Abstract
cis-Zeatin (cZ) is generally regarded as a cytokinin with little or no activity, compared with the highly active trans-zeatin (tZ). Although recent studies suggested possible roles for cZ, its physiological significance remains unclear. In our studies with rice (Oryza sativa), cZ inhibited seminal root elongation and up-regulated cytokinin-inducible genes, and its activities were comparable to those of tZ. Tracer experiments showed that exogenously supplied cZ-riboside was mainly converted into cZ derivatives but scarcely into tZ derivatives, indicating that isomerizations of cZ derivatives into tZ derivatives are a minor pathway in rice cytokinin metabolism. We identified three putative cZ-O-glucosyltransferases (cZOGT1, cZOGT2, and cZOGT3) in rice. The cZOGTs preferentially catalyzed O-glucosylation of cZ and cZ-riboside rather than tZ and tZ-riboside in vitro. Transgenic rice lines ectopically overexpressing the cZOGT1 and cZOGT2 genes exhibited short-shoot phenotypes, delay of leaf senescence, and decrease in crown root number, while cZOGT3 overexpressor lines did not show shortened shoots. These results propose that cZ activity has a physiological impact on the growth and development of rice.
Cytokinins, a class of phytohormones, are involved in the regulation of various biological processes, including organogenesis (Kurakawa et al., 2007; Pernisová et al., 2009; Marhavý et al., 2011), senescence (Gan and Amasino, 1995; Kim et al., 2006), and nutrient responses (Takei et al., 2004a; Sakakibara et al., 2006; Hirose et al., 2008; Ruffel et al., 2011). Natural cytokinins detected in plants are N6-substituted adenine derivatives and are structurally classified into isoprenoid cytokinins or aromatic cytokinins carrying an isoprene-derived or aromatic N6 side chain, respectively (Mok and Mok, 2001; Sakakibara et al., 2006). Both groups of cytokinins include members with minor modifications of the N6 side chain, such as hydroxylations. Isoprenoid cytokinins include N6-(Δ2-isopentenyl)adenine (iP), trans-zeatin (tZ), dihydrozeatin, and cis-Zeatin (cZ), whereas aromatic cytokinins include 6-benzyladenine, ortho-topolin (oT), and meta-topolin. Isoprenoid cytokinins have been detected in all plant species examined so far, while aromatic cytokinins have been found only in a subset of these (Strnad, 1997; Sakakibara, 2006).
The initial products of isoprenoid cytokinin biosynthesis in plants are iP-ribotides formed by adenosine phosphate-isopentenyltransferase (Kakimoto, 2001; Takei et al., 2001). After transhydroxylation of the prenyl side chain (Takei et al., 2004b), the tZ- and iP-ribotides are converted into the active free-base forms tZ and iP, respectively (Kurakawa et al., 2007; Kuroha et al., 2009; Tokunaga et al., 2012). The cZ biosynthesis pathway has not been fully elucidated, but a major step in Arabidopsis (Arabidopsis thaliana) is the release of precursors in the course of tRNA degradation (Miyawaki et al., 2006). For inactivation, the free-base cytokinins may be degraded by cytokinin oxidase (Houba-Hérin et al., 1999; Werner et al., 2001; Bartrina et al., 2011) or conjugated by glucosyltransferases (Dixon et al., 1989; Martin et al., 1999, 2001; Mok et al., 2000; Mok and Mok, 2001; Haberer and Kieber, 2002; Veach et al., 2003; Hou et al., 2004; Wang et al., 2011) and converted to cytokinin-ribotides in the purine salvage pathway (Moffatt et al., 1991; Allen et al., 2002). A pathway to convert cZ or cZ-riboside (cZR) into tZ or tZ-riboside (tZR) was also proposed following partial purification of a putative cis-trans-isomerase from endosperm of Phaseolus vulgaris (Bassil et al., 1993).
The relationship between side chain variation and activity has been investigated by classical bioassays and the characterization of cytokinin signaling components. For instance, in callus growth assays using tobacco (Nicotiana tabacum) and Phaseolus lunatus, tZ showed the highest activity followed by iP and cZ (Leonard et al., 1971; Schmitz et al., 1972; Mok et al., 1978). In Arabidopsis, the promoter activity of the cytokinin-inducible ARABIDOPSIS RESPONSE REGULATOR5 gene was efficiently up-regulated by lower concentrations of tZ than of cZ (Gajdošová et al., 2011). The first identified cytokinin receptor, CRE1/AHK4, complemented a yeast sln1 mutant lacking an endogenous osmosensing histidine kinase (HK) in the presence of iP and tZ but not cZ (Inoue et al., 2001). Thus, while tZ is considered a highly active cytokinin and has been the main focus of cytokinin research, cZ is regarded as a derivative of low activity.
However, there are several lines of evidence suggesting that cZ could be an active cytokinin in maize (Zea mays), which contains abundant cZ derivatives (Veach et al., 2003). For instance, two maize cZ-O-glucosyltransferases (cZOGTs), cisZOG1 and cisZOG2, preferentially catalyzed O-glucosylation in the N6 side chain of cZ rather than tZ using UDP-Glc as a Glc donor in vitro (Martin et al., 2001; Veach et al., 2003), although P. lunatus ZOG1 specifically catalyzed tZ-O-glucosylation (Dixon et al., 1989; Martin et al., 1999). Cytokinin O-glucosides have been assumed to represent reversibly inactivated storage forms (Pineda Rodó et al., 2008). Thus, the discovery of cisZOGs raises the possibility that cZ derivatives may play a more important role in cytokinin homeostasis than previously recognized (Martin et al., 2001). Furthermore, in a bacterial system, the maize cytokinin receptor ZmHK1 responded to cZ and tZ with comparable affinities (Yonekura-Sakakibara et al., 2004; Lomin et al., 2011). cZ and its derivatives are abundant not only in maize but also in other species, including chickpea (Cicer arietinum; Emery et al., 1998) and rice (Oryza sativa; Takagi et al., 1985, 1989; Kojima et al., 2009). In addition, a recent comprehensive screen showed that higher levels of cZ than tZ derivatives can be found in species across the complete evolutionary tree of land plants (Gajdošová et al., 2011). However, the physiological significance of cZ in plants is still poorly understood.
In this study, we demonstrate that cZ itself can act as an active cytokinin in rice in the physiological concentration range. We identified rice genes encoding cisZOGs (cZOGTs) and characterized transgenic rice plants overexpressing the cZOGT genes to gain further insights into the role of cZ. The transgenic plants exhibited developmental phenotypes with modified cytokinin profiles. In summary, we suggest that cZ is an active cytokinin playing a role in normal growth and development in rice.
RESULTS
Cytokinin Activity of Exogenously Supplied cZ in Rice
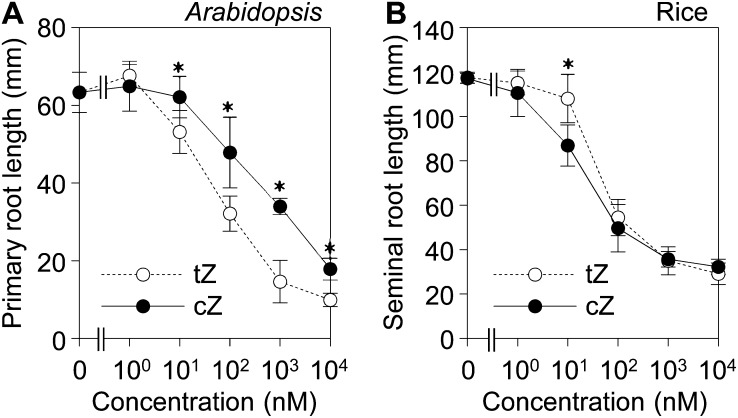
Exogenously supplied cytokinins inhibit root elongation via cytokinin receptors in a dose-dependent manner (Inoue et al., 2001; Higuchi et al., 2004). To test whether cZ is an active cytokinin in rice, bioactivities of cZ and tZ were compared in root growth assays using Arabidopsis and rice seedlings. In Arabidopsis, the inhibitory effect of cZ was significantly weaker than that of tZ above 10 nm (Fig. 1A). On the other hand, the effects of the two inhibitors were comparable in rice (Fig. 1B). This result suggested that exogenously supplied cZ has cytokinin activity, at least in long-term treatments.
Figure 1.
Comparison of the bioactivities of cZ and tZ in a root growth assay. Primary root length of Arabidopsis (A) and seminal root length of rice (B) were measured at 11 and 7 d, respectively, after sowing on agar medium containing various concentrations of tZ (white circles) and cZ (black circles). Asterisks indicate significant differences (Student’s t test; P < 0.01) between like concentrations of tZ and cZ. Mean values ± sd of at least 11 (Arabidopsis) and six (rice) plants are shown.
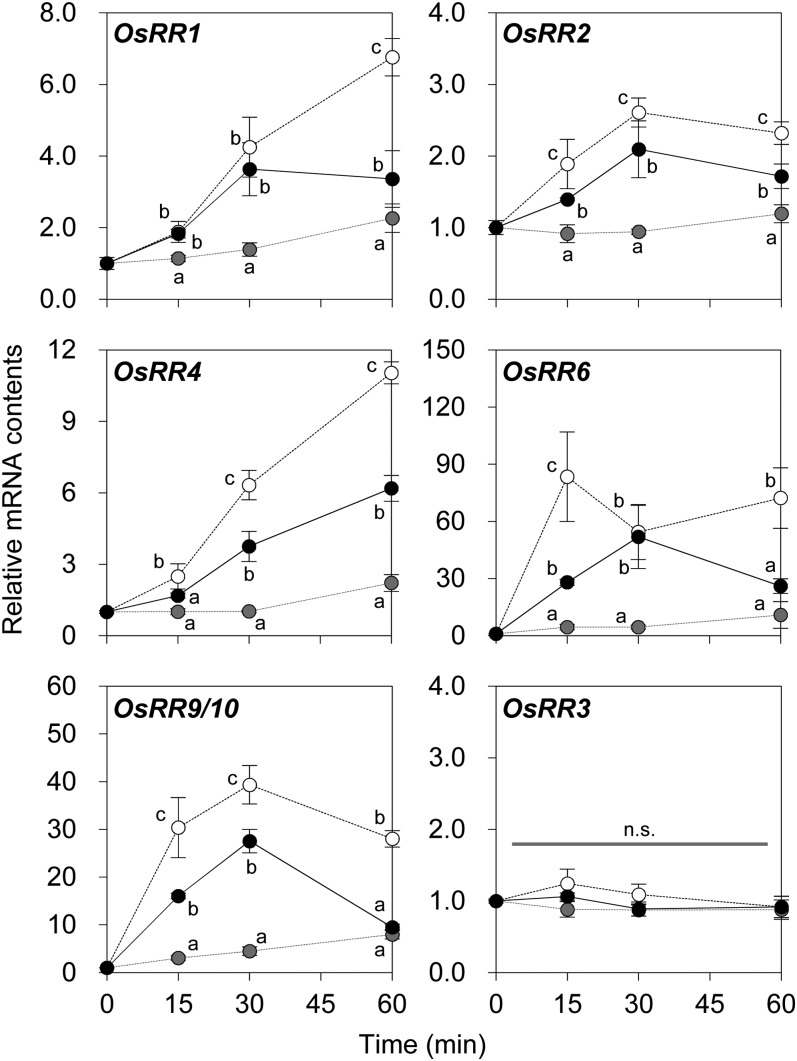
To further examine the cytokinin activity of cZ in rice, mRNA accumulation of type A response regulator genes (OsRRs; Ito and Kurata, 2006; Jain et al., 2006; Pareek et al., 2006) was analyzed in seedling roots. Quantitative reverse transcription (qRT)-PCR analysis showed that the levels of OsRR1, OsRR2, OsRR6, and OsRR9/10 mRNA were significantly increased within 15 min after exposure to 100 nm cZ or 100 nm tZ (Fig. 2). This immediate response of OsRRs indicated that cZ also exhibits cytokinin activity in the short term.
Figure 2.
Changes in mRNA levels of response regulator genes in response to zeatin isomers in roots of rice seedlings. Plants were grown in hydroponics for 2 weeks after sowing and then transferred to treatment solutions containing 100 nm tZ (white circles), cZ (black circles), or no cytokinin (gray circles). The roots were harvested before (0 min) and 15, 30, and 60 min after the transfer and submitted to qRT-PCR analysis for OsRR1 (top left), OsRR2 (top right), OsRR4 (middle left), OsRR6 (middle right), OsRR9 and/or OsRR10 (OsRR9/10; bottom left), and OsRR3 (bottom right). Because of their high degree of identity (Jain et al., 2006), mRNAs of OsRR9 and OsRR10 could not be separated. OsRR3, a gene not responsive to cytokinin, was used as an internal control (Jain et al., 2006). The mRNA contents were normalized to the value at 0 min. Mean values ± sd of at least three plants are shown. Different lowercase letters indicate statistically significant differences at a given time (Tukey-Kramer test following one-way ANOVA; P < 0.01). n.s., Not significant.
Isomerization of cZ and tZ Derivatives in Rice Seedlings
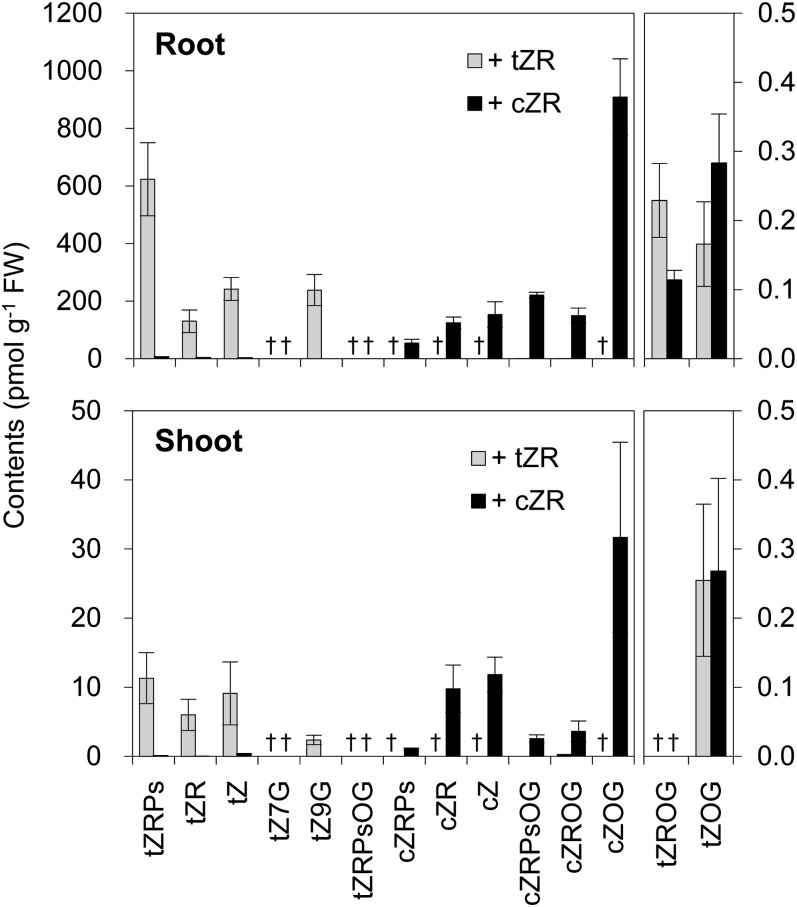
Since isomerization from cZ or cZR to tZ or tZR was proposed to occur (Bassil et al., 1993), the possible conversion of cZ to tZ should be considered when evaluating cytokinin activity. Therefore, we carried out tracer experiments using [1013C,515N]cZR and [1013C,515N]tZR [denoted cZR(+15) and tZR(+15), respectively, hereafter]. In roots fed with cZR(+15), cZ-O-glucoside (cZOG) was the major derivative containing the isotope labels, followed by cZ-ribotides-O-glucoside (cZRPsOG), cZ, cZR-O-glucoside (cZROG), cZR, and cZ-ribotides (cZRPs) at 1 h after feeding (Fig. 3). Although isotope labels were detected also in tZ-ribotides (tZRPs), tZR, tZ, tZR-O-glucoside (tZROG), and tZ-O-glucoside (tZOG), cZ derivatives represented 99.1% of all cytokinins derived from cZR(+15). On the other hand, in roots fed with tZR(+15), tZRPs were the major derivatives containing isotope labels, followed by tZ, tZ-9-N-glucoside, tZR, and tZRPs-O-glucoside (Fig. 3). Label was also detected in cZRPsOG and cZROG, but more than 99.9% of all cytokinins derived from tZR(+15) were tZ derivatives. Similar results were obtained from shoots fed with cZR(+15) and tZR(+15): cZ derivatives and tZ derivatives amounted to 98.8% and 99.1% of all cytokinins derived from cZR(+15) and tZR(+15), respectively. When we monitored the isotope labels until 24 h after feeding, the proportions remained essentially unchanged (Supplemental Tables S1 and S2). These results suggested that isomerization, if any occurs, is negligible compared with other reactions of cytokinin metabolism such as glucosylation and phosphorylation. Furthermore, isomerization can be neglected when rice seedlings are treated with zeatin derivatives at concentrations in the micromolar range for short periods. In combination with the results of the expression analysis of OsRR genes (Fig. 2), these findings led us to conclude that cZ functions as an active cytokinin in rice roots.
Figure 3.
Isomerization of tZ and cZ derivatives in rice seedlings. One micromolar [1013C,515N]tZR (+tZR; gray bars) or [1013C,515N]cZR (+cZR; black bars) was supplied to 2-week-old rice seedlings. After 1 h, roots (top panel) and shoots (bottom panel) were separately harvested. tZ and cZ derivatives containing isotope labels in the purine ring and the ribosyl group (total 15 D heavier than their authentic counterparts) and derivatives carrying label only in the purine ring (10 D heavier than their authentic counterparts) were quantified by UPLC-MS/MS. Means ± sd of five plants are shown. †, Not determined. FW, Fresh weight.
In Silico Exploration of Candidate Rice Genes Encoding Putative cZOGTs
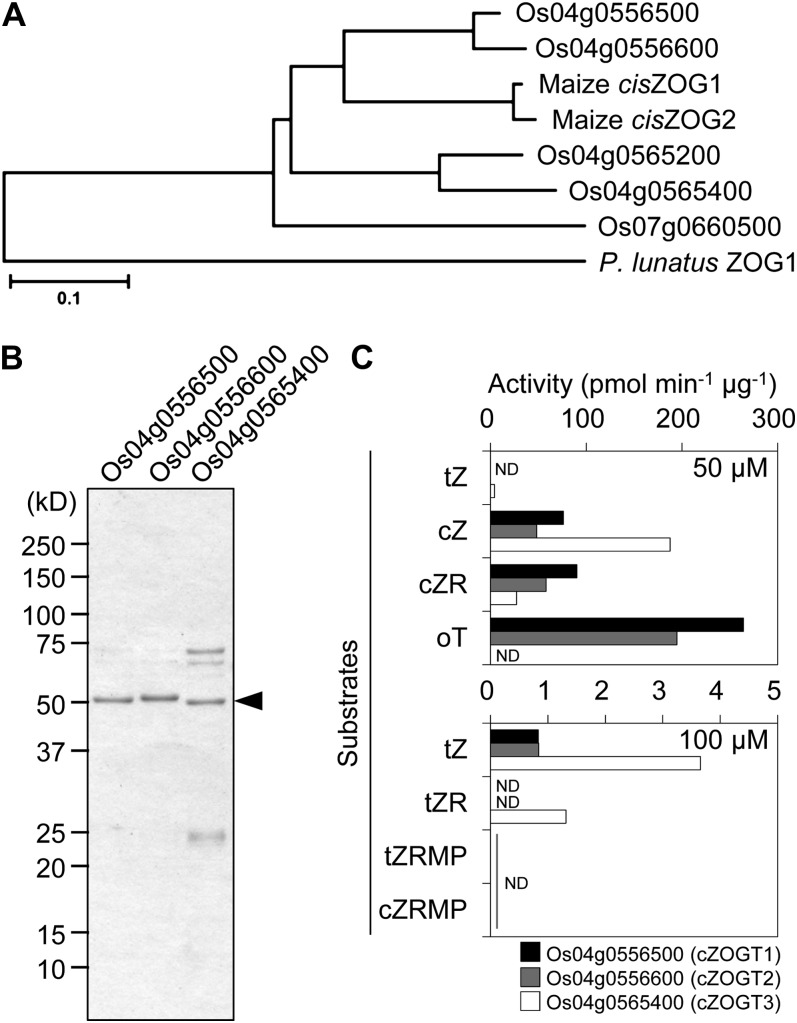
BLAST searches in rice genome databases (the Rice Annotation Project Database [RAP-DB; http://rapdb.dna.affrc.go.jp/] and the Rice Genome Annotation Project [http://rice.plantbiology.msu.edu/]) using amino acid sequences of maize cisZOGs as queries revealed six nuclear genes (RAP-DB identifiers Os04g0556400, Os04g0556500, Os04g0556600, Os04g0565200, Os04g0565400, and Os07g0660500) potentially encoding cZOGTs. All of the putative cZOGT genes seemed to lack introns. The complete predicted amino acid sequences for the cZOGT genes were highly similar to maize cisZOGs (Supplemental Fig. S1). A C-terminal conserved glycosyltransferase domain was predicted by National Center for Biotechnology Information Conserved Domain Search analysis (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; Supplemental Fig. S1). However, the protein encoded by Os04g0556400 seemed to lack glucosyltransferase activity due to a stop codon in the glycosyltransferase domain (Supplemental Fig. S1). All of the putative cZOGTs have been classified into the UDP-glucuronosyltransferase (UGT) family (GT1) in the Rice GT Database (http://ricephylogenomics.ucdavis.edu/cellwalls/gt/; Cao et al., 2008). We analyzed sequence similarities using the entire amino acid sequences of the rice putative cZOGTs, maize cisZOGs, and P. lunatus ZOG1 (Martin et al., 1999). The maize cisZOGs were nested within the rice proteins, but no cZOGT was closely related to P. lunatus ZOG1 (Fig. 4A). Os04g0556500 and Os04g0556600 were particularly closely related to the maize cisZOGs. When P. lunatus ZOG1 was used as the query in BLAST searches of rice databases, no counterparts were found.
Figure 4.
Identification of rice genes encoding cZOGTs. A, Phylogenetic relations of P. lunatus ZOG1, maize cisZOG1 and cisZOG2, and putative rice cZ-O-glucosyltransferases. The deduced amino acid sequences of the proteins were obtained from GenBank (accession nos. AF101972 for P. lunatus ZOG1, AF318075 for maize cisZOG1, and AY082660 for cisZOG2 [http://www.ncbi.nlm.nih.gov/genbank/]) and the RAP-DB (locus identifiers Os04g0556500, Os04g0556600, Os04g0565200, Os04g0565400, and Os07g0660500 for putative rice cZ-O-glucosyltransferases [http://rapdb.dna.affrc.go.jp/]). The phylogenetic relations were analyzed with the MEGA 4 program (http://www.megasoftware.net/) using the neighbor-joining method. The bar represents 0.1 amino acid substitutions per site. B, SDS-PAGE analysis of the purified recombinant Os04g0556500 (55.1 kD), Os04g0556600 (55.4 kD), and Os04g0565400 (52.2 kD) proteins. The gel was stained with Coomassie Brilliant Blue; the arrowhead points to the recombinant proteins, and molecular mass markers (kD) are indicated at the left. C, O-Glucosylation activities of the recombinant cZOGT1 (black bars), cZOGT2 (gray bars), and cZOGT3 (white bars) for 50 μm tZ, cZ, cZR, and oT (top panel) and for 100 μm tZ, tZR, tZRMP, and cZRMP (bottom panel). ND, Not detected.
In Vitro Activity of Rice cZOGTs
To examine O-glucosylation activity of the putative cZOGTs, poly-His tagged proteins were expressed in Escherichia coli cells, purified by metal-chelate affinity chromatography (Fig. 4B), and used for in vitro assays. Preliminary tests to determine activities were carried out using tZ, tZR, tZR 5′-monophosphate (tZRMP), cZ, cZR, cZR 5′-monophosphate (cZRMP), and oT as Glc acceptors (Fig. 4C). The recombinant proteins originating from Os04g0556500, Os04g0556600, and Os04g0565400, which were later named cZOGT1, cZOGT2, and cZOGT3, respectively, showed significant activities for cZ (Fig. 4C). Interestingly, cZOGT1 and cZOGT2 showed comparable activities for cZR and cZ, while cZOGT3 showed weaker activity for cZR than for cZ (Fig. 4C). cZOGT1 and cZOGT2 also exhibited notable activity for oT, but cZOGT3 did not (Fig. 4C). While cZOGT1 and cZOGT2 were weakly active for tZ but not tZR, cZOGT3 showed activities for both tZ and tZR (Fig. 4C). In other tested combinations of a recombinant protein and a Glc acceptor, no significant O-glucosylation was detected. Substrate specificity for sugar donors, requirements of Mg2+ and ATP, and optimum pH were also examined. The results suggested that cZOGTs catalyze O-glucosylation in a Mg2+-dependent and ATP-independent manner, specifically using UDP-Glc as a Glc donor, at optimum pH values of 6.0 for cZOGT1 and cZOGT3 and of 7.0 for cZOGT2 (Supplemental Fig. S2).
The kinetic parameters of the recombinant cZOGTs were determined for the substrates cZ and cZR (Table I; Supplemental Fig. S3). The Km values of rice cZOGTs for cZ were a little higher than the previously reported Km of maize cisZOGs (cisZOG1, 46 μm; cisZOG2, 96 μm; Veach et al., 2003). The affinity and specificity constant (kcat/Km) for cZR were similar to those for cZ in cZOGT1 and cZOGT2. The kinetic parameters of cZOGT3 for cZR and tZ could not be determined because of insufficient affinities.
Table I. Enzymatic properties of recombinant cZOGTs with various cytokinin substrates.
Kinetic parameters were calculated from Hanes-Woolf plots using values obtained from technical triplicates.
Expression Profiles of cZOGT Genes in Rice
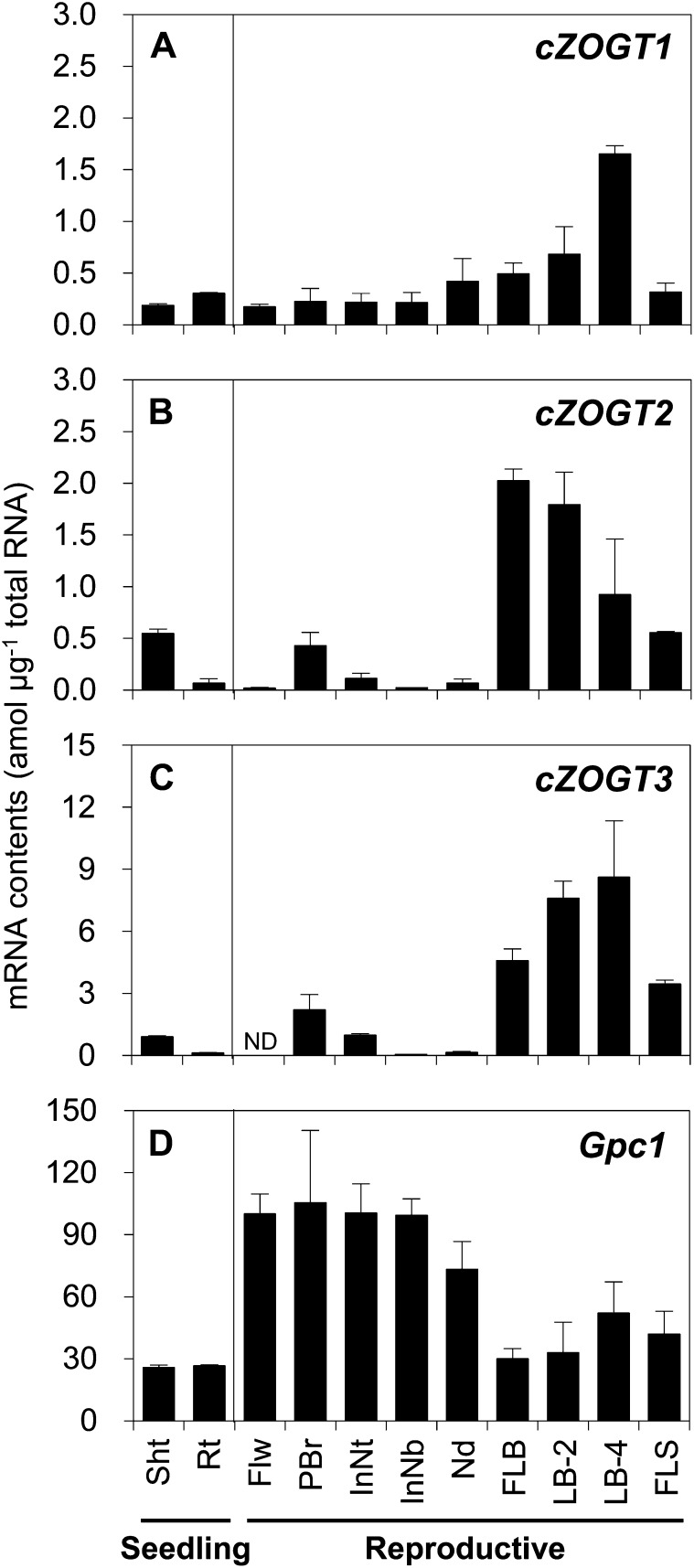
The accumulation of mRNAs of cZOGT genes was analyzed by qRT-PCR in various rice organs: total shoots and roots at the seedling stage, flowers before anthesis, panicle branches, top and basal parts of internode I, node I, blade and sheath of the flag leaf, and blades of leaves 2 and 4 counted down from the flag leaf (LB-2 and LB-4, respectively) at the reproductive stage. The cZOGT1 mRNA was detected in all tested organs, with the maximum level found in LB-4 (Fig. 5A). cZOGT2 and cZOGT3 mRNAs were detected in almost all organs examined, with the highest levels in blades of flag leaves as well as LB-2 and LB-4 (Fig. 5, B and C).
Figure 5.
Accumulation of cZOGT1 (A), cZOGT2 (B), cZOGT3 (C), and Gpc1 (D) mRNAs in various organs of rice. Total RNAs were extracted from shoots (Sht) and roots (Rt) of 2-week-old seedlings and from flowers (Flw), panicle branches (PBr), top and basal parts of internode I (InNt and InNb, respectively), node I (Nd), leaf blades of flag leaves (FLB), and blades of leaves 2 and 4 below the flag leaf (LB-2 and LB-4, respectively) of older plants. Total RNAs were subjected to qRT-PCR using gene-specific primers. The mRNA levels are indicated as amounts per total RNA without normalization by an internal control gene. The Gpc1 gene encoding glyceraldehyde-3-phosphate dehydrogenase is presented as an unrelated gene used to control for expression in the tissues. Mean values ± sd of three plants are shown. ND, Not detected.
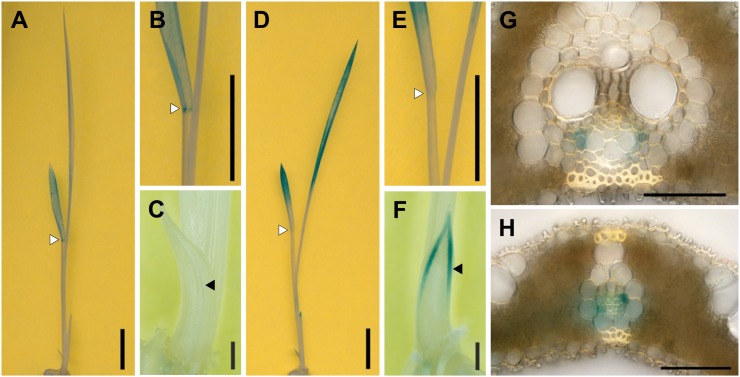
To reveal tissue specificities of cZOGT expression, promoter-reporter analysis was carried out. DNA fragments of 3,052, 3,952, and 3,089 bp upstream of the starting Met codon of cZOGT1, cZOGT2, and cZOGT3 (designated cZOGT1pro, cZOGT2pro, and cZOGT3pro, respectively) were fused with the GUS coding sequence. The cZOGT1pro-GUS, cZOGT2pro-GUS, and cZOGT3pro-GUS constructs were transformed into rice, and the T1 seedlings were stained for GUS activity. Unfortunately, no GUS activity was detected in cZOGT3pro-GUS seedlings. In cZOGT1pro-GUS and cZOGT2pro-GUS seedlings, significant GUS activity was reproducibly detected in leaf blades (Fig. 6, A and D) but not in roots. In cZOGT1pro-GUS seedlings, more or less uniform GUS activity was observed in leaf blades, with a somewhat stronger signal at the laminar joints (Fig. 6, A and B). In cZOGT2pro-GUS seedlings, GUS activity was stronger in the leaf tips and disappeared toward the laminar joints (Fig. 6, D and E). In cZOGT2pro-GUS but not in cZOGT1pro-GUS seedlings, GUS activity was detected also in vascular bundles of coleoptiles (Fig. 6, C and F). To analyze the tissue distribution of the promoter activities, cross-sections of fully expanded leaf blades were prepared from mature plants and stained for GUS activity. Tissues in which cZOGT1pro-GUS was active could not be specified because the signals were too weak. In cZOGT2pro-GUS plants, however, GUS activity appeared localized to phloem parenchyma cells of large and small vascular bundles (Fig. 6, G and H).
Figure 6.
Distribution of GUS activity under the control of the 5′ upstream region of the cZOGT1 (A–C) and cZOGT2 (D–H) genes. A to F, Whole shoots of the transformants were stained for GUS activity. Overviews (A and D) and closeups of the laminar joint of the second leaves (B and E) and coleoptiles (C and F) are shown. Bars = 10 mm for A, B, D, and E and 1 mm for C and F. White and black arrowheads indicate laminar joints and vascular bundles of coleoptiles, respectively. G and H, Cross-sections of mature leaf blades; a large (G) and a small (H) vascular bundle are shown. Bars = 50 μm.
Visible Phenotypes of Transgenic Rice Overexpressing cZOGT Genes
We had demonstrated that cZOGT1, cZOGT2, and cZOGT3 preferentially catalyzed O-glucosylation of cZ and cZR rather than tZ in vitro and that cZOGT1 and cZOGT2 also catalyzed O-glucosylation of oT (Fig. 4C). However, we had not detected oT, oT-riboside, and their O-glucosides in any sample (data not shown). In order to better understand the physiological significance of these findings, we perturbed cZ metabolism by generating transgenic rice in which cZOGT genes were ectopically overexpressed under the control of a rice actin promoter (McElroy at al., 1990; Supplemental Fig. S4). Because the transgenic lines for overexpression of cZOGT3 (cZOGT3-ox) were generated later than those for cZOGT1 and cZOGT2 (ZOGT1-ox and cZOGT2-ox, respectively), the cZOGT1-ox and cZOGT2-ox lines in the T2 or T3 generation and cZOGT3-ox lines in the T1 generation were compared with vector control (VC) lines in the corresponding generations.
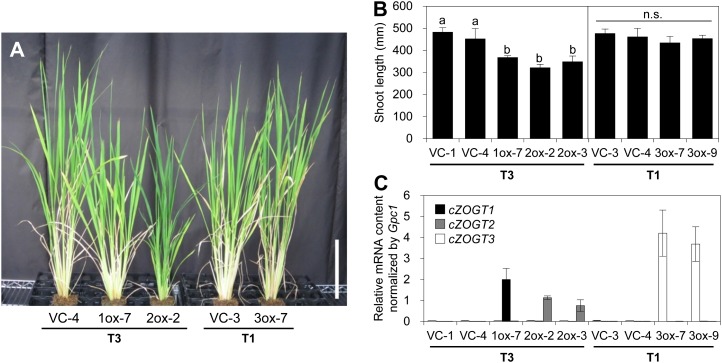
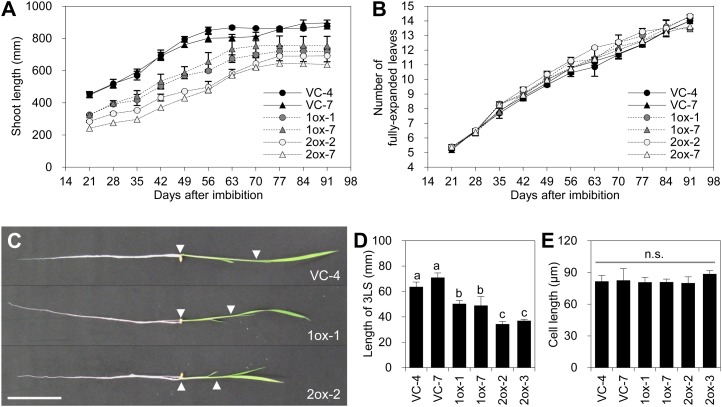
Significantly reduced shoot growth was observed in cZOGT1-ox and cZOGT2-ox but not in cZOGT3-ox, although the transgenes were overexpressed in all lines (Fig. 7). Growth analysis in plants of the T2 generation showed that cZOGT1-ox and cZOGT2-ox plants remained smaller than the VCs throughout their lifetime while the plastochron was not altered (Fig. 8, A and B). This suggested that the phenotype was due to reduced elongation growth rather than a disturbed sequence of developmental events. For instance, the third leaf sheaths were approximately 30% and approximately 50% shorter in cZOGT1-ox and cZOGT2-ox, respectively, than in the VCs (Fig. 8, C and D). Epidermal cell lengths were similar in all plants (Fig. 8E), suggesting that decreased cell production was involved in causing the short-shoot phenotype. The short-shoot phenotype in cZOGT1-ox and cZOGT2-ox lines occurred independently of the offspring generation in T1, T2, and T3 plants (data not shown).
Figure 7.
The short-shoot phenotype of transgenic rice overexpressing cZOGT1 or cZOGT2. Transgenic rice lines that overexpressed cZOGT1 (1ox-7), cZOGT2 (2ox-2 and 2ox-3), and cZOGT3 (3ox-7 and 3ox-9) were grown on soil in parallel with VC plants (VC-1, VC-3, and VC-4). The generations of the transgenic plants analyzed are indicated underneath the names of the lines. A, Transgenic rice at 89 d after sowing. Bar = 200 mm. B, Shoot length of the transgenic rice lines at 22 d after sowing. Mean values ± sd of three plants are shown. Means were tested for significant differences by the Tukey-Kramer test (P < 0.05) following one-way ANOVA; different lowercase letters indicate statistically significant differences (P < 0.05). n.s., Not significant. C, Accumulation of cZOGT1 (black bar), cZOGT2 (gray bars), and cZOGT3 (white bars) mRNAs in the shoots of transgenic rice lines at 22 d after sowing. cZOGT mRNA contents were normalized to the content of Gpc1 mRNA. Values shown are means ± sd of three plants.
Figure 8.
Characterization of the short-shoot phenotype in transgenic rice overexpressing cZOGT1 and cZOGT2. A and B, Shoot growth in transgenic rice lines overexpressing cZOGT1 (1ox-1, gray circles; 1ox-7, gray triangles) and cZOGT2 (2ox-2, white circles; 2ox-7, white triangles) and the corresponding VCs (VC-4, black circles; VC-7, black triangles). Shoot length (A) and number of fully expanded leaves (B) on the main culms of transgenic plants (T2) grown in soil were determined from 21 to 91 d after seed imbibition every 7 d. Mean values ± sd of three plants are shown. C, Transgenic rice seedlings grown in hydroponics for 2 weeks after sowing. Arrowheads highlight the proximal and distal ends of the sheath of the third leaf. Bar = 50 mm. D, Length of the sheath of the third leaf (3LS; means ± sd of three plants). Different lowercase letters indicate statistically significant differences (P < 0.05) of mean values in the Tukey-Kramer test following one-way ANOVA. E, Epidermal cell lengths in the distal part of the sheath of the third leaf. Average lengths for each plant were determined from 185 individual values, and values from three plants were used to calculate the mean for each transgenic line. No significant differences (n.s.) were detected by one-way ANOVA (P < 0.05). [See online article for color version of this figure.]
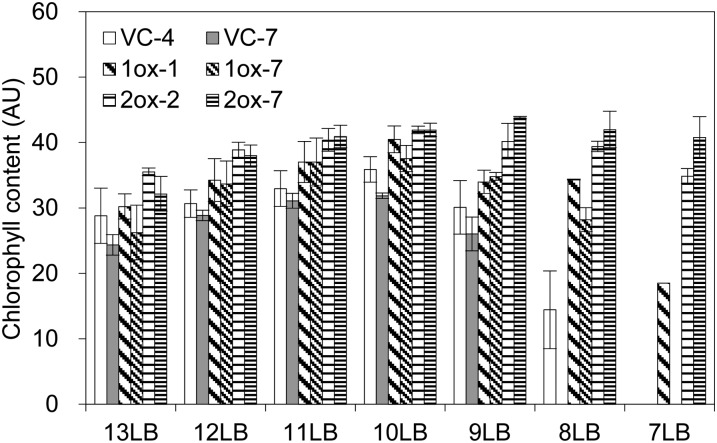
Mature cZOGT2-ox plants looked conspicuously dark green (Fig. 7A). Chlorophyll contents in leaf blades at various nodal positions remained high in aging cZOGT2-ox leaves at times when the corresponding VC began to die (Fig. 9). A similar trend was observed in cZOGT1-ox (Fig. 9). These results suggested that leaf senescence was delayed in cZOGT1-ox and cZOGT2-ox. The delay of leaf senescence in cZOGT2-ox was further confirmed by an analysis of the levels of the large subunit of Rubisco in leaf crude extracts (Supplemental Fig. S5).
Figure 9.
Chlorophyll retention in old leaf blades of cZOGT1- and cZOGT2-overexpressing rice lines. Relative chlorophyll contents in leaf blades at the ninth (9LB), 10th (10LB), 11th (11LB), 12th (12LB), and 13th (13LB) nodal positions of main culms were measured in transgenic lines that overexpressed cZOGT1 (1ox-1 and 1ox-7) or cZOGT2 (2ox-2 and 2ox-3) and in VC lines (VC-4 and VC-7); results are expressed in arbitrary units (AU). The plants were grown in soil for 77 d after seed imbibition. Mean values ± sd of three plants are given; no sd is shown where one or more leaves had started to wither.
Seminal roots of cZOGT1-ox were longer than those of VC, and fewer crown roots developed in cZOGT1-ox and cZOGT2-ox than in VC (Supplemental Fig. S6).
Endogenous Cytokinins in Transgenic Rice Overexpressing cZOGT Genes
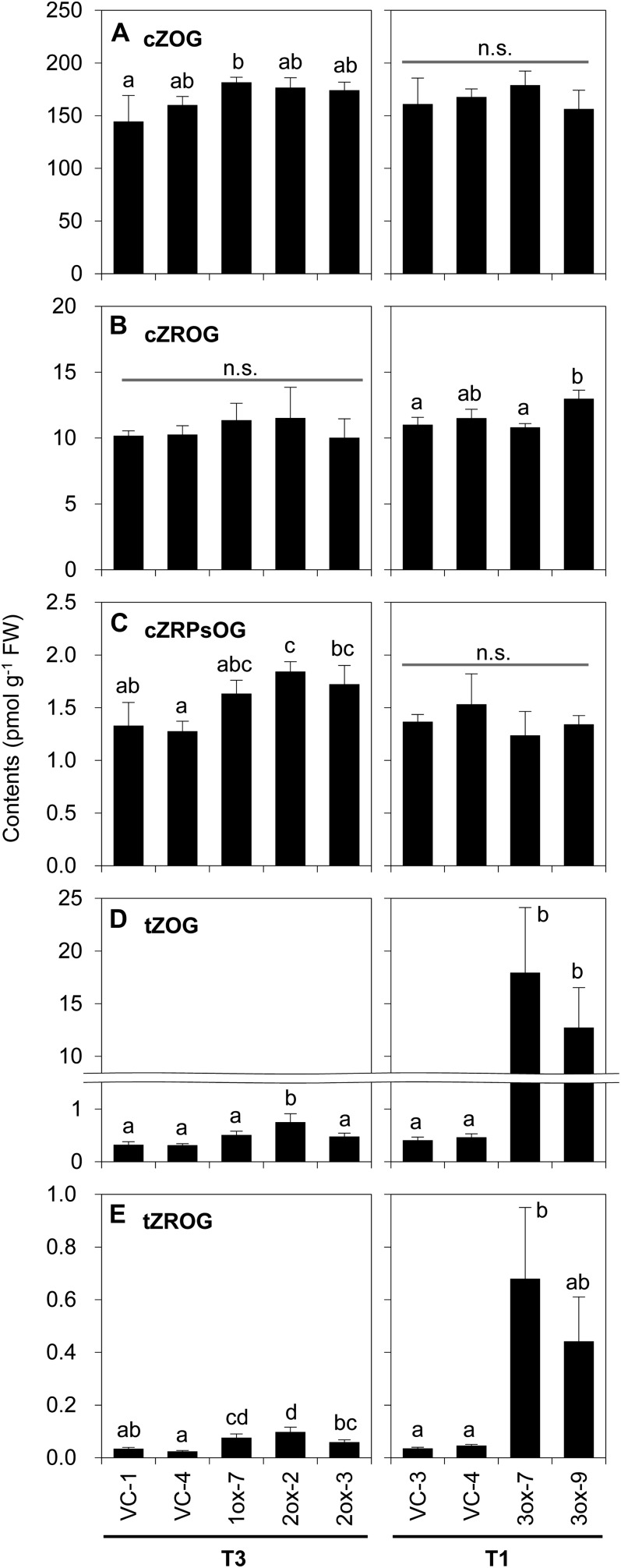
To investigate the effects of ectopic overexpression of cZOGT genes on the rice cytokinin profile, endogenous cytokinins were quantified in shoots of transgenic lines. For this analysis, the same plants were used from which the data presented as Figure 7, B and C, had been produced. Significant differences between cZOGT-ox and VC lines were found regarding the contents of cZRPsOG, cZROG, cZOG, tZROG, and tZOG but not with respect to the other cytokinins analyzed (Supplemental Table S3).
In all cZOGT1-ox and cZOGT2-ox lines examined, the contents of cZOG, cZRPsOG, tZOG, and tZROG were increased compared with those in the VC lines, although the differences were not always statistically significant (Fig. 10, A and C–E; Supplemental Table S3). On the other hand, in cZOGT3-ox lines, cZOG and cZRPsOG contents were unaffected, whereas tZOG and tZROG levels were dramatically increased (Fig. 10, A and C–E; Supplemental Table S3). No clear trends were observed in the cZROG contents of transgenic lines, even though a statistically significant difference was detected between a cZOGT3-ox line (3ox-9) and a VC line (VC-3; Fig. 10B; Supplemental Table S3).
Figure 10.
Contents of cZOG and tZOG and their precursors in cZOGT-overexpressing rice. Cytokinin derivatives were extracted from shoots of transgenic plants overexpressing cZOGT1 (1ox-7), cZOGT2 (2ox-2 and 2ox-3), and cZOGT3 (3ox-7 and 3ox-9) and from VC plants (VC-1, VC-3, and VC-4) grown in soil for 21 d after sowing (the same samples as in Fig. 7, B and C, were used) and were analyzed by UPLC-MS/MS. The contents of O-glucosides of cZ (cZOG; A), cZR (cZROG; B), cZRP (cZRPsOG; C), tZ (tZOG; D), and tZR (tZROG; E) are presented as means ± sd of three plants. The generations of transgenic plants are indicated underneath the names of the lines. Different lowercase letters indicate statistically significant differences in the same generation as detected by the Tukey-Kramer test (P < 0.05) following one-way ANOVA. FW, Fresh weight; n.s., not significant.
DISCUSSION
Based on studies with Arabidopsis, cZ has generally been thought to be a cytokinin derivative with no or low activity (Inoue et al., 2001; Spíchal et al., 2004; Romanov et al., 2006; Stolz et al., 2011). Although recent studies with nonmodel plants suggested possible roles for cZ (Vyroubalová et al., 2009; Goggin et al., 2010; Lomin et al., 2011), its physiological significance was yet to be clarified. Our root growth assay clearly demonstrated comparable activities of cZ and tZ in the inhibition of rice seminal root elongation (Fig. 1). The capacity of cZ to induce ZmRR1 gene expression in maize cultured cells has been reported to resemble that of tZ (Yonekura-Sakakibara et al., 2004). Our results further indicated that cZ has tZ-like cytokinin activity not only in the induction of cytokinin-responsive genes but also in the control of root growth at physiological concentration ranges. Thus, contrary to widespread assumptions, cZ plays a role as an active cytokinin in several aspects of rice growth and development. Previous biochemical studies on HK cytokinin receptors showed that the affinity to cZ was similar to that to tZ in maize HK (ZmHK1; Lomin et al., 2011), while cZ affinities were much lower than tZ affinities in Arabidopsis HKs (AHK2, AHK3, and CRE1/AHK4; Romanov et al., 2006; Stolz et al., 2011). Four cytokinin receptors have been identified in rice (OsHK3, OsHK4, OsHK5, and OsHK6; Ito and Kurata, 2006; Pareek et al., 2006; Du et al., 2007; Schaller et al., 2007). A recent study of ligand specificities of the OsHKs using a heterologous protoplast system showed that OsHK3 and OsHK4 have similar sensitivities to cZ and other cytokinins (Choi et al., 2012). These receptors may be involved in the cZ response in roots.
Our tracer experiments showed that cZR mostly was converted into O-glucosides, whereas tZR was initially converted into tZRPs and then into tZ-9-N-glucoside (Fig. 3; Supplemental Tables S1 and S2). However, rapid accumulation of cZRPs was also observed following the application of cZR, even though the cZRP levels reached were lower than the tZRP levels established following exposure to tZR (Supplemental Tables S1 and S2). Thus, tZR as well as cZR phosphorylation may occur, but the more rapid O-glucosylation may result in a lower accumulation of cZRPs. Interestingly, the accumulation of tZOG and tZROG after cZR application was comparable to that following tZR application, while the accumulation of tZ and tZR after the application of cZR was less pronounced than after tZR application (Fig. 3). These results suggest that isomerization may occur in the O-glucoside forms. However, our data indicate that isomerizations between cZ derivatives and tZ derivatives represent a minor pathway in rice cytokinin metabolism. No cis-trans-isomerization was observed in tobacco BY-2 cells and oat (Avena sativa) leaf segments exposed to radioisotope-labeled tZ or cZ (Gajdošová et al., 2011). The isomerization might occur predominantly in specific organs or tissues such as the endosperm of immature seeds (Bassil et al., 1993).
We identified three cZOGTs as putative cZOGTs in rice. Enzymatic characterization showed that the cZOGTs catalyzed not only cZ-O-glucosylation but also cZR-O-glucosylation. Although maize cisZOG1 did not recognize cZR as a substrate in a previous study (Martin et al., 2001), recombinant cisZOG1 catalyzed cZR-O-glucosylation in our tests (data not shown). The activities of rice cZOGT1 and cZOGT2 as well as maize cisZOG1 directed at the riboside form separate these enzymes from P. lunatus ZOG1, which does not accept tZR as a substrate (Dixon et al., 1989); the biological relevance of this difference, however, remains unclear. Maize cisZOG1 has been reported to catalyze oT-O-glucosylation (Mok et al., 2005); similarly, cZOGT1 and cZOGT2, but not cZOGT3, catalyzed oT-O-glucosylation in vitro (Fig. 4C). However, oT and its derivatives including oT-O-glucoside were not detected in cZOGT1-ox and cZOGT2-ox plants (Supplemental Table S3).
The enzymatic properties and phylogenetic analysis (Fig. 4A) indicated that rice cZOGT1 and cZOGT2 are cognates of maize cisZOGs. However, expression patterns suggested that cZOGT2 might have a different physiological function from that of maize cisZOGs at the seedling stage: cZOGT2 was predominantly expressed in shoots (Figs. 5 and 6), whereas cisZOG genes were active mostly in roots (Veach et al., 2003; Vyroubalová et al., 2009).
The observed alterations of O-glucoside contents in cZOGT1-ox and cZOGT2-ox lines were consistent with the in vitro activities targeting cZ but not with those targeting cZR and cZRMP (Figs. 4 and 10); this suggested that phosphorylation of cZROG might occur in rice. Although adenosine kinases have been reported to be involved in the phosphorylation of iP-ribotides and tZR (Chen and Eckert, 1977; von Schwartzenberg et al., 1998; Kwade et al., 2005; Schoor et al., 2011), no activity directed at O-glucosides has been described. Despite the higher activity of cZOGT3 for cZ than for tZ and tZR in vitro (Fig. 4C), cZOG contents remained unchanged in cZOGT3-ox, whereas tZOG and tZROG levels dramatically increased (Fig. 10). There is an example of a plant UGT whose actual substrate in vivo is determined by the substrate specificity of interacting enzymes that catalyze upstream reactions (Sorghum bicolor UGT85B1; Kahn et al., 1999; Nielsen et al., 2008). We can hypothesize that the cZOGTs interact with other proteins affecting the in vivo activity of cZOGTs.
The cZOGT1-ox and cZOGT2-ox plants exhibited conspicuous short-shoot phenotypes apparently caused by decreased cell numbers in affected organs (Fig. 8) and also a delay of leaf senescence (Fig. 9; Supplemental Fig. S5). These phenotypes are consistent with previous observations in cytokinin mutants (Riefler et al., 2006; Bartrina et al., 2011) and transgenic maize overexpressing P. lunatus ZOG1 (Pineda Rodó et al., 2008), suggesting that the action of cytokinins is perturbed in cZOGT1-ox and cZOGT2-ox. It should be noted that increased tZ and tZR O-glucosylation does not seem sufficient to cause the short-shoot phenotype because this phenotype was not found in cZOGT3-ox, which showed significantly increased tZOG and tZROG contents (Fig. 10). On the other hand, alteration of cZOG and cZRPsOG contents appeared to parallel the occurrence of the short-shoot phenotype (Figs. 7 and 10), suggesting that increased cZ O-glucosylation might be involved in the causation of the phenotype.
However, several issues need to be considered carefully. First, the Km values of rice cZOGTs for cZ and cZR (95–330 µm; Table I) seemed high compared with the endogenous cZ and cZR contents (around 1–3 nm; calculated from Supplemental Table S3). Second, the cZ content was not significantly decreased in cZOGT1-ox and cZOGT2-ox (Supplemental Table S3), and the increase of cZOG contents was rather moderate (Fig. 10). These results raise the question of whether there is another, unidentified substrate of cZOGTs controlling the phenotype. On the other hand, Km values in the micromolar range are not unusual for cytokinin metabolic enzymes, especially for glucosyltransferases. Examples are 50 µm in P. lunatus ZOG1 for tZ (Mok et al., 2005) and 70 to 240 µm in Arabidopsis N-glucosyltransferases UGT76C1 and UGT76C2 for iP and tZ (Hou et al., 2004). Moreover, substrate contents of cytokinin glucosyltransferases did not decrease in previously studied transgenic plants overexpressing P. lunatus ZOG1 (Pineda Rodó et al., 2008) and UGT76C2 (Wang et al., 2011), while glucosides increased. The moderate increase of cZOG in cZOGT1-ox and cZOGT2-ox may be explained by the natural abundance of cZOG in rice and/or the involvement of the O-glucoside in multiple metabolic pathways. However, we cannot exclude the possibility that cZOGTs are involved in additional mechanisms of plant growth and development. To fully clarify this issue, integrative metabolome and hormone analyses of the cZOGT-ox and cZOGT loss-of-function mutants will be required.
In conclusion, our evidence shown here suggests that cZ functions as an active cytokinin and that its metabolism including O-glucosylation possibly contributes to the control of cytokinin activity in rice. Further investigation into cZ metabolism, including the elucidation of O-glucoside metabolism and the identification of novel genes involved in its biosynthesis, will promote a better understanding of the physiological significance of cZ.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa ‘Nipponbare’) and Arabidopsis (Arabidopsis thaliana) ecotype Columbia were used as wild-type rice and Arabidopsis, respectively. Rice plants were grown under three conditions: sterile culture on agar plates, hydroponic culture, and soil culture. For sterile cultures, sterilized rice seeds were sown and germinated on one-half-strength Murashige and Skoog medium containing 1% (w/v) Suc and 1% (w/v) agar at 30°C for 1 to 2 d in the dark. The seedlings were further grown for 5 d in a growth chamber with a 13-/11-h day/night regime at 28°C/24°C. For hydroponic culture, rice seeds were germinated in distilled water at 30°C for 2 d in the dark. The germinated rice seeds were sown in tap water adjusted to pH 5.5 using HCl and grown for 1 week in a greenhouse with supplemental artificial light at 26°C/23°C. The seedlings were further grown for 1 week in one-quarter-strength nutrient solution (Makino et al., 1983). For soil culture, germinated rice seeds were sown on a synthetic soil (Mitsui-Toatsu No.3) and grown for 3 to 4 weeks in a greenhouse with irrigation and supplemental artificial light at 26°C/23°C. Then, the plants were transplanted with approximately 1 g of slow-release fertilizer per plant and grown in a greenhouse with irrigation and supplemental artificial light at 26°C/23°C until harvest of samples or the late grain-filling stage. Arabidopsis seedlings were aseptically cultured on one-half-strength Murashige and Skoog agar medium in a growth room with a 16/8-h day/night regime at 22°C for 13 d.
Oligonucleotide Primers
Oligonucleotide primers used in this study are shown in Supplemental Table S4.
Preparation of Total RNA and First-Strand cDNA and Quantitative PCR Analysis
Total RNA was extracted from plant samples using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Single-stranded complementary DNA (cDNA) was prepared from total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen) with oligo(dT)12-18 primers. Quantitative real-time PCR analysis was carried out with the StepOne Plus Real-Time PCR System (Applied Biosystems Japan) using gene-specific primers (Supplemental Table S1) and SYBR Premix Ex Taq II (Perfect Real Time; TaKaRa BIO). Purified plasmids containing the target sequences of the primers were used to obtain linear standard curves (r2 > 0.9985), and the mRNA contents were determined quantitatively within the linear range.
Assays for Cytokinin Activities
To prepare stock solutions, cytokinins were dissolved in dimethyl sulfoxide (DMSO) and added to agar medium or nutrient solution. In all conditions, the final concentration of DMSO was controlled to 0.02% (v/v). Root growth assays were carried out using plants in sterile culture as described above. The primary root length of Arabidopsis or seminal root length of rice was manually measured in seedlings that were grown in the presence of tZ or cZ at 0, 1, 10, 100, 1,000, or 10,000 nm. Expression analysis of OsRR genes was carried out in plants grown in hydroponic culture. The 2-week-old rice seedlings were pretreated with one-tenth-strength nutrient solution without nitrogen nutrient (1/10-N) for 1 d. Then, the seedlings were transferred to 1/10-N solution containing no cytokinin, 100 nm tZ, or 100 nm cZ. Whole roots were harvested at 15, 30, and 60 min after transfer. Roots harvested before transfer served as time-zero samples. Harvested roots were immediately frozen in liquid nitrogen and stored at −80°C until total RNA extraction. The expression levels of OsRR1, OsRR2, OsRR3, OsRR4, OsRR6, and OsRR9/10 (RAP-DB identifiers Os04g0442300, Os02g0557800, Os02g0830200, Os01g0952500, Os04g0673300, and Os11g0143300/Os12g0139400) were analyzed by qRT-PCR.
Synthesis of Stable Isotope-Labeled tZR and cZR
Stable isotope-labeled tZRMP was synthesized from [1013C,515N]AMP and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate with recombinant tZ secretion (Tzs) protein (Sugawara et al., 2008). Thus, cytokinins derived from this isotope-labeled tZRMP (labeled in the purine ring and ribosyl group; +15) have a molecular mass increased by 15 D. The tZRMP(+15) was dephosphorylated by alkaline phosphatase to form tZR(+15). To purify the tZR(+15), proteins were removed using Micropore-EZ columns (Amicon), and the resulting solution was loaded onto an octadecylsilyl reverse-phase column (Symmetry C18, 3.5 μm, 2.1 × 150 mm; Waters) in an HPLC system (Alliance 2695 system/2996 photodiode array detector; Waters). The products were separated at a flow rate of 1.0 mL min−1 with linear gradients of solvent A (2% acetic acid) and solvent B (acetonitrile) set as follows: 0 min, 99% A + 1% B; 1 min, 99% A + 1% B; 3 min, 93% A + 7% B; 11 min, 90% A + 10% B; 12 min, 60% A + 40% B; 13 min, 40% A + 60% B; 22 min, 40% A + 60% B; 23 min, 99% A + 1% B. Fractions containing tZR(+15) were collected and dried. Isomerization from tZR(+15) to cZR(+15) was carried out by nonenzymatic conversion (Bassil et al., 1993). Seventy microliters of 3 mm tZR(+15) in DMSO was mixed with 100 μL of 2-mercaptoethanol in a glass tube and incubated under artificial lights (D400 [Toshiba], MLBOC400C-U [Mitsubishi/Osram]) at 28°C for 30 min. The resulting solution was dried and dissolved in 2% acetic acid. The cZR(+15) was purified as described above for tZR(+15). The quantity and purity of the synthesized tZR(+15) and cZR(+15) were checked by ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analysis (Supplemental Fig. S7) as described previously (Kojima et al., 2009).
Tracer Experiments
Hydroponically grown rice seedlings (2 weeks old) were pretreated with 1/10-N solution for 1 d and then transferred into the 1/10-N solution containing 1 μm tZR(+15) or cZR(+15). Whole roots and shoots were separately harvested at 1 h after transfer. The harvested roots and shoots were immediately frozen in liquid nitrogen and stored at −80°C until cytokinin analysis (Kojima et al., 2009). Monitored ion transitions for the quantification of labeled cytokinin species were as follows: 367.2 > 230.2 for cZR(+15), 362.2 > 230.2 for cZR(+10), 230.2 > 146.1 for cZ(+10), 529.2 > 230.2 for tZROG(+15), 524.2 > 230.2 for tZROG(+10), 529.2 > 230.2 for cZROG(+15), 524.2 > 230.2 for cZROG(+10), 392.2 > 230.2 for tZOG(+10), and 392.2 > 230.2 for cZOG(+10). Other conditions for MS/MS analysis were as described previously (Kojima et al., 2009; Tokunaga et al., 2012).
Cloning of cDNAs, DNA Sequencing, and Phylogenetic Relationships
BLAST searches in the RAP-DB (http://rapdb.dna.affrc.go.jp/) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) Web sites were carried out using the deduced amino acid sequences of maize (Zea mays) cisZOG1 (AF318075) and cisZOG2 (AY082660) as queries. Predicted entire coding regions of Os04g0556400, Os04g0556500 (cZOGT1), Os04g0556600 (cZOGT2), Os04g0565200, Os04g0565400 (cZOGT3), and Os07g0660500 were amplified from first-strand cDNA by PCR and put into the pCR-Blunt II-TOPO vector (Invitrogen). DNA sequencing was carried out with a 3100 Genetic Analyzer (Applied Biosystems Japan). The phylogenetic tree was constructed by the neighbor-joining method with a bootstrap test using MEGA 4.0.2 software (http://www.megasoftware.net/).
Preparation of Recombinant Proteins
In vitro assays of cytokinin-O-glucosylation activities were performed with purified recombinant proteins expressed in Escherichia coli. The DNA fragments containing entire reading frames were inserted into the multicloning site of the pCold-I vector (TaKaRa BIO) to generate translational fusions with translation-enhanced elements and poly-His tags. The resulting expression plasmids were transformed into the E. coli strain BL21. Transfected BL21 cells were cultured and treated to express recombinant proteins according to the pCold-I manufacturer’s instructions. Crude soluble proteins were extracted from the BL21 cells by sonication in extraction buffer (50 mm NaH2PO4-NaOH, pH 8.0, 300 mm NaCl, 10 mm imidazole, and 1 mm dithiothreitol) containing a protease inhibitor cocktail (P8849; Sigma). The recombinant proteins were purified by nickel affinity chromatography using Ni-NTA Superflow (Qiagen) according to the manufacturer’s instructions. Purified recombinant proteins were stored at −80°C in the presence of 1 mm dithiothreitol and 15% (w/w) glycerol. Concentrations of recombinant proteins were determined with a Protein Assay kit (Bio-Rad) and by densitometry of SDS-polyacrylamide gels stained with Bio-safe Coomassie Brilliant Blue G-250 stain (Bio-Rad) using bovine serum albumin as a standard.
Enzyme Assay
The enzyme assay for cytokinin-O-glucosylation was based on a previous study of maize cisZOGs (Veach et al., 2003). One microgram of the purified recombinant protein was incubated in 198 μL of reaction premixture (50 mm Tris-acetate buffer, pH 5.0–7.0, or Tris-HCl, pH 7.0–10.0, 1.5 or 3.0 mm UDP-Glc, and 50 or 100 mm MgCl2) for at least 5 min at 30°C, and then reactions were started by the addition of 2 μL of cytokinin in DMSO as a substrate. To determine kinetic parameters, at least four different concentrations of a cytokinin substrate in the range of 10 to 500 µm were applied. Five minutes later, the reactions were stopped by the addition of 1 mL of cold ethanol, and the mixtures were incubated for at least 20 min at −30°C. After removal of the precipitates by centrifugation (20,400g, 20 min at 4°C), the supernatant was vacuum dried, dissolved in 60 μL of 2% (v/v) acetic acid, and loaded onto an octadecylsilyl column (Merck, Supersphere RP-select B, 4 mm × 250 mm) in an HPLC system. Compounds were separated at a flow rate of 1.0 mL min−1 with linear gradients of solvent A (2% acetic acid) and solvent B (acetonitrile) as follows: 0 min, 99% A + 1% B; 1 min, 93% A + 7% B; 6 min, 93% A + 7% B; 14 min, 90% A + 10% B; 38 min, 60% A + 40% B; 40 min, 50% A + 50% B; 55 min, 99% A + 1% B.
Promoter-GUS Analysis
A Gateway cassette (Invitrogen) was inserted into a unique HindIII site upstream of the GUS coding sequence of the pCAMBIA1390-GUS vector (Hirose et al., 2005) using the In-Fusion Advantage PCR Cloning Kit (Clontech). The resulting vector was designated as pCAMBIA1390-GW-GUS. Genomic sequences containing putative promoter regions of cZOGT1, cZOGT2, and cZOGT3 (−3,052, −3,952, and −3089 to −1 bp from the translational initiation codon, respectively) were designated as cZOGT1pro, cZOGT2pro, and cZOGT3pro and were put into the entry vector pDONR221 (Invitrogen) using the Gateway BP Clonase II Enzyme Mix (Invitrogen). Subsequently, to construct cZOGT1pro-GUS, cZOGT2pro-GUS, and cZOGT3pro-GUS, the putative promoter sequences were put into the pCAMBIA1390-GW-GUS vector using the Gateway LR Clonase II Enzyme Mix (Invitrogen). Transgenic rice plants were generated by the Agrobacterium tumefaciens-mediated method (Hirose et al., 2005) using the A. tumefaciens strain EHA105. Detection of GUS activity and histochemical analysis were carried out as described previously (Hirose et al., 2005).
Generation and Characterization of Transgenic Plants Overexpressing cZOGT Genes in Rice
The binary vector pActnos/Hmz (Sentoku et al., 2000; Hirose et al., 2007) was used to prepare the constructs for overexpression of rice cZOGT genes. The entire coding regions of the cZOGT cDNAs were inserted into an XbaI/SmaI site located between an Act1 promoter (Zhang et al., 1991) and a NOS terminator. Transformation of rice was carried out using A. tumefaciens as described above. The empty vector containing pActnos/Hmz served as a control. We generated four VC lines (VC-1, VC-3, VC-4, and VC-7), four cZOGT1-ox lines (1ox-1, 1ox-3, 1ox-4, and 1ox-7), four cZOGT2-ox lines (2ox-2, 2ox-3, 2ox-7, and 2ox-10), and four cZOGT3-ox lines (3ox-3, 3ox-7, 3ox-9, and 3ox-10); all of these were independent transgenic lines derived from different transformation events. The lengths of shoots and leaves were measured manually. To measure cell lengths of epidermal cells, seedlings were grown hydroponically, and 5 mm of the leaf sheaths was cut at their laminar joints. The leaf sheaths were stained with propidium iodide after fixation in 5% (v/v) formaldehyde solution containing 37% (w/v) formaldehyde, 5% (v/v) acetic acid, and 63% (v/v) ethanol. Propidium iodide fluorescence micrographs of epidermal cells were taken using a confocal laser-scanning microscope (FV1000; Olympus). Cell lengths were determined with ImageJ (http://rsbweb.nih.gov/ij/). Relative chlorophyll contents were analyzed in a noninvasive manner using a portable chlorophyll meter (SPAD-502Plus; Konica Minolta). The relative contents of each leaf were determined by three measurements at the tip, middle, and basal parts of the leaf. Crude proteins were extracted from leaf blades in extraction buffer (50 mm NaH2PO4-NaOH, pH 8.0, 1 mm EDTA, 0.1% [v/v] 2-mercaptoethanol, 0.1% [v/v] Tween 20, 5% [v/v] glycerol, and 1% [v/v] proteinase inhibitor cocktail III from Calbiochem). The homogenate was centrifuged at 20,000g for 20 min at 4°C, and the resulting supernatant was used as the crude protein extract. The protein content was determined by the Protein Assay kit (Bio-Rad) using bovine serum albumin as a standard. Cytokinin was quantified with a Xevo TQ-S (Waters) tandem quadrupole mass spectrometer; conditions for MS/MS analysis were as described previously (Kojima et al., 2009).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Os04g0556400 (NM_001060060), Os04g0556500 (NM_001060061), Os04g0556600 (NM_001060062), Os04g0565200 (NM_001060107), Os04g0565400 (NM_001060109), Os07g0660500 (NM_001067067), OsRR1 (AK072736), OsRR2 (AK070645), OsRR3 (AB249655), OsRR4 (AK101721), OsRR6 (AK059734), OsRR9 (AK058585), OsRR10 (AB249660), and Gpc1 (AK103777).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple alignment of deduced amino acid sequences of maize cisZOGs and putative rice cZOGTs.
Supplemental Figure S2. Preliminary tests to determine conditions for an enzymatic activity assay for rice cZOGTs.
Supplemental Figure S3. Plots of velocities of O-glucosylation catalyzed by cZOGT recombinant proteins against substrate concentrations.
Supplemental Figure S4. Overexpression of cZOGT genes in transgenic lines.
Supplemental Figure S5. Delay of leaf senescence in cZOGT2-overexpressing rice.
Supplemental Figure S6. Visible phenotypes in roots of cZOGT1- and cZOGT2-overexpressing rice at the seedling stage.
Supplemental Figure S7. UPLC-MS/MS analysis of synthesized [1013C,515N]tZR and [1013C,515N]cZR.
Supplemental Table S1. Time courses of cytokinin derivative contents of rice seedlings after application of [1013C,515N]tZR.
Supplemental Table S2. Time courses of cytokinin derivative contents of rice seedlings after application of [1013C,515N]cZ.
Supplemental Table S3. Cytokinin contents in shoots of cZOGT-overexpressing lines and VC lines.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. M. Matsuoka (Nagoya University) for providing us with pActnos/Hmz. We also thank Dr. K. Yonekura-Sakakibara (RIKEN) for providing sugar nucleotides and helpful suggestions for measuring glucosyltransferase activity.
Glossary
- iP
N6-(Δ2-isopentenyl)adenine
- tZ
trans-zeatin
- cZ
cis-zeatin
- oT
ortho-topolin
- cZR
cZ-riboside
- tZR
tZ-riboside
- qRT
quantitative reverse transcription
- cZOG
cZ-O-glucoside
- cZROG
cZR-O-glucoside
- cZRPsOG
cZ-ribotide-O-glucoside
- cZRP
cZ-ribotide
- tZRP
tZ-ribotide
- tZROG
tZR-O-glucoside
- tZOG
tZ-O-glucoside
- RAP-DB
Rice Annotation Project Database
- tZRMP
tZR 5′-monophosphate
- cZRMP
cZR 5′-monophosphate
- VC
vector control
- DMSO
dimethyl sulfoxide
- 1/10-N
one-tenth-strength nutrient solution without nitrogen nutrient
- UPLC
ultraperformance liquid chromatography
- MS/MS
tandem mass spectrometry
- cDNA
complementary DNA
References
- Allen M, Qin W, Moreau F, Moffatt B. (2002) Adenine phosphoribosyltransferase isoforms of Arabidopsis and their potential contributions to adenine and cytokinin metabolism. Physiol Plant 115: 56–68 [DOI] [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil NV, Mok D, Mok MC. (1993) Partial purification of a cis-trans-isomerase of zeatin from immature seed of Phaseolus vulgaris L. Plant Physiol 102: 867–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao PJ, Bartley LE, Jung KH, Ronald PC. (2008) Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol Plant 1: 858–877 [DOI] [PubMed] [Google Scholar]
- Chen CM, Eckert RL. (1977) Phosphorylation of cytokinin by adenosine kinase from wheat germ. Plant Physiol 59: 443–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Lee J, Kim K, Cho M, Ryu H, An G, Hwang I. (2012) Functional identification of OsHk6 as a homotypic cytokinin receptor in rice with preferential affinity for iP. Plant Cell Physiol 53: 1334–1343 [DOI] [PubMed] [Google Scholar]
- Dixon SC, Martin RC, Mok MC, Shaw G, Mok DW. (1989) Zeatin glycosylation enzymes in Phaseolus: isolation of O-glucosyltransferase from P. lunatus and comparison to O-xylosyltransferase from P. vulgaris. Plant Physiol 90: 1316–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Jiao F, Chu J, Jin G, Chen M, Wu P. (2007) The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics 89: 697–707 [DOI] [PubMed] [Google Scholar]
- Emery RJ, Leport L, Barton JE, Turner NC, Atkins CA. (1998) cis-Isomers of cytokinins predominate in chickpea seeds throughout their development. Plant Physiol 117: 1515–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdošová S, Spíchal L, Kamínek M, Hoyerová K, Novák O, Dobrev PI, Galuszka P, Klíma P, Gaudinová A, Źiźková E, et al. (2011) Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J Exp Bot 62: 2827–2840 [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM. (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988 [DOI] [PubMed] [Google Scholar]
- Goggin DE, Emery RJN, Powles SB, Steadman KJ. (2010) Initial characterisation of low and high seed dormancy populations of Lolium rigidum produced by repeated selection. J Plant Physiol 167: 1282–1288 [DOI] [PubMed] [Google Scholar]
- Haberer G, Kieber JJ. (2002) Cytokinins: new insights into a classic phytohormone. Plant Physiol 128: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H. (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539 [DOI] [PubMed] [Google Scholar]
- Hirose N, Makita N, Yamaya T, Sakakibara H. (2005) Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol 138: 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59: 75–83 [DOI] [PubMed] [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ. (2004) N-Glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279: 47822–47832 [DOI] [PubMed] [Google Scholar]
- Houba-Hérin N, Pethe C, d’Alayer J, Laloue M. (1999) Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J 17: 615–626 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kurata N. (2006) Identification and characterization of cytokinin-signalling gene families in rice. Gene 382: 57–65 [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. (2006) Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Fahrendorf T, Halkier BA, Møller BL. (1999) Substrate specificity of the cytochrome P450 enzymes CYP79A1 and CYP71E1 involved in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys 363: 9–18 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol 42: 677–685 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H. (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwade Z, Świątek A, Azmi A, Goossens A, Inzé D, Van Onckelen H, Roef L. (2005) Identification of four adenosine kinase isoforms in tobacco BY-2 cells and their putative role in the cell cycle-regulated cytokinin metabolism. J Biol Chem 280: 17512–17519 [DOI] [PubMed] [Google Scholar]
- Leonard NJ, Playtis AJ, Skoog F, Schmitz RY. (1971) A stereoselective synthesis of cis-zeatin. J Am Chem Soc 93: 3056–3058 [Google Scholar]
- Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. (2011) Ligand-binding properties and subcellular localization of maize cytokinin receptors. J Exp Bot 62: 5149–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Mae T, Ohira K. (1983) Photosynthesis and ribulose 1,5-bisphosphate carboxylase in rice leaves: changes in photosynthesis and enzymes involved in carbon assimilation from leaf development through senescence. Plant Physiol 73: 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Pařezová M, Petrášek J, Friml J, Kleine-Vehn J, et al. (2011) Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21: 796–804 [DOI] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Habben JE, Mok DW. (2001) A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. Proc Natl Acad Sci USA 98: 5922–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Mok DW. (1999) Isolation of a cytokinin gene, ZOG1, encoding zeatin O-glucosyltransferase from Phaseolus lunatus. Proc Natl Acad Sci USA 96: 284–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D, Rothenberg M, Wu R. (1990) Structural characterization of a rice actin gene. Plant Mol Biol 14: 163–171 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T. (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103: 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt B, Pethe C, Laloue M. (1991) Metabolism of benzyladenine is impaired in a mutant of Arabidopsis thaliana lacking adenine phosphoribosyltransferase activity. Plant Physiol 95: 900–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DWS, Martin RC, Shan X, Mok MC. (2000) Genes encoding zeatin O-glycosyltransferases. Plant Growth Regul 32: 285–287 [Google Scholar]
- Mok DWS, Mok MC. (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Mok MC, Martin RC, Dobrev PI, Vanková R, Ho PS, Yonekura-Sakakibara K, Sakakibara H, Mok DWS. (2005) Topolins and hydroxylated thidiazuron derivatives are substrates of cytokinin O-glucosyltransferase with position specificity related to receptor recognition. Plant Physiol 137: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok MC, Mok DW, Armstrong DJ. (1978) Differential cytokinin structure-activity relationships in Phaseolus. Plant Physiol 61: 72–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KA, Tattersall DB, Jones PR, Møller BL. (2008) Metabolon formation in dhurrin biosynthesis. Phytochemistry 69: 88–98 [DOI] [PubMed] [Google Scholar]
- Pareek A, Singh A, Kumar M, Kushwaha HR, Lynn AM, Singla-Pareek SL. (2006) Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol 142: 380–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisová M, Klíma P, Horák J, Válková M, Malbeck J, Souček P, Reichman P, Hoyerová K, Dubová J, Friml J, et al. (2009) Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc Natl Acad Sci USA 106: 3609–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda Rodó A, Brugière N, Vankova R, Malbeck J, Olson JM, Haines SC, Martin RC, Habben JE, Mok DWS, Mok MC. (2008) Over-expression of a zeatin O-glucosylation gene in maize leads to growth retardation and tasselseed formation. J Exp Bot 59: 2673–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN, Schmülling T. (2006) Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J Exp Bot 57: 4051–4058 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N. (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP, Kurata N, Mizuno T, Pareek A, Shiu SH, Wu P, et al. (2007) Nomenclature for two-component signaling elements of rice. Plant Physiol 143: 555–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RY, Skoog F, Playtis AJ, Leonard NJ. (1972) Cytokinins: synthesis and biological activity of geometric and position isomers of zeatin. Plant Physiol 50: 702–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoor S, Farrow S, Blaschke H, Lee S, Perry G, von Schwartzenberg K, Emery N, Moffatt B. (2011) Adenosine kinase contributes to cytokinin interconversion in Arabidopsis. Plant Physiol 157: 659–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Matsuoka M. (2000) Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev Biol 220: 358–364 [DOI] [PubMed] [Google Scholar]
- Spíchal L, Rakova NY, Riefler M, Mizuno T, Romanov GA, Strnad M, Schmülling T. (2004) Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol 45: 1299–1305 [DOI] [PubMed] [Google Scholar]
- Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T. (2011) The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J 67: 157–168 [DOI] [PubMed] [Google Scholar]
- Strnad M. (1997) The aromatic cytokinins. Physiol Plant 101: 674–688 [Google Scholar]
- Sugawara H, Ueda N, Kojima M, Makita N, Yamaya T, Sakakibara H. (2008) Structural insight into the reaction mechanism and evolution of cytokinin biosynthesis. Proc Natl Acad Sci USA 105: 2734–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Yokota T, Murofushi N, Ota Y, Takahashi N. (1985) Fluctuation of endogenous cytokinin contents in rice during its life cycle: quantification of cytokinins by selected ion monitoring using deuterium-labelled internal standards. Agric Biol Chem 49: 3271–3277 [Google Scholar]
- Takagi M, Yokota T, Murofushi N, Saka H, Takahashi N. (1989) Quantitative changes of free-base, riboside, ribotide and glucoside cytokinins in developing rice grains. Plant Growth Regul 8: 349–364 [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T. (2001) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276: 26405–26410 [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. (2004a) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. (2004b) Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J Biol Chem 279: 41866–41872 [DOI] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, Sakakibara H. (2012) Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J 69: 355–365 [DOI] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DWS, Malbeck J, Vankova R, Mok MC. (2003) O-Glucosylation of cis-zeatin in maize: characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol 131: 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwartzenberg K, Kruse S, Reski R, Moffatt B, Laloue M. (1998) Cloning and characterization of an adenosine kinase from Physcomitrella involved in cytokinin metabolism. Plant J 13: 249–257 [DOI] [PubMed] [Google Scholar]
- Vyroubalová S, Václavíková K, Turecková V, Novák O, Smehilová M, Hluska T, Ohnoutková L, Frébort I, Galuszka P. (2009) Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol 151: 433–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma XM, Kojima M, Sakakibara H, Hou BK. (2011) N-Glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol 52: 2200–2213 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T. (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. (2004) Molecular characterization of cytokinin-responsive histidine kinases in maize: differential ligand preferences and response to cis-zeatin. Plant Physiol 134: 1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, McElroy D, Wu R. (1991) Analysis of rice Act1 5′ region activity in transgenic rice plants. Plant Cell 3: 1155–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.