Abstract
Chloroplasts develop from proplastids in a process that requires the interplay of nuclear and chloroplast genomes, but key steps in this developmental process have yet to be elucidated. Here, we show that the nucleus-localized transcription factors GATA NITRATE-INDUCIBLE CARBON-METABOLISM-INVOLVED (GNC) and CYTOKININ-RESPONSIVE GATA1 (CGA1) regulate chloroplast development, growth, and division in Arabidopsis (Arabidopsis thaliana). GNC and CGA1 are highly expressed in green tissues, and the phytohormone cytokinin regulates their expression. A gnc cga1 mutant exhibits a reduction in overall chlorophyll levels as well as in chloroplast size in the hypocotyl. Ectopic overexpression of either GNC or CGA1 promotes chloroplast biogenesis in hypocotyl cortex and root pericycle cells, based on increases in the number and size of the chloroplasts, and also results in expanded zones of chloroplast production into the epidermis of hypocotyls and cotyledons and into the cortex of roots. Ectopic overexpression also promotes the development of etioplasts from proplastids in dark-grown seedlings, subsequently enhancing the deetiolation process. Inducible expression of GNC demonstrates that GNC-mediated chloroplast biogenesis can be regulated postembryonically, notably so for chloroplast production in cotyledon epidermal cells. Analysis of the gnc cga1 loss-of-function and overexpression lines supports a role for these transcription factors in regulating the effects of cytokinin on chloroplast division. These data support a model in which GNC and CGA1 serve as two of the master transcriptional regulators of chloroplast biogenesis, acting downstream of cytokinin and mediating the development of chloroplasts from proplastids and enhancing chloroplast growth and division in specific tissues.
Chloroplasts are organelles that convert light energy into chemical energy to sustain plant growth. Chloroplasts not only carry out photosynthesis but also play a pivotal role in plant metabolism, being involved in the biosynthesis of amino acids, fatty acids, and phytohormones (Neuhaus and Emes, 2000). Chloroplasts of vascular plants develop from a nonphotosynthetic progenitor, a proplastid, which is maintained in the meristematic cells. Proplastids are colorless and contain limited amounts of internal membranes, but they can differentiate into a variety of plastid types with specialized activities, such as amyloplasts in the roots for starch storage, leucoplasts for lipid storage, chromoplasts for pigment accumulation, etioplasts in dark-grown shoots, and chloroplasts in light-grown shoots. The different types of plastids are interconvertible in response to developmental and environmental changes (Mullet, 1988; López-Juez and Pyke, 2005). In the absence of light, proplastids can differentiate into etioplasts, which contain a semicrystalline structure called the prolamellar body with precursors of both chlorophyll and thylakoid membrane lipids (Armstrong et al., 1995). Upon illumination, genes associated with biogenesis of the mature chloroplast are expressed, and several subsequent activities, including import of nucleus-encoded chloroplast proteins, expression of plastid-encoded photosynthetic genes, biosynthesis of chlorophyll, assembly of the photosystems and the thylakoid network, and chloroplast division, occur in parallel to complete chloroplast development.
Multiple factors regulate chloroplast development, light being chief among these factors. Phytohormones, in particular cytokinins, can also modulate chloroplast development. For example, chloroplast maturation is induced by cytokinins in tobacco (Nicotiana tabacum) tissue cultures (Stetler and Laetsch, 1965), exogenous cytokinins induce regreening of senescent Nicotiana rustica leaves (Zavaleta-Mancera et al., 1999a, 1999b), and cytokinins activate chloroplast differentiation from proplastids in pumpkin (Cucurbita maxima) and watermelon (Citrullus vulgaris) cotyledons (Khokhlova, 1977; Longo et al., 1979). In addition, transcription of several photosynthetic genes encoded in the plastids of barley (Hordeum vulgare) is increased by exogenously applied cytokinins (Zubo et al., 2008). Similarly, seedling deetiolation is induced by cytokinins (Chory et al., 1994), and many proteins related to chloroplast biogenesis are up-regulated in response to cytokinins in Arabidopsis (Arabidopsis thaliana; Lochmanová et al., 2008). In spite of the significance of cytokinins for chloroplast biogenesis, the molecular mechanism of their actions on chloroplasts is largely unknown.
Chloroplast biogenesis requires the coordinated transcriptional regulation of genes encoded in the nuclear and chloroplast genomes. Thus, one might predict the existence of nuclear transcription factors that regulate the expression of genes required for chloroplast biogenesis. However, very few transcription factors functioning as positive regulators of chloroplast biogenesis have been identified. A notable exception is the GOLDEN TWO-LIKE (GLK) family of transcription factors (Fitter et al., 2002; Waters et al., 2009). The glk1 glk2 double mutant is pale green with a severe reduction in thylakoids, and the GLK transcription factors promote the expression of many nucleus-encoded photosynthetic genes that are associated with chlorophyll biosynthesis and light-harvesting functions (Waters et al., 2009). In addition, and tying back to the role of cytokinin in regulating chloroplast biogenesis, overexpression of the cytokinin-regulated transcription factor CYTOKININ RESPONSE FACTOR2 (CRF2; Rashotte et al., 2006) increases the chloroplast division rate by up-regulating the protein level of PLASTID DIVISION2 (PDV2; Okazaki et al., 2009).
GATA NITRATE-INDUCIBLE CARBON-METABOLISM-INVOLVED (GNC) and CYTOKININ-RESPONSIVE GATA1 (CGA1) were first identified based on their induction by nitrate (Wang et al., 2003; Price et al., 2004; Scheible et al., 2004; Bi et al., 2005). More recent studies indicate that expression of this family is also induced by light and cytokinin (Naito et al., 2007) and is repressed during flower development (Mara and Irish, 2008). Functionally, they have been shown to antagonize signaling by gibberellins, acting downstream of DELLA proteins (Richter et al., 2010). Effects on greening have also been observed in genetic studies, with the gnc cga1 mutant exhibiting reduced greening compared with the wild type and overexpression of either gene resulting in enhanced greening and chloroplast production (Bi et al., 2005; Naito et al., 2007; Mara and Irish, 2008; Richter et al., 2010; Hudson et al., 2011; Köllmer et al., 2011). The chloroplast-localized gene GLUTAMATE SYNTHASE (GLU1/Fd-GOGAT), a key gene involved in nitrogen assimilation, has been identified as a potential transcriptional target of GNC and CGA1 (Hudson et al., 2011). However, thus far, a detailed picture of the physiological function of this GATA family of transcriptional regulators is lacking. We hypothesize that these transcription factors are part of a pathway that links light and cytokinin signaling with the differentiation of photosynthetically active chloroplasts from proplastids and their proliferation throughout plant development. To characterize these two proteins functionally and to place them into the pathway that mediates chloroplast development, we performed an extensive physiological analysis of both loss- and gain-of-function lines. This approach allowed us to address what aspects of chloroplast biogenesis are modulated by the GNC/CGA1 family. Our study indicates that GNC and CGA1 mediate the development of chloroplasts from proplastids, enhance chloroplast growth and division, and serve as targets for cytokinin regulation of these processes. We propose a model in which GNC and CGA1 serve as key positive regulators of chloroplast biogenesis, integrating both light and cytokinin inputs to mediate chloroplast development, growth, and division.
RESULTS
GNC and CGA1 Are Highly Expressed in the Shoots and Are Regulated by Cytokinin
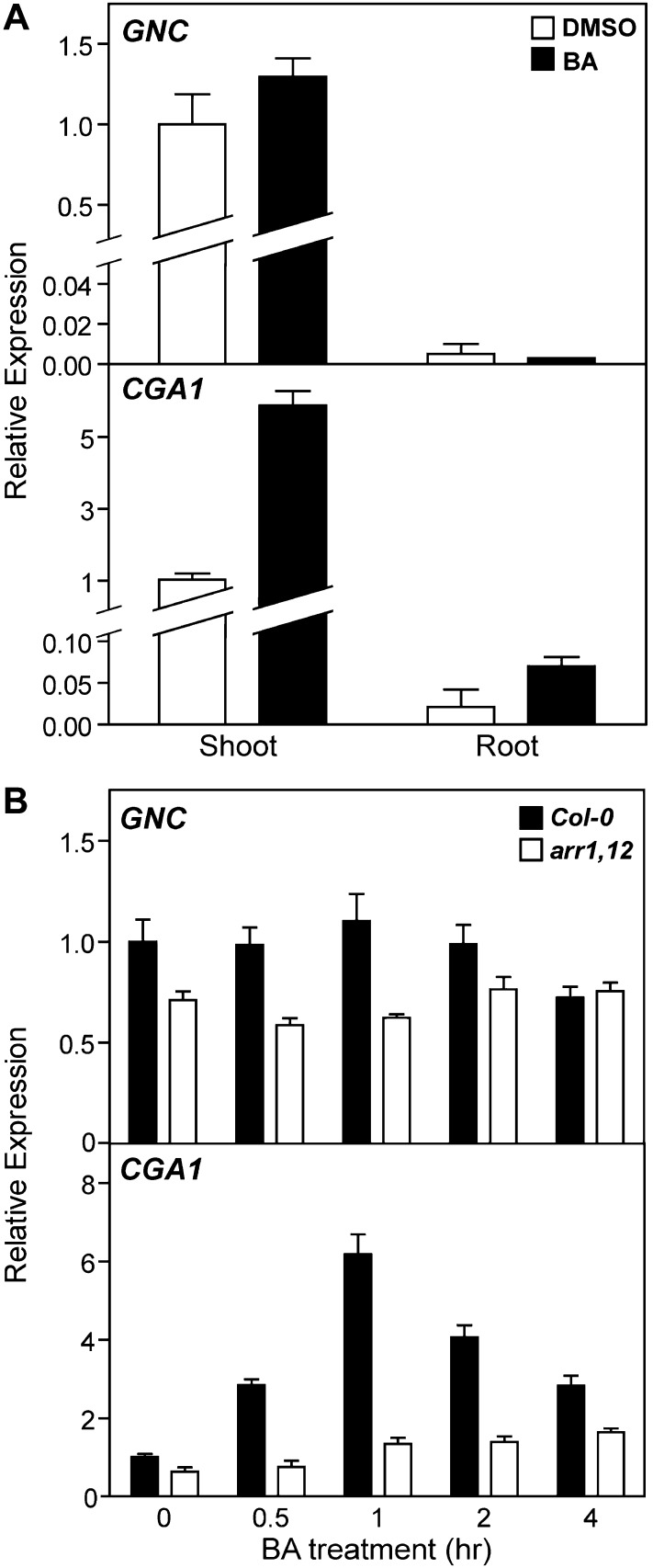
We used real-time quantitative reverse transcription (qRT)-PCR to examine the tissue specificity for expression of GNC and CGA1. The expression of both GNC and CGA1 is substantially higher in the shoot than in the root of light-grown seedlings (Fig. 1A), consistent with a role in greening (Naito et al., 2007; Mara and Irish, 2008; Richter et al., 2010). We also quantified the effect of exogenous cytokinin on the two mRNAs, since CGA1 message is reported to be induced by exogenous cytokinin treatment (Brenner et al., 2005; Kiba et al., 2005; Rashotte et al., 2006; Naito et al., 2007). A 1-h treatment with the cytokinin benzyladenine (BA) induced CGA1 expression 6-fold in the shoot and 3-fold in the root. In contrast, cytokinin had little effect on the expression of GNC in either the shoot or the root.
Figure 1.
Effect of cytokinin on GNC and CGA1 expression in shoots and roots. A, Fourteen-day-old Col-0 seedlings were treated with either 10 µm BA or a DMSO control for 1 h, and the expression of GNC and CGA1 was analyzed in shoots or roots by qRT-PCR. Error bars indicate sd. B, Fourteen-day-old Col-0 and arr1-3 arr12-1 seedlings were treated with 10 µm BA or a DMSO control for the times indicated, and the expression of GNC and CGA1 was analyzed in the shoots excised from the treated seedlings by qRT-PCR. Error bars indicate sd.
The transcriptional response to cytokinin is mediated by the type-B Arabidopsis response regulators (ARRs; Sakai et al., 2001; Mason et al., 2005; Argyros et al., 2008), with ARR1, ARR10, and ARR12 regulating the majority of this transcriptional response (Argyros et al., 2008). To determine if the cytokinin-induced expression of CGA1 requires the type-B ARRs, the arr1-3 arr12-1 double mutant was examined because this mutant exhibits a similar morphology to the wild type but significantly reduced sensitivity to cytokinin (Mason et al., 2005). Wild-type and arr1-3 arr12-1 seedlings were treated with 10 µm BA or a dimethyl sulfoxide (DMSO) vehicle control for up to 4 h, and the expression of both GNC and CGA1 was analyzed in the shoot by qRT-PCR (Fig. 1B). The expression of CGA1 in wild-type seedlings peaked at 1 h after cytokinin treatment and then declined. Cytokinin-induced expression of CGA1 was substantially reduced in the arr1-3 arr12-1 mutant (Fig. 1B), consistent with a similar loss of sensitivity to cytokinin for induction of CGA1 in a cytokinin receptor mutant (Naito et al., 2007). In addition, the basal expression levels of both GNC and CGA1 are decreased in the arr1-3 arr12-1 mutant compared with the wild type, which indicates that endogenous cytokinin levels regulate their expression.
We examined the spatial pattern of expression of GNC and CGA1 using promoter-GUS fusions (Supplemental Fig. S1). In mature embryos at 12 d post anthesis (West and Harada, 1993), the GUS staining of pGNC::GUS was observed in the cotyledons and hypocotyl, whereas the staining of pCGA1::GUS was largely restricted to the cotyledons. The microarray data from the Arabidopsis eFP Browser confirm that both GNC and CGA1 are expressed in the embryonic cotyledons, with a peak at the heart and torpedo stages, respectively, but neither is strongly expressed in roots (Casson et al., 2005; Winter et al., 2007). In 7-d-old seedlings, both pGNC::GUS and pCGA1::GUS plants exhibit staining in shoot tissues, including the tip, circumference, and vasculature of the cotyledons, the emerging leaves, the meristematic region, and the basal part of the hypocotyl. In addition, some weaker staining is found in the primary roots. In 14-d-old seedlings, staining for both GNC and CGA1 became more pronounced in the green shoot tissues, consistent with the strong expression identified in shoots by qRT-PCR (Fig. 1A). Based on the normalized microarray data from the Arabidopsis eFP Browser, both genes have higher expression in the vegetative rosette (GNC, 58.38 ± 5.82; CGA1, 29.5 ± 1.01) than in the roots (GNC, 0.6 ± 0.25; CGA1, 4.31 ± 1.9; Schmid et al., 2005; Winter et al., 2007).
To determine the subcellular localization of GNC and CGA1, the genomic sequences of these genes were translationally fused to GFP and transiently expressed from the cauliflower mosaic virus (CaMV) 35S promoter in Arabidopsis mesophyll protoplasts. In agreement with their roles as transcription factors, both GNC and CGA1 colocalized with a histone-2B-RFP fusion protein in the nucleus (Supplemental Fig. S2).
GNC and CGA1 Regulate Chlorophyll Production
To characterize the functions of GNC and CGA1, transferred DNA (T-DNA) insertion lines were obtained from the Arabidopsis Biological Resource Center. Both gnc (SALK_001778) and cga1 (SALK_003995) homozygous lines resulted in undetectable levels of full-length transcripts (Supplemental Fig. S3B). In agreement with previous reports that gnc and cga1 mutants have defects in greening (Bi et al., 2005; Mara and Irish, 2008; Richter et al., 2010), the gnc single mutant and gnc cga1 double mutant displayed significantly reduced chlorophyll levels in the shoots (Supplemental Fig. S4). The pale-green phenotype of the gnc single mutant and the higher basal levels of GNC transcripts compared with CGA1 transcripts (Naito et al., 2007) suggest that GNC plays the more dominant role in regulating chlorophyll production.
The decreased chlorophyll levels in the gnc cga1 mutant suggest a role of this gene family in a chloroplast function. However, this pale-green phenotype alone does not indicate a normal role in the control of chloroplast development, because loss of any gene critical to chloroplast function (e.g. genes required for the photosynthetic apparatus) will result in reduced chloroplast production and decreased chlorophyll levels (Ishizaki et al., 2005; Ma et al., 2007; Waters and Langdale, 2009). To get direct insight into the function of the GNC/CGA1 family, we expressed GNC and CGA1 as GFP fusion proteins in stable transgenic plants using the CaMV 35S promoter, which results in high-level, ubiquitous expression in Arabidopsis. The lines overexpressing either of these genes exhibited small dark-green cotyledons as well as green hypocotyls and roots, the latter being tissues in which chlorophyll is not produced at high levels in wild-type plants under these growth conditions (Fig. 2; Supplemental Fig. S5). The enhanced greening found in the CaMV 35S-driven GNC and CGA1 lines indicates, even though some expression is normally observed in these tissues based on GUS fusion analysis (Supplemental Fig. S1), that the native expression level of the GNC/CGA1 family is limiting or that there are differences in regulation between the endogenous and ectopically produced transcription factors. Overexpression lines in which the genes were expressed without the GFP tag exhibited similar greening phenotypes, indicating that the fused GFP does not alter the function of these transcription factors (Supplemental Fig. S5). For the remainder of this study, we focused on two independent GNC overexpression lines (OX-1 and OX-2), because the loss-of-function analysis supports a more substantial role for GNC compared with CGA1 in greening. However, CGA1 overexpression lines exhibited similar phenotypes to those found with GNC (Supplemental Figs. S5 and S6).
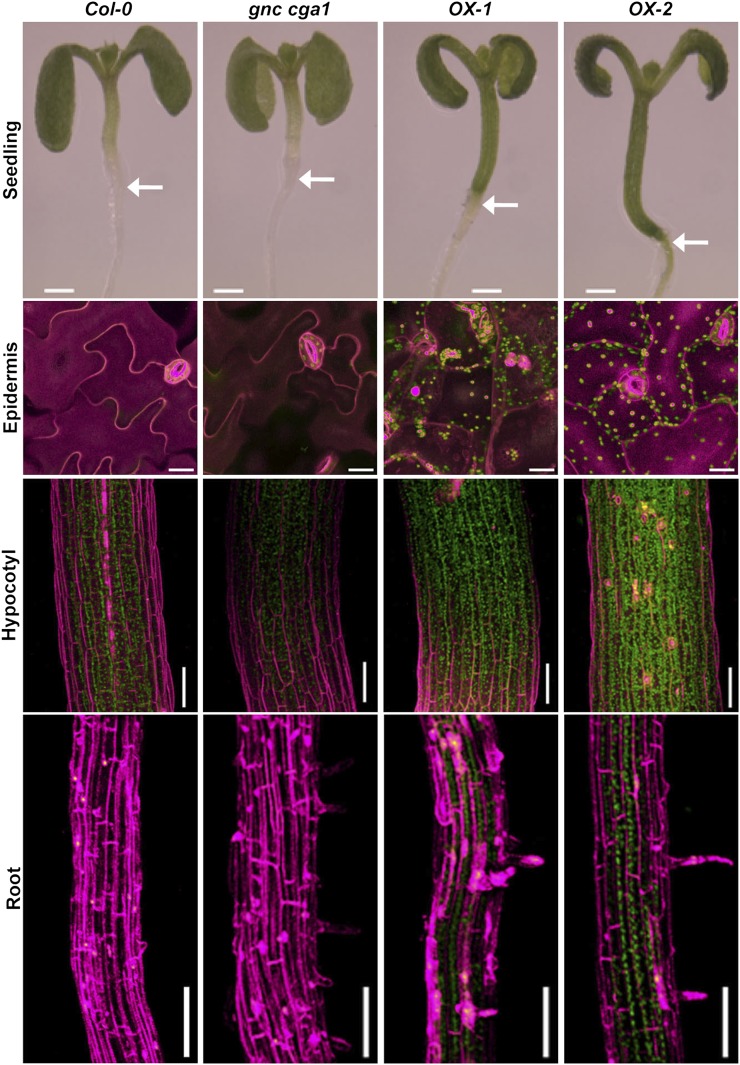
Figure 2.
Overexpression of GNC induces chloroplast biogenesis in the cotyledon epidermis, hypocotyls, and roots. First row, 7-d-old seedlings of Col-0, gnc cga1, and two independent GNC overexpression lines (OX-1 and OX-2) grown under constant white light. Bars = 0.5 mm. Arrows show the hypocotyl-root junction. Second row, detection of chlorophyll autofluorescence in the cotyledon adaxial epidermis of 14-d-old seedlings grown under constant white light. Bars = 20 µm. Third and fourth rows, detection of chlorophyll autofluorescence in the hypocotyls of 7-d-old seedlings (third row) and in the roots of 10-d-old seedlings (fourth row) grown under constant white light. Bars = 0.1 mm. Chlorophyll autofluorescence and cell wall staining by propidium iodide were false colored in green and magenta, respectively.
To further characterize the greening in cotyledons, hypocotyls, and roots, chlorophyll autofluorescence was examined by confocal microscopy. Interestingly, we observed chlorophyll fluorescence in the epidermal pavement cells of cotyledons, a cell type in which chloroplasts are typically not produced (Fig. 2; Supplemental Videos S1 and S2). Furthermore, the normal jigsaw-puzzle shape of cotyledon pavement cells was changed to a more rounded mesophyll-like shape in the overexpression lines, indicating that overexpression of the GNC/CGA1 family may lead to an alteration of cell fate. To our knowledge, these effects from the ectopic expression of GNC on chloroplast production and the shape of pavement cells have not previously been reported. Stronger chlorophyll fluorescence was detected in the hypocotyls of overexpression lines compared with those of the wild type (Fig. 2). In contrast, the gnc cga1 double mutant exhibited weaker chlorophyll fluorescence than the wild type (Fig. 2). We also observed that the hypocotyls of overexpression lines were longer compared with those of the wild type and the gnc cga1 double mutant (Fig. 2). Significantly, the overexpression lines exhibited chlorophyll fluorescence in the roots, whereas no fluorescence was detected in wild-type and gnc cga1 double mutant roots (Fig. 2), consistent with chlorophyll biosynthesis normally being repressed in the roots under these growth conditions. In all tissues and cells examined, the chlorophyll fluorescence was punctate in appearance, consistent with its localization to chloroplasts.
Ectopic Overexpression of GNC or CGA1 Increases Chloroplast Biogenesis in the Hypocotyl
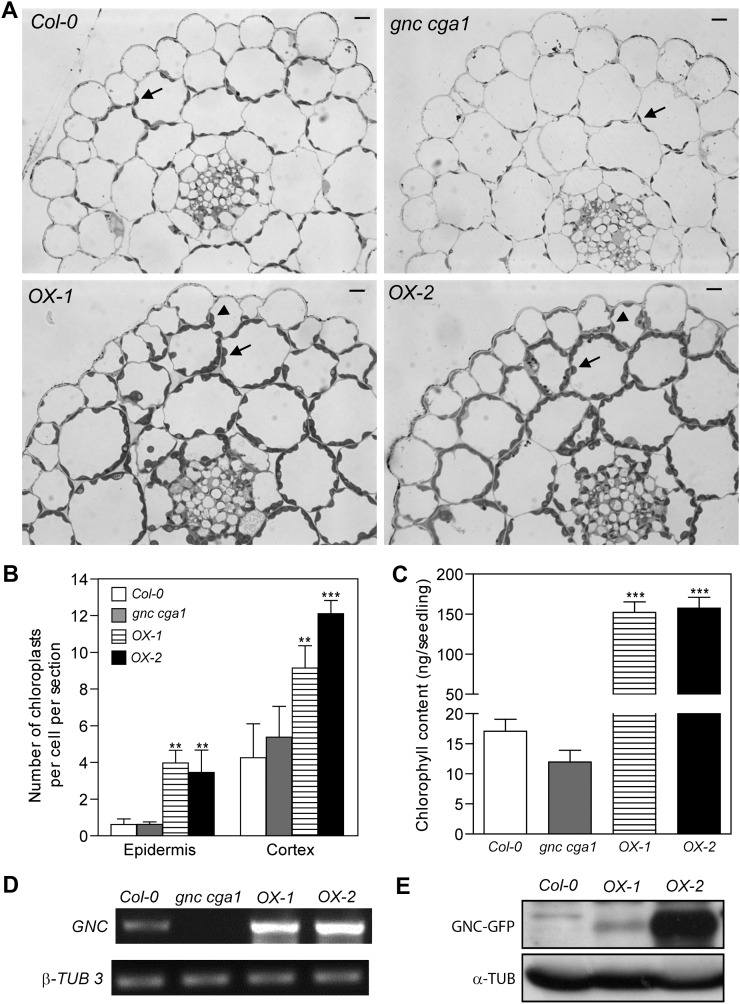
To determine whether the overexpression of GNC induces the biogenesis of functional chloroplasts, we examined the number and distribution of plastids in cross-sections of hypocotyls. We found two key differences between the overexpression lines and the wild type. First, chloroplast biogenesis in the cortical cell layers of the overexpression lines is significantly enhanced (Fig. 3, A and B). Quantification revealed that the chloroplast number per cell was increased more than 2-fold in the cortical cells of overexpression lines compared with the wild type or the gnc cga1 double mutant. The largest number of chloroplasts was found in the line OX-2 (Fig. 3B), which has the highest level of GNC protein (Fig. 3E), even though the transcript levels of GNC in the lines OX-1 and OX-2 were similar (Fig. 3D). There was no significant difference between the wild type and the gnc cga1 double mutant in the number of chloroplasts in hypocotyl cortical cells (Fig. 3B). These results are consistent with the analysis of chlorophyll content in the hypocotyls (Fig. 3C), in which the overexpression lines exhibited a 10-fold increase in chlorophyll levels compared with the wild type, but there was no significant difference in chlorophyll levels between the wild type and the gnc cga1 double mutant.
Figure 3.
Overexpression of GNC results in increased chloroplast number and expanded zones of chloroplast production. A, Cross-sections of fixed hypocotyls of 7-d-old Col-0, gnc cga1 mutant, OX-1, and OX-2 seedlings grown under constant white light. Bars = 0.01 mm. Arrows indicate chloroplasts in the cortical cells. Arrowheads indicate chloroplasts in the epidermal cells. B, Quantification of chloroplasts in all epidermal cells or outer cortical cells of hypocotyl cross-sections. Cells from three individual hypocotyls of each line were quantified. Error bars indicate sd (Dunnett’s multiple comparison test was performed by using Col-0 of each condition as a reference group after one-way ANOVA [**P < 0.01, ***P < 0.001]). C, Chlorophyll content of hypocotyls from 7-d-old seedlings grown under 100 µmol m−2 s−1 constant white light. Each sample represents the mean of seven replicates of 10 hypocotyls each. Error bars indicate se (Dunnett’s multiple comparison test was performed by using Col-0 as a reference group after one-way ANOVA [***P < 0.001]). D, RT-PCR and immunoblot analyses showing the levels of GNC transcript and protein. Total RNA extracted from 10-d-old seedlings was used for RT-PCR to examine the transcript of GNC. β-TUBULIN3 (β-TUB 3) was used as the internal control. E, Total protein extracted from 10-d-old seedlings was analyzed by anti-GFP antibodies. α-Tubulin as a loading control was detected by anti-tubulin antibodies.
The second difference between the overexpression lines and the wild type was in the production of chloroplasts in the hypocotyl epidermal cells. In the wild type and the gnc cga1 double mutant, the average number in a cross-section is about 0.6 chloroplasts per epidermal cell (Fig. 3B), consistent with epidermal cells not normally producing fully developed chloroplasts (Dupree et al., 1991). In contrast, overexpression of GNC results in enhanced chloroplast biogenesis, with about four chloroplasts per epidermal cell in a cross-section, representing an increase of greater than 6-fold. The ectopic production of chloroplasts in the hypocotyl epidermis is consistent with what we observed in the cotyledon pavement cells of the overexpression lines (Fig. 2) and indicates that ectopic expression of GNC expands the zones of chloroplast production. Cross-sections of hypocotyls from CGA1 overexpression lines revealed similar effects on the distribution and number of chloroplasts in cortical and epidermal cells to those seen in the GNC overexpression lines (Supplemental Fig. S6).
GNC and CGA1 Positively Regulate Chloroplast Growth in the Hypocotyl
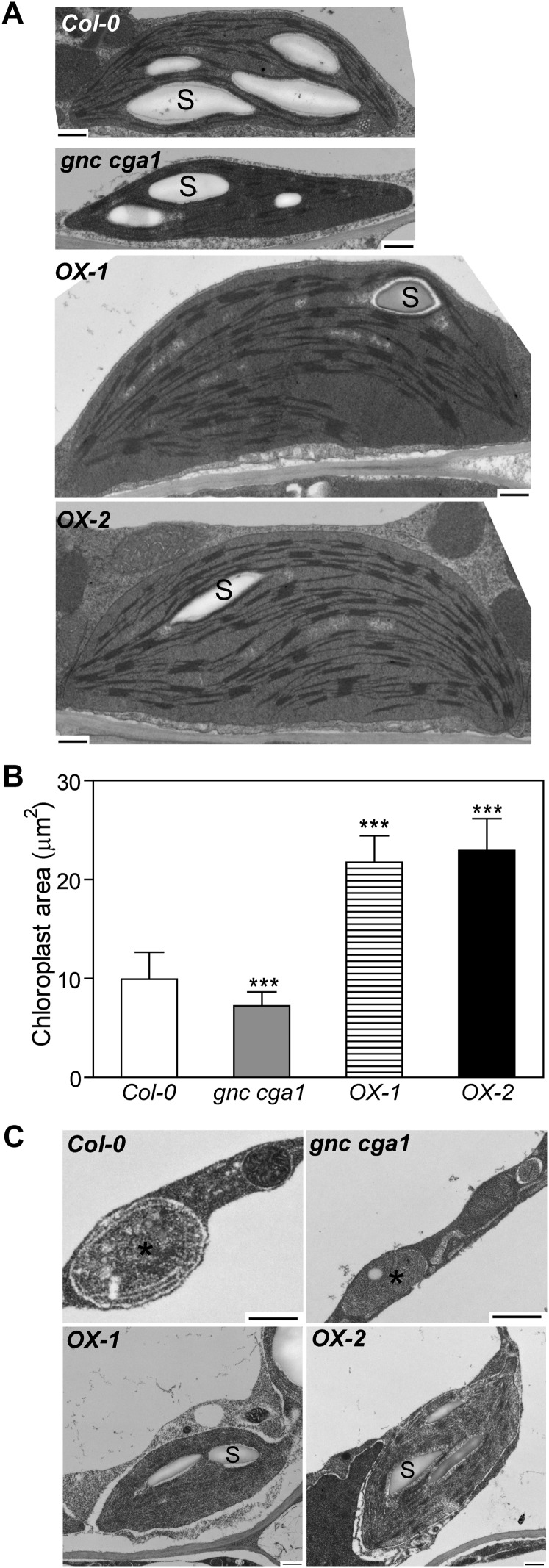
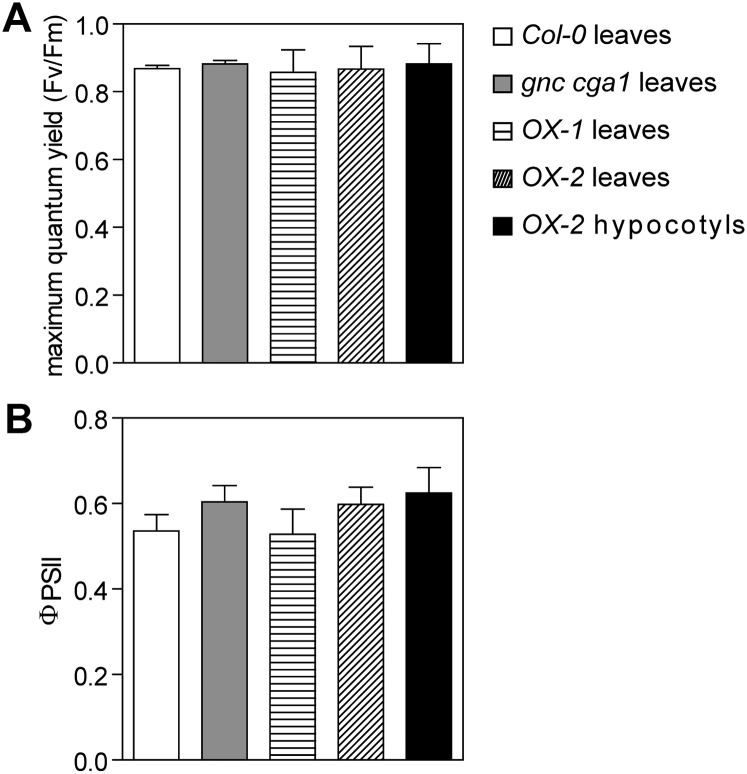
We examined the ultrastructure of chloroplasts in the hypocotyls by transmission electron microscopy (TEM). Interestingly, in spite of there being no significant difference in hypocotyl chloroplast number between the wild type and the gnc cga1 double mutant (Fig. 3B), the size of chloroplasts differs (Fig. 4, A and B). The cross area of chloroplasts in the cortical cells showed a 27% reduction in the gnc cga1 double mutant, whereas chloroplasts in the overexpression lines were over 2-fold larger than those in the wild type (Fig. 4B). It should be noted, however, that the chloroplasts in the hypocotyls of overexpression lines are not larger than those in the wild-type leaf (5–10 µm in diameter; López-Juez and Pyke, 2005), suggesting that GNC overexpression enhanced chloroplast growth in the hypocotyls where chloroplast development is normally retarded. These results demonstrate that GNC and CGA1 positively regulate chloroplast growth. In spite of the smaller chloroplasts in the gnc cga1 double mutant, no obvious defects of the thylakoid membranes or grana were observed (Fig. 4A). In addition, chloroplasts from the gnc cga1 double mutant and overexpression lines produced starch granules resembling those of wild-type chloroplasts (Fig. 4A). Photosynthetic efficiency analysis showed that the maximum quantum yield of PSII (Fv/Fm) and the flux of electrons through PSII (ΦPSII) in the hypocotyls of overexpression lines are similar to those in the leaves of the wild type, the gnc cga1 double mutant, and the overexpression lines, indicating that the chloroplasts from all these tissues exhibit normal photosynthetic activity (Fig. 5; Supplemental Fig. S7).
Figure 4.
GNC promotes chloroplast development in the hypocotyls and roots. A, Representative chloroplasts from hypocotyls of Col-0, gnc cga1, OX-1, and OX-2 seedlings grown under constant white light for 7 d. B, Chloroplast size of cortical cells in hypocotyls. The quantification was performed on 20 chloroplasts in the cortical cells from each hypocotyl. Three individual hypocotyls of each line were quantified. Error bars indicate sd (Dunnett’s multiple comparison test was performed by using Col-0 as a reference group after one-way ANOVA [n = 60; ***P < 0.001]). C, Ectopic chloroplast production in the roots overexpressing GNC. Proplastids are from Col-0 or gnc cga1 roots, and chloroplasts are from the roots of OX-1 and OX-2. Bars = 500 nm. S, Starch granules. Asterisks indicate plastids.
Figure 5.
Chloroplasts from the gnc cga1 mutant and GNC overexpression lines exhibit normal photosynthetic activity. A and B, Seedlings were grown in the dark for 3.5 d to elongate the hypocotyls and then transferred to a 16-h-light/8-h-dark cycle for 7 d. Fv/Fm (A) and ΦPSII (B) were measured at an actinic light intensity of 150 µE. Only OX-2 gives a strong enough signal to measure the photosynthetic parameters in the hypocotyls. Error bars indicate sd (n ≥ 20 for leaves and n = 11 for hypocotyls).
GNC Overexpression Promotes Chloroplast Biogenesis in the Root
Because the overexpression lines exhibit chlorophyll fluorescence in the roots, we examined the distribution of plastids in root cross-sections. The most pronounced difference between the GNC overexpression lines as compared with the wild type and the gnc cga1 mutant was in the proliferation of chloroplasts in the pericycle and cortex. The ability of the root pericycle to produce chloroplasts has been noted previously (López-Juez, 2007); we observed a few chloroplasts in the wild-type and gnc cga1 mutant pericycle, but these were reduced in number, smaller, and with fewer thylakoid membranes than found in the overexpression lines. Interestingly, we did not observe any proliferation of chloroplasts within the endodermis following overexpression of GNC, although chloroplasts were occasionally noted in the wild-type endodermis (one or two chloroplasts per cross-section). The size of the root cortical cells allowed for accurate quantification of chloroplast production. No chloroplasts were observed in the cortical cells of the wild type or the gnc cga1 mutant; in contrast, the GNC overexpression lines OX-1 and OX-2 contained 4.22 (sd = 1.58) and 4.54 (sd = 1.44) chloroplasts per cortical cell, respectively (n = 50).
We examined the ultrastructure of plastids in roots by TEM to shed light on the mode of GNC action in chloroplast biogenesis. Proplastids, between 0.2 and 1 µm in diameter with very limited membrane structures, were observed in the cortex of wild-type and gnc cga1 double mutant roots (Fig. 4C). As proplastids are colorless and nonphotosynthetic, these results are consistent with the lack of chlorophyll fluorescence in the roots of the wild type and the gnc cga1 double mutant (Fig. 2). However, most strikingly, regular lens-shaped chloroplasts with thylakoids and starch granules developed in the roots of the overexpression lines. The values of Fv/Fm and ΦPSII in the roots of the overexpression line OX-2 (Fv/Fm = 0.874 ± 0.008; ΦPSII = 0.651 ± 0.02) were similar to those in the leaves of the wild type (Fv/Fm = 0.914 ± 0.005; ΦPSII = 0.769 ± 0.025), supporting their functionality. These results suggest that overexpression of GNC bypasses the signals that retard chloroplast biogenesis in the roots.
GNC Overexpression Enhances Chloroplast Development in the Dark
Light is normally a prerequisite for chlorophyll biosynthesis and the completion of chloroplast development (Armstrong et al., 1995; Waters and Langdale, 2009). To determine whether overexpression of GNC enhances chloroplast development in the dark, we examined plastid morphology in the hypocotyls of dark-grown seedlings by TEM. Hypocotyls were used, as we observed pronounced effects of GNC on chloroplast production in this tissue. In 7-d-old dark-grown seedlings, proplastids were not yet differentiated into plastids either in wild-type or in gnc cga1 double mutant hypocotyls (Fig. 6A). In contrast, most plastids in the hypocotyls of the overexpression lines had transformed into etioplasts, with their distinctive prolamellar bodies and prothylakoids (Fig. 6A).
Figure 6.
Overexpression of GNC promotes chloroplast development in dark-grown hypocotyls and enhances the deetiolation process when plants are exposed to light. A, Chloroplast development in dark-grown hypocotyls. Cross-sections of hypocotyls from 7-d-old dark-grown seedlings were examined by TEM. Plastids from Col-0, the gnc cga1 mutant, and OX-1 are shown. M, Mitochondria; P, proplastids; PLB, prolamellar body in an etioplast. Bars = 500 nm. B, Overexpression of GNC enhances deetiolation. Seven-day-old dark-grown seedlings were transferred to 50 µmol m−2 s−1 white light for the times indicated. Bars = 0.1 cm. C, Chlorophyll content of mutant lines following 6 h of deetiolation. Seedlings were grown in the dark for 5 d and then illuminated for 6 h with 100 µmol m−2 s−1 white light before measuring chlorophyll content. Four replicates with 30 seedlings each were used for each line. Error bars represent sd. Dunnett’s multiple comparison test was performed by using Col-0 of each condition as a reference group after one-way ANOVA (*P < 0.05, ***P < 0.001). FW, Fresh weight.
To examine whether the etioplasts in the overexpression lines are readily converted to mature chloroplasts, we performed a light-induced deetiolation experiment. For this purpose, 7-d-old dark-grown seedlings were transferred to white light for different periods of time. Before illumination, the seedlings displayed etiolated phenotypes with closed yellow cotyledons and elongated hypocotyls (Fig. 6B). After 24 h of light exposure, wild-type seedlings showed yellow cotyledons with newly emerging green true leaves, while the gnc cga1 double mutant only exhibited yellow cotyledons. Interestingly, unlike the wild type and the gnc cga1 double mutant, the overexpression lines exhibited expanded green cotyledons with newly emerging green true leaves within 24 h after illumination. The strongest response was found in OX-2, with the highest GNC protein level.
To determine the immediate effect of the loss- and gain-of-function mutations on deetiolation, dark-grown seedlings were moved to light for 6 h and deetiolation was assessed by measuring chlorophyll levels in the seedlings (Fig. 6C). The gnc cga1 mutant exhibited reduced chlorophyll levels compared with the wild type. Significantly, the overexpression lines exhibited higher accumulation of chlorophyll than the wild type. These results suggest that overexpression of GNC enhances greening by promoting the development of etioplasts from proplastids in the dark and that these etioplasts are promptly converted to chloroplasts upon illumination.
Increases in Chloroplast Biogenesis Can Be Regulated Postembryonically
Many of the greening phenotypes we and others have associated with ectopic expression of the GNC/CGA1 family occur in the cotyledon, hypocotyl, and root (Richter et al., 2010; Hudson et al., 2011; Köllmer et al., 2011), all of which are embryonically derived. This is of significance because chloroplasts are present in these tissues during the early stages of embryo development, including being found within the epidermal cell layer (Tejos et al., 2010). During later stages of embryo development, chloroplasts undergo dedifferentiation to form basal-state plastids as seeds develop. These basal-state plastids can then redifferentiate to chloroplasts or other plastids upon germination, suggesting that postembryonic factors in concert with embryonic factors regulate chloroplast differentiation during plant development (Whatley, 1978; Zhao and Sack, 1999). Thus, a possible explanation for the increased levels and expanded zones of chloroplast production is that overexpression of the GNC/CGA1 family prevents the chloroplasts in the embryos from the dedifferentiation or increases levels of some embryonically derived factor such that plastids are more likely to take on a chloroplast identity following germination.
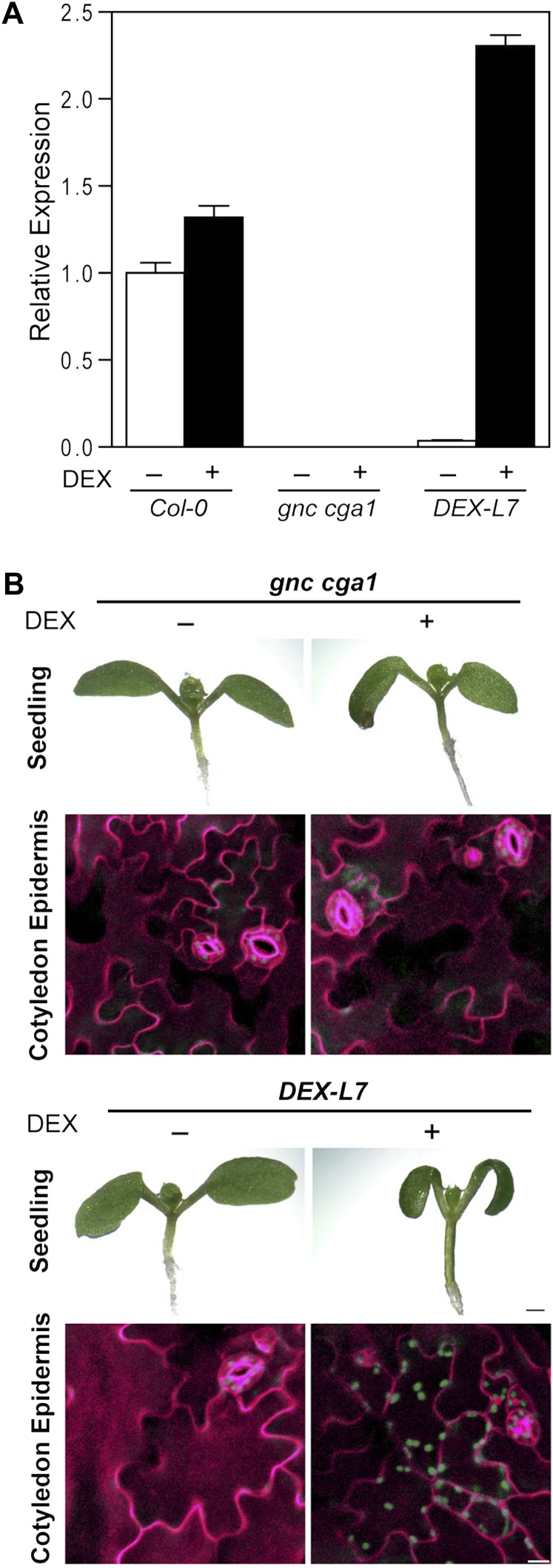
To determine whether the striking phenotypes in the embryonically derived tissues require the expression of GNC during embryogenesis, we generated transgenic lines in a gnc cga1 mutant background in which the expression of GNC can be induced by the application of a steroid hormone dexamethasone (DEX; Aoyama and Chua, 1997). The induction of GNC by DEX treatment was confirmed by a 4-h induction period with 10 µm DEX, following which we observed induction of GNC in the DEX-inducible line (DEX-L7) to twice the wild-type level; no detectable expression of GNC occurred in the absence of DEX treatment (Fig. 7A). To examine chloroplast production, DEX-inducible GNC seedlings were grown on the plates containing 1 µm DEX or a DMSO vehicle control under constant white light for 7 d. DEX-L7 seedlings exhibited long green hypocotyls only when DEX was supplemented on the plates (Fig. 7B), the same phenotype we observed when GNC was ectopically expressed (Fig. 2). Significantly, we also observed chlorophyll fluorescence in the epidermal pavement cells of cotyledons of DEX-L7 grown in the presence of DEX (Fig. 7B). In contrast, the DEX-L7 line grown in the absence of DEX as well as the gnc cga1 mutant control exhibited no chlorophyll fluorescence in the epidermal pavement cells of cotyledons (Fig. 7B). These results indicate that GNC can play a postembryonic role in the development of chloroplasts, even in those tissues that are derived from the embryo.
Figure 7.
Inducible expression of GNC simulates postembryonic chloroplast production in seedlings. A, GNC expression following 10 μm DEX induction for 4 h as determined by qRT-PCR. Expression was determined for Col-0, the gnc cga1 mutant, and a gnc cga1 line carrying DEX-inducible GNC (DEX-L7). Error bars indicate sd. B, Effect of GNC induction on chloroplast biogenesis. Seven-day-old seedlings of the gnc cga1 mutant and DEX-L7 were grown in the presence or absence of 1 µm DEX under constant white light. Bar = 0.5 mm. Detection of chlorophyll autofluorescence in the cotyledon abaxial epidermis of 7-d-old seedlings is also shown. Chlorophyll autofluorescence and cell wall staining by propidium iodide were false colored in green and magenta, respectively. Bar = 20 µm.
Effect of the GNC/CGA1 Family on Chlorophyll Levels, Chloroplast Size, and Chloroplast Number in Cotyledons and Leaves
The cotyledons, hypocotyls, and roots of seedlings overexpressing GNC or CGA1 were darker green than those of the wild type, but the cotyledons of the overexpression lines were smaller than those of the wild type (Fig. 2; Supplemental Fig. S5), raising the question of whether the total pigment content of the cotyledons actually differs between the mutant and wild-type lines. Therefore, we measured the levels of chlorophyll a in isolated cotyledons and normalized the chlorophyll measurement per cotyledon and per cotyledon area (Supplemental Fig. S8). Based on this analysis, chlorophyll levels are reduced in the gnc cga1 mutant and increased in the GNC overexpression lines, indicating an increased level of chlorophyll production in the overexpression lines that is independent of the cotyledon size. Analysis of chloroplast size and number per cell in mesophyll cells from cotyledons of the mutant lines indicated that the gnc cga1 mutant is similar to the wild type but that the GNC overexpression lines exhibit a decrease in chloroplast area with a compensating increase in the chloroplast number per cell (Fig. 8, A and B).
Figure 8.
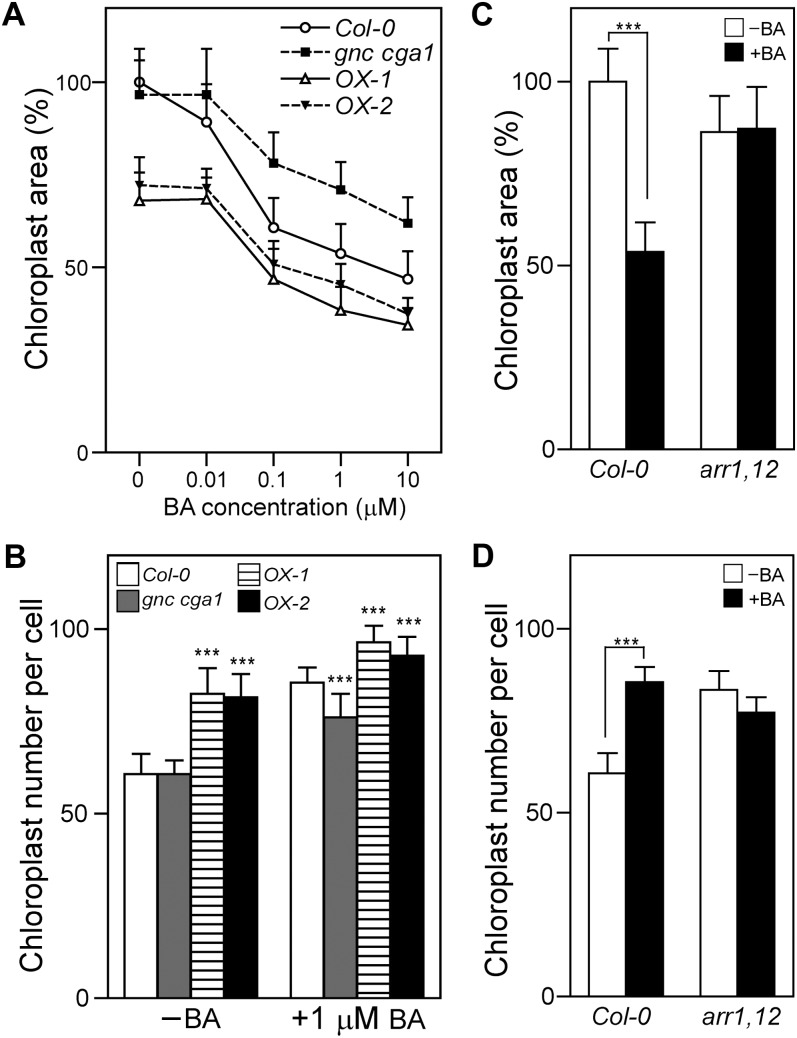
Effects of cytokinin on chloroplast division are altered in GNC/CGA1 and type B ARR mutants. A and B, Cytokinin effects on chloroplast size and number are altered in the GNC/CGA1 mutants. Seedlings were grown under white light in the presence of the indicated cytokinin concentrations for 11 d. Chloroplast size (A) and number (B) in the cotyledons of Col-0, the gnc cga1 mutant, OX-1, and OX-2 were determined. Error bars represent sd. Dunnett’s multiple comparison test was performed by using Col-0 of each condition as a reference group after one-way ANOVA (***P < 0.001). C and D, Cytokinin effects on chloroplast size and number are reduced in the arr1-3 arr12-1 (arr1,12) mutant. Chloroplast size (C) and number (D) in the cotyledons of Col-0 and the arr1-3 arr12-1 mutant grown in the presence of 1 µm cytokinin or a DMSO vehicle control for 11 d were determined. A Bonferroni test was performed after two-way ANOVA (***P < 0.001). For chloroplast size, 100 chloroplasts from seven seedlings were analyzed for each line, with average area normalized to the DMSO-treated wild-type control. For chloroplast number, chloroplasts of 30 cells from five seedlings were quantified for each line.
Both loss- and gain-of-function mutations in the GNC/CGA1 family negatively impact plant growth, based on the reduction in biomass of 5-week-old plants (Supplemental Fig. S9A), these results for the gnc cga1 mutant and the GNC overexpression lines being consistent with results from Mara and Irish (2008) and Richter et al. (2010), respectively. Leaves of the gnc cga1 mutant exhibit a decrease in levels of chlorophyll; conversely, the strongly expressing GNC overexpression line OX-2 exhibits an increase in levels of chlorophyll, although this is only apparent when normalized to leaf fresh weight (Supplemental Fig. S9, B and C). The loss- and gain-of-function mutants also affect cell size and chloroplast size (Supplemental Fig. S10). Both mesophyll cell area and chloroplast area are significantly increased in the gnc cga1 mutant compared with the wild type (Supplemental Fig. S10, B and C). Conversely, both mesophyll cell area and chloroplast area are significantly decreased in both the GNC overexpression lines (Supplemental Fig. S10, B and C). Even though both mesophyll cell area and chloroplast area are affected in the mutants, the actual number of chloroplasts per cell does not change due to the similar direction of changes (Supplemental Fig. S10). The effects of GNC/CGA1 on cell expansion are likely related to their role in the repression of GA signaling (Richter et al., 2010), while their effects on chloroplast size suggest a role in the regulation of chloroplast division (Okazaki et al., 2009). The effect of GNC on chloroplast size and division provides an explanation, at least in part, for the greater number of chloroplasts reported in leaves of GNC overexpression lines (Hudson et al., 2011). Chloroplast autofluorescence was decreased in the gnc cga1 mutant and increased in the strongly expressing GNC overexpression line OX-2 compared with the wild type, consistent with the results from Hudson et al. (2011), providing a basis for the altered chlorophyll levels observed in leaves (Supplemental Fig. S9). Chloroplasts were observed in the epidermal pavement cells of leaves when GNC was ectopically expressed from either the CaMV 35S promoter or following induction from a DEX-inducible promoter, although at substantially reduced levels compared with what we observed in the epidermal pavement cells of cotyledons (Fig. 2).
Effect of Cytokinin on Chloroplast Division in Mutants of the GNC/CGA1 Family
Cytokinin stimulates chloroplast division in cotyledons, resulting in a smaller size but increased numbers of chloroplasts (Okazaki et al., 2009). Therefore, we examined the effects of cytokinin on chloroplast size and number in the wild type, the gnc cga1 mutant, and the GNC overexpression lines (Fig. 8, A and B). Growth of the wild type with exogenous BA results in a reduction in chloroplast size, such that at 10 µm BA the chloroplast cross-sectional area in the wild type is approximately 50% of that found in untreated seedlings (Fig. 8A). The chloroplast size of the gnc cga1 mutant is similar to that of the wild type in the untreated sample but exhibits reduced sensitivity to cytokinin, resulting in significantly larger chloroplasts than the wild type at all cytokinin concentrations examined (Fig. 8A; P < 0.001 based on Dunnett’s multiple comparison test). Overexpression of GNC results in a significantly smaller chloroplast size in the untreated sample and a significantly enhanced reduction in size in all the cytokinin-treated samples (Fig. 8A; P < 0.001 based on Dunnett’s multiple comparison test). We also examined the cytokinin effect on chloroplast number per cell. Growth of the wild type with 1 µm BA results in a 1.41-fold increase in the chloroplast number per cell (Fig. 8B), consistent with a cytokinin effect on chloroplast division (Okazaki et al., 2009). Although the chloroplast number per cell of the gnc cga1 mutant is similar to that of the wild type in the untreated sample, it exhibits reduced sensitivity to cytokinin, resulting in significantly lower numbers of chloroplasts than the wild type in the presence of 1 µm BA (1.25-fold cytokinin-dependent increase in chloroplast number per cell for gnc cga1 compared with 1.41-fold increase for the wild type; Fig. 8B). Furthermore, overexpression of GNC results in an increase of chloroplast number per cell in the untreated sample (1.34-fold higher numbers of chloroplasts per cell for both OX-1 and OX-2 compared with the wild type) and an enhanced increase in numbers in the cytokinin-treated samples (1.13- and 1.08-fold higher numbers of chloroplasts per cell for OX-1 and OX-2, respectively, compared with the wild type; Fig. 8B). These results are all consistent with the GNC/CGA1 family playing a positive role in mediating the effect of cytokinin on chloroplast division.
The expression of CGA1 and potentially also of GNC is positively regulated by cytokinin through action of the type B ARRs, in particular ARR1 and ARR12 (Fig. 1). Therefore, we examined whether the cytokinin-mediated regulation of chloroplast division also requires ARR1 and ARR12 by examining the cytokinin responsiveness of the arr1-3 arr12-1 double mutant for this phenotype (Fig. 8, C and D). Whereas wild-type chloroplasts decrease in size and increase in number when grown with 1 µm BA, the arr1-3 arr12-1 mutant exhibits no difference in chloroplast size or number following cytokinin treatment, indicating that the type B ARRs are required for cytokinin effects on chloroplast division. These results support a general role for the GNC/CGA1 family in chloroplast division and, more specifically, indicate that cytokinin mediates its effects on chloroplast division through GNC and CGA1 in a type B ARR-dependent manner. The finding that the gnc cga1 mutant still exhibits some response to cytokinin indicates that other factors, such as the CRF family of transcription factors (Okazaki et al., 2009), functionally overlap with the GNC/CGA1 family in this response.
DISCUSSION
In this study, we characterized the role of GNC and CGA1 in plant development, focusing on several aspects of chloroplast biogenesis. Our study contributes novel evidence supporting roles for GNC and CGA1 in mediating the development of chloroplasts from proplastids, in enhancing chloroplast growth and division, and in serving as targets for cytokinin regulation of these processes. Below we discuss our results within the context of (1) chloroplast biogenesis, (2) cytokinin signaling, and (3) the overall transcriptional control of chloroplast development. A model situating the GNC/CGA1 family within this regulatory context is given in Figure 9.
Figure 9.
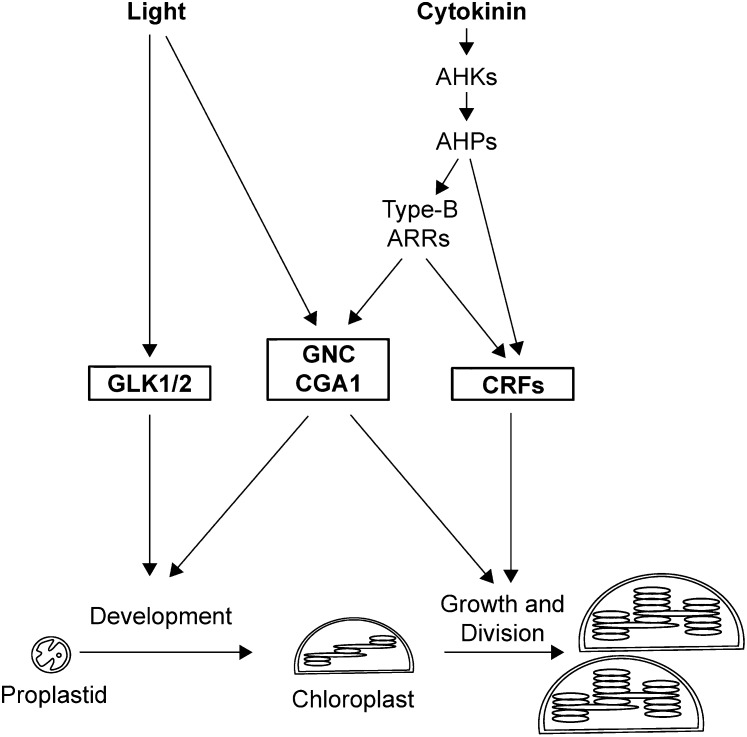
Model for the roles of GNC and CGA1 in the transcriptional regulation of chloroplast biogenesis. GNC and CGA1 are positive regulators of chloroplast development, growth, and division. The GLK and CRF families of transcription factors also play roles in these processes. Light and cytokinin induce the expression of these transcription factors as indicated. Cytokinin acts through a signaling pathway that involves cytokinin receptors (AHKs), His-containing phosphotransfer proteins (AHPs), and the type B ARR family of transcription factors.
Key Role of the GNC/CGA1 Family in Chloroplast Biogenesis
Our results demonstrate that the GNC/CGA1 family regulates multiple aspects of chloroplast biogenesis. In particular, the GNC/CGA1 family regulates chloroplast development from proplastids and, once chloroplasts are formed, their proliferation through growth and division (Fig. 9). A role in chloroplast development is supported by the finding that ectopic overexpression of the GNC/CGA1 family induces chloroplast production in hypocotyls and roots and, perhaps most significantly, expands chloroplast production to the epidermis of the hypocotyls and the epidermal pavement cells of the cotyledons. The presence of chloroplasts in the epidermal cells is not simply a holdover of embryonically derived chloroplasts (Tejos et al., 2010), because controlled induction of GNC in germinating seedlings was able to recapitulate this phenotype. We also observed that the shape of cotyledon pavement cells in the overexpression lines was more rounded and mesophyll like, suggesting that their cell fate may be altered by the overexpression of the GNC/CGA1 family at the stage of embryogenesis. Alternatively, a retrograde signal from the ectopic production of chloroplasts may alter the cell shape and/or cell fate. The accelerated deetiolation of the overexpression lines, including accelerated leaf emergence, could be separate manifestations of either phenomenon.
Overexpression of the GNC/CGA1 family phenocopies photomorphogenic mutants in the ability to promote the development of chloroplasts from proplastids. For example, like the GNC/CGA1 overexpression lines, the constitutive photomorphogenic mutants cop1 and cop9 contain pseudochloroplasts with partially formed thylakoid membranes under dark-grown conditions (Deng and Quail, 1992; Wei and Deng, 1992). Furthermore, like the GNC/CGA1 overexpression lines, the cop1 and det1 mutants also result in ectopic production of chloroplasts in the roots (Chory and Peto, 1990; Deng and Quail, 1992; Wei and Deng, 1992). These results suggest that the GNC/CGA1 family acts downstream of or in parallel with the COP1 complex in regulating chloroplast development. According to this model, overexpression of the GNC/CGA1 family overrides the repressive activity of the COP1 complex and promotes chloroplast biogenesis and photomorphogenesis. It is possible that activation of the GNC/CGA1 family circumvents other factors and pathways that repress chloroplast biogenesis or photomorphogenesis, one possibility being the PHYTOCHROME-INTERACTING FACTORS (PIFs). PIFs repress chloroplast development and photomorphogenesis (Leivar et al., 2008; Shin et al., 2009; Stephenson et al., 2009), and PIF3 was recently found to negatively regulate the expression of the GNC/CGA1 family by direct binding to their promoters (Richter et al., 2010). Thus, increased expression of the GNC/CGA1 family may counteract the repressive activity of PIFs on chloroplast development and photomorphogenesis.
Although overexpression of GNC and CGA1 induces chloroplast development in multiple cell types and tissues, the effect was not universal, being noticeably absent in flower petals. Petal cells of flowers contain chromoplasts differentiated from proplastids or chloroplasts, so the possibility for initiation of the pathway for converting proplastids to chloroplasts exists (Weston and Pyke, 1999). In addition, expression of the GNC/CGA1 family is suppressed in the floral tissue by APETALA3 (AP3) and PISTILLATA (PI) suggesting that the repression of chloroplast biogenesis in petals involves reducing transcription from the GNC/CGA1 family (Mara and Irish, 2008). However, we did not observe greening in the petals of overexpression lines, indicating that other factors required for greening are absent or that the activity of the GNC/CGA1 family is regulated posttranscriptionally.
The GNC/CGA1 family not only regulates the development of chloroplasts from proplastids but also, once the chloroplasts are formed, regulates their growth and division. A role in chloroplast growth is supported by the smaller chloroplasts found in hypocotyl cortex cells of the gnc cga1 mutant compared with the wild type, even though the absolute number of chloroplasts did not differ in the cells. Also consistent with an effect on growth is the finding that overexpression of GNC and CGA1 results in the opposite phenotype, chloroplasts of the hypocotyl cortex cells being larger than those found in the wild type. A role in chloroplast division is supported by our analysis of cytokinin-regulated chloroplast division in cotyledons. Evidence that division is altered in other tissues comes from our finding that overexpression of GNC and CGA1 results in increased numbers of chloroplasts in epidermal and cortical cells of the hypocotyl as well as the finding that GNC/CGA1 mutants alter the numbers of chloroplasts in leaves (Hudson et al., 2011). Chloroplasts multiply by binary fission (Possingham and Lawrence, 1983; Kuroiwa et al., 1998), and thus an increase in chloroplast number indicates an acceleration of chloroplast division.
Role of the GNC/CGA1 Family in Cytokinin Signaling
There is a long history of research implicating cytokinins in the control of chloroplast development and maintenance. For example, cytokinins induce chloroplast maturation in tobacco tissue cultures (Stetler and Laetsch, 1965), induce regreening of senescent Nicotiana spp. leaves (Zavaleta-Mancera et al., 1999a, 1999b), promote chloroplast differentiation from proplastids (Khokhlova, 1977; Longo et al., 1979; Chory et al., 1994), regulate the biosynthesis of chloroplast proteins as well as pigments (Zavaleta-Mancera et al., 1999a, 1999b; Yaronskaya et al., 2006), and enhance chloroplast division (Okazaki et al., 2009). Genetic analysis of the cytokinin signal transduction pathway is also consistent with cytokinin regulating chloroplast development, because mutation of the cytokinin receptors (ahk2 ahk3 ahk4 triple mutant) or the type B ARRs (arr1 arr10 arr12 triple mutant) results in reduced chlorophyll levels (Riefler et al., 2006; Argyros et al., 2008). In spite of the significance of cytokinin for chloroplast biogenesis, the molecular mechanism of its action on chloroplasts is largely unknown. Our results suggest that the GNC/CGA1 family serves as one of the transcriptional outputs by which cytokinin modulates chloroplast development (Fig. 9).
GNC was first identified based on its transcriptional induction by nitrate (Wang et al., 2003; Price et al., 2004; Scheible et al., 2004; Bi et al., 2005), which in retrospect provided a first link with cytokinin because of the well-established role for nitrogen in regulating cytokinin biosynthesis and translocation (Samuelson and Larsson, 1993; Takei et al., 2001, 2004; Miyawaki et al., 2004). CGA1 expression is strongly affected by exogenous cytokinin (Brenner et al., 2005; Kiba et al., 2005; Rashotte et al., 2006; Naito et al., 2007), and our quantitative data now demonstrate that both CGA1 and GNC are misregulated in arr1 arr12 mutants. These observations firmly place CGA1 and GNC in a cytokinin-dependent signaling pathway. Our genetic analysis of GNC/CGA1 function demonstrates that changes in their expression modulate chloroplast biogenesis, and thus the effects of cytokinin on their expression are predicted to similarly modulate various aspects of chloroplast development.
Our data support a role for the GNC/CGA1 family in mediating the effects of cytokinin on chloroplast division. Exogenously applied cytokinins or overexpression of the cytokinin-regulated transcription factor CRF2 (Rashotte et al., 2006) induce chloroplast division by up-regulating the protein level of PDV2, suggesting that increased cytokinin signaling accelerates chloroplast division through PDV2 (Okazaki et al., 2009). By using chloroplast size and numbers as readouts for the rate of division, we found that the gnc cga1 mutant is less responsive than the wild type to cytokinin, whereas overexpression of GNC mimics the effects of cytokinin on chloroplast division. Although less responsive, the gnc cga1 mutant did not abolish the effects of cytokinin on chloroplast division, indicating that other factors also play a role in this process, the cytokinin-regulated transcription factor CRF2 being a likely candidate (Okazaki et al., 2009). We did not observe increased expression of PDV1 or PDV2 upon cytokinin treatment or in the GNC overexpression lines (data not shown), suggesting that an increase in PDV2 protein by cytokinin is mediated posttranscriptionally and/or that overexpression of GNC may regulate chloroplast division through other plastid division proteins.
The GNC/CGA1 family may also play a role in the molecular mechanism by which cytokinin mediates deetiolation. Exogenously applied cytokinins partially mimic the effect of light on dark-grown seedlings, enhancing the deetiolation of dark-grown seedlings based on their induction of chloroplast development, cotyledon expansion, and leaf development and their inhibition of hypocotyl elongation (Chory et al., 1994). Overexpression of the GNC/CGA1 family phenocopies the effect of cytokinin on plastid differentiation in the dark, resulting in the production of etioplasts with prolamellar bodies and similarly enhancing chlorophyll production when dark-grown seedlings are moved to the light.
The GNC/CGA1 Family and the Transcriptional Control of Chloroplast Biogenesis
Three transcription factor families (GNC/CGA1, GLK, and CRF) are now implicated in directly and positively regulating chloroplast biogenesis (Bi et al., 2005; Naito et al., 2007; Mara and Irish, 2008; Waters et al., 2008, 2009; Okazaki et al., 2009; Richter et al., 2010; Hudson et al., 2011; Köllmer et al., 2011; Fig. 9). Several lines of evidence indicate that these transcription factor families act in concert, with both overlapping and unique functions. Both the GNC/CGA1 and GLK families play roles in chloroplast development. Like the gnc cga1 double mutant, the glk1 glk2 double mutant exhibits a decrease in chlorophyll levels (Fitter et al., 2002), the defect in the glk1 glk2 mutant being primarily owed to a decreased abundance of thylakoid membranes and grana (Waters et al., 2008). Overexpression of both GNC/CGA1 and GLK family members leads to an increase of chlorophyll content compared with wild-type plants (Waters et al., 2008; Hudson et al., 2011), but the ectopic production of chloroplasts in the epidermis and roots has only been reported in the GNC/CGA1 overexpression lines. However, overexpression of rice GLK1 (OsGLK1) induces chloroplast development in nongreen rice callus cells (Nakamura et al., 2009), consistent with both GLK and GNC/CGA1 transcription factors serving to positively regulate chloroplast development from proplastids, although the conditions under which this has been observed differ. It will be of interest to determine if these transcription factor families play roles in other systems where ectopic chloroplast development has been observed, such as in green curd cauliflower (Brassica oleracea; Zhou et al., 2011).
The GNC/CGA1 family also functions after chloroplast development in the regulation of chloroplast growth and division, and at this point it appears to overlap with the function of the CRF family (Fig. 9). Unlike GNC/CGA1 overexpression lines, neither ectopic production of chloroplasts nor increased chlorophyll content has been reported from CRF overexpression lines (Okazaki et al., 2009). However, like the GNC/CGA1 overexpression lines, overexpression of CRF2 mimics the effect of cytokinin on chloroplast division, resulting in increased numbers of smaller chloroplasts (Okazaki et al., 2009).
Based on their roles in the regulation of chloroplast biogenesis, it is perhaps not too surprising that expression of these transcription factors is regulated by key signals implicated in the control of chloroplast development, in particular light and cytokinin (Fig. 9). The GNC/CGA1 and GLK families are both transcriptionally up-regulated by light (Fitter et al., 2002; Naito et al., 2007), supporting an increased role under conditions where chloroplasts are actively produced. In addition, both the GNC/CGA1 and CRF families are transcriptionally up-regulated by cytokinin (Rashotte et al., 2006; Naito et al., 2007). We find that CGA1 is strongly and transiently up-regulated in response to treatment with exogenous cytokinin. GNC is less responsive to exogenous cytokinin but, like CGA1, exhibits reduced expression in a type B arr mutant background, consistent with the expression of both being affected by changes in endogenous cytokinin signal output. Although the expression of the GLK family does not appear to be regulated by cytokinin, the GLK transcription factors have a DNA-binding domain similar to that of type B ARRs (Fitter et al., 2002), implying that their overlapping role in chloroplast biogenesis may be mediated in part by both binding to similar promoter sequences. Taken together, these data support a transcriptional network in which the GNC/CGA1 family acts in concert with at least two other families of transcription factors to control chloroplast development, growth, and division and is responsive to inputs by both light and cytokinin to modulate these processes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
T-DNA insertion lines for the Arabidopsis (Arabidopsis thaliana) genes GNC (SALK_001778) and CGA1 (SALK_003995) were obtained from the Arabidopsis Biological Resource Center (Bi et al., 2005; Naito et al., 2007; Mara and Irish, 2008). The double gnc cga1 mutant was generated by crossing. T-DNA insertion mutants were genotyped by PCR using the T-DNA left border and the gene-specific primers as listed in Supplemental Table S1.
Unless stated otherwise, seeds were surface sterilized and stratified for 3 d in the dark at 4°C before being moved into the light. Seedlings for molecular and physiological assays were grown on medium containing 0.8% (w/v) phytoagar (Research Products International), 1× Murashige and Skoog (MS) salts containing Gamborg’s vitamins (PhytoTechnology Laboratories), 1% (w/v) Suc, and 0.05% (w/v) MES (pH 5.7). Seeds were germinated and grown under continuous white light (50 µmol m−2 s−1) at 22°C, with white light being generated by standard fluorescent bulbs augmented with 18,000K fluorescent bulbs (Aqua-GLO; Rolf C. Hagen). Layers of black nylon mesh were used to reduce light levels as needed (Argyros et al., 2008). For gene expression analysis, seedlings were grown on filter paper for 14 d and treated with either 10 µm BA or a DMSO vehicle control as described (Argyros et al., 2008). The roots and shoots were then separated and frozen at −80°C before RNA extraction. For the light-induced deetiolation experiment, 7-d-old dark-grown seedlings were transferred to 50 µmol m−2 s−1 white light for the indicated periods of time. For analysis of adult phenotypes, plants were grown in pots under long-day conditions (16 h of light).
Plasmid Constructs
To generate the 35S::GNC-GFP, 35S::CGA1-GFP, 35S::CGA1, 35S::GNC, and 35S::CGA1-CFP constructs, genomic sequences of GNC or CGA1 were amplified from ecotype Columbia (Col-0) genomic DNA using the appropriate primers (Supplemental Table S1). The PCR products were cloned into pCR8/GW/TOPO/ (Invitrogen) to generate an entry clone and were sequenced to ensure that no mutations had occurred. The entry clones were recombined into pMDC32 (Curtis and Grossniklaus, 2003), pEarleygate103 (GFP tag), or pEarleygate102 (cyan fluorescent protein tag; Earley et al., 2006) Gateway-compatible destination binary vectors that confer hygromycin or Basta resistance to transformed plants.
For pGNC::GUS and pCGA1::GUS constructs, approximately 900-bp fragments upstream of the start codon were amplified from Col-0 genomic DNA using Primestar polymerase (TaKaRa Bio) and appropriate primers (Supplemental Table S1). The PCR products were cloned into pCR8/GW/TOPO/ and recombined into pMDC163, a Gateway-compatible plant destination binary vector that confers hygromycin resistance to transformed plants (Curtis and Grossniklaus, 2003).
For DEX-inducible constructs, the Gateway cassette with a yellow fluorescent protein-hemagglutinin (YFP-HA) fragment from pEarleygate101 (Earley et al., 2006) was cloned into pTA7001 vector (Aoyama and Chua, 1997) to generate a Gateway-compatible DEX-inducible vector, pTA-7001 GW-YFP-HA. The genomic fragments of CGA1 and GNC on the pCR8/GW/TOPO/ vector were recombined into pTA-7001 GW-YFP-HA, which confers hygromycin resistance to transformed plants. For stable transformation of Arabidopsis, binary vectors were transformed into Agrobacterium tumefaciens GV3101 or AGL1 and then introduced into Arabidopsis by the floral dip method (Clough and Bent, 1998).
Analysis of GUS Activity
Histochemical analysis of GUS activity in stably transformed lines of Arabidopsis was performed as described (Argyros et al., 2008). Embryo GUS staining was performed as described (Yadegari et al., 1994) with the following modification. Siliques were opened longitudinally and then incubated with GUS staining solution at 37°C overnight. Siliques were then fixed in an ethanol:acetic acid solution (1:1) for 3 h and cleared in a chloral hydrate:glycerol:water solution (8:1:2 , w/v/v) overnight. Dissected ovules were mounted in 50% glycerol with a few drops of chloral hydrate solution, and embryos were removed from the ovules by applying pressure with needles. The GUS-stained tissues were visualized and photographed using an EPSON scanner or Zeiss Axioplan 2 microscope under bright-field conditions.
Gene Expression Analysis
RNA isolation and qRT-PCR analyses were performed as described (Argyros et al., 2008). Briefly, total RNA was isolated by using the RNeasy plant kit according to the manufacturer, with the incorporation of a DNase treatment (Qiagen). First-strand complementary DNA synthesis for reverse transcription (RT)-PCR and qRT-PCR were performed using SuperScript III with oligo(dT) primers (Invitrogen). For RT-PCR, gene-specific primers (Supplemental Table S1) were used to examine the full-length transcripts of GNC or CGA1 with HS-Taq polymerase (TaKaRa Bio). qRT-PCR was performed using SYBR Premix Ex Taq II (TaKaRa Bio) and the primer sets listed in Supplemental Table S1. Average cycle threshold values were generated and analyzed by SDS software version 1.4, which uses the comparative cycle threshold method (Livak and Schmittgen, 2001). β-TUBULIN3 (AT5G62700) was used as a control for both RT-PCR and qRT-PCR.
Immunoblot Analysis
Ten-day-old seedlings were ground in liquid nitrogen and treated with homogenization buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10 mm EDTA, 0.1% Nonidet P-40, and plant protease inhibitor cocktail (Sigma-Aldrich). The mixture was centrifuged at 12,000g for 15 min, the supernatant was isolated, and protein concentration was determined using the bicinchoninic acid protein assay (Thermo Scientific). Proteins were separated on 10% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes as described (Gamble et al., 2002). GFP fusion proteins were detected with a horseradish peroxidase-conjugated monoclonal anti-GFP antibody (Santa Cruz Biotechnology). The loading control α-tubulin was detected with a mouse monoclonal anti-tubulin antibody (Sigma-Aldrich) and a secondary goat anti-mouse antibody (Santa Cruz Biotechnology).
Chlorophyll Measurements
Total chlorophyll levels were determined by extraction of plant material with 95% (v/v) ethanol at 4°C in the dark and shaking for 16 h, with spectrophotometric determination of chlorophyll content made as described (Lichtenthaler, 1987) with the following equations: chlorophyll a (µg mL−1) = (13.36 × OD664) − (5.19 × OD648); chlorophyll b (µg mL−1) = (27.43 × OD648) − (8.12 × OD664), where OD represents optical density at the given value. For measurements of chlorophyll a in individual cotyledons, the cotyledons were incubated with 90% (v/v) ethanol for 24 h at 4°C in the dark with shaking, and the extracted chlorophyll a levels were determined with a fluorometer (model 10-AU; Turner Designs). Dunnett’s multiple comparison test was performed by using the wild type as a reference group after one-way ANOVA.
For deetiolation, seeds were stratified and illuminated under 100 µmol m−2 s−1 white light for 1 h to coordinate germination. Seedlings were grown on 1× MS medium for 5 d in the dark and illuminated with 100 µmol m−2 s−1 white light for 6 h to induce deetiolation. Chlorophyll content was measured as described (Lichtenthaler, 1987). Four replicates with 30 seedlings each were used for each line. Dunnett’s multiple comparison test was performed by using the wild type of each condition as a reference group after one-way ANOVA.
Chloroplast Visualization
For visualization of chloroplasts by autofluorescence in living tissues, hypocotyls from 7-d-old light-grown seedlings and roots from 10-d-old light-grown seedlings were excised, stained with propidium iodide (10 µg mL−1), mounted in water, and visualized using a Leica TCS SP UV confocal microscope at an excitation wavelength of 488 nm. Chlorophyll autofluorescence was detected between 631 and 729 nm. Propidium iodide emission was detected between 562 and 612 nm. Image series taken in the z plane were processed by Imaris (Bitplane Scientific Software) to obtain the maximum intensity projections. For visualization in cotyledons, 14-d-old light-grown seedlings were used, mounted adaxial side uppermost in water, and visualized using a Nikon A1 confocal microscope at the excitation and emission wavelengths given above. For visualization of chlorophyll autofluorescence in the palisade mesophyll cells of leaves, a Nikon A1 confocal microscope was used with quantitative analysis of chlorophyll autofluorescence performed using NIS-Elements AR Analysis 4.00.03 software.
For visualization of chloroplast autofluorescence from DEX-inducible lines, cotyledons from 7-d-old seedlings grown in the presence or absence of 1 µm DEX were used. Samples were prepared as described above, mounted abaxial side uppermost in water, and visualized using a Nikon A1 confocal microscope at the excitation and emission wavelengths given above.
For visualization of chloroplasts in the palisade mesophyll cells of leaves, a Zeiss Axioplan 2 microscope was used. The areas of the chloroplasts as well as of the palisade cells were measured using ImageJ software (version 1.32; National Institutes of Health).
For visualization of chloroplasts in fixed tissues, 1-mm-long hypocotyl segments at the root-hypocotyl junction were dissected from 7-d-old light-grown seedlings, 2-mm-long hypocotyl segments were dissected under dim green light from 4-d-old dark-grown seedlings, and 2-mm-long root segments were dissected at 5 mm away from the root-hypocotyl junction of 7-d-old light-grown seedlings. Preparation of the samples was performed as described (Härtel et al., 1998). Samples were fixed by vacuum infiltration with 3% (v/v) glutaraldehyde, 1% (w/v) paraformaldehyde, and 0.1 m sodium phosphate buffer, pH 7.2. The fixative was replaced three times over 8 h, and the samples were left under vacuum overnight. Samples were postfixed with 2% (w/v) OsO4 in 0.1 m sodium phosphate buffer (pH 7.2), dehydrated through a graded series of ethanol and propylene oxide, and embedded in an Epon LX112 Kit (Ladd Research). For visualization under bright-field conditions, thick cross-sections (500 nm) were prepared and stained with 0.5% (w/v) methylene blue in 0.5% (w/v) sodium borate and 0.5% (w/v) Azure II. Sections were visualized using a Zeiss Axioplan 2 microscope. For visualization by TEM, ultrathin sections (70–90 nm) were stained with methanolic uranyl acetate for 20 min and Reynold’s lead citrate for 3 min. All TEM images were taken at 100 kV on a JEOL TEM 1010 apparatus equipped with a XR-41B AMT digital camera.
For quantification of chloroplast number in epidermal or cortical cells of hypocotyls, cross-sections of fixed hypocotyls of each line were prepared as described above. Chloroplasts in all epidermal or outer cortical cells of the cross-sections were quantified. Only one cross-section from one individual hypocotyl was quantified. Each sample represents the mean on a per cell basis of three individual hypocotyls. The same approach was used to quantify chloroplast number in cortical cells of the root. For the measurements of chloroplast size in fixed hypocotyls, the area of chloroplasts from the cross-sections was measured by using ImageJ software (version 1.32; National Institutes of Health). The quantification was performed on 20 chloroplasts in the outer cortical cells from each of three hypocotyls. Dunnett’s multiple comparison test was performed by using the wild type as a reference group after one-way ANOVA.
Photochemical Efficiency Analysis
For chlorophyll fluorescence analysis of hypocotyls and cotyledons, plants were grown on vertical plates in 0.6% agar-solidified (Sigma-Aldrich) one-half-strength MS medium (Caisson Laboratories) supplemented with 1% Suc (Sigma-Aldrich). After stratification, germination was induced by a 6-h illumination. Plants were then kept in the dark for 3.5 d followed by 7 d in the light (60 µmol m−2 s−1) with a 16-h-light/8-h-dark cycle at 23°C. Prior to chlorophyll fluorescence imaging analysis, plants were dark adapted for 30 min. Analysis was performed as described (Cohu et al., 2009). The Fv/Fm and ΦPSII were calculated as described (Maxwell and Johnson, 2000).
Cytokinin Effects on Chloroplast Division
For chloroplast size measurement, chloroplasts were isolated from the cotyledon tips of 11-d-old seedlings grown on 1× MS medium supplied with 1% (w/v) Suc and the indicated concentrations of BA or a DMSO vehicle control. Cotyledon tips were chopped by a razor blade in 0.33 m sorbitol, 2 mm EDTA, and 50 mm Tris-HCl (pH 8.0) to release chloroplasts. Chloroplasts were visualized using a Zeiss Axioplan 2 microscope, and size was measured using ImageJ software (version 1.32; National Institutes of Health), with 100 chloroplasts from seven seedlings analyzed for each line. For measurement of chloroplast number, the cotyledon tips were mounted in 0.33 m sorbitol, 2 mm EDTA, and 50 mm Tris-HCl (pH 8.0), and cells were released by gently squeezing the cotyledon tips between coverslips and slides. For statistical analysis, the cells were visualized using a Zeiss Axioplan 2 microscope, and chloroplasts were counted in 30 cells from five seedlings for each line. For Figure 8, A and B, Dunnett’s multiple comparison test was performed by using the wild type of each condition as a reference group after one-way ANOVA. For Figure 8, C and D, the Bonferroni test was performed after two-way ANOVA (***P < 0.001).
Subcellular Localization of GNC and CGA1
Arabidopsis leaf mesophyll protoplasts were isolated from wild-type mature leaves and transfected with 35S::GNC-GFP or 35S::CGA1-GFP constructs with a histone-2B-RFP nuclear marker construct as described (Kovtun et al., 2000). Subcellular localization of fusion proteins was observed using a Zeiss Axioplan 2 fluorescence microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT5G56860 (GNC), AT4G26150 (CGA1), gnc (SALK_001778), and cga1 (SALK_003995).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Tissue-specific expression of GNC and CGA1.
Supplemental Figure S2. GNC and CGA1 transcription factors localize to the nucleus.
Supplemental Figure S3. Characterization of the gnc and cga1 T-DNA insertion alleles.
Supplemental Figure S4. Altered levels of chlorophyll in the gnc cga1 mutant.
Supplemental Figure S5. Overexpression of GNC and CGA1 induces chloroplast biogenesis in the hypocotyls.
Supplemental Figure S6. Overexpression of GNC and CGA1 results in increased chloroplast number and expanded zones of chloroplast production.
Supplemental Figure S7. Photosynthetic activity of Col-0, the gnc cga1 mutant, and GNC overexpression lines (OX-1 and OX-2).
Supplemental Figure S8. Chlorophyll a content in cotyledons of the gnc cga1 mutant and GNC overexpression lines is altered compared with the wild type.
Supplemental Figure S9. Effects of gnc cga1 mutation and GNC overexpression on rosette weight and chlorophyll accumulation in leaves.
Supplemental Figure S10. Effects of gnc cga1 mutation and GNC overexpression on chloroplast autofluorescence, size of mesophyll cells, chloroplast size, and number of chloroplasts per cell.
Supplemental Table S1. DNA primers used in this study.
Supplemental Video S1. Detection of chlorophyll autofluorescence in pavement and guard cells of the cotyledon epidermis in the GNC overexpression line (OX-2).
Supplemental Video S2. Detection of chlorophyll autofluorescence in pavement and guard cells of the cotyledon epidermis in the wild type.
Supplementary Material
Glossary
- qRT
reverse transcription
- BA
benzyladenine
- DMSO
dimethyl sulfoxide
- CaMV
cauliflower mosaic virus
- TEM
transmission electron microscopy
- Fv/Fm
maximum quantum yield of PSII
- ΦPSII
flux of electrons through PSII
- DEX
dexamethasone
- MS
Murashige and Skoog
- Col-0
ecotype Columbia
- T-DNA
transferred DNA
References
- Aoyama T, Chua N-H. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang Y-H, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE. (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong GA, Runge S, Frick G, Sperling U, Apel K. (1995) Identification of NADPH:protochlorophyllide oxidoreductases A and B: a branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol 108: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y-M, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S. (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44: 680–692 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K. (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42: 111–123 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA. (1990) Mutations in the DET1 gene affect cell-type-specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc Natl Acad Sci USA 87: 8776–8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M. (1994) A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Plant Physiol 104: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohu CM, Abdel-Ghany SE, Gogolin Reynolds KA, Onofrio AM, Bodecker JR, Kimbrel JA, Niyogi KK, Pilon M. (2009) Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: regulation and unexpected phenotypes in an Arabidopsis mutant. Mol Plant 2: 1336–1350 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Quail PH. (1992) Genetic and phenotypic characterization of Cop-1 mutants of Arabidopsis thaliana. Plant J 2: 83–95 [Google Scholar]
- Dupree P, Pwee K-H, Gray JC. (1991) Expression of photosynthesis gene-promoter fusions in leaf epidermal cells of transgenic tobacco plants. Plant J 1: 115–120 [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE. (2002) Mutational analysis of the ethylene receptor ETR1: role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol 128: 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Essigmann B, Lokstein H, Hoffmann-Benning S, Peters-Kottig M, Benning C. (1998) The phospholipid-deficient pho1 mutant of Arabidopsis thaliana is affected in the organization, but not in the light acclimation, of the thylakoid membrane. Biochim Biophys Acta 1415: 205–218 [DOI] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi Y-M, Rothstein SJ. (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. (2005) A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J 42: 133–144 [DOI] [PubMed] [Google Scholar]
- Khokhlova VA. (1977) Effect of cytokinin on formation of plastids in isolated pumpkin cotyledons under light and in dark. Soviet Plant Physiol 24: 956–961 [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T. (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry. Plant Cell Physiol 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Köllmer I, Werner T, Schmülling T. (2011) Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J Plant Physiol 168: 1320–1327 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J. (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. (1998) The division apparatus of plastids and mitochondria. Int Rev Cytol 181: 1–41 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lochmanová G, Zdráhal Z, Konecná H, Koukalová Š, Malbeck J, Souček P, Válková M, Kiran NS, Brzobohatý B. (2008) Cytokinin-induced photomorphogenesis in dark-grown Arabidopsis: a proteomic analysis. J Exp Bot 59: 3705–3719 [DOI] [PubMed] [Google Scholar]
- Longo GP, Pedretti M, Rossi G, Longo CP. (1979) Effect of benzyladenine on the development of plastids and microbodies in excised watermelon cotyledons. Planta 145: 209–217 [DOI] [PubMed] [Google Scholar]
- López-Juez E. (2007) Plastid biogenesis, between light and shadows. J Exp Bot 58: 11–26 [DOI] [PubMed] [Google Scholar]
- López-Juez E, Pyke KA. (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49: 557–577 [DOI] [PubMed] [Google Scholar]
- Ma J, Peng L, Guo J, Lu Q, Lu C, Zhang L. (2007) LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mara CD, Irish VF. (2008) Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Mullet JE. (1988) Chloroplast development and gene-expression. Annu Rev Plant Physiol Plant Mol Biol 39: 475–502 [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T. (2007) Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Muramatsu M, Hakata M, Ueno O, Nagamura Y, Hirochika H, Takano M, Ichikawa H. (2009) Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol 50: 1933–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ. (2000) Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51: 111–140 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Kabeya Y, Suzuki K, Mori T, Ichikawa T, Matsui M, Nakanishi H, Miyagishima SY. (2009) The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possingham JV, Lawrence ME. (1983) Controls to plastid division. Int Rev Cytol 84: 1–56 [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC. (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Samuelson ME, Larsson C-M. (1993) Nitrate regulation of zeation riboside levels in barley roots: effects of inhibitors of N assimilation and comparison with ammonium. Plant Sci 93: 77–84 [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee C-H, Lee D, Choi G. (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ. (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler DA, Laetsch WM. (1965) Kinetin-induced chloroplast maturation in cultures of tobacco tissue. Science 149: 1387–1388 [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. (2001) Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol 42: 85–93 [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H. (2004) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Tejos RI, Mercado AV, Meisel LA. (2010) Analysis of chlorophyll fluorescence reveals stage specific patterns of chloroplast-containing cells during Arabidopsis embryogenesis. Biol Res 43: 99–111 [PubMed] [Google Scholar]
- Wang RC, Okamoto M, Xing XJ, Crawford NM. (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Langdale JA. (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA. (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56: 432–444 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. (1992) COP9: a new genetic locus involved in light-regulated development and gene expression in Arabidopsis. Plant Cell 4: 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Harada JJ. (1993) Embryogenesis in higher plants: an overview. Plant Cell 5: 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston EL, Pyke KA. (1999) Developmental ultrastructure of cells and plastids in the petals of wallflower (Erysimum cheiri). Ann Bot (Lond) 84: 763–769 [Google Scholar]
- Whatley JM. (1978) Suggested cycle of plastid developmental interrelationships. New Phytol 80: 489–502 [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Paiva G, Laux T, Koltunow AM, Apuya N, Zimmerman JL, Fischer RL, Harada JJ, Goldberg RB. (1994) Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 6: 1713–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaronskaya E, Vershilovskaya I, Poers Y, Alawady AE, Averina N, Grimm B. (2006) Cytokinin effects on tetrapyrrole biosynthesis and photosynthetic activity in barley seedlings. Planta 224: 700–709 [DOI] [PubMed] [Google Scholar]
- Zavaleta-Mancera HA, Franklin KA, Ougham HJ, Thomas H, Scott IM. (1999a) Regreening of senescent Nicotiana leaves. I. Reappearance of NADPH-protochlorophyllide oxidoreductase and light-harvesting chlorophyll a/b-binding protein. J Exp Bot 50: 1677–1682 [Google Scholar]
- Zavaleta-Mancera HA, Thomas BJ, Thomas H, Scott IM. (1999b) Regreening of senescent Nicotiana leaves. II. Redifferentiation of plastids. J Exp Bot 50: 1683–1689 [Google Scholar]
- Zhao L, Sack FD. (1999) Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am J Bot 86: 929–939 [PubMed] [Google Scholar]
- Zhou X, Fei Z, Thannhauser TW, Li L. (2011) Transcriptome analysis of ectopic chloroplast development in green curd cauliflower (Brassica oleracea L. var. botrytis). BMC Plant Biol 11: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, Shakirova FM, Avalbaev AM, Kudryakova NV, Zubkova NK, Liere K, Kulaeva ON, Kusnetsov VV, et al. (2008) Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.