Abstract
Glycosylation is the most abundant and complex posttranslational modification to be considered for recombinant production of therapeutic proteins. Mucin-type (N-acetylgalactosamine [GalNAc]-type) O-glycosylation is found in eumetazoan cells but absent in plants and yeast, making these cell types an obvious choice for de novo engineering of this O-glycosylation pathway. We previously showed that transient implementation of O-glycosylation capacity in plants requires introduction of the synthesis of the donor substrate UDP-GalNAc and one or more polypeptide GalNAc-transferases for incorporating GalNAc residues into proteins. Here, we have stably engineered O-glycosylation capacity in two plant cell systems, soil-grown Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum) Bright Yellow-2 suspension culture cells. Efficient GalNAc O-glycosylation of two stably coexpressed substrate O-glycoproteins was obtained, but a high degree of proline hydroxylation and hydroxyproline-linked arabinosides, on a mucin (MUC1)-derived substrate, was also observed. Addition of the prolyl 4-hydroxylase inhibitor 2,2-dipyridyl, however, effectively suppressed proline hydroxylation and arabinosylation of MUC1 in Bright Yellow-2 cells. In summary, stably engineered mammalian type O-glycosylation was established in transgenic plants, demonstrating that plants may serve as host cells for the production of recombinant O-glycoproteins. However, the present stable implementation further strengthens the notion that elimination of endogenous posttranslational modifications may be needed for the production of protein therapeutics.
Posttranslational modifications (PTMs) of recombinant protein therapeutics are important, with glycosylation being by far the most important PTM to address. N- and O-linked glycosylation are often essential for biological activity and pharmacokinetic properties (Li and d’Anjou, 2009). Recombinant production of glycoprotein therapeutics has so far generally aimed to mimic the glycosylation found on naturally produced human glycoproteins, primarily to avoid adverse immunological reactivity and improve circulatory half-life. Considerable efforts have thus been devoted to the design and optimization of host cells aimed for recombinant expression of biologics. Significant success has been achieved with designed N-glycosylation in higher eukaryotes such as plants (Strasser et al., 2008; Gomord et al., 2010), lower eukaryotes such as yeast (Gerngross, 2004), and even extended to some prokaryotes (Hug et al., 2011). In contrast, limited progress has been made with respect to O-glycosylation. O-Glycosylation is one of the most complex regulated PTMs of proteins and also one of the least understood (Bennett et al., 2012). O-Glycosylation is initiated by a family of up to 20 UDP-GalNAc polypeptide N-acetylgalactosaminyltransferases (GalNAc-Ts), which allows for huge variability of O-glycan attachments to proteins in different cells, a feature not found for most other types of protein glycosylation. Engineering glycosylation in host cells may also aim to produce aberrant truncated glycoforms that mimic those found in disease with the intent, for example, to target proteins to antigen-presenting cells and augment immunity in vaccine strategies. Therefore, it may be desirable to build glycosylation capacity in host cells de novo in order to attain complete design freedom without an extensive requirement for gene elimination. This is particularly desirable for mammalian O-glycosylation, where cell-specific repertoires of the many GalNAc-Ts direct the glycosylation capacity.
Most glycoprotein therapeutics are produced in Chinese hamster (Cricetulus griseus) ovary (CHO) cells (Durocher and Butler, 2009). CHO cells produce monosialylated and disialylated core 1 (Galβ1-3GalNAcα1 -O-Ser/Thr) O-glycan structures (Sasaki et al., 1987). Recently, the repertoire of GalNAc-Ts expressed in CHO cells was shown to include only four isoforms (GalNAc-T2, -T7, -T11, and -T19; Xu et al., 2011), thus demonstrating that CHO cells have limited capacity for O-glycosylation. Insect cells produce immature O-glycans generally without sialic acids and have an as yet unknown GalNAc-T repertoire (Thomsen et al., 1990). Yeast does not produce GalNAc-type O-glycosylation but O-mannosylates similar Ser/Thr residues as GalNAc O-glycosylation in higher eukaryotes. The yeast Pichia pastoris was previously used as a model system to engineer the first step of human (Homo sapiens) O-glycosylation from scratch (Amano et al., 2008). However, since endoplasmic reticulum-localized O-mannosylation precedes GalNAc O-glycosylation in the Golgi apparatus, considerable O-mannosylation was found on glycoproteins expressed in the engineered cells. This competing O-mannosylation was partly blocked by coculturing in rhodanine-3-acetic acid, but complete elimination of O-mannosylation is presumably not possible, as knockout of three or more of the seven O-mannosyltransferases is lethal (Strahl-Bolsinger et al., 1999).
Plants are emerging as an attractive alternative platform for the expression of glycoproteins (Cox et al., 2006; Sainsbury and Lomonossoff, 2008; Pogue et al., 2010). Several studies have demonstrated that plants can efficiently produce complex mammalian proteins, such as monoclonal antibodies with humanized N-glycan structure (Cox et al., 2006; Strasser et al., 2008; Castilho et al., 2010; Pogue et al., 2010). Whereas plants and mammalian cells have identical core N-linked glycosylation, plants do not perform GalNAc O-glycosylation (Gomord and Faye, 2004; Saint-Jore-Dupas et al., 2007; Bennett et al., 2012). Plants, however, produce another type of O-glycosylation, which is primarily found in the Hyp-rich glycoprotein superfamily, where, for example, arabinogalactan and arabinosides are attached to Hyp residues of arabinogalactan proteins (AGPs) and extensins, respectively. This type of O-glycosylation requires the initial action of prolyl 4-hydroxylases (P4Hs), and although a consensus motif for this type of modification has not been defined, it appears to occur mainly in regions with a high density of Pro residues (Shimizu et al., 2005; Estévez et al., 2006; Jamet et al., 2008; Lamport et al., 2011). The Hyp contiguity hypothesis predicts arabinosylation of contiguous Hyp residues (two or more) with extensin-type β-linked arabinofuranosides β-Arafs) of up to four residues in length and galactoarabinosylation of clustered noncontiguous Hyp residues in AGPs (Shpak et al., 1999; Zhao et al., 2002; Tan et al., 2010). Several small peptide hormones and plasma membrane kinases involved in cell wall sensing have also been shown to be arabinosylated (for review, see Ringli, 2010; Matsubayashi, 2011). While small peptide hormones and plasma membrane kinases invariably carry the Hyp-(β-Araf)3 side chain, extensin β-Araf side chains may be shorter or additionally capped with a α-linked arabinofuranosides. The amino acid sequence SerProProPro(Pro) directs arabinosylation in extensins (Kieliszewski et al., 2011), but a glycosylation motif has not yet been identified in the plasma membrane kinases or the small peptide hormones. Plants also produce O-linked galactosylation of Ser residues, which is found on some vacuolar proteins such as sweet potato (Ipomoea batatas) sporamin of the Solanaceae family (solanaceous lectin-type O-galactosylation) and in the plant-specific extensin SerProProPro(Pro) motif repeat of cell wall extensins (Lamport et al., 1973; Yong-Pill and Chrispeels, 1976; Matsuoka et al., 1995; Kieliszewski and Shpak, 2001). The restricted occurrence or sequence specificity of these plant-specific types of O-glycosylation should make plants an ideal candidate cell system for the custom design and buildup of O-glycosylation from scratch with respect to both sites of O-glycan attachment and O-glycan structures.
Daskalova et al. (2010) initially started engineering plants for mammalian O-glycosylation and suggested that GalNAc O-glycosylation in Nicotiana benthamiana plants required a Glc(NAc) C4-epimerase, a UDP-Gal(NAc) transporter, and a human GalNAc-T. We subsequently tested a variety of design strategies for the introduction of GalNAc O-glycosylation in plants using transient expression of the glycogenes in N. benthamiana and found that O-glycosylation capacity was achieved by the introduction of only the Pseudomonas aeruginosa Glc(NAc) C4-epimerase (WbpP; Creuzenet et al., 2000) and a human GalNAc-T. The apparent discrepancy with regard to the requirement for the introduction of a UDP-GalNAc transporter is not fully clarified, but the study by Daskalova et al. (2010) was solely based on lectin labeling, and the authors subsequently demonstrated that the lectins used detect endogenous plant proteins (Reaves et al., 2011), causing serious concerns with regard to the original findings. Our study included the production of recombinant glycoproteins in quantities allowing for a detailed characterization of the introduced glycosylation by mass spectrometry (Yang et al., 2012). However, stoichiometry and subcellular localization of the introduced glycoenzymes and subcellular substrate pools are increasingly difficult to control as the engineering effort increases in complexity. Stable engineering of especially the glycosylation machineries is thus generally needed at an early point, as seen in the engineering efforts to humanize plants with respect to N-linked protein glycosylation (Strasser et al., 2008; Castilho et al., 2010; Kaulfürst-Soboll et al., 2011; Nagels et al., 2011; Schoberer and Strasser, 2011; Yin et al., 2011).
Here, we have established stable O-glycosylation capacity in two plant cell systems, soil-grown Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) suspension culture cells. In agreement with our previous studies using transient expression (Yang et al., 2012), we found that efficient O-glycosylation capacity was obtained without the introduction of a UDP-Gal(NAc) transporter in Arabidopsis and tobacco BY-2 suspension cells. We also found that Pro hydroxylation and Hyp-linked arabinosylation produced by the endogenous plant PTM machinery were considerably more pronounced in stable engineered cells compared with our transiently engineered cells. Interestingly, for suspension cell cultures, the presence of a P4H inhibitor resulted in a marked reduction of the endogenous PTMs. In summary, plants were stably engineered with efficient O-glycosylation machineries, but further host cell engineering aimed at eliminating Pro hydroxylation in O-glycoproteins prone for this endogenous modification appear to be needed.
RESULTS
Engineering GalNAc O-Glycosylation in Plants
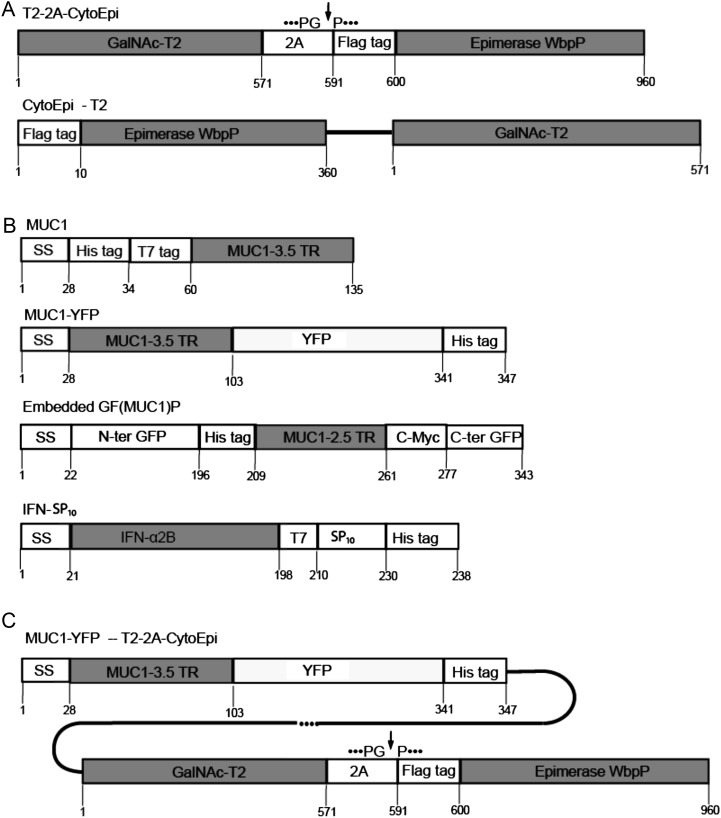
Previously, using transient expression in N. benthamiana, we identified optimal two-component GalNAc O-glycosylation machinery designs (Fig. 1), which conferred efficient O-glycosylation of coexpressed reporter substrates (Yang et al., 2012). Of the earlier transiently tested O-glycosylation machinery constructs, T2-2A-CytoEpi and CytoEpi-T2 (Fig. 1A), expressing Golgi-targeted GalNAc-T2 and cytoplasmic epimerase as a single polyprotein interspaced by the 2A self-cleavage sequence or as separate proteins, respectively, conferred efficient O-glycosylation in the stable implementations.
Figure 1.
Construct designs for engineering O-glycosylation. A, Depiction of O-glycosylation machinery construct designs. Designs included Flag-tagged cytoplasm-targeted P. aeruginosa C4-epimerase WbpP (CytoEpi) and Golgi-targeted polypeptide GalNAc-T2 (T2) expressed as a single polycistronic protein interspaced by the 2A self-cleaving sequence (cleavage site indicated by the arrow; T2-2A-CytoEpi) or as separate transcriptional units (CytoEpi-T2). B, Depiction of the O-glycosylation reporter constructs used: MUC1-3.5TR (MUC1) with and without a C-terminal fusion to YFP (MUC1-YFP); MUC1 embedded into GFP [GF(MUC1)P]; Full-coding IFNα2B cytokine with a C-terminal T7 and SP10 glycomodule (IFN-SP10). All reporter constructs included N- or C-terminal His6 tags and N-terminal signal peptide sequences (SS). C, Depiction of a single combined construct with reporter MUC1-YFP and the 2A-linked O-glycosylation machinery under the control of two promoters (MUC1-YFP-T2-2A-CytoEpi).
Stably introduced O-glycosylation reporter constructs included the following: (1) the approximately 11-kD 3.5-tandem repeat (TR) sequence of the cancer-associated mucin MUC1 (MUC1-3.5TR) N terminally fused to a His6 and T7 tag (MUC1); (2) MUC1-3.5TR C terminally fused to yellow fluorescent protein (YFP; MUC1-YFP); (3) MUC1-2.5TR embedded into GFP [GF(MUC1)P]; (4) the full-length secreted IFNα2B cytokine tagged with a T7 tag and an AGP type SP10 glycomodule (IFN-SP10; Fig. 1B); and (5) 1.2TR of MUC16 tagged with T7 and His6 tags (MUC16; Supplemental Fig. S1B). All reporter constructs included an N-terminal signal peptide sequence for direction into the secretory pathway.
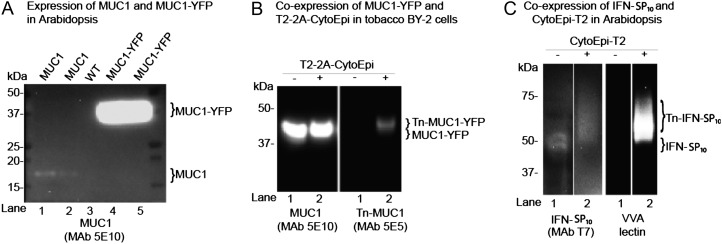
We first tested the expression and stability of reporter substrates. Expression of the small approximately 11-kD MUC1 3.5TR (MUC1) reporter in Arabidopsis and water-grown Lemna minor conferred only weak accumulation (Fig. 2A, lanes 1 and 2; Supplemental Fig. S1A) but was not detectable in BY-2 cells (data not shown), as evidenced by SDS-PAGE western-blot analyses. Expression of the fusion reporter substrate MUC1-YFP (Fig. 1B), however, resulted in agreement with the transient implementations (Yang et al., 2012) in an approximately 20-fold increased accumulation in Arabidopsis (Fig. 2A, lanes 4 and 5) as compared with expression of MUC1 alone (Fig. 2A, lanes 1 and 2). Expression of MUC1-YFP in BY-2 cell callus also resulted in detectable intracellular accumulation (Fig. 2B, lane 1). Intracellular O-glycosylation of MUC1-YFP in BY-2 cells was achieved by coexpression of the O-glycosylation machinery T2-2A-CytoEpi, as evidenced by western-blot analysis using a GalNAc-MUC1 glycoform (Tn-MUC1)-specific monoclonal antibody (MAb), 5E5 (Fig. 2B, lane 2). However, neither MUC1-YFP nor Tn-MUC1-YFP was detectable in the BY-2 cell medium (data not shown). Expression of another mucin-type substrate, MUC16, also conferred accumulation in the BY-2 medium. The MUC16 reporter construct design contains three potential N-glycosylation sites and up to 30 O-glycosylation sites, and peptide N-glycosidase F treatment of MUC16-containing medium fractions resulted in mobility shifts to an apparent slightly lower molecular weight on SDS-PAGE western blots, in agreement with the presence of N-glycosylation sites that are utilized by the endogenous plant N-glycosylation machinery (Supplemental Fig. S1B).
Figure 2.
Implementation of O-glycosylation in Arabidopsis and tobacco BY-2 suspension cells. A, SDS-PAGE western-blot analysis of four Arabidopsis lines expressing either MUC1 (lanes 1 and 2) or MUC1-YFP (lanes 4 and 5) alone. The contrast of the image was adjusted to visualize very weak bands for MUC1 (lanes 1 and 2). Lane 3 shows results for the wild type (WT). B, SDS-PAGE western-blot analysis of two tobacco BY-2 suspension cell lines expressing MUC1-YFP alone (lane 1) and MUC1-YFP together with the 2A-linked O-glycosylation machinery T2-2A-CytoEpi (lane 2). The absence (−) or presence (+) of O-glycosylation machinery is indicated above the lanes. C, An Arabidopsis line transgenic for T7-tagged full-coding secreted IFNα2B expressed alone (−) or coexpressed with the O-glycosylation machinery CytoEpi-T2 (+). The glycosylation of His tag-purified IFNα2B was detected by VVA lectin-blot analysis. Total protein extracts from transgenic Arabidopsis leaves or BY-2 cell callus were loaded and blots reacted with MUC1-specific MAbs 5E10 and 5E5, where 5E5 is specific for GalNAc-glycosylated MUC1 (Tn-MUC1) and does not react with nonglycosylated MUC1. Approximately 30 μg of total protein was loaded in each lane.
The secreted form of IFNα2B tagged with a SP10 glycomodule (IFN-SP10) was also tested as an O-glycosylation substrate in Arabidopsis. Coexpression with the O-glycosylation machinery CytoEpi-T2 resulted in a mobility shift and reactivity with the Vicia villosa (VVA) lectin (Fig. 2C). IFNα2B only has a single O-glycosylation site (GVGVT132ETPLM137), which is known to be O-glycosylated specifically by GalNAc-T2 (DeFrees et al., 2006). The observed mobility shift and VVA labeling clearly indicate that the protein was O-glycosylated (Fig. 2C), although mass spectrometry analysis is needed for confirmation of the glycosylation site.
Finally, using transient expression of the glycogenes, prior introduction of the glycosylation machinery appeared to enhance the full occupancy of the three GalNAc-T2-specific O-glycosylation sites in the MUC1 TR from approximately 5% to approximately 40% (Supplemental Fig. S4), perhaps pointing to the significance of providing ample amounts of donor substrate and transferase enzyme.
MUC1 of MUC1-YFP Is Degraded in BY-2 Cell Medium
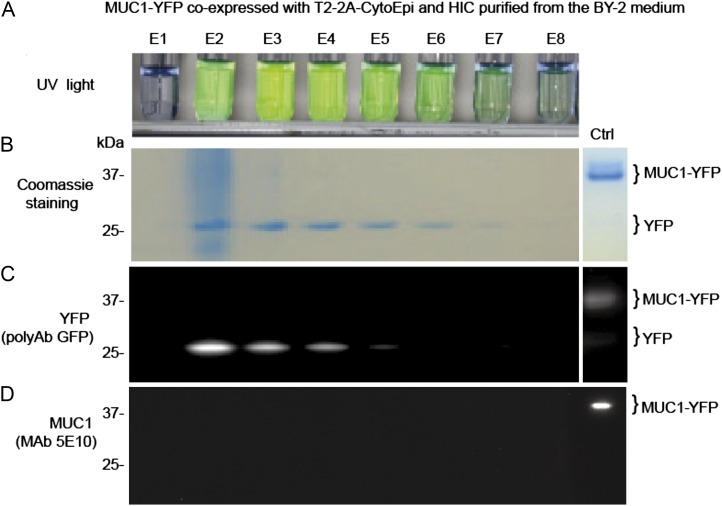
Repeated failed attempts to detect MUC1 expressed alone or in fusion with the carrier YFP in the BY-2 medium fractions prompted us to look further into the fate of MUC1-YFP. A BY-2 transgenic cell line stably expressing MUC1-YFP and the O-glycosylation machinery T2-2A-CytoEpi failed to produce detectable levels of the MUC1-YFP protein (37 kD) in BY-2 medium, as evidenced by MUC1-specific western-blot analysis (data not shown). His tag affinity purification of MUC1-YFP from spent medium of the BY-2 cell line was also not possible (data not shown). Concentration by hydrophobic interaction chromatography (HIC), however, revealed the presence of substantial amounts of fluorescent protein (Fig. 3A), which migrated as a 25-kD protein on SDS-PAGE gels and reacted with a GFP polyclonal antibody (Fig. 3, B and C) but not with a MUC1-specific MAb (Fig. 3D). The yield was estimated to be approximately 25 mg L−1, as judged from Coomassie blue-stained SDS-PAGE gels (Fig. 3B). Based on this, we concluded that MUC1-YFP was partially degraded in the BY-2 medium, resulting in release of the C-terminal His tag and the N-terminal MUC1 sequence. A construct expressing a fusion of an AGP-type SP10 glycomodule (Xu et al., 2010) to the C terminus of MUC1 also failed to confer the accumulation of MUC1 in the BY-2 medium (data not shown).
Figure 3.
MUC1-YFP is expressed and glycosylated in BY-2 cells, but MUC1 is degraded in the medium. A, UV image of fractions (E1–E8) from HIC of MUC1-YFP secreted from BY-2 cells coexpressing T2-2A-CytoEpi. B, SDS-PAGE Coomassie blue-stained analysis of fractions. C, Western blot with anti-GFP antibody. D, Western blot with anti-MUC1 (5E10). Ten microliters of 2-mL eluate fractions was applied in each lane. [See online article for color version of this figure.]
Embedding and Glycosylation Stabilize Secreted MUC1 in BY-2 Medium
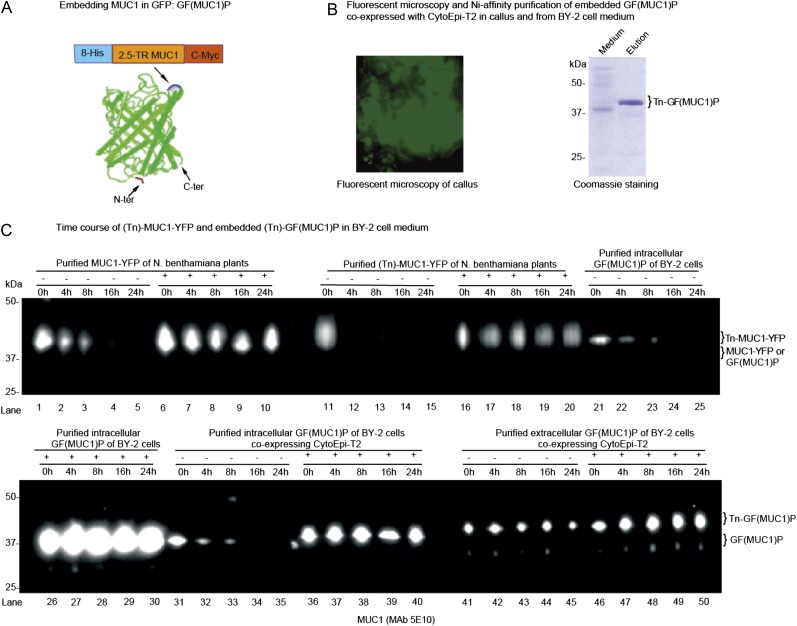
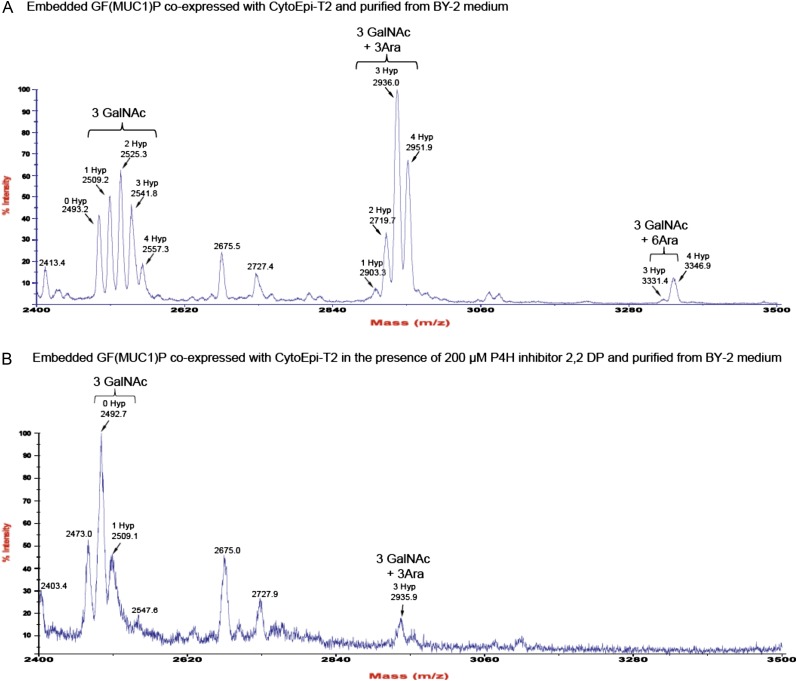
Previous studies have shown that embedding of small otherwise labile proteins in GFP may confer increased stabilization (Kobayashi et al., 2008). Accordingly, we prepared an embedded MUC1 construct, designated GF(MUC1)P, where a MUC1-2.5TR sequence flanked C terminally with a His8 tag and N terminally with a Myc tag was embedded in the loop between Asp-196 and Gly-197 of GFP (Fig. 4A). A transgenic BY-2 cell line expressing the embedded GF(MUC1)P substrate together with the O-glycosylation machinery CytoEpi-T2 appeared to accumulate fluorescence in callus cells (Fig. 4B, left panel). Isolation of GF(MUC1)P from the medium by nickel-affinity chromatography was now feasible, and the isolated product migrated as a single band (approximately 40 kD) as judged by Coomassie blue-stained SDS-PAGE gels (Fig. 4B, right panel), suggesting that MUC1 was stabilized by embedding it into GFP. Endoproteinase Asp mediated release of the MUC1 TR 20-mer peptide from the purified product with subsequent matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) confirmed a uniform incorporation of three GalNAc residues (Fig. 5A). However, evidence was also found for the modification of Pro to Hyp and of Hyp with three (approximately 45%) and less frequently six (approximately 5%) Ara residues attached (Fig. 5A). The MUC1 repeat contains five Pro residues, of which up to four were hydroxylated.
Figure 4.
O-Glycosylation and embedding in GFP stabilize MUC1 in BY-2 cell culture. A, Construct design for embedding MUC1-2.5TR into GFP [GF(MUC1)P], and corresponding barrel structure showing the loop into which MUC1-2.5 TR flanked with His8 and C-Myc tags was inserted after Asp-196 in the loop between the β-strands (blue) located opposite the N and C termini. This figure was adapted from Kobayashi et al. (2008). B, Fluorescence microscopy of a stable BY-2 transgenic line coexpressing GF(MUC1)P with the O-glycosylation machinery CytoEpi-T2 (left panel), and SDS-PAGE Coomassie blue staining of nickel chromatography-purified secreted GalNAc-glycosylated GF(MUC1)P [Tn-GF(MUC1)P] of this line (right panel). C, Analysis of the degradation of MUC1-YFP and GF(MUC1)P in BY-2 cell culture medium by SDS-PAGE western blotting: (1) MUC1-YFP (lanes 1–10) and GalNAc-glycosylated MUC1-YFP (Tn-MUC1-YFP; lanes 11–20) transiently produced in leaves of N. benthamiana plants; (2) intracellularly embedded GF(MUC1)P (lanes 21–30) purified from a stably transformed BY-2 cell line expressing GF(MUC1)P alone; (3) intracellularly (lanes 31–40) and extracellularly (lanes 41–50) embedded (Tn)-GF(MUC1)P purified from a transgenic BY-2 cell line additionally coexpressing CytoEpi-T2. The isolated proteins were added to 7-d-old unboiled (−) or boiled (+) wild-type BY-2 medium fractions, which were further incubated for up to 24 h under the same conditions. Approximately 5 μg of purified proteins was added to 1 mL of BY-2 medium fractions, of which approximately 15 μL was loaded and blots probed with anti-MUC1 MAb 5E10. The corresponding Tn-MUC1-specific western blot using MAb 5E5 is presented in Supplemental Figure S3. [See online article for color version of this figure.]
Figure 5.
Inhibition of Pro hydroxylation of MUC1 in BY-2 cells. A, MALDI-TOF-MS analysis of endoproteinase Asp-released MUC1 tandem repeats (DTRPAPGSTAPPAHGVTSAP; indicated by arrows) from Tn-GF(MUC1)P expressed in BY-2 cells coexpressing CytoEpi-T2. The analysis revealed the presence of 3 mol of attached GalNAc (i.e. complete GalNAc-T2-mediated GalNAc occupancy of sites), with varying Pro hydroxylations of up to 4 mol per tandem repeat. Hydroxylated MUC1 peptides were further modified by the attachment of mainly three, and less frequently of six, Ara residues. B, MALDI-TOF-MS analysis of MUC1 tandem repeats (indicated by arrows) of GF(MUC1)P coexpressed with CytoEpi-T2 in BY-2 cells grown in the presence of a P4H inhibitor. Growth medium was exchanged at day 7 with fresh medium including 200 µm 2,2-DP, followed by incubation in similar growth conditions. [See online article for color version of this figure.]
O-Glycosylation Further Stabilizes MUC1
An in vitro degradation model was used to evaluate the effect of O-glycosylation on MUC1 stability in BY-2 medium. Purified GF(MUC1)P of BY-2 cell lysates, purified glycosylated Tn-GF(MUC1)P of BY-2 medium, and purified MUC1-YFP and Tn-MUC1-YFP, transiently produced in N. benthamiana plants, were added to 7-d-old BY-2 wild-type medium with and without prior boiling (Fig. 4). While all MUC1 substrates were stable in boiled medium (Fig. 4C), only the Tn-GF(MUC1)P was stable in nonboiled medium after 24 h of incubation (Fig. 4C, lanes 41–45), as evidenced by western-blot analysis with the MUC1-specific MAb 5E10. For the corresponding Tn-MUC1 glycoform-specific western blot, see Supplemental Figure S3.
2,2-Dipyridyl Inhibits Pro Hydroxylation and Arabinosylation in BY-2 Cells
In order to block or reduce formation of the observed endogenous PTMs on the GF(MUC1)P reporter substrate, three inhibitors of Pro hydroxylase activity were selected (ethyl-3,4-dihydroxybenzoate, 3,4-dehydroproline, and 2,2-dipyridyl [2,2-DP]; Schmidt et al., 1991; Velasquez et al., 2011) and added to the BY-2 medium at various concentrations. Addition of all three inhibitors reduced the accumulated level of GF(MUC1)P in the medium as well as cell growth rates (Supplemental Fig. S5). Only 2,2-DP was able to efficiently suppress Pro hydroxylation of MUC1 TRs and the accompanying attachment of pentose sugars (Fig. 5B; Supplemental Fig. S6). The larger arabinogalactans and the short arabinofuranosides are the only known Hyp-linked glycans in plants (Lamport et al., 2011). In the transient expression system, addition of the Pro hydroxylase inhibitors conferred a minor reduction in the Pro hydroxylation level, from approximately 20% to approximately 10%, without an accompanying reduced level of expressed MUC1-YFP (Supplemental Fig. S7).
MUC1-YFP Is Glycosylated and Hyp Is Arabinosylated in Glycoengineered Arabidopsis
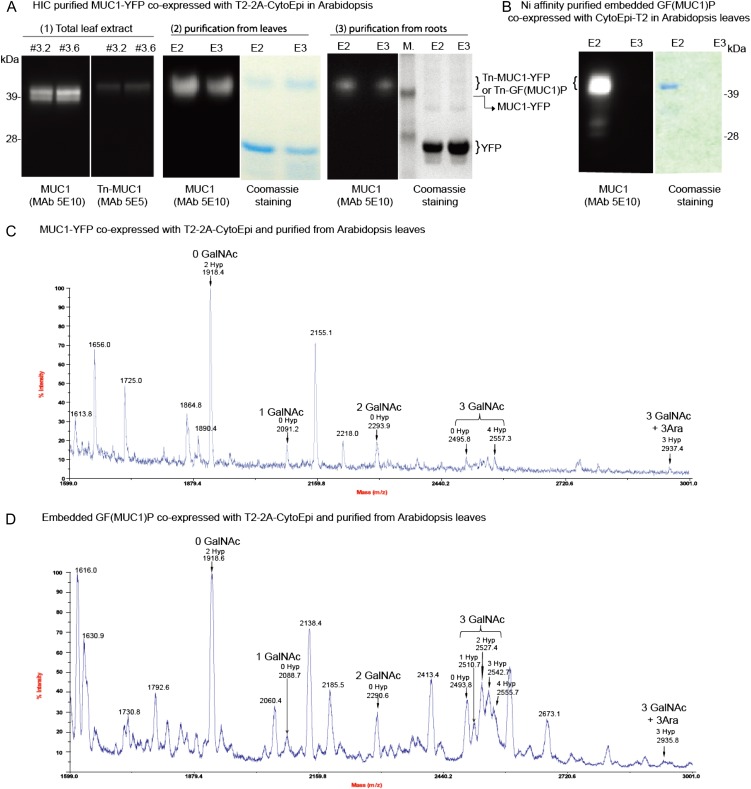
Expression of the MUC1 constructs and their use as glycosylation targets were also tested in soil-grown Arabidopsis. SDS-PAGE western-blot analysis of Arabidopsis lines expressing a single construct encoding combined expression of MUC1-YFP and the O-glycosylation machinery T2-2A-CytoEpi (Fig. 1C) revealed the presence of two closely migrating bands when probed with MAb 5E10 specific to MUC1 (Fig. 6A, 1). Only the upper of these bands reacted with MAb 5E5, which is specific to Tn-MUC1 (Fig. 6A, 1). The MUC1-YFP was purified from leaves and roots by HIC, and O-glycosylation was confirmed by MALDI-TOF-MS analysis. Coomassie blue staining of purified MUC1-YFP from Arabidopsis leaves revealed that the major product was YFP without the MUC1 entity (Fig. 6A, 2). MUC1-YFP of the roots was almost completely degraded, as judged by Coomassie blue-stained SDS-PAGE gels (Fig. 6A, 3, right panel), with the MUC1-YFP being only detectable in HIC-enriched elution fractions (Fig. 6A, 3, left panel). Nickel-affinity purification of embedded GF(MUC1)P coexpressed with CytoEpi-T2 revealed very little degradation (Fig. 6B).
Figure 6.
MUC1-YFP and GF(MUC1)P expressed with or without O-glycosylation machinery in Arabidopsis. A, SDS-PAGE analysis of a transgenic Arabidopsis line expressing a combined construct (MUC1-YFP-T2-2A-CytoEpi) comprising both the reporter MUC1-YFP and the O-glycosylation machinery T2-2A-CytoEpi. Shown are MUC1-specific and Tn-MUC1-specific western-blot analysis of total leaf extracts (1) and MUC1-YFP purified from leaves (2) and roots (3) by HIC as visualized by Coomassie blue-stained SDS-PAGE gels and MUC1-specific MAb 5E10 western blots of the eluates. B, His tag purification of embedded GF(MUC1)P coexpressed with the O-glycosylation machinery CytoEpi-T2 as visualized by Coomassie blue-stained SDS-PAGE gels and MUC1-specific MAb 5E10 western blot. C and D, MALDI-TOF-MS analysis of MUC1 tandem repeats (DTRPAPGSTAPPAHGVTSAP; indicated by arrows) from His tag-purified Asp-N-digested MUC1-YFP (C) and GF(MUC1)P (D) coexpressed with the O-glycosylation machineries T2-2A-CytoEpi and CytoEpi-T2, respectively. [See online article for color version of this figure.]
Both MUC1-YFP (Fig. 6C) and embedded GF(MUC1)P (Fig. 6D) coexpressed with T2-2A-CytoEpi and CytoEpi-T2, respectively, were subjected to Asp-N digestion for release of the MUC1 TR for subsequent MALDI-TOF-MS analysis. Approximately 30% and 70% of MUC1-YFP (Fig. 6C) and embedded GF(MUC1)P (Fig. 6D), respectively, were O-glycosylated. The dominant peak was found to be nonglycosylated MUC1-1TR with two Pro hydroxylations (mass-to-charge ratio of 1,918; Fig. 6, C and D). Peaks corresponding to Tn-MUC1-1TR with one to three GalNAc attachments and one to four Pro hydroxylations were observed (Fig. 6, C and D). Minor additional products, corresponding to the addition of three Ara residues, were also identified (Fig. 6C).
DISCUSSION
We previously established mammalian GalNAc O-glycosylation capacity transiently in plants. GalNAc O-glycosylation capacity required the introduction of a two-component O-glycosylation machinery basically consisting of a C4-epimerase and a GalNAc-T (Yang et al., 2012). Here, we have established O-glycosylation capacity stably in two different plant cell systems: (1) tobacco BY-2 suspension cells, which offers potential for the secretion of products (Hellwig et al., 2004) and has low reproductive constraints with regard to the propagation and manipulation of transgenic lines; and (2) soil-grown Arabidopsis, which has a comprehensive transferred DNA knockout repertoire (http://signal.salk.edu/) enabling, for example, the manipulation of endogenous-derived PTMs. A previous report proposed that a three-component machinery (additionally including a UDP-GalNAc-T) was required for the introduction of O-glycosylation in plants (Daskalova et al., 2010), but our results using transient (Yang et al., 2012) and now also stable introduction of O-glycosylation machineries clearly demonstrate that the UDP-GalNAc-T is not required.
We did find unexpected differences in efficiency of the glycosylation machinery designs when introduced stably compared with transiently. The two most efficient designs in the transient implementations (HA-T2-2A-CytoEpi and CytoEpi-2A-T2) failed to express the enzyme proteins and glycosylate reporter substrates in stable implementations (Supplemental Fig. S2A). Surprisingly, removal of the N-terminal hemagglutinin tag in HA-T2-2A-CytoEpi markedly improved production of the two functional enzymes in stable implementations (Fig. 2B). Degradation of all 2A-derived polyproteins was observed, as evidenced by western-blot analysis (Supplemental Fig. S2B). Also, the one gene-one promoter O-glycosylation machinery design (CytoEpi-T2), which in transient implementations proved markedly less efficient than the 2A-linked designs, conferred a similar O-glycosylation efficiency to T2-2A-CytoEpi in a stable implementation in BY-2 cells (Fig. 4C, lanes 41–46). There are, to our knowledge, few reports of stably integrated 2A-linked implementations where the two proteins are destined for different subcellular localizations. Our results suggest that the design of 2A-linked polyprotein constructs with respect to the order of proteins may be important.
We previously observed that the MUC1 reporter substrate was somewhat unstable when expressed transiently in plants (Yang et al., 2012). Here, we observed even more severe problems with the stability of the small MUC1 reporter substrate, whether expressed alone (MUC1) or in fusion with the carrier YFP (MUC1-YFP; Figs. 1 and 3). We also showed that incubation of nonglycosylated and glycosylated MUC1-YFP in BY-2 medium conferred a complete protease-mediated breakdown of MUC1, resulting in the trimmed resistant stable barrel YFP structure. Therefore, we tested an alternative strategy where we embedded 2.5 TRs of MUC1 with tags into GFP (Fig. 4A), as proposed previously (Kobayashi et al., 2008). Those authors demonstrated that 67-amino acid residues could be inserted into a loop between the β-strands located on the opposite side of the N and C termini of the GFP without substantial loss of fluorescence capacity. Coexpression of the embedded construct GF(MUC1)P with the O-glycosylation machinery CytoEpi-T2 resulted in the secretion of GalNAc-glycosylated GF(MUC1)P in the BY-2 medium (Fig. 5A). MALDI-TOF-MS analysis revealed that GF(MUC1)P was uniformly glycosylated with three GalNAc residues, which corresponds to the substrate specificity of GalNAc-T2 (Bennett et al., 1998). Approximately 50% of the GF(MUC1)P in the BY-2 medium further contained up to four Hyp residues as well as three and six Ara residues (Fig. 5A). Introduction of a single construct for the expression of GF(MUC1)P and the O-glycosylation machinery CytoEpi-2A-T2 resulted in moderate accumulation of GF(MUC1)P (approximately 5 μg g−1 fresh weight) in Arabidopsis (Fig. 6B), which was modified with zero to three GalNAc residues, zero to four Pro hydroxylations, and minor arabinosides (Fig. 6D).
In agreement with our previous transient implementation (Yang et al., 2012), introduction of the mature secreted form of the cytokine IFNα2B in Arabidopsis resulted in expression of the intact product, which was glycosylated when coexpressed with the O-glycosylation machinery CytoEpi-T2 (Fig. 2C). Likewise, 1.2 TRs of MUC16, which also realizes a full structural unit with three potential N-glycosylation sites and up to 30 O-glycosylation sites, was expressed and shown to be N-glycosylated by the endogenous N-glycosylation machinery in BY-2 cells (Supplemental Fig. S1). Explanations for the apparent instability of the small MUC1-3.5TR may include a lack of proper folding and a lack of protective O-glycosylation, where native mammalian mucins are large (greater than 200 kD) heavily O-glycosylated glycoproteins abundant in mucus layers, where they hydrate, lubricate, and protect cells from proteases as well as from pathogens. In plants, hundreds of genes encode proteins involved in proteolytic processes (Rawlings et al., 2008), with an estimated 1,900 genes in Arabidopsis being implicated directly or indirectly in peptide bond hydrolysis (Schaller, 2004; Smalle and Vierstra, 2004; Benchabane et al., 2008). The high protease activity in the BY-2 medium (Robertson et al., 1997; Okushima et al., 2000) may require, for the production of some pharmaceutical proteins, tailored knockout of specific proteases. It is well established that N- and O-linked glycosylation may enhance protein stability, probably by acting as a physical barrier to protease-mediated degradation (Li and d’Anjou, 2009). The finding that O-linked GalNAc glycosylation protected the MUC1 protein offers an alternative approach to combating protease-mediated degradation.
The plant-specific Hyp O-glycosylation is an obvious concern for the recombinant production of human therapeutics in plants. Pro hydroxylation is initiated by a family of P4Hs, and the resulting Hyp residues may subsequently be glycosylated with arabinogalactan or shorter arabinosides. P4Hs are membrane-bound type II proteins found in the endoplasmic reticulum and extending into the Golgi apparatus (Yuasa et al., 2005). Thirteen putative P4Hs have been identified in Arabidopsis (Liang et al., 2010), and our understanding of the functions of these in terms of peptide/protein substrate specificities and sequence contexts (which proteins and where) is still limited. This is especially true for the transgene expression of proteins. Hyp residues, for example, were identified on human erythropoietin (Weise et al., 2007) and IgA1 (Karnoup et al., 2005) recombinantly expressed in Physcomitrella patens and maize (Zea mays), respectively, and Hyp-linked O-glycosylation was additionally identified on IgA1, specifically in the Pro/Ser/Thr-rich hinge region where GalNAc O-glycosylation occurs in humans (Karnoup et al., 2005). More recently, Pinkhasov et al. (2011) found Hyp formation (Pro-11 or Pro-12) and Hyp substituted with three arabinofuranoside residues in MUC1 TRs transiently expressed in plants. This finding was confirmed in our studies with an additional interesting observation, namely that the expression of MUC1 from stably integrated constructs markedly increased the level of Pro hydroxylation and subsequent Hyp-linked arabinosides. Kieliszewski et al. (2011) recently summarized plant Hyp-O-glycosylation, with noncontiguous Hyp residues being glycosylated exclusively with arabinogalactan polysaccharides and contiguous Hyp residues being glycosylated exclusively with arabinosides We demonstrated that substantial reduction in Hyp modifications could be achieved by addition of the 2,2-DP inhibitor, but clearly, targeted knockout of one or more P4Hs acting on the expressed proteins of interest would be more efficient. Several reports suggest the involvement of the three P4Hs, AtP4H2, AtP4H5, and AtP4H13, in the hydroxylation of contiguous Pro motifs in particular (Hieta and Myllyharju, 2002; Tiainen et al., 2005; Keskiaho et al., 2007; Velasquez et al., 2011), perhaps pointing to these enzymes as first targets for engineering plants with knockout of plant-specific Pro hydroxylation and Hyp-linked arabinosylation. The clinical significance of Pro hydroxylations and Hyp-linked arabinosylations is unknown or at best speculative (Petersen et al., 1998; Leonard et al., 2005; Altmann, 2007; Manduzio et al., 2012, Pinkhasov et al., 2011). In relation to the present glycoengineering, these endogenously derived PTMs did not appear to affect the GalNAc-T2-mediated O-glycosylation of MUC1.
Our interest in designing plant cell factories with engineered human-style O-glycosylation stems from a need for the production of MUC1 and other vaccines with aberrant O-glycosylation. We have previously identified cancer-associated immunodominant GalNAc-glycopeptide epitopes in MUC1 mucin that are not covered by immunological tolerance (Sørensen et al., 2006; Sabbatini et al., 2007; Tarp et al., 2007). We have also shown that chemoenzymatically produced Tn-MUC1 glycopeptides induce IgG antibodies in cancer patients (Sabbatini et al., 2007), and spontaneous IgG antibodies to the same epitope are found in some cancer patients at the time of diagnosis (Wandall et al., 2010; Blixt et al., 2011). The absence of GalNAc-type O-glycosylation makes plants an ideal cell system for engineering O-glycosylation de novo and the production of aberrant O-glycosylation glycoforms, as shown in this study.
The absolute requirement of a carrier protein for stabilization of the small MUC1-2.5TR and MUC1-3.5TR proteins, which in the BY-2 cell system needed to be embedded into the carrier GFP and glycosylated, plus the add-on presence of endogenous PTMs render production of this particular small vaccine candidate in plants nonfeasible or at best cumbersome. In summary, stably engineered plants with the capacity to perform mammalian O-glycosylation on recombinantly expressed secreted target proteins were established. Problems with degradation and plant-specific PTMs were identified, highlighting the need for additional engineering for plants to become versatile production platforms of therapeutics. Although in its infancy, engineering O-glycosylation capacities in plants may eventually be combined with engineered human N-glycosylation (Strasser et al., 2008; Kaulfürst-Soboll et al., 2011; Nagels et al., 2011; Schoberer and Strasser, 2011) to generate more general platforms for the expression of glycoprotein pharmaceuticals in plants.
MATERIALS AND METHODS
Transformation and Plant Growth Conditions
Tobacco (Nicotiana tabacum) BY-2 suspension cells were cultivated by shaking (25°C, 130 rpm) in the dark. Transformation and fermentation of BY-2 cells are described by Mayo et al. (2006). Agrobacterium tumefaciens strain C58C1 pGV3850 was used for both stable transformation and transient A. tumefaciens-mediated expression. Transformation of Arabidopsis (Arabidopsis thaliana) was performed as described (Horsch et al., 1986). Growth conditions and transformation of transgenic Arabidopsis plants were as described (Egelund et al., 2007). Transformation and maintenance of Lemna minor were done as described (Rival et al., 2008). A. tumefaciens-mediated expression was done essentially as described in our previous study (Yang et al., 2012).
DNA Constructs for Plant Transformation
Open-source vectors used for A. tumefaciens-mediated expression and transformation are pBI121 (GenBank accession no. AY781296), pCAMBIA 2300 (GenBank accession no. AF234315), and pCAMBIA 1302 (GenBank accession no. AF234298). For the legacy of open-source pCAMBIA binary vectors, see http://www.cambia.org. pPS48 is an intermediate Escherichia coli-only vector (Odell et al., 1985) that contains the cauliflower mosaic virus 35S promoter followed by the 35S terminator interspaced by a multiple cloning site, into which genes of interest (goi) were cloned. Entire transcriptional units (35S-Pro-goi-35S-term) were excised using XbaI or HindIII and ligated into the multiple cloning site of the pCAMBIA-derived plant expression plasmids. The tobacco ubiquitin promoter and terminator were synthesized by GenScript.
The designs of expression constructs used are depicted in Figure 1. Secreted reporter constructs for O-glycosylation were designed with either of the following signal peptide sequences, (1) NtSP derived from tobacco Pro-rich protein 3 (UniProt accession no. Q40502; sequence, MGKMASLFASLLVVLVSLSLA); or (2) PpSP derived from Physcomitrella patens aspartic protease (EMBL accession no. AJ586914, sequence, MGASRSVRLAFFLVVLVVLAALAEA), and these were generated as described previously (Yang et al., 2012). A novel construct with a 2.5-TR sequence of MUC1 embedded in GFP [GF(MUC1)P] was prepared as follows (Fig. 1). The MUC1-2.5TR sequence was inserted into GFP after Asp-196 into the NcoI site of modified GFP (Kobayashi et al., 2008), followed by PCR-mediated insertion of the signal peptide for secretion. The primer sets were SSGFPfor (5′-GAGCTCCATGGGTAAGACTAATCTTTTTCTCTTTCTCATCTTTTCACTTCTCCTATCATTATCCTCGGCCGAGCAAGTGAGCAAGGGCGAGGAGCT-3′) and GFPrev (5′-CATATGCTTGTACAGCTCGTCCATG-3′), and the resulting PCR fragments were cloned into the SacI site of pPS48 under the control of the ubiquitin promoter and the ubiquitin terminator. Finally, the whole expression cassette was inserted into the HindIII site of pCAMBIA2300, yielding pC2300D-UbiPro-NtSP-GF(MUC1-2.5TR)P-UbiTerm [GF(MUC1)P].
Design of the O-glycosylation machinery was essentially as described previously (Yang et al., 2012; Fig. 1, A and C) and consisted of a cytosolic Pseudomonas aeruginosa Glc(NAc) C4-epimerase (CytoEpi) WbpP (Creuzenet et al., 2000; GenBank accession no. AAF23998.1) and human (Homo sapiens) polypeptide GalNAc-T2 (UniProt accession no. X85019). The O-glycosylation machinery was implemented from a single construct either with each gene driven by a single promoter or a polycistronic transcript encoding two genes interspaced by nucleotides encoding the FMDV 2A autocleavable sequence (El Amrani et al., 2004; Szymczak et al., 2004). Construction was carried out as follows. Constructs encoding the epimerase and the GalNAc-T2 were assembled by inserting the XbaI-35SPro-CytoEpi-35STerm-XbaI of construct CytoEpi into the XbaI site of construct T2, resulting in pC1302D-35SPro-T2-35STerm;35SPro-CytoEpi-35STer (CytoEpi-T2). This construct was also made as a 2A-linked polycistronic construct. A modified version of pC1302-35SPro-HA-T2-2A-CytoEpi-35STerm (HA-T2-2A-CytoEpi; Yang et al., 2012) without a hemagglutinin tag at the N terminus of GalNAc-T2 was also prepared as follows. T2-2A-CytoEpi was cloned by PCR using the T2For (5′-CCATGGCTATGCGGCGGCGCTCGCGGAT-3′) and PFwbppRev (5′-GAGCTCTCACTTGTCGTCGTCGTCCTTGTAATCTTTCAAAAACATGATGTACCAAG-3′) primer set, followed by insertion into the SacI site of pPS48 vector. The entire fragment containing 35SPro-T2-2A-CytoEpi-35STerm was inserted into the HindIII site of pCAMBIA1302, yielding pC1302-35SPro-T2-2A-CytoEpi-35STerm (T2-2A-CytoEpi). The 2A linker region of T2-2A-CytoEpi encodes GSGQTLNFDLLKLAGDVESNPG↓PM, where italics designate the 2A sequence and the arrow indicates the autocleavage site (El Amrani et al., 2004; Szymczak et al., 2004). The XbaI-digested fragment of 35SPro-PpSP-MUC1-3.5TR-YFP(His)6-35STerm was inserted into T2-2A-CytoEpi, yielding pC1302-35SPro-PpSP-MUC1-3.5TR-YFP(His)6-35STerm;35SPro-T2-2A-CytoEpi-35STerm (MUC1-YFP-T2-2A-CytoEpi; Fig. 1C).
Preparation of Total Protein Extracts
Approximately 1 g of freshly harvested leaves or roots was frozen in liquid N2 and comminuted using a pestle and mortar with 2 mL of extraction buffer A (50 mm NaPO4, 250 mm NaCl, and 5 mm imidazole, pH 8.0) containing Complete Proteinase Inhibitor (Roche) and 1 mm phenylmethanesulfonyl fluoride. The sample was incubated for 10 min on ice, insoluble material was pelleted by centrifugation (20,000g) for 10 min, the supernatant was recovered and stored at −20°C.
SDS-PAGE Western Blotting
SDS-PAGE western-blot analysis was performed as described previously (Petersen et al., 2009). MAbs to T7 tag and MUC16 (M11) were obtained from Invitrogen and Dako, respectively. MAbs to MUC1 and GalNAc-MUC1 (Tn-MUC1), 5E10 and 5E5, respectively, were described previously (Tarp et al., 2007). Horseradish peroxidase-conjugated Vicia villosa (VVA) lectin (EY Laboratories) was also used for the detection of GalNAc glycoforms.
HIC
MUC1-YFP was purified by HIC using the ÄKTA FPLC System with a HiTrap HIC column (5 mL; GE) prepacked with Phenyl Sepharose 6 Fast Flow. BY-2 cell medium fraction (100 mL), additionally adjusted to include 3 m NaCl and 20 mm Tris buffer (pH 8.0), was centrifuged at 25,000g for 20 min. The supernatant was loaded (2 mL min−1) to the preequilibrated HIC column and then washed with 20 mL of wash buffer (2 m NaCl in 20 mm Tris-buffer, pH 8.0). MUC1-YFP was eluted by water in 10 fractions of 2 mL.
Nickel Chromatography
Cleared supernatants (50 mL, 20,000g for 30 min) were incubated with 0.5 mL of nickel-nitrilotriacetic acid agarose beads (Qiagen) for 2 h (4°C) under gentle rolling. Beads were washed for 10 min with 20 mL of wash buffer (50 mm NaPO4, 250 mm NaCl, and 20 mm imidazole, pH 8.0) and eluted with 5 × 1 mL of elution buffer (50 mm NaPO4, 250 mm NaCl, and 250 mm imidazole, pH 8.0).
Degradation Study of MUC1-YFP and Embedded GF(MUC1)P in BY-2 Medium
Culture medium (50 mL) of wild-type BY-2 cells grown for 7 d was collected by centrifugation at 10,000g for 5 min (wild-type BY-2 medium fraction). Approximately 5 μg of purified MUC1-YFP (transiently produced in leaves of Nicotiana benthamiana plants with/without O-glycosylation machinery) and embedded GF(MUC1)P (purified from intracellular or extracellular BY-2 cells with/without O-glycosylation machinery) was added to 7-d-old 1 mL of unboiled or boiled (for 15 min) wild-type BY-2 medium fractions, which were further incubated at 25°C under shaking (130 rpm) for an additional 24 or 48 h under the same conditions and ultimately subjected to MUC1- and Tn-MUC1-specific (MAbs 5E10 and 5E5, respectively) western-blot analysis.
Endoproteinase Asp Digestion of MUC1-TR and Sample Purification
Purified MUC1-YFP (approximately 25 µg) was incubated with 1 µg of endoproteinase Asp-N from Pseudomonas fragi (Roche) in 300 µL of 100 mm Tris-HCl (pH 8.0) for 16 h at 37°C under shaking (600 rpm). In the case of embedded GF(MUC1)P, in-gel digestion (Shevchenko et al., 2006) was used to release MUC1 tandem repeats from the GFP, as in-gel protein digestion of embedded GF(MUC1)P was found to give a better recovery compared to the “in-solution” method. Both Asp-N (10 ng μL−1) and trypsin (10 ng μL−1) were added into the digestion buffer (25 mm NH4HCO3 and 10% acetonitrile). The digest was cleaned on a C18 Zip-Tip (Millipore). Briefly, the digestion mixture was dissolved in 20 μL of 0.1% trifluoroacetic acid and drawn through the column, desalted with 0.5% formic acid, and eluted with 100% acetonitrile.
MALDI-TOF-MS
Lyophilized peptides were dissolved in 20 μL of water. The MALDI matrix 2,5-dihydroxybenzoic acid (Sigma-Aldrich) was dissolved (25 g L−1) in a 1:1 mixture of water and methanol. Samples were prepared for analysis by placing 0.5 μL of sample solution on a MALDI target, followed by 0.5 μL of matrix solution, which was then air dried. All mass spectra were acquired in the linear mode on a Voyager-Elite MALDI time-of-flight mass spectrometer (Perseptive Biosystem) equipped with delayed extraction; external calibration was used.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of the mucin type O-glycoproteins MUC1 and MUC16 in L. minor and BY-2 cells, respectively.
Supplemental Figure S2. SDS-PAGE western-blot analysis of 2A-linked O-glycosylation machinery.
Supplemental Figure S3. Time course of Tn-MUC1 and (Tn)-GF(MUC1)P in BY-2 cell medium.
Supplemental Figure S4. Prior introduction of O-glycosylation machinery enhanced the glycosylation level of MUC1-YFP in a transient expression system.
Supplemental Figure S5. Chemical inhibitors of P4Hs reduce extracellular accumulation of GF(MUC1)P and inhibit growth of BY-2 cells.
Supplemental Figure S6. Chemical inhibition of proline hydroxylation of MUC1 expressed in BY-2 cells.
Supplemental Figure S7. Transient expression of MUC1-YFP with coinfiltration of different C4 P4Hs in N. benthamiana.
Supplementary Material
Acknowledgments
pET23-WbpP was a generous gift from J.S. Lam (University of Guelph). Takashi Murayama (Juntendo University School of Medicine) is acknowledged for sharing the construct modified GFP. Dr. Jose Estevez (Universidad de Buenos Aires) is thanked for helpful suggestions with relation to Pro hydroxylation inhibition experiments.
Glossary
- PTM
posttranslational modification
- GalNAc-T
UDP-GalNAc polypeptide N-acetylgalactosaminyltransferase
- CHO
Chinese hamster ovary
- AGP
arabinogalactan protein
- P4H
prolyl 4-hydroxylase
- β-Araf
β-linked arabinofuranoside
- BY-2
Bright Yellow-2
- YFP
yellow fluorescent protein
- MAb
monoclonal antibody
- VVA
Vicia villosa
- HIC
hydrophobic interaction chromatography
- MALDI
matrix-assisted laser desorption ionization
- TOF
time-of-flight
- MS
mass spectrometry
- TR
tandem repeat
- 2,2-DP
2,2-dipyridyl
References
- Altmann F. (2007) The role of protein glycosylation in allergy. Int Arch Allergy Immunol 142: 99–115 [DOI] [PubMed] [Google Scholar]
- Amano K, Chiba Y, Kasahara Y, Kato Y, Kaneko MK, Kuno A, Ito H, Kobayashi K, Hirabayashi J, Jigami Y, et al. (2008) Engineering of mucin-type human glycoproteins in yeast cells. Proc Natl Acad Sci USA 105: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchabane M, Goulet C, Rivard D, Faye L, Gomord V, Michaud D. (2008) Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol J 6: 633–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, van Kessel AG, et al. (1998) Cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J Biol Chem 273: 30472–30481 [DOI] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22: 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Bueti D, Burford B, Allen D, Julien S, Hollingsworth M, Gammerman A, Fentiman I, Taylor-Papadimitriou J, Burchell JM. (2011) Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res 13: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho A, Strasser R, Stadlmann J, Grass J, Jez J, Gattinger P, Kunert R, Quendler H, Pabst M, Leonard R, et al. (2010) In planta protein sialylation through overexpression of the respective mammalian pathway. J Biol Chem 285: 15923–15930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KM, Sterling JD, Regan JT, Gasdaska JR, Frantz KK, Peele CG, Black A, Passmore D, Moldovan-Loomis C, Srinivasan M, et al. (2006) Glycan optimization of a human monoclonal antibody in the aquatic plant Lemna minor. Nat Biotechnol 24: 1591–1597 [DOI] [PubMed] [Google Scholar]
- Creuzenet C, Belanger M, Wakarchuk WW, Lam JS. (2000) Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J Biol Chem 275: 19060–19067 [DOI] [PubMed] [Google Scholar]
- Daskalova SM, Radder JE, Cichacz ZA, Olsen SH, Tsaprailis G, Mason H, Lopez LC. (2010) Engineering of N. benthamiana L. plants for production of N-acetylgalactosamine-glycosylated proteins: towards development of a plant-based platform for production of protein therapeutics with mucin type O-glycosylation. BMC Biotechnol 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFrees S, Wang ZG, Xing R, Scott AE, Wang J, Zopf D, Gouty DL, Sjoberg ER, Panneerselvam K, Brinkman-Van der Linden EC, et al. (2006) GlycoPEGylation of recombinant therapeutic proteins produced in Escherichia coli. Glycobiology 16: 833–843 [DOI] [PubMed] [Google Scholar]
- Durocher Y, Butler M. (2009) Expression systems for therapeutic glycoprotein production. Curr Opin Biotechnol 20: 700–707 [DOI] [PubMed] [Google Scholar]
- Egelund J, Obel N, Ulvskov P, Geshi N, Pauly M, Bacic A, Petersen BL. (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol Biol 64: 439–451 [DOI] [PubMed] [Google Scholar]
- El Amrani A, Barakate A, Askari BM, Li X, Roberts AG, Ryan MD, Halpin C. (2004) Coordinate expression and independent subcellular targeting of multiple proteins from a single transgene. Plant Physiol 135: 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez JM, Kieliszewski MJ, Khitrov N, Somerville C. (2006) Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiol 142: 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerngross TU. (2004) Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat Biotechnol 22: 1409–1414 [DOI] [PubMed] [Google Scholar]
- Gomord V, Faye L. (2004) Posttranslational modification of therapeutic proteins in plants. Curr Opin Plant Biol 7: 171–181 [DOI] [PubMed] [Google Scholar]
- Gomord V, Fitchette AC, Menu-Bouaouiche L, Saint-Jore-Dupas C, Plasson C, Michaud D, Faye L. (2010) Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol J 8: 564–587 [DOI] [PubMed] [Google Scholar]
- Hellwig S, Drossard J, Twyman RM, Fischer R. (2004) Plant cell cultures for the production of recombinant proteins. Nat Biotechnol 22: 1415–1422 [DOI] [PubMed] [Google Scholar]
- Hieta R, Myllyharju J. (2002) Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana: effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J Biol Chem 277: 23965–23971 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Klee HJ, Stachel S, Winans SC, Nester EW, Rogers SG, Fraley RT. (1986) Analysis of Agrobacterium tumefaciens virulence mutants in leaf discs. Proc Natl Acad Sci USA 83: 2571–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug I, Zheng B, Reiz B, Whittal RM, Fentabil MA, Klassen JS, Feldman MF. (2011) Exploiting bacterial glycosylation machineries for the synthesis of a Lewis antigen-containing glycoprotein. J Biol Chem 286: 37887–37894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet E, Albenne C, Boudart G, Irshad M, Canut H, Pont-Lezica R. (2008) Recent advances in plant cell wall proteomics. Proteomics 8: 893–908 [DOI] [PubMed] [Google Scholar]
- Karnoup AS, Turkelson V, Anderson WH. (2005) O-Linked glycosylation in maize-expressed human IgA1. Glycobiology 15: 965–981 [DOI] [PubMed] [Google Scholar]
- Kaulfürst-Soboll H, Rips S, Koiwa H, Kajiura H, Fujiyama K, von Schaewen A. (2011) Reduced immunogenicity of Arabidopsis hgl1 mutant N-glycans caused by altered accessibility of xylose and core fucose epitopes. J Biol Chem 286: 22955–22964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskiaho K, Hieta R, Sormunen R, Myllyharju J. (2007) Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 19: 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA, Tan L, Cannon MC. (2011) Hydroxyproline-Rich Glycoproteins: Form and Function. P Ulvskov, ed, Annual Plant Reviews: Plant Polysaccharides, Biosynthesis and Bioengineering. Wiley-Blackwell, Oxford, pp 321–342 [Google Scholar]
- Kieliszewski MJ, Shpak E. (2001) Synthetic genes for the elucidation of glycosylation codes for arabinogalactan-proteins and other hydroxyproline-rich glycoproteins. Cell Mol Life Sci 58: 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Morone N, Kashiyama T, Oyamada H, Kurebayashi N, Murayama T. (2008) Engineering a novel multifunctional green fluorescent protein tag for a wide variety of protein research. PLoS ONE 3: e3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DT, Katona L, Roerig S. (1973) Galactosylserine in extensin. Biochem J 133: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DTA, Kieliszewski MJ, Chen Y, Cannon MC. (2011) Role of the extensin superfamily in primary cell wall architecture. Plant Physiol 156: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard R, Petersen BO, Himly M, Kaar W, Wopfner N, Kolarich D, van Ree R, Ebner C, Duus JO, Ferreira F, et al. (2005) Two novel types of O-glycans on the mugwort pollen allergen Art v 1 and their role in antibody binding. J Biol Chem 280: 7932–7940 [DOI] [PubMed] [Google Scholar]
- Li H, d’Anjou M. (2009) Pharmacological significance of glycosylation in therapeutic proteins. Curr Opin Biotechnol 20: 678–684 [DOI] [PubMed] [Google Scholar]
- Liang Y, Faik A, Kieliszewski M, Tan L, Xu WL, Showalter AM. (2010) Identification and characterization of in vitro galactosyltransferase activities involved in arabinogalactan-protein glycosylation in tobacco and Arabidopsis. Plant Physiol 154: 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manduzio H, Fitchette AC, Hrabina M, Chabre H, Batard T, Nony E, Faye L, Moingeon P, Gomord V. (2012) Glycoproteins are species-specific markers and major IgE reactants in grass pollens. Plant Biotechnol J 10: 184–194 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011) Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol 52: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Watanabe N, Nakamura K. (1995) O-Glycosylation of a precursor to a sweet potato vacuolar protein, sporamin, expressed in tobacco cells. Plant J 8: 877–889 [DOI] [PubMed] [Google Scholar]
- Mayo KJ, Gonzales BJ, Mason HS. (2006) Genetic transformation of tobacco NT1 cells with Agrobacterium tumefaciens. Nat Protoc 1: 1105–1111 [DOI] [PubMed] [Google Scholar]
- Nagels B, Van Damme EJM, Pabst M, Callewaert N, Weterings K. (2011) Production of complex multiantennary N-glycans in Nicotiana benthamiana plants. Plant Physiol 155: 1103–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH. (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Koizumi N, Kusano T, Sano H. (2000) Secreted proteins of tobacco cultured BY2 cells: identification of a new member of pathogenesis-related proteins. Plant Mol Biol 42: 479–488 [DOI] [PubMed] [Google Scholar]
- Petersen A, Schramm G, Schlaak M, Becker WM. (1998) Post-translational modifications influence IgE reactivity to the major allergen Phl p 1 of timothy grass pollen. Clin Exp Allergy 28: 315–321 [DOI] [PubMed] [Google Scholar]
- Petersen BL, Egelund J, Damager I, Faber K, Jensen JK, Yang Z, Bennett EP, Scheller HV, Ulvskov P. (2009) Assay and heterologous expression in Pichia pastoris of plant cell wall type-II membrane anchored glycosyltransferases. Glycoconj J 26: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkhasov J, Alvarez ML, Rigano MM, Piensook K, Larios D, Pabst M, Grass J, Mukherjee P, Gendler SJ, Walmsley AM, Mason HS. (2011) Recombinant plant-expressed tumour-associated MUC1 peptide is immunogenic and capable of breaking tolerance in MUC1.Tg mice. Plant Biotechnol J 9: 991–1001 [DOI] [PubMed] [Google Scholar]
- Pogue GP, Vojdani F, Palmer KE, Hiatt E, Hume S, Phelps J, Long L, Bohorova N, Kim D, Pauly M, et al. (2010) Production of pharmaceutical-grade recombinant aprotinin and a monoclonal antibody product using plant-based transient expression systems. Plant Biotechnol J 8: 638–654 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. (2008) MEROPS: the peptidase database. Nucleic Acids Res 36: D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves ML, Lopez LC, Daskalova SM. (2011) Non-canonical interactions between plant proteins and lectins cause false positives in lectin blots. Research in Plant Biology 1: 49–54 [Google Scholar]
- Ringli C. (2010) Monitoring the outside: cell wall-sensing mechanisms. Plant Physiol 153: 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rival S, Wisniewski JP, Langlais A, Kaplan H, Freyssinet G, Vancanneyt G, Vunsh R, Perl A, Edelman M. (2008) Spirodela (duckweed) as an alternative production system for pharmaceuticals: a case study, aprotinin. Transgenic Res 17: 503–513 [DOI] [PubMed] [Google Scholar]
- Robertson D, Mitchell GP, Gilroy JS, Gerrish C, Bolwell GP, Slabas AR. (1997) Differential extraction and protein sequencing reveals major differences in patterns of primary cell wall proteins from plants. J Biol Chem 272: 15841–15848 [DOI] [PubMed] [Google Scholar]
- Sabbatini PJ, Ragupathi G, Hood C, Aghajanian CA, Juretzka M, Iasonos A, Hensley ML, Spassova MK, Ouerfelli O, Spriggs DR, et al. (2007) Pilot study of a heptavalent vaccine-keyhole limpet hemocyanin conjugate plus QS21 in patients with epithelial ovarian, fallopian tube, or peritoneal cancer. Clin Cancer Res 13: 4170–4177 [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Lomonossoff GP. (2008) Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol 148: 1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C, Faye L, Gomord V. (2007) From planta to pharma with glycosylation in the toolbox. Trends Biotechnol 25: 317–323 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Bothner B, Dell A, Fukuda M. (1987) Carbohydrate structure of erythropoietin expressed in Chinese hamster ovary cells by a human erythropoietin cDNA. J Biol Chem 262: 12059–12076 [PubMed] [Google Scholar]
- Schaller A. (2004) A cut above the rest: the regulatory function of plant proteases. Planta 220: 183–197 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Datta K, Marcus A. (1991) Peptidyl proline hydroxylation and the growth of a soybean cell culture. Plant Physiol 96: 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Strasser R. (2011) Sub-compartmental organization of Golgi-resident N-glycan processing enzymes in plants. Mol Plant 4: 220–228 [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Shimizu M, Igasaki T, Yamada M, Yuasa K, Hasegawa J, Kato T, Tsukagoshi H, Nakamura K, Fukuda H, Matsuoka K. (2005) Experimental determination of proline hydroxylation and hydroxyproline arabinogalactosylation motifs in secretory proteins. Plant J 42: 877–889 [DOI] [PubMed] [Google Scholar]
- Shpak E, Leykam JF, Kieliszewski MJ. (1999) Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc Natl Acad Sci USA 96: 14736–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Sørensen AL, Reis CA, Tarp MA, Mandel U, Ramachandran K, Sankaranarayanan V, Schwientek T, Graham R, Taylor-Papadimitriou J, Hollingsworth MA, et al. (2006) Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology 16: 96–107 [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Gentzsch M, Tanner W. (1999) Protein O-mannosylation. Biochim Biophys Acta 1426: 297–307 [DOI] [PubMed] [Google Scholar]
- Strasser R, Stadlmann J, Schähs M, Stiegler G, Quendler H, Mach L, Glössl J, Weterings K, Pabst M, Steinkellner H. (2008) Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol J 6: 392–402 [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DAA. (2004) Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol 22: 589–594 [DOI] [PubMed] [Google Scholar]
- Tan L, Varnai P, Lamport DT, Yuan C, Xu J, Qiu F, Kieliszewski MJ. (2010) Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J Biol Chem 285: 24575–24583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarp MA, Sørensen AL, Mandel U, Paulsen H, Burchell J, Taylor-Papadimitriou J, Clausen H. (2007) Identification of a novel cancer-specific immunodominant glycopeptide epitope in the MUC1 tandem repeat. Glycobiology 17: 197–209 [DOI] [PubMed] [Google Scholar]
- Thomsen DR, Post LE, Elhammer AP. (1990) Structure of O-glycosidically linked oligosaccharides synthesized by the insect cell line Sf9. J Cell Biochem 43: 67–79 [DOI] [PubMed] [Google Scholar]
- Tiainen P, Myllyharju J, Koivunen P. (2005) Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J Biol Chem 280: 1142–1148 [DOI] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-Glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Wandall HH, Blixt O, Tarp MA, Pedersen JW, Bennett EP, Mandel U, Ragupathi G, Livingston PO, Hollingsworth MA, Taylor-Papadimitriou J, et al. (2010) Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res 70: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A, Altmann F, Rodriguez-Franco M, Sjoberg ER, Bäumer W, Launhardt H, Kietzmann M, Gorr G. (2007) High-level expression of secreted complex glycosylated recombinant human erythropoietin in the Physcomitrella Delta-fuc-t Delta-xyl-t mutant. Plant Biotechnol J 5: 389–401 [DOI] [PubMed] [Google Scholar]
- Xu J, Okada S, Tan L, Goodrum KJ, Kopchick JJ, Kieliszewski MJ. (2010) Human growth hormone expressed in tobacco cells as an arabinogalactan-protein fusion glycoprotein has a prolonged serum life. Transgenic Res 19: 849–867 [DOI] [PubMed] [Google Scholar]
- Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, et al. (2011) The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol 29: 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Drew DP, Jørgensen B, Mandel U, Bach SS, Ulvskov P, Levery SB, Bennett EP, Clausen H, Petersen BL. (2012) Engineering mammalian mucin-type O-glycosylation in plants. J Biol Chem 287: 11911–11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin B-J, Gao T, Zheng N-Y, Li Y, Tang S-Y, Liang L-M, Xie Q. (2011) Generation of glyco-engineered BY2 cell lines with decreased expression of plant-specific glycoepitopes. Protein Cell 2: 41–47 [Google Scholar]
- Yong-Pill C, Chrispeels MJ. (1976) Serine-O-galactosyl linkages in glycopeptides from carrot cell walls. Phytochemistry 15: 165–169 [Google Scholar]
- Yuasa K, Toyooka K, Fukuda H, Matsuoka K. (2005) Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J 41: 81–94 [DOI] [PubMed] [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DT, Kieliszewski MJ. (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31: 431–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.