The ubiquitin-proteasome system (UPS) plays a role in nearly every aspect of plant biology. The wealth of emerging data demonstrates that regulated protein degradation rivals the well-studied area of transcriptional regulation for importance in cellular regulation. In this Update, we will highlight current research findings that illustrate the contributions of the UPS to phytohormone signaling. We will also consider outstanding questions and discuss possible experimental approaches that will increase our understanding of regulated protein degradation.
UPS activity involves a three-step enzymatic cascade between E1, E2, and E3 enzymes that results in the covalent transfer of ubiquitin to target proteins. This process can result in different outcomes, including (1) proteolytic degradation by the 26S proteasome or reversible, nonproteolytic regulatory events. Current plant genome annotations indicate that most species can produce over 1,000 different UPS components; E3 ligase family proteins are the most abundant (Du et al., 2009; Vierstra, 2009). Thus, the potential scope of the UPS is extensive. E3 ligases provide specificity by directly controlling the transfer of ubiquitin to the target substrate and have been classified into four groups based on their complex composition and modes of action: HECT, RING, U-box, and cullin-RING ligases (CRLs; Vierstra, 2009). CRLs are multisubunit ligases that can be further subdivided into four groups based on a variable target recognition module: (1) F-box proteins in the case of S-phase kinase-associated protein1-cullin1-F-box (SCF) ligases; (2) bric-a-brac-tramtrak-broad complex (BTB) proteins that define the BTB group of E3s; (3) the DNA damage-binding (DDB) class that utilizes WD40 domain-containing DWD proteins; and (4) the anaphase-promoting complex category that contains several interchangeable recognition proteins (Vierstra, 2009). Altogether, these enzymatic modules are incredibly diverse, and many of their targets are currently unknown.
The phytohormones are small molecule regulators that collectively impact every aspect of plant growth and development. They regulate genetically determined aspects of development such as organ development as well as pathogen response, integration of environmental cues such as light and temperature, and regulation of circadian clock output (Ho et al., 2008; Vierstra, 2009; Lee and Lee, 2012). Recent discoveries highlight the particular importance of UPS action in phytohormone signaling. In fact, UPS-mediated protein degradation has been implicated or demonstrated for every plant hormone, including abscisic acid (ABA), auxin, brassinosteroid (BR), cytokinin, ethylene, GA, jasmonic acid (JA), and strigolactone (SL). Notably, the UPS is also very important for steroid hormone signaling in humans (Lee and Lee, 2012). While some plant hormones undergo long-distance transport and thus have different sites of synthesis and action (e.g. auxin, SL), others appear to be synthesized and function in the same tissue (e.g. BR). As we will discuss in more detail below, one of the major functions of UPS pathways in hormone signaling is the selective destruction of proteins whose concentrations must vary with time and alterations in the state of the cell.
REGULATING THE REGULATORS: KNOWN E3-TARGET INTERACTIONS
UPS action appears to regulate hormone biosynthesis, transport, and perception and thus provides a direct mechanism to control the magnitude and duration of hormone signaling (Figs. 1 and 2). Detailed studies of UPS function in phytohormone pathways have revealed a number of patterns. First, most of the hormone-related UPS targets described to date are proteins associated with transcription. Furthermore, many of the known target transcription factors are classified as repressors and contain ETHYLENE RESPONSE FACTOR-associated amphiphilic repression domains. This includes the auxin/indole-3-acetic acid (Aux/IAA; auxin), jasmonate-ZIM domain (JAZ; JA), and BRASSINAZOLE RESISTANT1 (BZR1; BR) proteins (Kagale et al., 2010). Second, a number of hormones directly control ligase-substrate interactions (e.g. auxin, JA, and GA). The protein degradation mechanisms involved in auxin, JA, and GA signaling are strikingly similar (Fig. 1, A, D, and E). Additionally, in the case of both auxin and JA, the F-box proteins responsible for target proteolysis are themselves part of the hormone receptor complex. UPS components controlling BR and ethylene signaling also appear to be superficially similar. We will describe the types of E3 ligases and their target proteins (when known) that are active in Arabidopsis (Arabidopsis thaliana) in more detail below. The hormones are organized with the aim of highlighting the mechanistic similarities and differences of UPS action. Conservation of these pathways in other plant species is discussed separately at the end of the article.
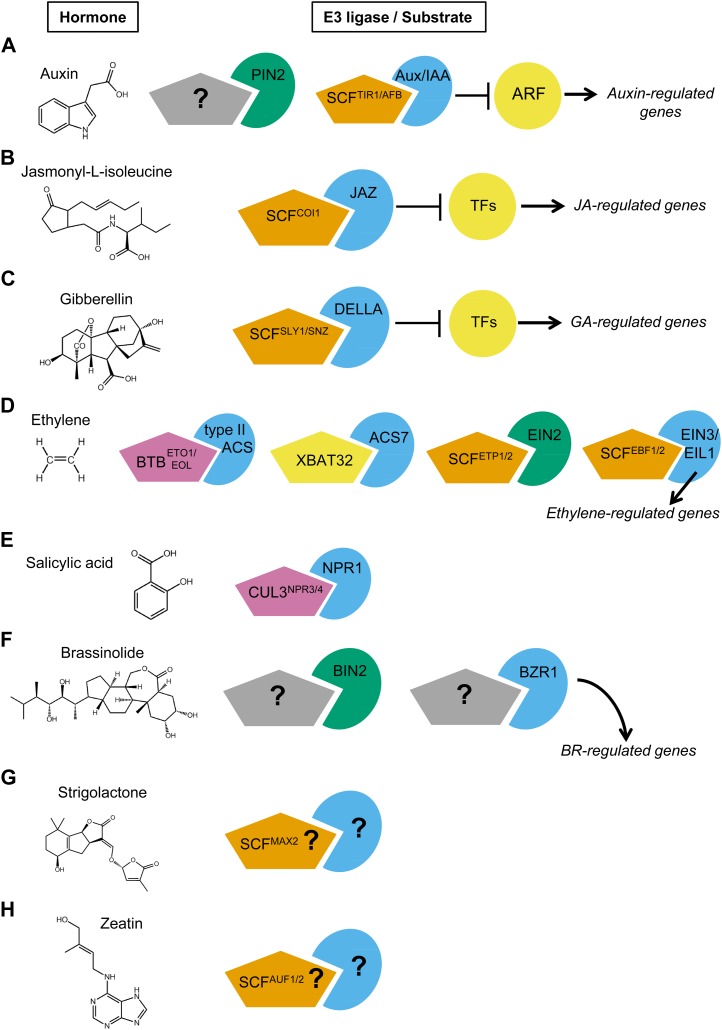
Figure 1.
Conserved and diverse UPS degradation mechanisms control hormone signaling (ABA is considered separately due to the abundance of UPS components; see Fig. 2). SCF-type E3 ligases are shown as orange pentagons, RING-type ligases are shown as yellow pentagons, and CRL3-based ligases are depicted as pink pentagons. Unknown ligases are in gray, with a question mark. Known (or unknown) target proteins are shown in blue (transcription factors) or green (kinases and transporters) pie wedges. Transcription factors that are directly bound by SCF target substrates are shown as yellow circles. A, An auxin efflux carrier, PIN2, is degraded in a ubiquitin-dependent manner. In the presence of high auxin concentrations, SCFTIR1/AFB E3 ligases direct the degradation of Aux/IAA transcriptional repressors, releasing bound ARF transcription factors and thus inducing auxin-mediated transcription. B, In the presence of JA-Ile, SCFCOI1 directs the degradation of JAZ transcriptional repressors, releasing bound transcription factors and thus inducing JA-mediated transcription. C, Nucleus-localized growth-repressive DELLA proteins are targeted for degradation by SCFSLY1/SNZ E3 ligases in response to GA perception. D, Ethylene biosynthesis enzymes are degraded by the proteasome. Type-II ACS proteins are targeted by BTBETO1/EOL, while ACS7 is ubiquitylated by XBAT32. In the absence of ethylene, ETP1/2 directs the degradation of the membrane-bound protein EIN2. Degradation of the ETHYLENE INSENSITIVE3 (EIN3) transcription factor is controlled by EBF1 and EBF2 and is inversely correlated with ethylene levels. Thus, as ethylene levels rise, both EIN2 and EIN3 accumulate and ethylene-regulated transcription is stimulated. E, In the absence of SA, the transcription factor NPR1 is targeted for degradation by the NPR4 CUL3-based ligase. The presence of SA reduces the affinity of CUL3NPR4 for NPR1 and thus allows the accumulation of NPR1 and induced resistance. F, Two key components of BR signaling, BIN2 kinase and the transcription factor BZR1, are targets of regulated proteolysis in response to BR via unknown ligase activity. G, The F-box protein MAX2 has been implicated in SL signaling. H, AUF1 may control levels of a cytokinin (represented here by kinetin) response regulator.
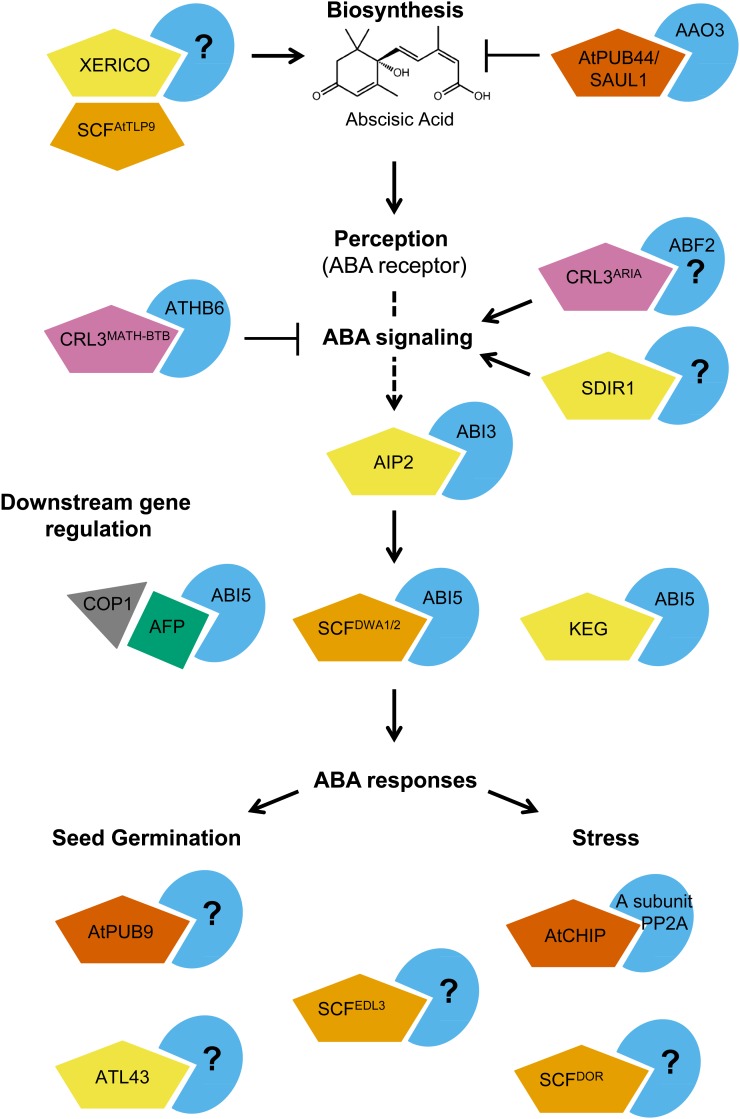
Figure 2.
E3 ligases involved in ABA signaling. The schematic of known UPS components in ABA signaling illustrates that there is regulation of a single substrate (ABI5) by multiple E3 ligases and a number of different classes of E3s act at multiple levels in the ABA pathway, including biosynthesis and downstream gene regulation. RING-type E3 ligases are shown as yellow pentagons, U-box-type E3 ligases as dark orange pentagons, CRL E3 ligases as orange pentagons, and CRL3-based E3 ligases as pink pentagons. AFP is depicted is a green square, and COP1 is a gray triangle. Known (or unknown) ligase substrates are blue pie wedges.
Auxin
Almost every aspect of plant growth and development is controlled by auxin signaling (Stewart and Nemhauser, 2010). The role of UPS activity in the auxin response has been extensively studied and is well established (Santner et al., 2009; Shan et al., 2012). A small family of six F-box proteins, TRANSPORT INHIBITOR RESPONSE1 (TIR1) and AUXIN SIGNALING F-BOX1 (AFB1) to AFB5, act as auxin receptors (Dharmasiri et al., 2005; Greenham et al., 2011). In the presence of auxin, members of the TIR1/AFB1 to -5 family can direct the polyubiquitylation and proteasomal degradation of Aux/IAA transcriptional repressors. There are 29 Aux/IAA proteins in Arabidopsis, and 23 of these are thought to be substrates of SCFTIR1/AFB (Fig. 1A; Calderón Villalobos et al., 2012). Auxin is directly bound by both TIR1/AFB and an Aux/IAA protein simultaneously and thus creates a “coreceptor” complex (Calderón Villalobos et al., 2012). Additionally, the binding affinity of auxin appears to be predominantly controlled by the Aux/IAA proteins and not the TIR1/AFB proteins (Calderón Villalobos et al., 2012). A 13-amino acid motif known as the degron located within domain II of Aux/IAA proteins (Ramos et al., 2001) contributes to substrate stability and turnover rates that range from approximately 10 to 80 min (Dreher et al., 2006). The varied protein-protein interaction affinities between TIR1/AFB proteins and Aux/IAA proteins (Calderón Villalobos et al., 2012) combined with distinct spatiotemporal patterns of accumulation (Parry et al., 2009; Vernoux et al., 2011) may contribute to the broad role of auxin in diverse growth processes.
Under low-auxin conditions, members of the Aux/IAA family participate in repressive transcriptional complexes containing an AUXIN RESPONSE FACTOR (Mansfield et al., 2004) transcription factor and TOPLESS (TPL) corepressor protein (Szemenyei et al., 2008). When auxin levels increase, Aux/IAA proteins are targeted for proteasomal degradation. Thus, the specific degradation of the Aux/IAA protein by SCFTIR1/AFB1-5 will result in the disassembly of the repressive complex. It is worth noting that degradation of ARF1 by the proteasome occurs independently of SCFTIR1/AFB1-5, suggesting that ARF protein levels may also be tightly controlled (Salmon et al., 2008).
Auxin transport is also regulated by the UPS through proteasomal degradation of the auxin efflux carrier protein PIN-FORMED2 (PIN2; Leitner et al., 2012). Ubiquitylation of PIN2 leads to endocytosis and vacuolar targeting of PIN2 in response to auxin and gravity, which affects auxin distribution within the root (Leitner et al., 2012). It will be interesting to determine which E3 ligase(s) catalyzes the ubiquitylation of PIN2; often, endocytic sorting of a target protein involves multiple E3s (Polo, 2012).
Jasmonates
Recent studies indicate that UPS action during the jasmonate response parallels what occurs during auxin signaling. JA-Ile (Fig. 1B) is a lipid-derived hormone known to regulate plant development and response to environmental stresses. The F-box protein CORONATINE-INSENSITIVE1 (COI1) acts as a JA-Ile receptor and directs the degradation of JAZ proteins, a small family of transcriptional repressors (Fig. 1B; Shan et al., 2012). A number of JAZ proteins (e.g. JAZ1) contain a degron motif that facilitates binding to COI1 (Grunewald et al., 2009; Pauwels et al., 2010; Sheard et al., 2010). Similar to the situation for auxin, COI and the JAZ protein act as coreceptors for JA-Ile; thus, JA-Ile directly stabilizes SCFCOI1 interactions with some JAZ proteins (Sheard et al., 2010). Some JAZ proteins can form complexes with TPL through the adapter protein NOVEL INTERACTOR OF JAZ (Pauwels et al., 2010), whereas the ETHYLENE RESPONSE FACTOR-associated amphiphilic repression domain-containing JAZ proteins (e.g. JAZ8) interact directly with TPL or other corepressors to inhibit JA responses (Shyu et al., 2012). This JAZ-based complex acts to repress the activity of transcription factors such as MYC2 and MYB21/24 (Pauwels et al., 2010; Song et al., 2011). In the presence of JA-Ile, JAZ proteins are degraded, JAZ-mediated repression is relieved, and early JA-responsive genes are rapidly expressed (Pauwels et al., 2010). Similar to the Aux/IAA proteins, sequence variation in the degron motif alters the stability and repressive action of JAZ proteins (Shyu et al., 2012).
GA
Similar SCF-mediated regulation has been shown for GA signaling (Fig. 1C; Gao et al., 2011). The effects of GA include promotion of seed germination, stimulation of organ elongation, and induction of flowering. GA is perceived by a protein called GIBBERELLIN INSENSITIVE DWARF1 (GID1). GA binding to GID1 results in binding to nucleus-localized growth repressors called DELLA proteins (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006; Willige et al., 2007). This tripartite GID-GA-DELLA complex is subsequently targeted for ubiquitylation by SCFSLY1/SNZ E3 ligases, resulting in degradation of the DELLAs (Gao et al., 2011). This scenario is reminiscent of UPS action during auxin and JA-Ile signaling, because DELLA proteins bind and repress the activity of transcription factors such as the PHYTOCHROME INTERACTING FACTORs (de Lucas et al., 2008; Feng et al., 2008). Thus, this scenario is reminiscent of UPS action during auxin and JA signaling because hormone perception results in the degradation of proteins involved in transcriptional repression. However, while protein degradation is directly involved in GA perception, it is different compared with auxin and JA receptor systems, where the receptors are themselves part of E3 ligase complexes.
Ethylene
Ethylene functions as a critical growth regulator and is also important for biotic and abiotic stress responses (Schaller, 2012). Many aspects of ethylene signaling are tightly controlled by the UPS (Fig. 1D). First, a number of enzymes involved in ethylene biosynthesis are targeted for proteasomal degradation. This includes (1) type-2 1-aminocyclopropane-1-carboxylic acid synthase proteins (ACS4, ACS5, and ACS9), which are ubiquitylated by ETO1 and ETO-like1/2 BTB ligases (Wang et al., 2004; Yoshida et al., 2005; Christians et al., 2009), and (2) ACS7, a type-3 ACS enzyme that is ubiquitylated by the RING-type E3 ligase XBAT32 (Lyzenga et al., 2012). These degradation mechanisms provide a rapid way to change ethylene concentrations in planta.
In the absence of ethylene, a pair of F-box proteins, EIN2-TARGETING PROTEIN1 (ETP1) and ETP2, promote the degradation of the ethylene signaling protein ETHYLENE INSENSITIVE2 (EIN2), reducing the ethylene response (Qiao et al., 2009). In the presence of ethylene, ETP expression is repressed, allowing the accumulation of EIN2. Downstream of EIN2 lies EIN3 and EIN3-like1 (EIL1), transcription factors that directly target ethylene-responsive genes. At low ethylene levels, EIN3 and EIL1 are targeted for ubiquitylation and degradation by another pair of F-box proteins, EIN3-BINDING F-BOX1 (EBF1) and EBF2, which are also subject to proteasomal degradation (Guo and Ecker, 2003; Potuschak et al., 2003; An et al., 2010). As ethylene levels increase, the stability of EBF1/2 decreases, leading to a buildup of EIN3 and EIL1, thus inducing transcription (An et al., 2010).
Salicylic Acid
Salicylic acid (SA) is a secondary metabolite produced by a wide range of organisms, and salicylates have been used by humans for pain relief for centuries (An and Mou, 2011). In plants, SA functions as a plant hormone required for innate immunity (Vlot et al., 2009). Two paralogous receptors were recently identified for SA, NPR1-LIKE PROTEIN3 (NPR3) and NPR4, which are BTB-CUL3 ligases (Fu et al., 2012). CUL3NPR3/4 direct the degradation of NONEXPRESSOR OF PR GENES1 (NPR1), a master transcriptional regulator of plant defense (Fu et al., 2012). NPR1 interacts with other transcription factors in the nucleus to mediate pathogen resistance (Fu et al., 2012). Thus, the perception of SA (Fig. 1E) parallels other UPS-controlled hormone signaling mechanisms (Fig. 1).
Other Hormones
To what extent (and how) UPS activity plays a role in BR, SL, and cytokinin signaling is not clear at present (Fig. 1, F–H). A couple of key components of BR signaling appear to be degraded by the 26S proteasome in response to BR by unknown ligases (Fig. 1F). Specifically, the BRASSINOSTEROID INSENSITIVE2 (BIN2) kinase is regulated by proteasome-mediated protein degradation (Peng et al., 2008). Additionally, the transcription factor BZR1 is phosphorylated by BIN2 and rapidly degraded by the proteasome to mediate the feedback inhibition of several BR biosynthetic genes (He et al., 2002). A recent report also demonstrated that the endoplasmic reticulum-associated protein degradation pathway controls levels of the BR receptor BIN1 via a stress-induced ubiquitin conjugation enzyme, UBC32 (Cui et al., 2012). UPS activity is implicated in SL signaling because MORE AXILLARY BRANCHES2 (MAX2) encodes an F-box protein (Stirnberg et al., 2002). A direct role for the UPS in cytokinin action has not been described. However, the recent identification of AUXIN UP-REGULATED F-BOX PROTEIN1 (AUF1) implicates SCFAUF1/2 in mediating interactions between cytokinin and auxin (Zheng et al., 2011). Given the extent of protein degradation for other hormones, the identification of MAX2 and AUF1/2 targets should prove to be quite interesting.
ABA
The number of E3 ligases involved (or implicated) in ABA signaling is more than for any other hormone to date. As a result, many aspects of ABA biology are controlled by protein degradation (Fig. 2). ABA plays important roles in many physiological processes, including seed germination and stress responses, both biotic and abiotic. The current understanding of ABA signaling, from receptors to responses, includes many intermediate steps and is quite complex (Cutler et al., 2010; Kim, 2012).
At least one ligase has been shown to directly impact ABA biosynthesis. SENESCENCE-ASSOCIATED E3 UBIQUITIN LIGASE1/ARABIDOPSIS THALIANA PLANT U-BOX44 (AtPUB44) regulates the levels of ABSCISIC ALDEHYDE OXIDASE3, an enzyme that converts abscisic aldehyde to ABA (Raab et al., 2009). Another gene that has been shown to affect ABA levels is XERICO, which encodes a RING-H2 domain-containing protein that can interact with UPS components in yeast, including the F-box TUBBY-LIKE PROTEIN9 (Ko et al., 2006).
The levels of ABA INSENSTITIVE5 (ABI5), a basic Leu zipper transcription factor, are regulated by at least two different classes of E3 ligases, including KEEP ON GOING, a RING-type E3, and DDB-BINDING WD40 PROTEIN1 (DWA1) and DWA2, CUL4-based CRLs (Stone et al., 2006; Lee et al., 2010). The nucleus-localized ABI FIVE BINDING PROTEIN (AFP) family appears to promote ABI5 degradation in nuclear bodies in concert with the RING protein CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), although it is not exactly clear how this process occurs (Lopez-Molina et al., 2003). Additionally, posttranslational modifications such as phosphorylation and sumoylation of ABI5 may also play a role in proteasome-mediated degradation of ABI5 (Lopez-Molina et al., 2001, 2003; Miura et al., 2009). The regulation of a single substrate by multiple E3 ligases has been well documented in animal systems. For example, the transcription factor p53 was targeted for ubiquitylation and degradation by numerous E3 ligases (Benkirane et al., 2010). Such a mechanism may rapidly integrate different inputs to control the level of a key transcription factor leading to the appropriate response(s). Additionally, a RING-type E3, ABI3-INTERACTING PROTEIN2, acts to control ABI3 transcription factor abundance and modulate downstream ABA signaling (Zhang et al., 2005).
A small family of CUL3-based E3 ligases designated CRLBPM have recently been found to play a key role in ABA signaling by directing the proteasomal degradation of the class I homeobox-Leu zipper (HD-ZIP) transcription factor ATHB6 (Lechner et al., 2011). Furthermore, all six MATH-BTB proteins can interact with three different HD-ZIPI transcription factors (ATHB5, ATHB6, and ATHB16), suggesting that this family of E3 ligases may regulate other processes as well (Lechner et al., 2011). A yeast two-hybrid (Y2H) screen with ABSCISIC ACID RESPONSIVE ELEMENTS-BINDING FACTOR2 (ABF2) identified an arm repeat protein interacting with ABF2 (Brentani et al., 2003), which may act as part of a CUL3-based ligase (Kim et al., 2004). Additionally, the E3 ligase AtCHIP can modulate the activity of an A subunit of protein phosphatase 2A by ubiquitylation, leading to altered ABA-induced stomatal closing and inhibition of seed germination (Luo et al., 2006).
A number of other classes of E3 ligases have been shown to function during ABA signaling, but their targets (and direct roles in protein degradation) have not been established yet. These include the F-box proteins Drought Tolerance Repressor (Zhang et al., 2008) and EID1-LIKE PROTEIN3 (Koops et al., 2011), AtPUB9 (Samuel et al., 2008), and two RING-type E3s, SALT- AND DROUGHT-INDUCED RING FINGER1 (Zhang et al., 2007) and ATL43 (Serrano et al., 2006).
INTERACTIONS BETWEEN HORMONES
Plant growth and development involves the simultaneous integration of many different hormone signals at both the cellular and organ levels. With respect to UPS activity and the hormones, there are a number of emerging examples illustrating how such cross talk may occur. For instance, TPL proteins are general corepressors that affect both auxin and JA signaling pathways through the interaction with specific transcription factors (e.g. Aux/IAA and JAZ proteins). There are also examples of one hormone inducing an E3 ligase that acts on a target active in another hormone signaling pathway. For example, AUF1 is expressed in response to auxin and affects ARR1 levels (Zheng et al., 2011). The physical interaction of JAZ and DELLA proteins is an example of how protein-protein interactions directly link two distinct hormone pathways (Hou et al., 2010; Arabidopsis Interactome Mapping Consortium, 2011; Kazan and Manners, 2012; Yang et al., 2012). As protein-protein interactions continue to be characterized in plants, we expect the degree of connectedness to increase among hormones.
EVOLUTIONARY CONSIDERATIONS
The sheer number of UPS components in plants suggests that the ubiquitin pathway has a broad role in cellular regulation (Vierstra, 2009). Given what is already known from Arabidopsis, it is likely that many pathways will be conserved in other species. However, there are also likely to be some differences. In fact, all of the UPS components and protein-protein interactions involved in auxin signaling, including the F-box coreceptors and TPL-Aux/IAA-ARF interactions, appear to be conserved in the moss Physcomitrella patens (Prigge et al., 2010; Causier et al., 2012). Over 30 plant genomes are now available at Phytozome (Goodstein et al., 2012), providing ample opportunities for in silico identification of UPS components for further study. Additionally, a curated database of UPS proteins, called plantsUPS database, is available online at http://bioinformatics.cau.edu.cn/plantsUPS/ (Du et al., 2009).
FUTURE DIRECTIONS
How many proteins are modified by ubiquitin and degraded by the proteasome? This is currently a major outstanding question in plant biology. Initial descriptions of ubiquitylated proteins in Arabidopsis have identified about 100 proteins that are modified by ubiquitin, including known E3 target proteins such as EIN3 and JAZ6 (Maor et al., 2007; Saracco et al., 2009). The human ubiquitinome (ubiquitin-modified proteome) was recently characterized using a unique ubiquitin-based antibody specific for ubiquitin-conjugated proteins (Kim et al., 2011). Because this antibody recognizes signature di-Gly fragments on ubiquitin conjugates following trypsin digestion, a similar strategy could be employed in plants to capture ubiquitylated proteins for identification by mass spectrometry. Such data could help identify target substrates for known ubiquitin ligases (Figs. 1 and 2) and also identify new proteins that are under UPS control. Combining these techniques with hormone treatments and receptor mutants could help place the role of the UPS in hormone-specific contexts. It is worth noting that a three-step immunoprecipitation workflow combined with tandem mass spectrometry successfully identified 357 proteins modified by SUMO (for small ubiquitin-like modifier) in Arabidopsis (Miller and Vierstra, 2011). Most of the SUMOylated proteins are involved in transcription and chromatin modification, suggesting that attachment of SUMO may be an important posttranslational modification for gene regulation.
While the degradation motifs are known for Aux/IAA and JAZ proteins, we do not know what other degron(s) contribute to ligase-substrate interactions and protein stability. These domains are short and cannot be easily identified in silico due to a lack of sequence conservation and because the attachment of ubiquitin to substrates may not be restricted to Lys residues (Dreher et al., 2006; Hochstrasser et al., 2008). In fact, we have very poor knowledge of ubiquitylation sites on substrates altogether, even for well-studied proteins like the Aux/IAAs. The use of immunoprecipitation strategies combined with mass spectrometry is one approach to identifying such residues. Additionally, degron motifs may be useful sensors of hormone action within cells and tissues. A fluorescently tagged version of Aux/IAA domain II (DII-VENUS), which contains the canonical degron for this family of SCFTIR1/AFB1-5 substrates, has been used to quantitatively image auxin in the root and shoot (Band et al., 2012; Brunoud et al., 2012). Another potential area of investigation involves determining the half-lives for E3 targets. Aux/IAA and JAZ proteins exhibit rapid degradation within the range of approximately 1 to 80 min (Dreher et al., 2006; Pauwels et al., 2010), but we do not know the stability of other target proteins.
Proteomics studies from various organisms have shown a very poor, slightly positive correlation between mRNA levels and protein abundance (Taniguchi et al., 2010; Maier et al., 2011; Schwanhäusser et al., 2011; Vogel and Marcotte, 2012). Additionally, it is clear that many proteins are under posttranslational control, and we do not currently know to what extent posttranslational modifications (such as phosphorylation) play a role in UPS-mediated processes in plants. For instance, only 56% (78 out of 138) of the hormone- and UPS-related proteins discussed here (Figs. 1 and 2) are present in the current proteomics databases (MASCP Gator search; Joshi et al., 2011). Thus, we need to develop new techniques and approaches in order to determine protein abundance and posttranslational modifications in planta more routinely.
A number of protein-protein assays have been used to identify ligase-substrate pairs, but most E3 ligases in Arabidopsis are currently orphans. A high-throughput Y2H assay could be utilized to identify new ubiquitylation targets (network evolution in an Arabidopsis interactome map; Arabidopsis Interactome Mapping Consortium, 2011). Considering that a large number of known E3 targets are transcription factors (Figs. 1 and 2), screening with a transcription factor-specific library (Pruneda-Paz et al., 2009; Brady et al., 2011; Ou et al., 2011) could be very fruitful. Such screens would also be informative for describing the activities of UPS targets. For instance, new protein-protein interactions were described based on a Y2H assay between a sunflower (Helianthus annuus) Aux/IAA protein (HaIAA27) and a heat shock transcription factor, HaHSFA9 (Carranco et al., 2010). Once target proteins are identified, a number of assays can be used to confirm that they are ubiquitylated by ligases. Efficient ubiquitylation assays have been recently developed using tobacco (Nicotiana tabacum) leaves (Liu et al., 2010) and UBIQapture columns (Lechner et al., 2011).
It truly is an exciting time to be studying ubiquitylation and protein degradation in plants, as there are still many questions and new tools being developed to help us find answers. Although DNA and RNA molecules provide the templates, proteins are the keystones of cells. Further insights into the regulation of protein abundance, especially with respect to plant hormone signaling, have strong potential to positively impact many areas of plant agriculture.
Glossary
- UPS
ubiquitin-proteasome system
- CRL
cullin-RING ligase
- ABA
abscisic acid
- BR
brassinosteroid
- JA
jasmonic acid
- SL
strigolactone
- SA
salicylic acid
- Y2H
yeast two-hybrid
References
- An C, Mou Z. (2011) Salicylic acid and its function in plant immunity. J Integr Plant Biol 53: 412–428 [DOI] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Larrieu A, Sun J, Middleton AM, French AP, Brunoud G, Sato EM, Wilson MH, Péret B, et al. (2012) Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc Natl Acad Sci USA 109: 4668–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Sardet C, Coux O. (2010) Lessons from interconnected ubiquitylation and acetylation of p53: think metastable networks. Biochem Soc Trans 38: 98–103 [DOI] [PubMed] [Google Scholar]
- Brady SM, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, Liu W, Zeng A, Taylor-Teeples M, Kim D, et al. (2011) A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentani H, Caballero OL, Camargo AA, da Silva AM, da Silva WA, Jr, Dias Neto E, Grivet M, Gruber A, Guimaraes PE, Hide W, et al. (2003) The generation and utilization of a cancer-oriented representation of the human transcriptome by using expressed sequence tags. Proc Natl Acad Sci USA 100: 13418–13423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranco R, Espinosa JM, Prieto-Dapena P, Almoguera C, Jordano J. (2010) Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc Natl Acad Sci USA 107: 21908–21913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Lloyd J, Stevens L, Davies B. (2012) TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signal Behav (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hansen M, Binder BM, Kieber JJ, Vierstra RD. (2009) The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J 57: 332–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Liu L, Zhao Q, Zhang Z, Li Q, Lin B, Wu Y, Tang S, Xie Q. (2012) Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Li L, Su Z. (2009) plantsUPS: a database of plants’ ubiquitin proteasome system. BMC Genomics 10: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XH, Xiao SL, Yao QF, Wang YJ, Fu XD. (2011) An updated GA signaling ‘relief of repression’ regulatory model. Mol Plant 4: 601–606 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, Santner A, Castillejo C, Mooney S, Sairanen I, Ljung K, Estelle M. (2011) The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr Biol 21: 520–525 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Grunewald W, Vanholme B, Pauwels L, Plovie E, Inzé D, Gheysen G, Goossens A. (2009) Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep 10: 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MS, Ou C, Chan YR, Chien CT, Pi H. (2008) The utility F-box for protein destruction. Cell Mol Life Sci 65: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M, Deng M, Kusmierczyk AR, Li X, Kreft SG, Ravid T, Funakoshi M, Kunjappu M, Xie Y. (2008) Molecular genetics of the ubiquitin-proteasome system: lessons from yeast. Ernst Schering Foundation Symposium Proceedings, Vol 2. Springer-Verlag, Heidelberg, Germany, pp 41–66. [DOI] [PubMed]
- Hou X, Lee LY, Xia K, Yan Y, Yu H. (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Joshi HJ, Hirsch-Hoffmann M, Baerenfaller K, Gruissem W, Baginsky S, Schmidt R, Schulze WX, Sun Q, van Wijk KJ, Egelhofer V, et al. (2011) MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol 155: 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17: 22–31 [DOI] [PubMed] [Google Scholar]
- Kim S, Choi HI, Ryu HJ, Park JH, Kim MD, Kim SY. (2004) ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol 136: 3639–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH. (2012) Plant stress surveillance monitored by ABA and disease signaling interactions. Mol Cells 33: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH. (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47: 343–355 [DOI] [PubMed] [Google Scholar]
- Koops P, Pelser S, Ignatz M, Klose C, Marrocco-Selden K, Kretsch T. (2011) EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J Exp Bot 62: 5547–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E, Leonhardt N, Eisler H, Parmentier Y, Alioua M, Jacquet H, Leung J, Genschik P. (2011) MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev Cell 21: 1116–1128 [DOI] [PubMed] [Google Scholar]
- Lee JH, Lee MJ. (2012) Emerging roles of the ubiquitin-proteasome system in the steroid receptor signaling. Arch Pharm Res 35: 397–407 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO, Deng XW. (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J, Petrasek J, Tomanov K, Retzer K, Parezova M, Korbei B, Bachmair A, Zazimalova E, Luschnig C. (2012) Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci USA 109: 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q. (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61: 893–903 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Shen G, Yan J, He C, Zhang H. (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J 46: 649–657 [DOI] [PubMed] [Google Scholar]
- Lyzenga WJ, Booth JK, Stone SL. (2012) The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J 71: 23–34 [DOI] [PubMed] [Google Scholar]
- Maier T, Schmidt A, Güell M, Kühner S, Gavin AC, Aebersold R, Serrano L. (2011) Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol 7: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, et al. (2004) MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 36: 1079–1083 [DOI] [PubMed] [Google Scholar]
- Maor R, Jones A, Nühse TS, Studholme DJ, Peck SC, Shirasu K. (2007) Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol Cell Proteomics 6: 601–610 [DOI] [PubMed] [Google Scholar]
- Miller MJ, Vierstra RD. (2011) Mass spectrometric identification of SUMO substrates provides insights into heat stress-induced SUMOylation in plants. Plant Signal Behav 6: 130–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Ou B, Yin KQ, Liu SN, Yang Y, Gu T, Wing Hui JM, Zhang L, Miao J, Kondou Y, Matsui M, et al. (2011) A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol Plant 4: 546–555 [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P, Yan Z, Zhu Y, Li J. (2008) Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol Plant 1: 338–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S. (2012) Signaling-mediated control of ubiquitin ligases in endocytosis. BMC Biol 10: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M. (2010) Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol 20: 1907–1912 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR. (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab S, Drechsel G, Zarepour M, Hartung W, Koshiba T, Bittner F, Hoth S. (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J 59: 39–51 [DOI] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon J, Ramos J, Callis J. (2008) Degradation of the auxin response factor ARF1. Plant J 54: 118–128 [DOI] [PubMed] [Google Scholar]
- Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, Goring DR. (2008) Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol 147: 2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Calderon-Villalobos LI, Estelle M. (2009) Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol 5: 301–307 [DOI] [PubMed] [Google Scholar]
- Saracco SA, Hansson M, Scalf M, Walker JM, Smith LM, Vierstra RD. (2009) Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE. (2012) Ethylene and the regulation of plant development. BMC Biol 10: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Serrano M, Parra S, Alcaraz LD, Guzmán P. (2006) The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J Mol Evol 62: 434–445 [DOI] [PubMed] [Google Scholar]
- Shan X, Yan J, Xie D. (2012) Comparison of phytohormone signaling mechanisms. Curr Opin Plant Biol 15: 84–91 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C, Figueroa P, Depew CL, Cooke TF, Sheard LB, Moreno JE, Katsir L, Zheng N, Browse J, Howe GA. (2012) JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24: 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. (2011) The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JL, Nemhauser JL. (2010) Do trees grow on money? Auxin as the currency of the cellular economy. Cold Spring Harb Perspect Biol 2: a001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. (2010) Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13: 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR. (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. (2005) Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH. (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu W, Li Z, Deng XW, Wu W, Xue Y. (2008) F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis. Plant Physiol 148: 2121–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Miller ND, Lewis DR, Christians MJ, Lee KH, Muday GK, Spalding EP, Vierstra RD. (2011) AUXIN UP-REGULATED F-BOX PROTEIN1 regulates the cross talk between auxin transport and cytokinin signaling during plant root growth. Plant Physiol 156: 1878–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]