Abstract
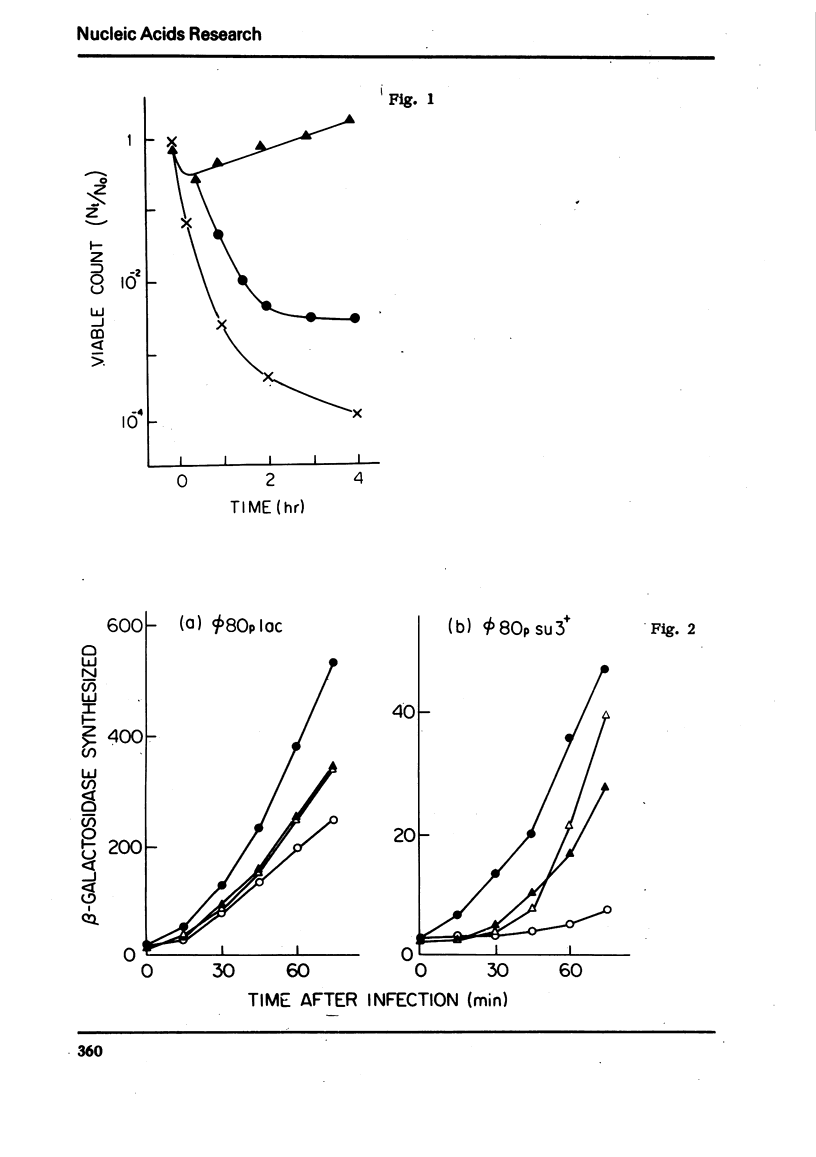
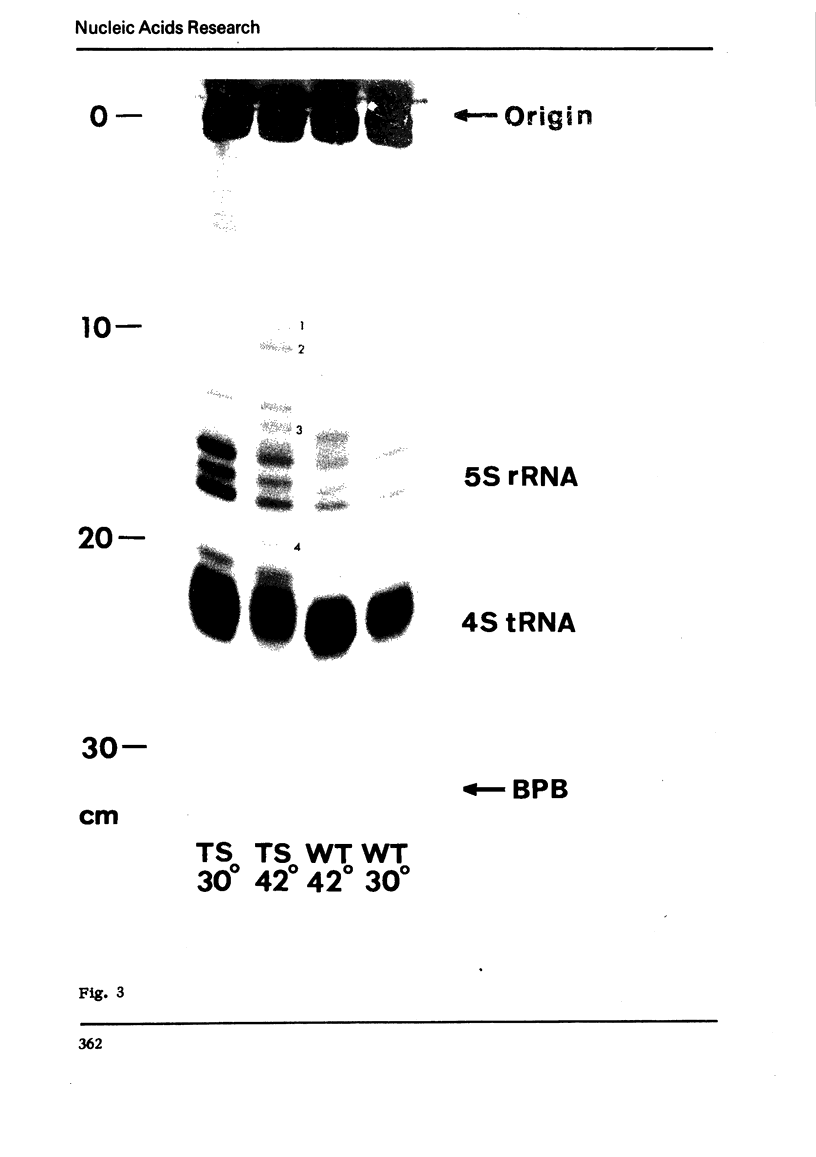
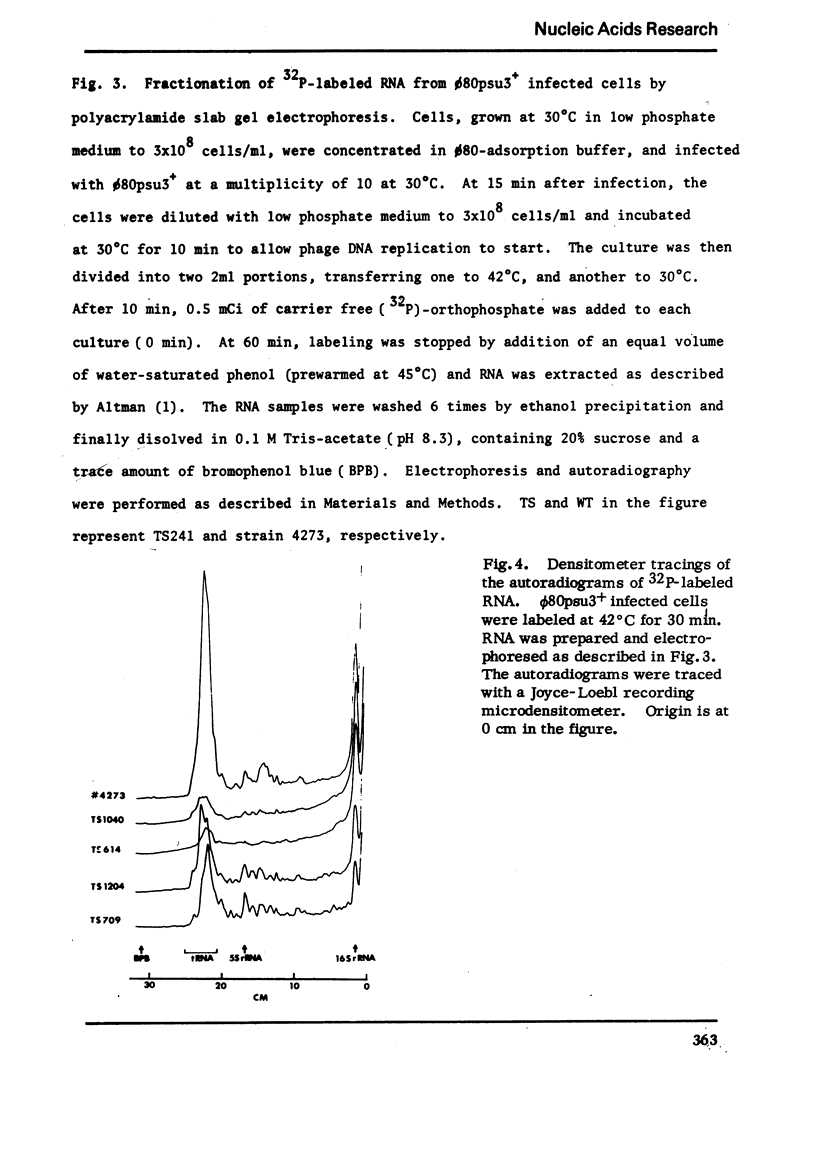
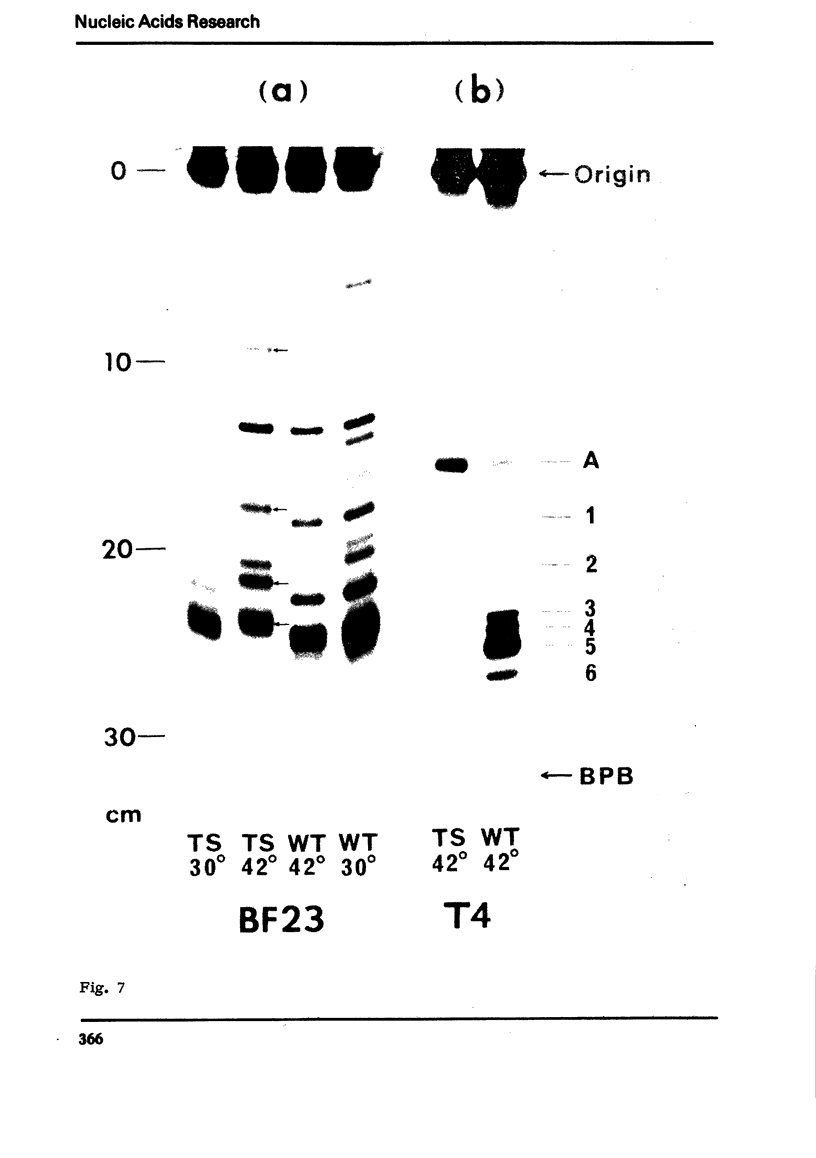
An efficient method was devised to isolate temperature sensitive mutants of E. coli defective in tRNA biosynthesis. Mutants were selected for their inability to express suppressor activity after su3+-transducing phage infection. In virtually all the mutants tested, temperature sensitive synthesis of tRNATyr was demonstrated. Electrophoretic fractionation of 32P labeled RNA synthesized at high temperature showed in some mutants changes in mobility of the main tRNA band and the appearance of slow migrating new species of RNA. Temperature sensitive function of mutant cells was also evident in tRNA synthes: directed by virulent phage T4 and BF23. We conclude that although the mutants show individual differences, many are temperature sensitive in tRNA maturation functions.
In spite of much information on the structure and function of transfer RNA (tRNA), our knowledge concerning the biosynthesis of tRNA is relatively poor. It is generally assumed that complete tRNA molecules are made via a series of processing steps from the original transcription products of tRNA genes which are presumably unmodified and longer than mature tRNA molecules. In the case of tyrosine suppressor tRNA of su3+, an unmodified precursor RNA carrying additional residues at the 3′ and 5′ ends has been isolated (1,2), and an endonuclease cleaving at the 5′ side of this precursor has been identified in E. coli (3). In the case of T4 encoded tRNA, a large precursor molecule for several tRNA's has been reported (4). Some enzymes that catalyze the modifications have also been described (5). However, the over-all picture and the precise mechanisms of tRNA maturation are as yet largely unkown.
For study of tRNA biosynthesis in E. coli, a genetic approach may prove useful, as has been the case in other biosynthetic pathways. In order to obtain mutants blocked in any of the intermediary steps of tRNA synthesis, we have developed an efficient selection system that enriches these mutants. Since any mutational block in tRNA biosynthesis might well be lethal, we looked for conditional lethal mutants in which the defect in tRNA synthesis occurs only at high temperature. In this selection system, the su3 gene carried by a temperate phage was newly introduced into cells(su−) and those cells incapable of synthesizing su3+ tRNA at high temperature were selected. Such mutants were easily enriched by using conditions in which cells expressing suppressor activity were killed by two virulent phages. In this communication, we report the method for isolation of mutants and some characterization of tRNA synthesis in these mutants. Recently, Schedl and Primakoff (6) have independently isolated thermosensitive mutants of E. coli defective in tRNA synthesis which may or may not be different types from ours.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S. Isolation of tyrosine tRNA precursor molecules. Nat New Biol. 1971 Jan 6;229(1):19–21. doi: 10.1038/newbio229019a0. [DOI] [PubMed] [Google Scholar]
- Altman S., Smith J. D. Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol. 1971 Sep 8;233(36):35–39. doi: 10.1038/newbio233035a0. [DOI] [PubMed] [Google Scholar]
- Andoh T., Ozeki H. Suppressor gene Su3+ of E. coli, a structural gene for tyrosine TRNA. Proc Natl Acad Sci U S A. 1968 Mar;59(3):792–799. doi: 10.1073/pnas.59.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., Dahlberg J. E. Small ribonucleic acids of Escherichia coli. I. Characterization by polyacrylamide gel electrophoresis and fingerprint analysis. J Biol Chem. 1973 Jul 25;248(14):5024–5032. [PubMed] [Google Scholar]
- Landy A., Abelson J., Goodman H. M., Smith J. D. Specific hybridization of tyrosine transfer ribonucleic acids with DNA from a transducing bacteriophage phi-80 carrying the amber suppressor gene su 3. J Mol Biol. 1967 Nov 14;29(3):457–471. doi: 10.1016/0022-2836(67)90112-x. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guthrie C., Barrell B. G. Eight transfer RNAs induced by infection of Escherichia coli with bacteriophage T4. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3703–3707. doi: 10.1073/pnas.69.12.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierlich D. P., Lamfrom H., Sarabhai A., Abelson J. Transfer RNA synthesis in vitro. Proc Natl Acad Sci U S A. 1973 Jan;70(1):179–182. doi: 10.1073/pnas.70.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisioka T., Ozeki H. Early abortive lysis by phage BF23 in Escherichia coli K-12 carrying the colicin Ib factor. J Virol. 1968 Nov;2(11):1249–1254. doi: 10.1128/jvi.2.11.1249-1254.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Altman S., Smith J. D. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. J Biol Chem. 1972 Aug 25;247(16):5243–5251. [PubMed] [Google Scholar]
- Russell R. L., Abelson J. N., Landy A., Gefter M. L., Brenner S., Smith J. D. Duplicate genes for tyrosine transfer RNA in Escherichia coli. J Mol Biol. 1970 Jan 14;47(1):1–13. doi: 10.1016/0022-2836(70)90397-9. [DOI] [PubMed] [Google Scholar]
- Schedl P., Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherberg N. H., Weiss S. B. Detection of bacteriophage T4- and T5-coded transfer RNAs. Proc Natl Acad Sci U S A. 1970 Nov;67(3):1164–1171. doi: 10.1073/pnas.67.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., Abelson J. N. Bacteriophage T4 transfer RNA. II. Mutants of T4 defective in the formation of functional suppressor transfer RNA. J Mol Biol. 1972 Aug 14;69(1):57–73. doi: 10.1016/0022-2836(72)90023-x. [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Ozeki H. Comparative study of thermal inactivation of phage phi 80 and lambda. Virology. 1972 May;48(2):316–322. doi: 10.1016/0042-6822(72)90042-6. [DOI] [PubMed] [Google Scholar]