Abstract
Chitin is commonly found in fungal cell walls and is one of the well-studied microbe/pathogen-associated molecular patterns. Previous studies showed that lysin motif (LysM)-containing proteins are essential for plant recognition of chitin, leading to the activation of plant innate immunity. In Arabidopsis (Arabidopsis thaliana), the LYK1/CERK1 (for LysM-containing receptor-like kinase1/chitin elicitor receptor kinase1) was shown to be essential for chitin recognition, whereas in rice (Oryza sativa), the LysM-containing protein, CEBiP (for chitin elicitor-binding protein), was shown to be involved in chitin recognition. Unlike LYK1/CERK1, CEBiP lacks an intracellular kinase domain. Arabidopsis possesses three CEBiP-like genes. Our data show that mutations in these genes, either singly or in combination, did not compromise the response to chitin treatment. Arabidopsis also contains five LYK genes. Analysis of mutations in LYK2, -3, -4, or -5 showed that LYK4 is also involved in chitin signaling. The lyk4 mutants showed reduced induction of chitin-responsive genes and diminished chitin-induced cytosolic calcium elevation as well as enhanced susceptibility to both the bacterial pathogen Pseudomonas syringae pv tomato DC3000 and the fungal pathogen Alternaria brassicicola, although these phenotypes were not as dramatic as that seen in the lyk1/cerk1 mutants. Similar to LYK1/CERK1, the LYK4 protein was also localized to the plasma membrane. Therefore, LYK4 may play a role in the chitin recognition receptor complex to assist chitin signal transduction and plant innate immunity.
In nature, plants are confronted with a great variety of fungal pathogens as well as other types of pathogens. In response, plants have evolved various defense mechanisms to thwart or limit these infections. One such mechanism involves the recognition of nonself microbe-associated molecular patterns (MAMPs; synonymously termed “pathogen-associated molecular patterns”), such as chitin (Bent and Mackey, 2007; Boller and Felix, 2009), by pattern recognition receptors (PRRs) to activate so-called MAMP-triggered immunity (Jones and Dangl, 2006). Chitin is a linear polymer of β-1,4-linked GlcNAc units and a major component of fungal cell walls. The integrity of fungal cell walls is important for pathogenesis, since fungal pathogens with less chitin deposition are less virulent (Madrid et al., 2003; Michielse et al., 2009; Lenardon et al., 2010). A number of publications have documented that plants activate a variety of defense responses when challenged with chitin, including the production of reactive oxygen species, the activation of defense genes, and the accumulation of phytoalexins (Shibuya and Minami, 2001).
Recently, lysin motif (LysM)-containing proteins were shown to be involved in plant chitin recognition. The LysM domain was initially identified in bacterial enzymes involved in binding and degrading the bacterial cell wall component peptidoglycan (Joris et al., 1992; Steen et al., 2003; Buist et al., 2008), which is structurally similar to chitin. In rice (Oryza sativa), CEBiP (for chitin elicitor-binding protein) was shown to be important in the activation of plant innate immunity upon chitin addition (Kaku et al., 2006). This protein has an extracellular domain containing two LysMs and a single transmembrane domain. The analysis of mutants identified LYK1/CERK1 (for LysM-containing receptor-like kinase1/chitin elicitor receptor kinase1) as the primary PRR for chitin recognition in Arabidopsis (Arabidopsis thaliana; Miya et al., 2007; Wan et al., 2008a; Shimizu et al., 2010). The LYK proteins have an extracellular domain containing one to three LysMs, a single transmembrane domain, and an intracellular Ser/Thr kinase domain. This protein family appears to be found exclusively in plants (Zhang et al., 2007, 2009). In addition to these plant receptors, fungal pathogens appear to employ secreted LysM domain-containing proteins to either compete for binding chitin with the plant chitin receptors or to coat the fungal cell wall to prevent the release of elicitor-active chitin fragments by plant chitinases (van den Burg et al., 2006; de Jonge and Thomma, 2009; Stergiopoulos and de Wit, 2009; de Jonge et al., 2010). Collectively, these plant and fungal studies point to a central role for LysM proteins in chitin recognition and modulating plant innate immunity in response to fungal infection.
The Arabidopsis genome encodes five LYKs: LYK1/CERK1 and LYK2 to LYK5 (Zhang et al., 2007; Wan et al., 2008a). As shown previously, a knockout mutation in lyk1/cerk1 blocked the induction of virtually all chitin-responsive genes (CRGs), the production of reactive oxygen species, and the activation of mitogen-activated protein kinases (MPKs), and enhanced susceptibility to fungal (Miya et al., 2007; Wan et al., 2008a) and bacterial (Gimenez-Ibanez et al., 2009a, 2009b) pathogens. Recent biochemical studies confirmed that both CEBiP and LYK1/CERK1 proteins can directly bind chitin (Iizasa et al., 2010; Petutschnig et al., 2010). A protein similar to LYK1/CERK1, called OsCERK1, was also shown to be involved in chitin signaling in rice and was proposed to function together with CEBiP (Kaku et al., 2006; Shimizu et al., 2010). Proteins similar to rice CEBiP (i.e. CEBiP-like1, -2, and -3) are also encoded by the Arabidopsis genome, but so far their role in chitin signaling has not been established.
Interestingly, in legumes, LYK proteins were shown to be the essential receptors for the perception of the lipochitooligosaccharide nodulation factors (NFs), produced by rhizobia and essential for establishment of the nitrogen-fixing symbiosis (Limpens et al., 2003; Madsen et al., 2003; Mulder et al., 2006; Radutoiu et al., 2007; Smit et al., 2007). Therefore, the LysM domain appears to be capable of recognizing/binding a sugar backbone of β-1,4-linked aminosugars, such as chitin, peptidoglycan, and NFs. Surprisingly, the different LysM domains found in the various LYKs were shown to specifically recognize only their cognate signal, which, in the case of NF recognition, was mapped to only a few, critical amino acid residues (Radutoiu et al., 2007).
Although the chitin receptors have been identified from both Arabidopsis and rice, relatively little is known about the downstream signaling pathway, especially how the chitin signal is transduced into intracellular responses leading to plant innate immunity. In rice, Chen et al. (2010) showed that Hsp90 and its cochaperone Hop/Sti1 interacted with OsCERK1 in the endoplasmic reticulum and were critical for the efficient transport of OsCERK1 from the endoplasmic reticulum to the plasma membrane (PM). Furthermore, Hop/Sti1 and Hsp90 were also localized in a complex at the PM with the plant-specific ρ-type GTPase OsRac1, and Hop/Sti1 were required for chitin-triggered immunity and resistance to rice blast fungus. These data suggest that in rice, the Hop/Sti1-Hsp90 chaperone complex may play an important role in transducing the perceived chitin signal to downstream components in the chitin signaling pathway (Chen et al., 2010).
In Arabidopsis, a simple pathway has been proposed that starts with the perception of chitin by LYK1/CERK1, followed by activation of the MPK pathway, leading to the activation of a variety of transcription factors and, ultimately, the induction of genes involved in pathogen defense (Wan et al., 2004, 2008a; Libault et al., 2007). An analysis of the genes induced by various MAMPs, including chitin, flagellin, and EF-Tu, showed that although each is recognized by a unique PRR, these pathways converge on a common set of genes (Wan et al., 2008a, 2008b).
In this work, we examined the question of whether other LysM proteins in Arabidopsis, in addition to LYK1/CERK1, are involved in chitin perception. Additionally, some of these genes were also induced by chitin, for instance, LYK4 and LYK5, as well as LYK1 (Miya et al., 2007; Wan et al., 2008a), indicating their potential involvement in chitin signaling. Furthermore, recently, Petutschnig et al. (2010) found that several LysM proteins (i.e. LYK1/CERK1, LYK4, LYK5, and CEBiP-like1) can be pulled down from an Arabidopsis cellular extract by chitin magnetic beads, supporting the potential involvement of other LysM proteins in chitin perception.
In order to test the role of the three CEBiP-like proteins (i.e. CEBiP-like1, -2, and -3), we identified mutations in each of the encoding genes and also generated a triple mutant totally lacking in CEBiP expression. However, unlike the case in rice, mutations in one or all of the genes encoding CEBiP-like proteins showed no significant effects on the plant response to chitin addition.
We also obtained mutants for the other four LYK genes. Only the mutations in the LYK4 gene (At2g23770) resulted in a significant reduction in the plant chitin response, including reducing the induction of CRGs and chitin-induced calcium signaling and enhancing susceptibility to both fungal and bacterial pathogens. Similar to LYK1/CERK1, the LYK4 protein is also localized to the PM. Therefore, LYK4 is partially, but clearly, involved in chitin signaling, likely functioning in a chitin recognition receptor complex.
RESULTS
Analysis of CEBiP-Like Genes
To investigate whether CEBiP-like genes are involved in chitin signaling in Arabidopsis, we obtained homozygous mutants for each of the three genes, CEBiP-like1, -2, and -3, and also generated the triple mutant for all three genes through crossing. We then used reverse transcription (RT)-PCR to investigate whether the expression levels of CRGs were changed in these mutants after treatment with chitin. The results showed that the mutants responded in a similar fashion to the wild type (Supplemental Fig. S1), suggesting that, contrary to the case of the rice OsCEBiP (Kaku et al., 2006), these genes are not involved in chitin signaling in Arabidopsis. Consistent with this, expression of the CEBiP-like genes was not induced by chitin treatment, as shown by our previous microarray experiments (Ramonell et al., 2005; Wan et al., 2008a). Therefore, although OsCEBiP and OsCERK1 appear to function as part of a chitin receptor complex in rice, it is unlikely that CEBiP-like proteins play a similar role in Arabidopsis.
The Expression Pattern of LYK4
The Arabidopsis genome encodes five LYK proteins, and one of them, LYK1/CERK1, was previously shown to be involved in chitin perception (Miya et al., 2007; Wan et al., 2008a). To examine the potential involvement of the remaining four LYKs in chitin recognition, we obtained knockout mutants for each of these four genes, LYK2 to LYK5. Interestingly, only mutations in LYK4 affected the chitin response (see Fig. 5 below), whereas mutants of LYK2, LYK3, or LYK5 (Zhang et al., 2007; Wan et al., 2008a) as well as the triple mutant showed no significant effect on chitin signaling based on the induction of CRGs (e.g. WRKY53; Supplemental Fig. S2). Hence, we focused on further characterization of LYK4 as described below.
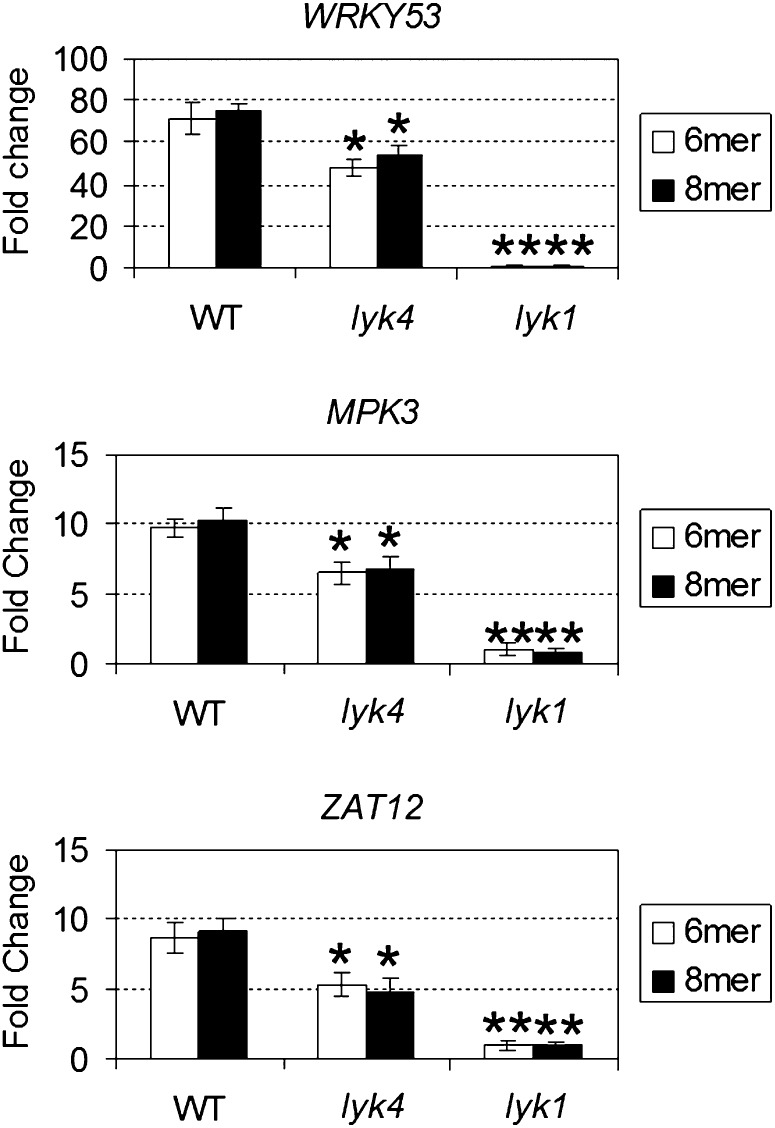
Figure 5.
Analysis of chitin-responsive genes in the lyk4 mutant. Seedlings were treated with the purified chitin oligomers, chitohexaose (6-mer) and chitooctaose (8-mer), at a final concentration of 1 μm for 30 min. The relative fold change ± se of the chitin-responsive genes (WRKY53, MPK3, and ZAT12) in a particular genotype was obtained from the comparison between the chitin-treated plants and the mock-treated plants after normalization with the reference gene SAND. Asterisks indicate statistically significant differences with respect to the wild type (WT): *P < 0.05, **P < 0.01.
The LYK4 gene is 1,839 bp long and lacks introns. The coding sequence is annotated to encode a protein of 612 amino acids, with a signal peptide, an extracellular LysM domain (containing three LysMs), a transmembrane domain, and an intracellular Ser/Thr kinase domain (Fig. 1A). This protein appears to be evolutionarily related to NF receptors and the chitin receptor LYK1/CERK1 (Zhang et al., 2007).
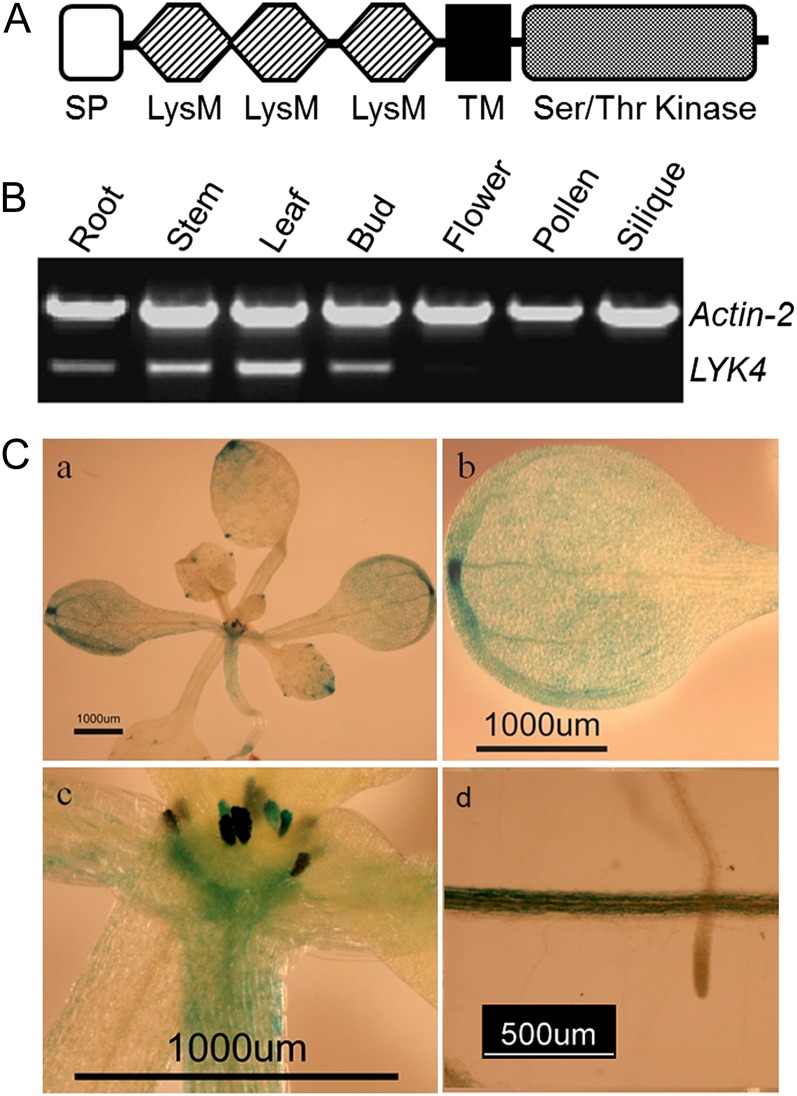
Figure 1.
Expression patterns of LYK4. A, Schematic representation of the LYK4 protein (not drawn to scale). SP, Signal peptide; TM, transmembrane domain. B, Semiquantitative RT-PCR analysis of LYK4 gene expression in different tissues. Actin2 was included as an internal control. C, LYK4 promoter-GUS transgenic plants: a, whole seedling; b, leaf; c, apical meristem and stipules; d, root.
To examine the expression pattern of LYK4, we first analyzed the mRNA level in various tissues using RT-PCR. These data showed measurable expression of LYK4 in root, stem, leaf, and flower bud, with relatively higher expression in leaf, stem, and root. Little or no LYK4 expression was detected in older flowers, pollen, or siliques (Fig. 1B).
We further examined the expression pattern of the LYK4 gene by analyzing the expression of a LYK4 promoter-GUS fusion in transgenic plants. The results showed that LYK4 is predominantly expressed in leaf (predominantly in hydathodes), stem, and root, roughly in agreement with the RT-PCR results (Fig. 1C), suggesting that LYK4 may function in these tissues. Interestingly, a higher expression level was found in the apical meristem and stipule (Fig. 1C), suggesting a possible spatial regulation of LYK4 expression in different tissues and, perhaps, a role for LYK4 in growth and development. However, lyk4 mutants showed no obvious growth defects.
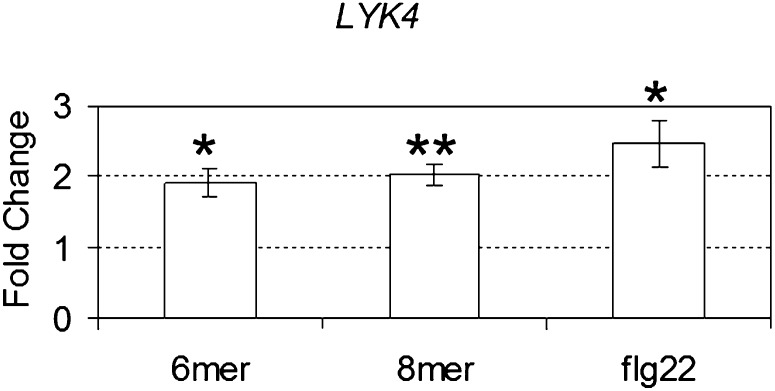
In addition, we examined the expression of LYK4 in response to chitin and flagellin. As shown in Figure 2, LYK4 was moderately induced by the purified chitin oligomers chitohexaose (6-mer) and chitooctaose (8-mer). A similar induction pattern was seen with LYK1 and LYK5, but not LYK2 or LYK3, in response to chitooctaose (Ramonell et al., 2005; Wan et al., 2008a). Interestingly, LYK4 was also moderately induced by the addition of flg22, an elicitor-active epitope of 22 amino acids derived from flagellin (Felix et al., 1999; Fig. 2), as was seen with LYK1 (Zipfel et al., 2004), suggesting that the expression of both may respond to more than one MAMP.
Figure 2.
LYK4 expression in response to chitin and flagellin. Plants were treated with chitohexaose (6-mer), chitooctaose (8-mer), and the flagellin-derived peptide flg22 at a final concentration of 1 μm for 30 min. The relative fold change ± se of LYK4 gene expression was obtained using quantitative RT-PCR from a comparison between the chitin- or flg22-treated plants and the mock-treated plants after normalization with the reference gene SAND. Average results from three independent experiments are reported. Asterisks indicate statistically significant differences with respect to the wild type: *P < 0.05, **P < 0.01.
Subcellular Localization of LYK4
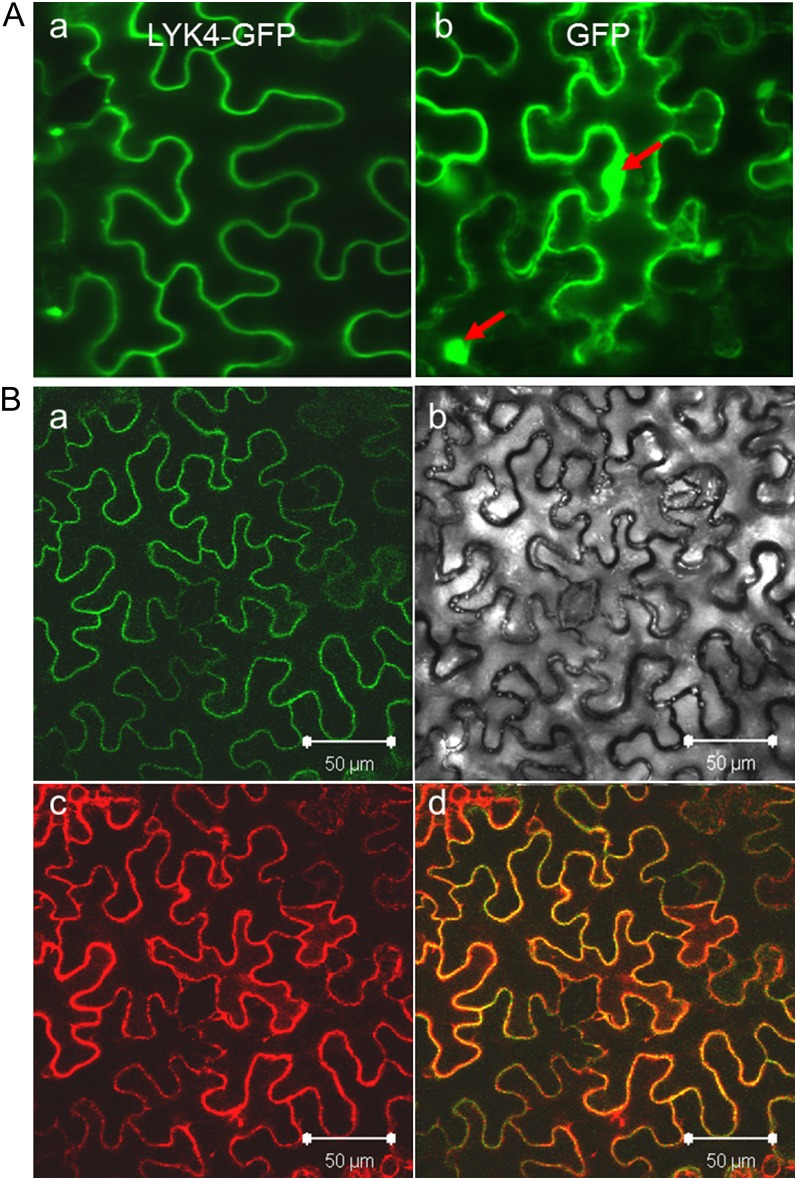
To investigate the subcellular localization of LYK4, we generated transgenic Arabidopsis plants expressing a translational fusion of LYK4 with GFP under the control of the strong cauliflower mosaic virus (CaMV) 35S promoter (Benfey and Chua, 1990). However, the signal from the LYK4-GFP fusion was very weak in both seedling leaves and roots compared with that from the GFP alone, also driven by the CaMV 35S promoter. The signal was still weak even after chitin or flg22 treatment. We then tested the transient expression of the same LYK4-GFP construct in tobacco (Nicotiana benthamiana) using the pressure-mediated Agrobacterium tumefaciens infiltration approach (Kapila et al., 1997). In this system, the expression of LYK4-GFP appeared to be localized at the PM of leaf cells, although the GFP alone appeared to be present in both the cytoplasm and nucleus (Fig. 3A). To further support this localization, the LYK4-GFP construct was coinfiltrated with a specific PM marker, the Arabidopsis PM aquaporin AtPIP2A fused with the red fluorescent protein mCherry (Nelson et al., 2007). After merging the images obtained from the green and red channels, both LYK4-GFP and AtPIP2A-mCherry appeared to colocalize (Fig. 3B), supporting that LYK4 is located at the PM, in agreement with the prediction that LYK4 is primarily located at the PM using the program WoLF PSORT developed by Horton et al. (2007).
Figure 3.
Subcellular localization of LYK4. A, Epifluorescence images of tobacco leaf cells infiltrated with Agrobacterium harboring the LYK4-GFP fusion construct (a) or the GFP construct (b). The red arrows indicate nuclei. B, Epifluorescence images of tobacco leaf cells coinfiltrated with Agrobacterium harboring the LYK4-GFP fusion construct and the PM marker-mCherry construct: a, green channel image; b, bright-field image; c, red channel image; d, merged image of the green and red channels. Bars = 50 µm.
Mutation in the LYK4 Gene Shows Reduced Induction of Chitin-Responsive Gene Expression in Response to Chitin
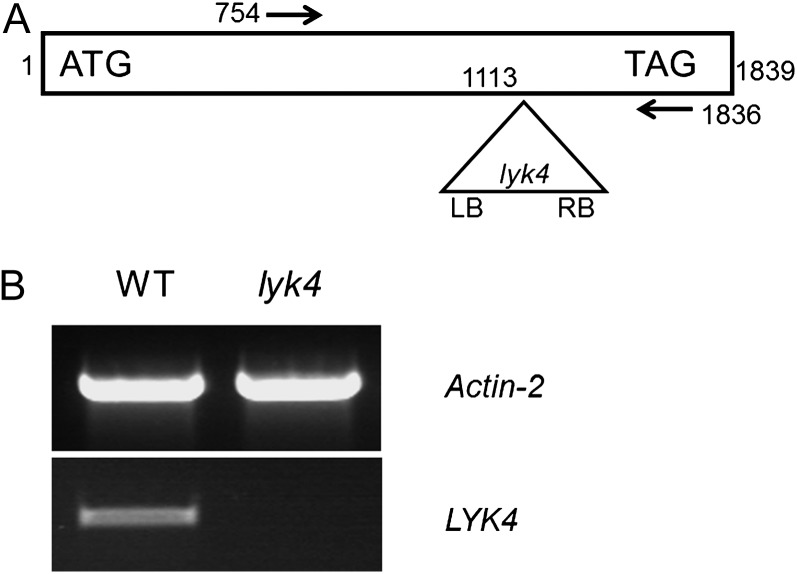
A knockout mutant of the LYK4 gene, as shown in Figure 4A, was used to examine the role of this gene in chitin signaling (see below). The mutant WiscDsLox297300_01C (lyk4) has a transferred DNA (T-DNA) insertion after the 1,113th nucleotide downstream from the A in the ATG start codon. RT-PCR analysis using primers flanking the insertion (arrows in Fig. 4A) failed to detect LYK4 mRNA expression in the mutant, which was easily detected in the wild-type plant (Fig. 4B).
Figure 4.
The lyk4 mutant. A, Schematic representation of the lyk4 mutant (not drawn to scale). Numbers indicate nucleotide positions. ATG, Start codon; LB, T-DNA left border; RB, T-DNA right border; TAG, stop codon. The arrows indicate the primers used in B. B, Semiquantitative RT-PCR analysis of LYK4 gene expression in the lyk4 mutant and wild-type (WT) plants. Actin2 was included as an internal control.
In order to test whether the lyk4 mutant is compromised in chitin signaling, we treated seedlings with chitin oligomers (i.e. chitohexaose and chitooctaose) and then examined the expression of selected CRG genes (Wan et al., 2004; Libault et al., 2007). As mentioned above, mutations in LYK2, -3, and -5 showed no effect on CRG expression (Supplemental Fig. S2). However, as shown in Figure 5, the induction of the CRG genes by chitin oligomers (chitohexaose and chitooctaose) appeared to be moderately reduced in the lyk4 mutant when compared with wild-type plants. As expected, the induction of these genes was totally blocked in the mutant lyk1 (Fig. 5), as reported previously (Miya et al., 2007; Wan et al., 2008a). A similar pattern of gene expression was found when mRNA levels in aerial tissues (mostly containing leaves) and root tissues were examined separately (Supplemental Fig. S3). Additionally, we generated complementation lines in which the LYK4 gene was expressed under the control of the CaMV 35S promoter in the lyk4 mutant background. As shown in Supplemental Figure S4, the induction of the CRG genes appeared to be normal in the complementation lines of the lyk4 mutant. Taken together, the attenuation of CRG expression caused in the lyk4 line suggests that LYK4 may play an auxiliary role in chitin signaling.
To see whether LYK4 is possibly involved in other signaling pathways, we examined the effect of the mutation in LYK4 on the induction of the genes WRKY53, MPK3, and ZAT12 in response to the treatment with flg22 or elf26. As shown in Supplemental Figure S5, the induction of these genes was normal in response to flg22 and elf26 in both the lyk4 and lyk1 mutants, suggesting that LYK4 functions specifically in chitin signaling.
The lyk4 Mutant Shows a Reduced Cytosolic Calcium Response to Chitin Treatment
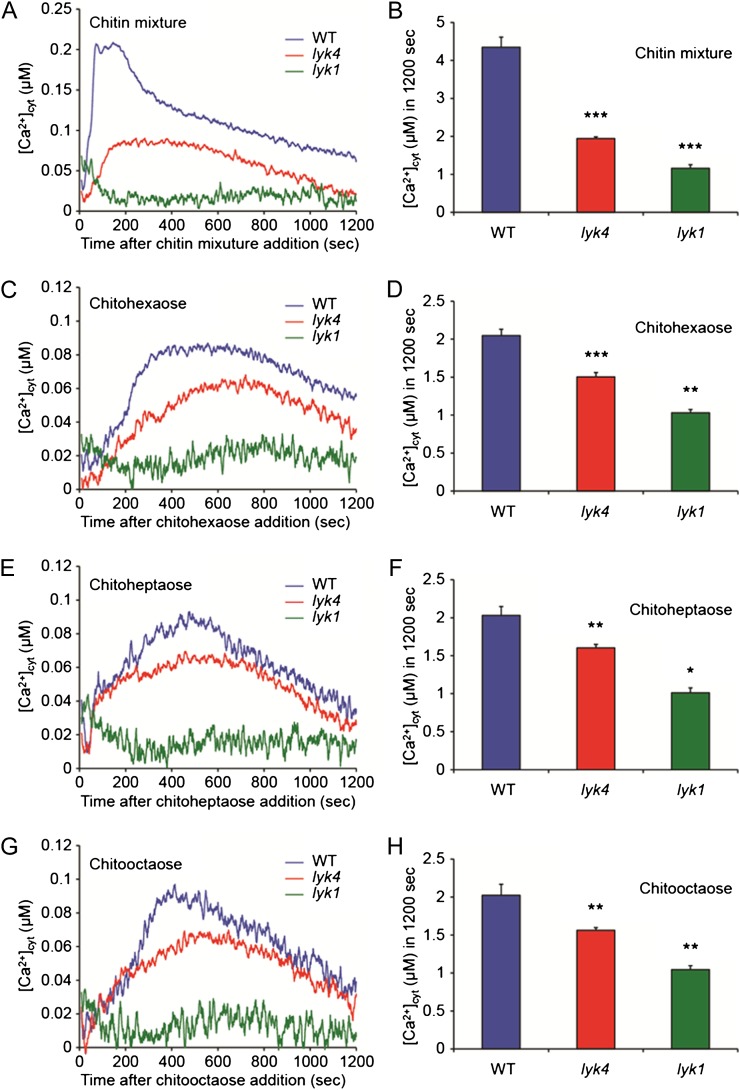
The elevation of cytosolic free calcium ion concentration ([Ca2+]cyt) is one of the plant cell responses to chitin elicitation (Kwaaitaal et al., 2011). To assess the chitin-induced [Ca2+]cyt changes in the lyk4 mutant, we generated a mutant line in the aequorin transgenic background by cross-pollination (see “Materials and Methods”). We also generated an aequorin-expressing lyk1 mutant line as a control. The individual mutants were treated with chitin to measure the chitin-induced [Ca2+]cyt response. As shown in Figure 6, the [Ca2+]cyt in the wild type was significantly elevated 0 to 150 s after addition of the chitin mixture or at 300 to 500 s after addition of the purified chitin oligomers. In contrast, the chitin-induced [Ca2+]cyt responses in the lyk4 mutant were delayed and moderately reduced in comparison with that in the wild type, while the responses were completely blocked in the lyk1 mutant (Fig. 6). The reduced chitin-induced [Ca2+]cyt responses were recovered in the complementation lines of the lyk4 mutant (Supplemental Fig. S6). Collectively, the results show that the mutation in the LYK4 gene significantly reduces the chitin-induced [Ca2+]cyt response but not to the same degree as found in the LYK1 mutant line.
Figure 6.
Effects of chitin treatments on the [Ca2+]cyt response in the lyk4 mutant. Five-day-old aequorin transgenic seedlings of the wild type (WT) and lyk4 and lyk1 mutants were treated with 10 μg mL−1 chitin mixture (A and B) or 1 µm chitohexaose (C and D), chitoheptaose (E and F), or chitooctaose (G and H). Line graphs show kinetic differences in chitin-induced [Ca2+]cyt responses. Histograms represent integrated [Ca2+]cyt values over 1,200 s after chitin treatments. Each value shows a mean of 12 seedlings ± se. Asterisks indicate statistically significant differences compared with the wild-type control: *P < 0.05, **0.001 < P < 0.01, ***P < 0.001.
Mutation in LYK4 Causes Enhanced Susceptibility to Fungal and Bacterial Pathogens
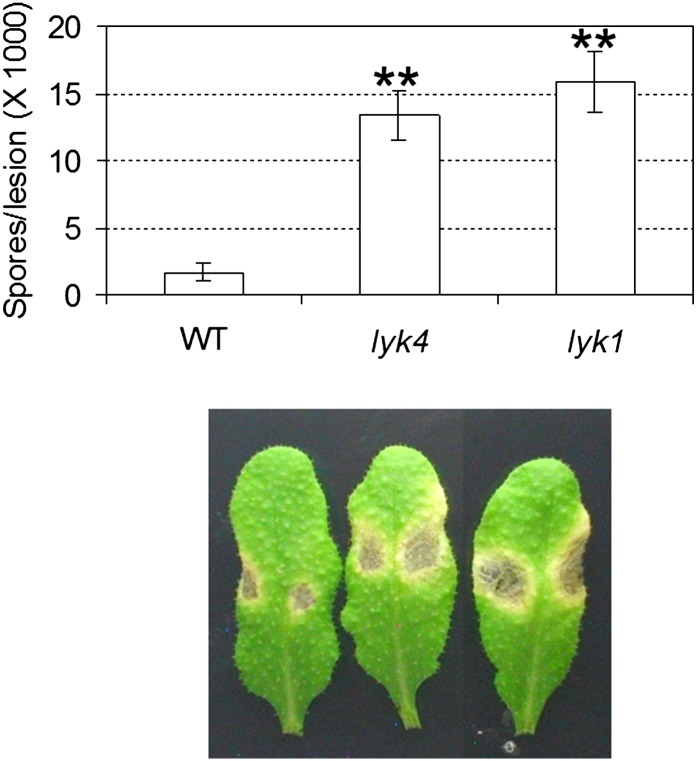
To investigate the potential role of LYK4 in plant defense, we performed disease assays with the necrotrophic fungal pathogen Alternaria brassicicola. The results clearly showed that the lyk4 plants were more susceptible to this fungal pathogen than wild-type plants, as exhibited by more severe symptoms and higher spore production per lesion (Fig. 7). These data suggest that LYK4 is involved in the plant defense pathway to fungal infection, likely mediated by chitin recognition.
Figure 7.
Response of the lyk4 mutant to a fungal pathogen. The fungal pathogen A. brassicicola was spot inoculated onto different plants at a concentration of 5 × 105 spores mL−1. Asterisks indicate statistically significant differences with respect to the wild type (WT) : **P < 0.01. The experiment was repeated three times with similar results. [See online article for color version of this figure.]
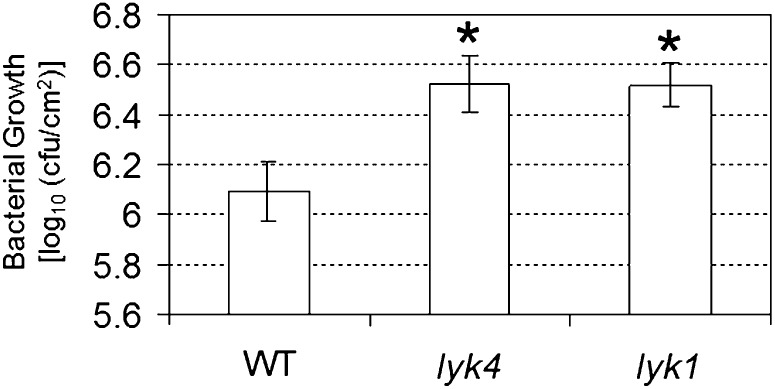
We also tested the lyk4 mutant to discern the susceptibility to infection with the hemibiotrophic bacterial pathogen Pseudomonas syringae pv tomato DC3000. As shown in Figure 8, both lyk4 and lyk1 mutants appeared to be significantly more susceptible to this bacterial pathogen than wild-type plants, as indicated by a higher bacterial growth rate (colony-forming units cm−2). These data suggest that LYK4 may also be involved in plant defense against bacterial pathogens.
Figure 8.
Response of the lyk4 mutant to a bacterial pathogen, P. syringae pv tomato DC3000. The pathogen was inoculated at a concentration of 5 × 104 colony-forming units (cfu) mL−1. Asterisks indicate statistically significant differences with respect to the wild type (WT): *P < 0.05. The experiment was repeated three times with similar results.
DISCUSSION
The LysM domain, first identified in enzymes involved in degrading peptidoglycan in the bacterial cell wall (Joris et al., 1992), has the capacity to recognize and bind sugar polymers composed of β-1,4-linked aminosugars, such as chitin, peptidoglycan, and the lipochitooligosaccharide NFs (Limpens et al., 2003; Madsen et al., 2003; Mulder et al., 2006; Miya et al., 2007; Radutoiu et al., 2007; Smit et al., 2007; Wan et al., 2008a). All sequenced plant genomes appear to have multiple LYK genes, with the possibility that each protein plays a different role in perceiving slightly different signals.
The Arabidopsis genome encodes three LysM domain-containing proteins (i.e. CEBiP-like1, -2, and -3), similar to OsCEBiP, which was shown to be critical for chitin perception in rice (Kaku et al., 2006). Interestingly, analyses of mutations in each gene or in combination of these genes indicate that they are not critical for chitin signaling in Arabidopsis. This difference suggests that different plants may employ different LysM proteins to perceive chitin; rice employs OsCERK1/OsCEBiP, and we suggest that Arabidopsis employs LYK1/LYK4 (see below).
The Arabidopsis genome encodes five LYKs (Zhang et al., 2007, 2009), of which LYK1/CERK1 appears to be the essential PRR involved in chitin perception and plant innate immunity (Miya et al., 2007; Wan et al., 2008a). In this work, we showed that LYK4 also plays an important role in chitin signaling, since a mutation in LYK4 led to diminished induction of CRGs and [Ca2+]cyt levels as well as enhanced susceptibility to both fungal and bacterial pathogens. Since mutations in LYK1 blocked every known response elicited by chitin, and LYK1 was shown to directly bind to chitin without apparent need of accessory proteins, this protein is clearly the major and essential PRR for chitin recognition. Given these findings, what is the role of LYK4?
One possibility is that LYK4 may function differentially in different parts of the plant, such as leaf and root. However, analysis of mRNA levels showed that LYK4 is expressed in both leaf and root as well as in other tissues (Fig. 1), and mutations in LYK4 had a similar effect on CRG expression in both leaf and root (Supplemental Fig. S3). Another possibility is that LYK4 may differ in its specificity for specific chitin oligomers, perhaps expanding the range of chitin oligomers that can be recognized by LYK1, which can recognize all known elicitor-active chitin oligomers (i.e. chitohexaose, chitoheptaose, and chitooctaose; Zhang et al., 2002; Miya et al., 2007; Wan et al., 2008a). However, our data indicate that lyk4 mutant plants were similarly defective in their response to both chitohexaose and chitooctaose in terms of gene regulation (Fig. 5). Similar results were seen in the calcium responses (Fig. 6). Therefore, LYK4 does not appear to be able to distinguish between chitohexaose and chitooctaose. Oligomers smaller than chitohexaose have only weak elicitor activity in wild-type, lyk4, and lyk1 plants. Additionally, we cannot rule out the possibility of recognition of other, unknown chitin-like molecules by LYK4.
Another possibility is that LYK4 may be a part of the receptor complex, in which it may interact with LYK1 to mediate chitin perception and signaling. In the case of rice, the data suggest that OsCEBiP likely mediates chitin recognition via interaction with the OsCERK1 protein (Shimizu et al., 2010). These proteins likely act as part of a protein complex that may also involve the Hop/Sti1, Hsp90, and OsRAC proteins (Chen et al., 2010). In the case of legumes, at least two LysM proteins appear to be involved in NF recognition. For example, in Lotus japonicus, two LYKs, LjNFR1 and LjNFR5, are postulated to interact and both are required for recognition of the lipochitooligosaccharide NF (Madsen et al., 2003; Radutoiu et al., 2003). Therefore, it is possible that LYK4 may interact directly or indirectly with LYK1 to aid in chitin recognition. However, direct evidence for this interaction is currently lacking. Recently, Petutschnig et al. (2010) showed that LYK4, LYK5, and CEBiP-like1 were pulled down together with LYK1 by chitin magnetic beads and were further eluted by chitohexaose from the beads (Petutschnig et al., 2010). We repeated this experiment and also found that LYK1, LYK4, LYK5, and CEBiP-like1 were pulled down by chitin magnetic beads and eluted by chitooctaose (Supplemental Table S1). Therefore, it remains a possibility that these proteins may constitute a chitin-recognition complex. However, only LYK1 appears to be essential for chitin recognition; hence, at best, LYK4 is serving to enhance the chitin response.
In the case of rice, it is presumed that OsCERK1 provides the necessary kinase function, since OsCEBiP does not have a kinase domain (Kaku et al., 2006; Miya et al., 2007; Chen et al., 2010; Shimizu et al., 2010; Shinya et al., 2010). In the case of LjNFR1 and LjNFR5, while both are LysM RLKs, only the LjNFR1 protein appears to have an active kinase (Radutoiu et al., 2003; Madsen et al., 2011). Hence, it is possible that chitin recognition requires two interacting LysM proteins, with at least one possessing the necessary kinase activity to initiate the downstream chitin signaling pathway. Interestingly, similar to LjNFR5, LYK4 also appears to be an inactive kinase, since the comparison of the LYK4 kinase domain with other typical kinases revealed that certain key residues are missing (Supplemental Fig. S7), especially those residues involved in aligning ATP for γ-phosphate group transfer in domain VII (Hanks and Hunter, 1995). Additionally, kinase assays using recombinant LYK4 protein isolated after expression in Escherichia coli failed to show phosphorylation of the common kinase substrate Myelin Basic Protein, although a kinase domain derived from a similar LYK was capable of phosphorylating the same substrate (Supplemental Fig. S8). Therefore, we speculate that, like LjNFR1 and LjNFR5 or OsCERK1 and OsCEBiP, Arabidopsis LYK1 and LYK4 may interact either directly or indirectly to form a chitin-receptor complex in which a single kinase domain, provided by LYK1, is the key step in downstream chitin signaling.
Our work here also suggests that LYK4 may be involved in plant defense against bacterial pathogens. These results are consistent with the observation that lyk1 mutant plants were more susceptible to P. syringae pv tomato DC3000 and that LYK1 is targeted by the bacterial effector protein (AvrPtoB) for degradation (Gimenez-Ibanez et al., 2009a, 2009b). Moreover, the mutations in LYK1 and LYK4 have a negligible impact on fls22 or elf26 signaling, based on the gene expression analysis (Supplemental Fig. S5). The findings that LYK1/CERK1 and LYK4 are involved in defense against both fungal and bacterial pathogens suggest that, in addition to chitin, they may be able to recognize and bind another unidentified elicitor from bacterial pathogens. Consistent with this hypothesis, a recent study demonstrated that the flagellin receptor FLS2 also mediates the perception of Xanthomonas AX21-secreted peptides to lead to defense against this pathogen (Danna et al., 2011). A likely candidate would be the cell wall peptidoglycan of pathogenic bacteria, as reported recently (Willmann et al., 2011), since the LysM domain has the potential to recognize and bind peptidoglycan (Buist et al., 2008).
CONCLUSION
This work has demonstrated that the mutation in LYK4 reduced the induction of CRGs in Arabidopsis and enhanced susceptibility to both a fungal and a bacterial pathogen. LYK4 was further shown to be induced by chitin and located at the PM. Therefore, LYK4 likely plays an auxiliary role in the chitin receptor complex to assist chitin signal transduction and plant innate immunity.
MATERIALS AND METHODS
Plants
Arabidopsis (Arabidopsis thaliana) Ds insertion mutants pst15072 and pst17581 were obtained for the CEBiP-like1 gene (At2g17120) and the CEBiP-like2 gene (At1g21880), respectively, from the RIKEN BioResource Center. One T-DNA insertion mutant, SALK_111212, was obtained for the CEBiP-like3 gene (At1g77630) from the Arabidopsis Biological Resource Center (ABRC). All the mutants were genotyped and raised to homozygosity. Additionally, crossing was done to generate a homozygous triple mutant for all three genes.
The lyk4 mutant, WiscDsLox297300_01C (also named CS850683 and CS863985 in the ABRC, with CS863985 derived from CS850683), was obtained from the Genome Center of Wisconsin (University of Wisconsin-Madison). Genomic DNA was isolated according to the protocol described by Edwards et al. (1991). The homozygous plants were identified by genotyping using the following gene-specific primers, 5′-CCACAATCGGTTTCTCCTCCTCCATTGTC-3′ and 5′-GTACGACGATTCTTCCCAGTTCTGCGTAG-3′, together with the T-DNA left border primer 5′-AACGTCCGCAATGTGTTATTAAGTTGTC-3′. These two primers were also used to detect whether the expression of the LYK4 gene was blocked in the mutant plants by RT-PCR (see below).
The mutant lines SALK_152226 for LYK2 (At3g01840) and SALK_140374 for LYK3 (At1g51940) were obtained from the ABRC, and CSHL_GT7089 for LYK5 (At2g33580) was obtained from Dr. Rob Martienssen. From these single mutants, a homozygous triple mutant was also made for all three genes through crossing.
Mutants of LYK4 and LYK1 in the Aequorin Transgenic Background and [Ca2+]cyt Measurement
The lyk1 (a Gabi-Kat line: 096F09; Wan et al., 2008a) and lyk4 mutants in the aequorin transgenic background were screened from F2 generation pools after cross-pollination between each mutant and the aequorin transgenic Arabidopsis line (kindly provided by Dr. Marc R. Knight). Homozygous T-DNA insertions were detected by PCR using the gene-specific primers and the T-DNA left border-specific primer (described above). The presence of the aequorin transgene was confirmed in the F2 generation by the detection of bioluminescence in its dissected cotyledon using reconstitution buffer and discharging solution (Tanaka et al., 2010); homozygosity was then determined in the F3 generation by the same method. [Ca2+]cyt measurement was performed as described previously (Tanaka et al., 2010).
Growth of Seedlings and Treatment with Chitin and flg22
Arabidopsis seedlings were grown in one-half-strength Murashige and Skoog liquid medium as described previously (Zhang et al., 2002). Approximately 10-d-old seedlings were treated with either chitohexaose or chitooctaose (Sigma) at a final concentration of 1 μm, or the flagellin-derived flg22 peptide, or elf26 peptide (GeneScript) at a concentration of 1 µm for 30 min. As a negative control, plants were similarly treated with an equivalent amount of solvents used to solubilize the elicitors. After treatment, samples were collected and frozen in liquid nitrogen for RNA isolation.
RNA Isolation and RT-PCR
Total RNA was isolated using the Trizol Reagent according to the manufacturer’s instructions (Invitrogen). The isolated RNA was further treated with Turbo DNase (Ambion) according to the manufacturer’s instructions to remove potential DNA contamination. Complementary DNA was synthesized using Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s instructions (Promega).
To analyze the expression of LYK4 in different tissues using RT-PCR, the following LYK4-specific primer pair was used, 5′-ATGATCTCGTTTTCATTTCATCTCCTC-3′ and 5′-GATACTTCACGCCATCTTCGTTGATC-3′, together with an internal control Actin2 (At3g18780) primer pair as follows: 5′-GACTAAGAGAGAAAGTAAGAGATAATCCAG-3′ and 5′-CAGCCTTTGATTTCAATTTGCATGTAAGAG-3′. RT-PCR was performed with both the LYK4-specific primer pair and the Actin2 primer pair under the following PCR conditions: 94°C for 3 min; 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min; and then 72°C for 3 min. The resultant PCR products were resolved on a 1% agarose gel for comparison. In the case of detecting LYK4 gene expression for comparison between the wild type and the lyk4 mutant, the cycles were increased from 25 to 39.
For quantitative RT-PCR, primer sequences for the following genes we tested were described previously (Libault et al., 2007): WRKY53 (At4g23810), MPK3 (At3g45640), ZAT12 (At5g59820), and a SAND gene (At2g28390). The SAND gene was used as an internal control to normalize gene expression across different samples. The reactions were conducted on a 7500 Real-Time PCR System (Applied Biosystems) using the SYBRGreen Master Mix (Applied Biosystems). The relative fold change of the target gene, normalized by the expression level of the SAND gene and relative to the gene expression in the control sample, was calculated as described before (Libault et al., 2007).
Generation of the CaMV 35S-LYK4, LYK4 Promoter-GUS, and LYK4-GFP Transgenic Plants
In order to express the LYK4 gene, the full-length coding sequence (1,839 bp) was amplified from the genomic DNA isolated from Arabidopsis ecotype Columbia (Col-0) using the following primer pair: 5′-acaaaGGTACCATCACGATGATCTCGTTTTC-3′ (with an engineered KpnI site underlined) and 5′-acaaaTCTAGATTAGTACGACGATTCTTCCCAG-3′ (with an underlined XbaI site). The amplified sequence was cloned into the modified binary vector pCAMBIA1200-35S under the control of the CaMV 35S promoter. The cloned LYK4 sequence was confirmed by sequencing. The final construct was electroporated into Agrobacterium tumefaciens AGL1. The resultant Agrobacterium was then used to transform Arabidopsis Col-0 plants via floral dipping (Clough and Bent, 1998). Transgenic plants were selected in the presence of hygromycin.
To make the LYK4 promoter-GUS fusion transgenic plants, the 2,046-bp sequence before the LYK4 start codon was amplified from the genomic DNA isolated from Arabidopsis Col-0 plants using the following primer pair: 5′-acaaaGTCGACGATCCGATTGTCACTCTCTG-3′ (with an underlined SalI site) and 5′-acaaaCCATGGCGTGATTCTGTAAGATTTGGT-3′ (with an underlined NcoI site). The amplified sequence was cloned into the binary vector pCAMBIA1391Z and confirmed by sequencing. The final construct was then used to generate transgenic plants using the previously mentioned methods. GUS staining was conducted as described (Jefferson et al., 1987).
To make the LYK4-GFP fusion transgenic plants, the LYK4 coding sequence (without the stop codon) was cloned into the binary vector pMDC83 to form a translational fusion with the GFP sequence via Gateway cloning strategies (Curtis and Grossniklaus, 2003), and the final construct was then used to generate transgenic plants using the previously mentioned methods. The GFP fluorescence in transgenic plants was observed with a confocal microscope (a Zeiss LSM 510 META NLO two-photon point-scanning confocal system).
Transient Expression of the LYK4-GFP Fusion Construct in Tobacco
The LYK4-GFP fusion construct, as well as the GFP construct, were electroporated into Agrobacterium tumefaciens GV3101. The different Agrobacterium strains were cultured overnight, pelleted, and resuspended in 10 mm MgCl2. After pretreatment with 40 µm acetosyringone (Sigma) for approximately 2 h at room temperature, the Agrobacterium strains harboring either the LYK4-GFP or GFP construct were mixed with Agrobacterium C58C1 expressing the silencing suppressor HC-Pro (Llave et al., 2000), with each strain adjusted to a final optical density of 0.3 at 600 nm. The bacterial mixture was coinfiltrated into leaves of 4-week-old tobacco (Nicotiana benthamiana) plants using a syringe to examine the transient expression of LYK4-GFP or GFP. Three days after infiltration, the infiltrated area was cut and observed using the previously mentioned confocal microscope.
Disease Assays
Disease assays with Alternaria brassicicola were conducted as described (van Wees et al., 2003; Veronese et al., 2006) with a spore suspension of 5 × 105 spores mL−1 by dot inoculating 5 µL of the spore solution onto leaves. The disease assays with Pseudomonas syringae pv tomato DC3000 carrying the luxCDABE operon (Fan et al., 2008; Bartels et al., 2009) were conducted at a bacterial concentration of 5 × 104 colony-forming units mL−1 as described (Fan et al., 2008; Bartels et al., 2009) using the Photek HRPCS4 photon detection camera system.
Expression of the LYK4 Recombinant Protein
To test the LYK4 chitin-binding capability, the coding sequence encoding the extracellular part (minus the signal peptide) of LYK4 was cloned into the expression vector pGEX-4T-3 to form the glutathione S-transferase (GST) fusion construct. The fusion protein was purified with Glutathione Sepharose 4B beads (GE Healthcare) and used in the chitin-binding assay.
To test the kinase activity of LYK4, the coding sequence encoding the intracellular part of LYK4 was cloned into pGEX-4T-3 to form the GST fusion construct. The fusion protein was purified and used in the in-gel kinase assay.
Chitin-Binding Assay and Pull-Down of Microsomal Fraction Proteins Using Chitin Magnetic Beads
In order to demonstrate protein binding to the chitin magnetic beads (New England Biolabs), 30 µg of the purified protein was inoculated with the chitin beads according to the method described by Petutschnig et al. (2010). The proteins pulled down by the beads were separated on a 12% SDS gel, transferred to a polyvinylidene difluoride membrane (Bio-Rad), and detected with the GST antibody from Sigma.
The pull-down of microsomal fraction proteins by chitin magnetic beads was conducted according to Petutschnig et al. (2010).
In Vitro Kinase Assay
The purified recombinant proteins were input in the in vitro kinase assay as described previously (Liu et al., 2011). The phosphorylated substrate was visualized by autoradiography after being separated on a 12.5% SDS-polyacrylamide gel.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of chitin-responsive genes in the CEBiP-like mutants.
Supplemental Figure S2. Induction of WRKY53 by chitin is not blocked in the mutants of the LYK2, LYK3, and LYK5 genes.
Supplemental Figure S3. Expression levels of chitin-responsive genes in the aerial tissue or root tissue of the lyk4 mutant seedlings.
Supplemental Figure S4. Analysis of chitin-responsive genes in complementation lines of the lyk4 mutant.
Supplemental Figure S5. Analysis of flg22- or elf26-induced gene expression in the lyk4 and lyk1 mutants.
Supplemental Figure S6. Effects of chitin treatments on the [Ca2+]cyt response in complementation lines of the lyk4 mutant.
Supplemental Figure S7. Comparison of the LYK4 kinase domain with other kinases.
Supplemental Figure S8. Kinase assay of the recombinant LYK4 protein.
Supplemental Table S1. Microsomal fraction proteins pulled down by chitin magnetic beads.
Supplementary Material
Acknowledgments
We thank Dr. Patrick J. Krysan (University of Wisconsin-Madison) for the lyk4 mutant WiscDsLox297300_01C, Dr. Rob Martienssen (Cold Spring Harbor Laboratory) for the lyk5 mutant CSHL_GT7089, Dr. Christopher Lawrence (Virginia Bioinformatics Institute) for Alternaria brassicicola, Dr. Chris Lamb (John Innes Centre) for P. syringae pv tomato DC3000 carrying the luxCDABE operon, Dr. Marc R. Knight (University of Oxford) for aequorin transgenic Arabidopsis, and Dr. Scott C. Peck (University of Missouri) for technical advice on using the Photek HRPCS4 photon detection camera.
Glossary
- LysM
lysin motif
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- CRG
chitin-responsive gene
- NF
nodulation factor
- PM
plasma membrane
- RT
reverse transcription
- CaMV
cauliflower mosaic virus
- [Ca2+]cyt
cytosolic free calcium ion concentration
- ABRC
Arabidopsis Biological Resource Center
- Col-0
ecotype Columbia
- GST
glutathione S-transferase
- T-DNA
transferred DNA
References
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R. (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21: 2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Chua NH. (1990) The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250: 959–966 [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok J, Kuipers OP. (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68: 838–847 [DOI] [PubMed] [Google Scholar]
- Chen L, Hamada S, Fujiwara M, Zhu T, Thao NP, Wong HL, Krishna P, Ueda T, Kaku H, Shibuya N, et al. (2010) The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 7: 185–196 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna CH, Millet YA, Koller T, Han SW, Bent AF, Ronald PC, Ausubel FM. (2011) The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc Natl Acad Sci USA 108: 9286–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, Thomma BP. (2009) Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol 17: 151–157 [DOI] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MH, Thomma BP. (2010) Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329: 953–955 [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Crooks C, Lamb C. (2008) High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J 53: 393–399 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. (2009a) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Ntoukakis V, Rathjen JP. (2009b) The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal Behav 4: 539–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9: 576–596 [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa EI, Mitsutomi M, Nagano Y. (2010) Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem 285: 2996–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Joris B, Englebert S, Chu CP, Kariyama R, Daneo-Moore L, Shockman GD, Ghuysen JM. (1992) Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol Lett 70: 257–264 [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapila J, De Rycke R, Van Montagu M, Angenon G. (1997) An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci 122: 101–108 [Google Scholar]
- Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R. (2011) Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem J 440: 355–365 [DOI] [PubMed] [Google Scholar]
- Lenardon MD, Munro CA, Gow NA. (2010) Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol 13: 416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20: 900–911 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZD, Coaker G. (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Carrington JC. (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA 97: 13401–13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid MP, Di Pietro A, Roncero MI. (2003) Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol Microbiol 47: 257–266 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Antolín-Llovera M, Grossmann C, Ye J, Vieweg S, Broghammer A, Krusell L, Radutoiu S, Jensen ON, Stougaard J, et al. (2011) Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J 65: 404–417 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Michielse CB, van Wijk R, Reijnen L, Cornelissen BJ, Rep M. (2009) Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large-scale insertional mutagenesis. Genome Biol 10: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L, Lefebvre B, Cullimore J, Imberty A. (2006) LysM domains of Medicago truncatula NFP protein involved in Nod factor perception: glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology 16: 801–809 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. (2010) The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem 285: 28902–28911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S. (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138: 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N, Minami E. (2001) Oligosaccharide signalling for defence responses in plant. Physiol Mol Plant Pathol 59: 223–233 [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T, Osada T, Desaki Y, Hatamoto M, Yamanaka Y, Hirano H, Takai R, Che FS, Kaku H, Shibuya N. (2010) Characterization of receptor proteins using affinity cross-linking with biotinylated ligands. Plant Cell Physiol 51: 262–270 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen A, Buist G, Leenhouts KJ, El Khattabi M, Grijpstra F, Zomer AL, Venema G, Kuipers OP, Kok J. (2003) Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem 278: 23874–23881 [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I, de Wit PJ. (2009) Fungal effector proteins. Annu Rev Phytopathol 47: 233–263 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Swanson SJ, Gilroy S, Stacey G. (2010) Extracellular nucleotides elicit cytosolic free calcium oscillations in Arabidopsis. Plant Physiol 154: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, de Wit PJ. (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19: 1420–1430 [DOI] [PubMed] [Google Scholar]
- van Wees SC, Chang HS, Zhu T, Glazebrook J. (2003) Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol 132: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang S, Stacey G. (2004) Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol Plant Pathol 5: 125–135 [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. (2008a) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Stacey G. (2008b) Chitin signaling and plant disease resistance. Plant Signal Behav 3: 831–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono JJ, Cullimore JV, et al. (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA 108: 19824–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Ramonell K, Somerville S, Stacey G. (2002) Characterization of early, chitin-induced gene expression in Arabidopsis. Mol Plant Microbe Interact 15: 963–970 [DOI] [PubMed] [Google Scholar]
- Zhang XC, Cannon SB, Stacey G. (2009) Evolutionary genomics of LysM genes in land plants. BMC Evol Biol 9: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, Cannon SB, Stacey G. (2007) Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol 144: 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.