E, Kretsch T. (2005) A new type of mutation in phytochrome A causes enhanced light sensitivity and alters the degradation and subcellular partitioning of the photoreceptor. Plant J 41: 146–161 [DOI] [PubMed] [Google Scholar]
- Dieterle M, Buche C, Schafer E, Kretsch T. (2003) Characterization of a novel non-constitutive photomorphogenic cop1 allele. Plant Physiol 133: 1557–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schäfer E, Funk M, Kretsch T. (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C. (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. (2003) HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J 34: 827–836 [DOI] [PubMed] [Google Scholar]
- Elfving N, Davoine C, Benlloch R, Blomberg J, Brännström K, Müller D, Nilsson A, Ulfstedt M, Ronne H, Wingsle Get al. (2011) The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc Natl Acad Sci USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Larner VS, Whitelam GC. (2005) The signal transducing photoreceptors of plants. Int J Dev Biol 49: 653–664 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Poppe C, Sweere U, Martin A, Schäfer E. (2001) Negative interference of endogenous phytochrome B with phytochrome A function in Arabidopsis. Plant Physiol 125: 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Poppe C, Unger S, Schäfer E. (1999) Control of hypocotyl elongation in Arabidopsis thaliana by photoreceptor interaction. Planta 208: 257–263 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13: 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Lisch DR, Quail PH. (2003) The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J 34: 453–471 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11: 550–558 [DOI] [PubMed] [Google Scholar]
- Iñigo S, Alvarez MJ, Strasser B, Califano A, Cerdán PD. (2012) PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J 69: 601–612 [DOI] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang SW, Yang JY, Chua NH. (2007) Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev 21: 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. (2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW. (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17: 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Cahill DM, Manners JM, Schenk PM, Kazan K. (2011) Diverse roles of the Mediator complex in plants. Semin Cell Dev Biol 22: 741–748 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kretsch T. (2010) Phenotypic characterization of photomorphogenic responses during plant development. Methods Mol Biol 655: 189–202 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C. (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco K, Zhou Y, Bury E, Dieterle M, Funk M, Genschik P, Krenz M, Stolpe T, Kretsch T. (2006) Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J 45: 423–438 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J. (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol 102: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW. (2000) The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol 124: 1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou B, Yin KQ, Liu SN, Yang Y, Gu T, Wing Hui JM, Zhang L, Miao J, Kondou Y, Matsui Met al. (2011) A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol Plant 4: 546–555 [DOI] [PubMed] [Google Scholar]
- Ouyang X, Li J, Li G, Li B, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, et al. (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke F, Kretsch T. (2011) Genome-wide analysis of light-dependent transcript accumulation patterns during early stages of Arabidopsis seedling deetiolation. Plant Physiol 155: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GIet al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW. (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Zhu D, Li J, Rubio V, Zhou Z, Shen Y, Hoecker U, Wang H, Deng XW. (2008) Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol Cell 31: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ. (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 97: 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH. (1991) The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3: 1263–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW. (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. (2005) Remembering winter: toward a molecular understanding of vernalization. Annu Rev Plant Biol 56: 491–508 [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F. (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW. (2002) Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma L, Habashi J, Li J, Zhao H, Deng XW. (2002) Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J 32: 723–733 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg AC, Strasser B, Cerdán PD, Amasino RM. (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol 148: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Jang IC, Henriques R, Chua NH. (2009) FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 21: 1341–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Casal JJ, Luppi JP. (1997) The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J 12: 659–667 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW. (2005) COP1: from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW. (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhou YC, Dieterle M, Büche C, Kretsch T. (2002) The negatively acting factors EID1 and SPA1 have distinct functions in phytochrome A-specific light signaling. Plant Physiol 128: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C. (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.