Abstract
Brassinosteroid (BR) signaling is essential for plant growth and development. In Arabidopsis (Arabidopsis thaliana), BRs are perceived by the BRASSINOSTEROID INSENSITIVE1 (BRI1) receptor. Root growth and hypocotyl elongation are convenient downstream physiological outputs of BR signaling. A computational approach was employed to predict root growth solely on the basis of BRI1 receptor activity. The developed mathematical model predicts that during normal root growth, few receptors are occupied with ligand. The model faithfully predicts root growth, as observed in bri1 loss-of-function mutants. For roots, it incorporates one stimulatory and two inhibitory modules, while for hypocotyls, a single inhibitory module is sufficient. Root growth as observed when BRI1 is overexpressed can only be predicted assuming that a decrease occurred in the BRI1 half-maximum response values. Root growth appears highly sensitive to variation in BR concentration and much less to reduction in BRI1 receptor level, suggesting that regulation occurs primarily by ligand availability and biochemical activity.
Brassinosteroids (BRs) play a key role in plant growth and development. In Arabidopsis (Arabidopsis thaliana), BRs are perceived by the plasma membrane-located receptor kinase BRASSINOSTEROID INSENSITIVE1 (BRI1). Detailed models (He et al., 2000; Wang et al., 2006; Jaillais et al., 2011a; Ye et al., 2011) of BR signaling describe the binding of BR to the extracellular leucine-rich repeat domain of BRI1 followed by activation of the receptor through autophosphorylation (Wang and Chory, 2006). Subsequently, BRI1 phosphorylates BRI1-KINASE INHIBITOR1 (BKI1), resulting in the release of BKI1 from the intracellular BRI1 kinase domain (Jaillais et al., 2011b). BKI1 restricts the association between BRI1 and another leucine-rich repeat receptor-like kinase, the BRI1-ASSOCIATED KINASE1 (BAK1; Li et al., 2002; Nam and Li, 2002). The release of BKI1 from the kinase domain of BRI1 enables transphosphorylation between BRI1 and BAK1 (Wang et al., 2008), initiating a phosphorylation-and-dephosphorylation cascade mediated by cytoplasmic BR signaling kinases (Kim et al., 2009). This cascade results in the transcriptional regulation of BR-responsive genes (Yin et al., 2002; Sun et al., 2010; Wu et al., 2011; Yu et al., 2011) mediated by the BRASSINAZOLE RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR-1 (BES1)/BZR2 transcriptional regulators (Wang et al., 2002; Yin et al., 2002). The PROTEIN PHOSPHATASE2A (PP2A) regulates at least two different steps in the BRI1 signaling pathway, dephosphorylation and degradation of BRI1 after activation (Wu et al., 2011) and dephosphorylation of BKI1, thereby promoting its translocation to the nucleus (Tang et al., 2011). Thus, both negative (BKI1 and PP2A) and positive (BAK1 and BR signaling kinase) regulators exist in the pathway (Supplemental Fig. S1). Disruption of BR signaling results in complex phenotypes such as extreme dwarfed stature, impaired photomorphogenesis, fertility defects, and impaired root growth (Clouse, 1996; Wang et al., 2001). BRs stimulate root growth at low ligand concentrations and are strongly inhibitory at high ligand concentrations (Müssig et al., 2003). Overexpression of BRI1, a reduction of BRI1 activity by mutation, and exogenously applied ligands all result in an arrest in cell cycle progression in the root meristem (González-García et al., 2011). Hence, BRI1-mediated BR signaling may not only affect root growth by promoting cell elongation (Szekeres et al., 1996; Müssig et al., 2003) but also by affecting cell division. In Arabidopsis roots, the meristem and elongation zone are very sensitive to changes in BR concentration, with the epidermis playing a major role in translating the BR signal into a root growth response (Hacham et al., 2011). Remarkably, BRI1 receptor density in wild-type roots remains almost constant in the meristematic region (van Esse et al., 2011). To help understand the complex cellular responses to BRI1-mediated BR signaling, a mathematical model is beneficial. Modeling was previously employed to describe the radial patterning of the vascular bundles in shoots, which is controlled by polar auxin transport and BR signaling (Ibañes et al., 2009; Fàbregas et al., 2010), and to help understand the role of the BREVIS RADIX (BRX) gene in BR and auxin signaling (Sankar et al., 2011).
The aim of this work is to develop a mathematical model to link BRI1 receptor activity directly to a downstream physiological response. Therefore, wild-type Arabidopsis root growth was used based on its reproducible and quantifiable response to BR signaling activity. Sufficient biochemical parameters such as the BRI1 dissociation constant (Wang et al., 2001) and the BRI1 concentration (van Esse et al., 2011) are now available for this purpose. The developed model includes one stimulatory and two inhibitory modules. For validation, roots were used in which BRI1 receptor activity was either increased by expressing an extra BRI1 copy or decreased by using strong and weak bri1 mutant alleles. Computer-generated model simulations show that the growth behavior of the bri1 mutant roots can be faithfully predicted in terms of BRI1 receptor occupancy level. Interestingly, a model structure with only one inhibitory response faithfully predicts the hypocotyl elongation response, previously reported to primarily rely on cell elongation (Gendreau et al., 1997). We conclude that mathematical modeling of the BRI1 plant signaling pathway helps to explain complex physiological responses in both roots and hypocotyls.
RESULTS
BRI1 Receptor Occupancy
Receptor occupancy is a widely used estimated parameter to describe signaling activity (Brent, 2009). To accurately predict BRI1 occupancy at physiological ligand concentrations, a good estimate of BRI1 receptor and BR ligand concentration is required. BRI1 is capable of binding the most active BR, 24-epibrassinolide (BL), but also 22-homobrassinolide (HBL) and the bioactive precursor castasterone (Wang et al., 2001), the latter two being approximately 5-fold less active. The endogenous level of castasterone is about 0.03 to 0.05 ng g−1 fresh weight (Bancoş et al., 2002; Shimada et al., 2003) or 0.06 to 0.1 nm, assuming an equal distribution throughout the inside and outside of the cells. In roots, only trace amounts of BL and HBL are detected (Bancoş et al., 2002; Shimada et al., 2003). It is not known where the natural pool of BRs exist; if this is exclusively outside of the cells, it is reasonable to assume that physiological levels can be about 10-fold higher (i.e. up to approximately 1 nm). To accurately predict BRI1 receptor occupancy at physiological ligand concentrations, all root growth experiments described here were performed using seedlings precultured in the presence of brassinazole (BRZ), a potent inhibitor of BR biosynthesis (Asami et al., 2000). To ensure that the stimulatory and inhibitory effects of BRs on root growth entirely depend on the exogenous supply of ligand, BRZ remained present in the medium throughout the experiments. With this experimental setup, we were able to monitor a clear stimulatory effect of exogenously applied BL on root growth with concentrations up to 1 nm (Fig. 1A). For comparison, the stimulatory effect of BL is hardly visible when wild-type roots are treated without the presence of BRZ (Supplemental Fig. S2), confirming previous reports (Müssig et al., 2003; González-García et al., 2011). To estimate BRI1 occupancy under these physiological ligand concentrations, several assumptions had to be made based on the literature. First, the highest affinity ligand is BL (Wang et al., 2001). Therefore, the model only considers BL binding to BRI1. Second, BL is bound at the island domain of BRI1 (He et al., 2000) in a ratio of one molecule to one monomer (Hothorn et al., 2011; She et al., 2011). At present, it is not clear whether monomers, homodimers, or heterodimers (Russinova et al., 2004) represent the active state. The third assumption is that the affinity of BRI1 for BL does not change between the monomeric, homodimeric, or heterodimeric state. For the BL dissociation constant, values ranging between 7.4 to 15 nm (Wang et al., 2001) and 55 nm (Caño-Delgado et al., 2004) have been reported, so simulations were done at a comparable range of values. Fourth, the BRI1 receptor concentration is estimated at 62 ± 4 nm in wild-type seedling roots (van Esse et al., 2011). The fifth assumption is that BRI1 receptor concentration remains constant during receptor activation and as long as required to record a downstream physiological effect (Geldner et al., 2007).
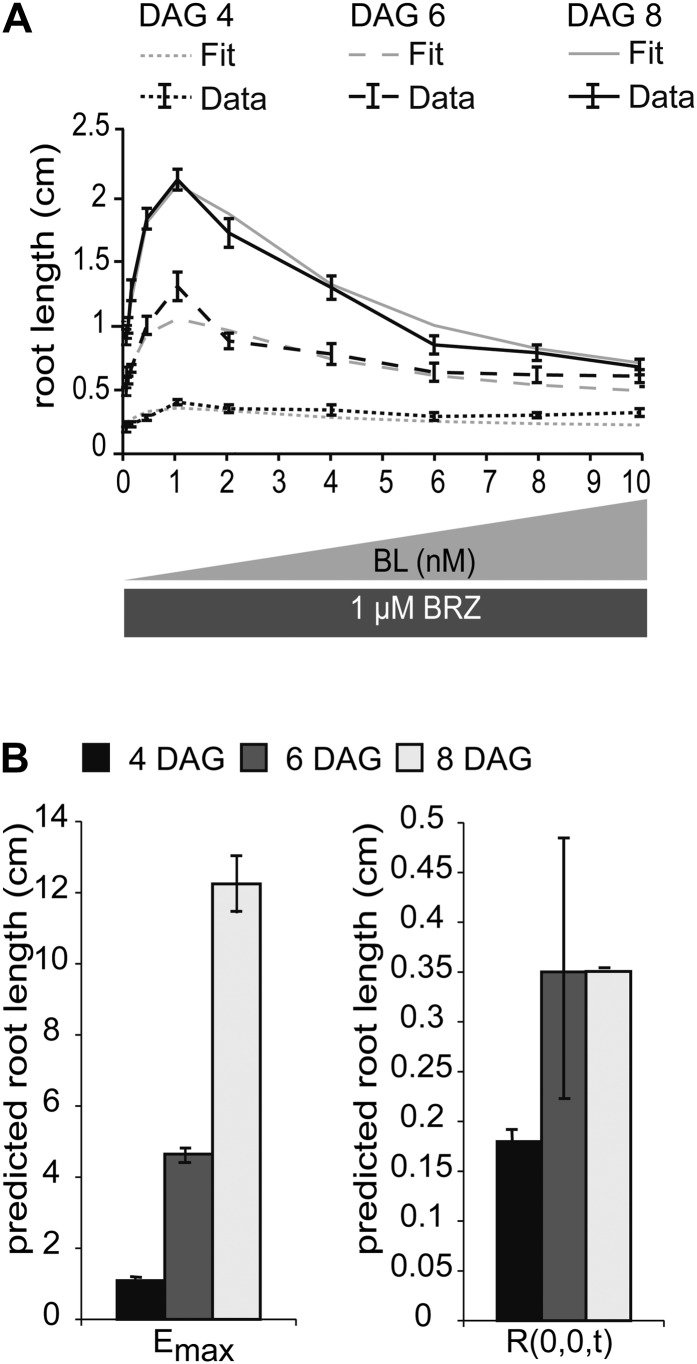
Figure 1.
BRI1 receptor occupancy is linked to root growth. A, Visualization of root-promoting and -inhibitory effects of BL on root growth at 4, 6, and 8 DAG. The medium was supplemented with 1 μm BRZ, after which increasing amounts of BL were added. Error bars represent se for 30 roots per data point measured in three independent replicates. The model is able to fit the predicted root length with the experimental data set. B, Model predictions of Emax and R(0,0,t) at 4, 6, and 8 DAG. The model was fit to the measured root length of seedlings at 4, 6, and 8 DAG.
With these assumptions, the concentration of ligand-occupied BRI1 receptors can be described as a steady-state equilibrium between free receptor and free ligand.
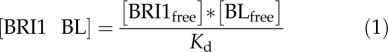 |
In accordance with the first three assumptions, this equilibrium is subject to the mass balance:
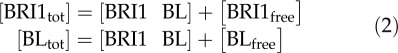 |
By combining Equations 1 and 2, and the condition that without ligand there is no signaling, the number of BRI1 molecules occupied by ligand can be calculated analytically as:
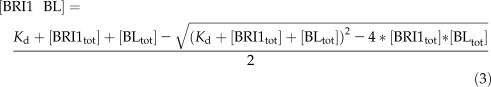 |
Here, [BRI1 BL] is the amount of BRI1 receptor occupied by ligand, Kd is the dissociation constant, [BRI1tot] is the total amount of BRI1, and [BLtot] is the total amount of BL. To calculate the BRI1 receptor occupancy, concentrations of 1 and 10 nm BL were used in the calculations, in line with the root growth stimulatory and inhibitory modes, respectively (Fig. 1A). From Equation 3, it follows that ligand occupancy of the BRI1 receptor at a physiological, stimulatory BL concentration is less than 1% and at full inhibitory mode is not more than 15% (Supplemental Fig. S3).
Relationship between BRI1 Receptor Occupancy and Root Growth
A model was developed to describe root growth in terms of ligand-dependent receptor occupancy. The model structure with the best predictive power has one stimulatory module and two inhibitory modules (Eq. 4). Which biological entities represent the inhibitory modules is not clear at present. As discussed in the previous section, the receptor ligand concentration is considered constant over time, as are the half-maximum response values in the pathway. However, BRI1-dependent and -independent root growth are an unknown function of time. This gives the following root growth model:
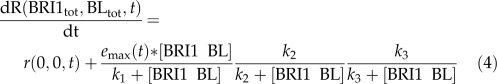 |
Here, r(0,0,t) is the root growth at time t in the absence of BRI1-mediated signaling, emax(t) is the maximum possible root growth when no inhibitory mechanisms are present, and k1, k2, and k3 are the half-maximum response values (k values). Integrating Equation 4 over time gives the root length at time t:
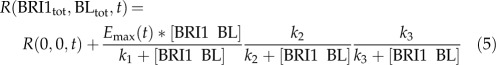 |
with
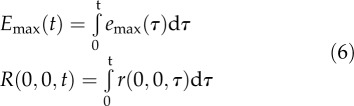 |
in which τ is the integration variable representing the time over which it is integrated, going from 0 to t. The model structure in Equation 5 can be fitted precisely to the biological data (Fig. 1A). Emax(t) and R(0,0,t) are regarded as free parameters that are changed over time (Fig. 1B). According to the Akaike (AIC) and Bayesian (BIC) information criteria (Klipp et al., 2009), several model structures explain the data equally well. To select the best model, the predictive power with respect to the bri-116 null mutant was also taken into account. Of all tested model structures, there was only one with a better prediction, although with higher AIC and BIC scores (Supplemental File S1; Supplemental Table S1; Supplemental Figs. S4–S6). Models with only one module or with a single inhibitory module had significantly higher AIC and BIC scores, clearly indicating a minimal necessary model complexity (Supplemental File S1; Supplemental Table S1).
Plants treated with BRZ display similar phenotypes to deetiolated2 (det2) mutants (Nagata et al., 2000). The det2 mutant has been reported to contain less than 10% of the wild-type levels of BRs (Fujioka et al., 1997). Therefore, the model assumes that there is still a small amount of endogenous BRs (0.1 nm or less in the extracellular compartment) present despite the BRZ treatment. However, the stimulatory effect of BL on root length is only observed in roots depleted from endogenous BL using BRZ. The model can explain this observation by fitting the root length of seedlings treated with BL only. Assuming a physiological BL level of 1 nm, only the inhibitory effect of BL on root growth is observed (Supplemental Fig. S7).
The model parameters R(0,0,t), Emax(t) at discrete t, as well as k1, k2, and k3 were calibrated with the actual measured root lengths in wild-type plants after treatment with BL in increasing amounts. Roots of different ages do not have the same sensitivity to BRs. Therefore, calibration was done on root lengths of wild-type seedlings at 4, 6, and 8 d after germination (DAG). For this, a controlled random search parameter-fitting procedure was conducted (Price, 1976) that found the same minima as a hybrid algorithm consisting of two runs of the Matlab Genetic Algorithm, a global search algorithm, followed by the Matlab gradient-based search algorithm lsqnonlin (Supplemental File S2; Supplemental Table S2).
Model Validation Reveals New Insights into BRI1-Mediated Root Growth
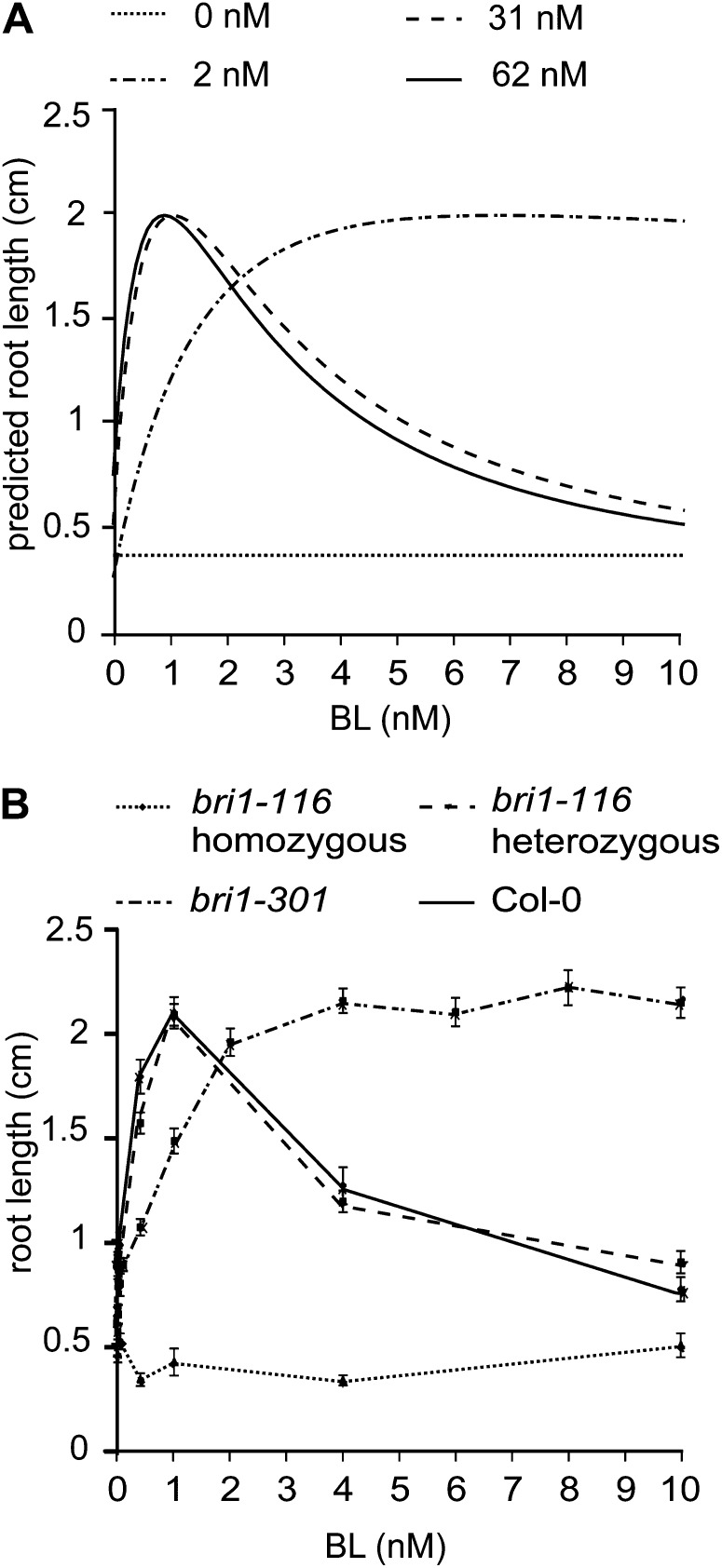
The bri1 mutant roots typically exhibit enhanced resistance toward BL. For modeling purposes, this is considered as a reduced concentration of active BRI1. Therefore, it was of interest to validate our model by varying the BRI1 receptor concentration. The model was first calibrated based on the wild-type root length assays, after which the BRI1 receptor concentration was theoretically reduced stepwise to 0 (Fig. 2A). The model predicts that a 2-fold reduction will not have a severe effect on root growth, either at BL concentrations below 1 nm or at a high BL concentration. This is obviously in line with the recessive nature of bri1 loss-of-function alleles, corroborated here by the experimental evidence showing no difference between wild-type seedlings and a segregating population of the bri1-116 mutant (Fig. 2B). Similar results were obtained for the bri1-201 mutant (Supplemental Fig. S8). Further theoretical reduction in receptor concentration to 0 nm BRI1 yields roots that are predicted to become completely insensitive toward BL (Fig. 2A). The model also predicts that the root length at 0 nm BRI1 will be about 20% of the wild-type value (Fig. 2A), which is exactly what is observed for real bri1-116 roots (Fig. 2B).
Figure 2.
Experimental validation of the mathematical model. A, Model predictions of the root length when only the BRI1 receptor concentration and ligand concentrations are altered. At 62 nm, the model predicts the Col-0 wild-type root length, 31 nm represents the heterozygous bri1-116 line, and 2 and 0 nm represent the bri1-301 and the homozygous bri1-116 lines, respectively. B, Experimental verification of the model predictions using Col-0, bri1-116 heterozygotes, bri1-116 homozygotes, and bri1-301 homozygotes. Error bars represent se for 30 roots per data point measured in three independent replicates.
Interestingly, the model predicts that at an active BRI1 receptor level of about 1% to 3% of the wild type, there is a more severe effect on the inhibition of root growth when compared with the stimulation of root growth. Although it requires much more ligand to obtain the stimulatory effect, eventually such roots will exhibit the same growth as the wild type, while at high ligand concentrations they exhibit insensitivity (Fig. 2A). This pattern is experimentally demonstrated using the weak bri1-301 mutant. bri1-301 harbors a two-nucleotide change (GG-AT) resulting in a Gly-989Ile conversion that causes reduced sensitivity toward BL (Xu et al., 2008). When bri1-301 is exposed to BL in the presence of BRZ, there is a stimulatory effect on root growth (Fig. 2B), while roots are insensitive to BL at higher BL concentrations. Assuming a 30-fold decrease in activity of the BRI1 receptor in bri1-301, the model output (Fig. 2A) agrees well with the experimental data (Fig. 2B). Similar results were obtained when the root lengths of seedlings at 4 and 8 DAG were predicted (Supplemental Fig. S9). A striking observation is that below 3 nm BL, wild-type root growth is highly sensitive to variation in BL concentration but not very sensitive to moderate reduction in the amount of BRI1 receptor. This suggests that under physiological conditions, BR signaling activity depends on ligand availability rather than on receptor density.
As a final validation of the model, the response of the roots toward the less potent ligand HBL was simulated by decreasing the Kd between BRI1 and BL. The resulting predictions indicate that the slope of the root growth curves and the ligand concentration at which maximum root growth is observed are altered, although the eventual root lengths are the same for both BL and HBL (Supplemental Fig. S10, A and B). The model predictions were experimentally corroborated using HBL (Supplemental Fig. S10, C and D).
BRI1 Kinetics Is Altered in BRI1-GFP Lines
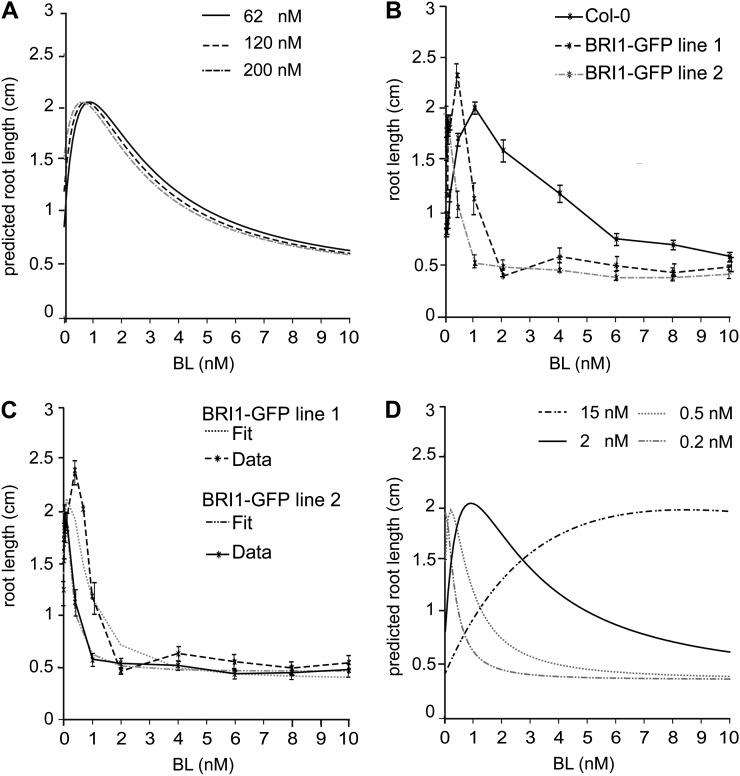
According to the model prediction, an increase of BRI1 receptor should only have a minor effect on root growth (Fig. 3A). To verify this, commonly used BRI1-GFP lines in the wild type were used. Surprisingly, actual root growth of BRI1-GFP line 1 and 2 is already inhibited at 0.4 and 0.1 nm BL, respectively (Fig. 3B). These lines have about two times (120 nm) and three times (200 nm) more BRI1 receptor when compared with the wild-type line (van Esse et al., 2011). In particular for BRI1-GFP line 2, hardly any stimulatory effect is seen, while even in the presence of BRZ the roots indeed grow faster than the wild type (Geldner et al., 2007). Alternative model structures (Supplemental File S2) were not able to explain the enhanced BL sensitivity observed in the BRI1-GFP lines. These results suggest that there may be an additional component in the pathway not detected in wild-type seedling roots. Most likely, upon introduction of another copy of the BRI1 receptor, a normally present rate-limiting component is out-titrated, making these roots almost constitutively sensitive to the ligand. In the model, this would result in a reduction of the k values. To test this hypothesis, we ran the model (Eq. 5) now calibrating the system to the BRI1-GFP lines instead of the wild type. A good fit was obtained between the model and the actual data set (Fig. 3C). However, this is only obtained when k values are decreased by at least a factor of 4 (Fig. 3D). Therefore, our model unexpectedly reveals that BR signaling in the BRI1-GFP lines is altered in a way that cannot simply be explained at the level of initial ligand-receptor interaction (Supplemental File S3; Supplemental Fig. S11). Interestingly, an increase in the k values to 15 nm results in a similar trend to that observed in the bri1-301 line (Figs. 2B and 3D). This suggests that the 30-fold reduction of receptor activity predicted for the bri1-301 line (Fig. 2, A and B) could also be due to a failure to interact with a positive regulator. In modeling terms, this is the same as a reduction in the total number of functional BRI1 receptors.
Figure 3.
The BRI1-GFP lines are more sensitive to BL. A, Model prediction for the behavior of a BRI1-GFP line. B, Root length assay of BRI1-GFP lines 1 and 2 containing two and three times more BRI1, respectively. The effect of the additional BRI1 copy is more severe than what the model predicts. All points in the graph are shown ± se for 30 roots per data point measured in three independent replicas. C, The model is able to fit the results of the BRI1-GFP lines. All parameters were fit to the BRI1-GFP lines and Col-0 wild-type lines separately. The BRI1 concentrations used for modeling are 62 nm for Col-0, 120 nm for BRI1-GFP line 1, and 200 nm for BRI1-GFP line 2. D, The behavior of BRI1-GFP lines can only be modeled when k1, k2, and k3 are altered when compared with the Col-0 lines. The following values for k1, k2, and k3 were used for modeling: 2 nm for Col-0, 0.5 nm for BRI1-GFP line 1, 0.2 nm for BRI1-GFP line 2, and 15 nm for bri1-301. The maximum root length and the minimum root length were 12 and 0.35 cm, respectively.
Modeling of Hypocotyl Elongation
To determine if the developed model is generally applicable to other BRI1-related developmental processes, hypocotyl elongation was used as an alternative physiological readout. In the presence of BRZ, a clear stimulatory effect of BL on hypocotyl elongation can be observed, while inhibition of hypocotyl elongation is observed at 1 µm BL (Fig. 4A; Supplemental Fig. S12). Different model structures (Supplemental File S2; Supplemental Table S3; Supplemental Fig. S13) were evaluated using the same criteria as described for the model comparison for root growth.
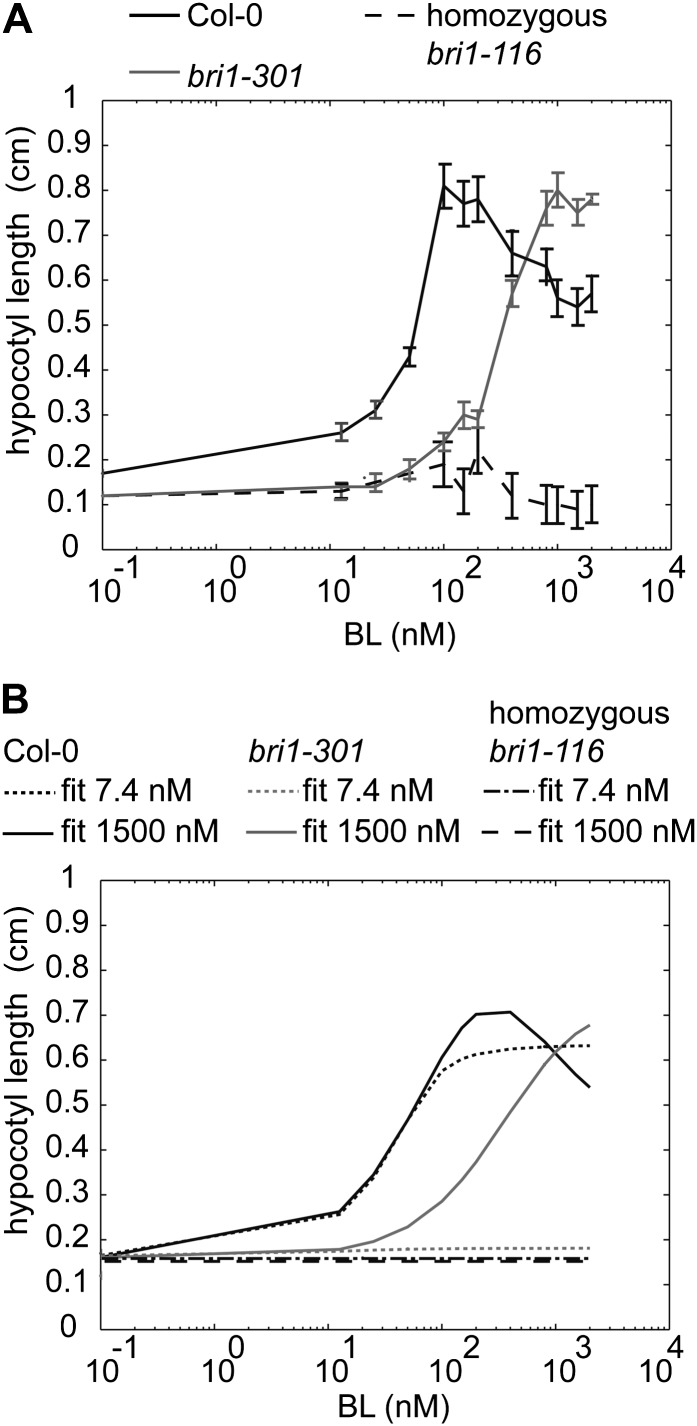
Figure 4.
BRI1 receptor occupancy linked to hypocotyl elongation. A, Hypocotyl lengths at different BL concentrations when 1 μm BRZ is added to the medium. Seedlings were grown for 5 d in the dark, after which the hypocotyl length was measured. All points in the graph are shown ± se for 15 hypocotyls per data point measured in three independent replicas. B, A good fit and prediction of the model described in Equation 7 was possible when assuming a Kd between 750 and 3,000 nm and total BRI1 concentrations of 62 nm for Col-0 and 0 nm for bri1-116. Whereas in bri1-301 roots, a 30-fold reduction in BRI1 activity was predicted, bri1-301 hypocotyls required only a 6-fold reduction. The graphs show a fit at Kd values of 7.4 and 1,500 nm, respectively.
Strikingly, only one inhibitory module was sufficient to explain the experimental data, in contrast with the root, where two modules are required. The following model structure was selected, based on low AIC and BIC scores as well as a good prediction with respect to the bri-116 null mutant:
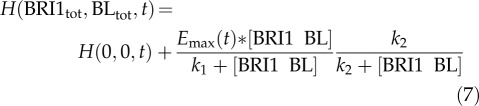 |
Here, H(0,0,t) is the measured hypocotyl length at time t in the absence of BRI1-mediated signaling, Emax(t) is the maximum possible hypocotyl length when no inhibitory mechanisms are present, and k1, k2, and k3 are the k values. Similar to the root, the model parameters H(0,0,t), Emax(t), k1, k2, and k3 were calibrated to the measured hypocotyl lengths in wild-type plants under variation of the BL level. The model was validated using homozygous bri1-116 and bri1-301 mutants (Fig. 4B). The BL concentration to which hypocotyls respond is about 10-fold higher when compared with roots. This is in agreement with previous observations showing that the endogenous BR concentration is higher in shoots (Shimada et al., 2003) and requires more exogenous BL (Gou et al., 2012). This implies that in hypocotyls, BRI1 receptor occupancy is much higher or the Kd of BRI1 is much lower when compared with the root. A good prediction of the hypocotyl length was only obtained when the Kd was increased from 7.4 nm to almost 1 µm (Fig. 4B; Supplemental Fig. S12), while a comparable BRI1 receptor density was found in leaf epidermal cells (van Esse et al., 2011). Thus, BRI1 has a much lower affinity for BL in the hypocotyl or accessibility to exogenous BL is impaired.
DISCUSSION
This work describes a mathematical approach in which BRI1 receptor activity, expressed as receptor occupancy, is used to model root growth and hypocotyl elongation in Arabidopsis. The model parameters employed were tuned on wild-type root growth and required a stimulatory module and two inhibitory modules. The in silico root growth predictions of bri1 mutants fitted well with the experimental data. A good prediction of bri1 mutant hypocotyl elongation required a stimulatory module and only a single inhibitory module besides an approximately 100-fold higher BRI1 dissociation constant. Both impaired and enhanced BRI1 signaling result in shorter roots and reduced hypocotyl elongation. For roots, it was recently proposed that this effect is due to a decrease in meristem size via the control of cell cycle progression as well as a reduction in elongation (González-García et al., 2011). This hypothesis is in line with our observation that the best fit and prediction of root growth are obtained when incorporating one stimulatory module and two inhibitory modules. It is attractive to propose that these two inhibitory modules reflect cell expansion and cell division, but no evidence is available at present to support this idea. Hypocotyl elongation is thought to be affected only through cell expansion (Gendreau et al., 1997). Therefore, it is of interest to note that the use of a single inhibitory module gave the best fit between BRI1-mediated BR signaling and hypocotyl elongation. Clearly, our model is applicable to BR-controlled responses in general.
In Arabidopsis, there are various mathematical models that describe plant architecture in terms of auxin signaling (Grieneisen et al., 2007; Laskowski et al., 2008; Ibañes et al., 2009; Vernoux et al., 2011). These models are used to study the effect of polarized auxin transport resulting in local auxin maxima to evaluate root growth (Grieneisen et al., 2007; Laskowski et al., 2008) or auxin-mediated signaling on the shoot apex (Vernoux et al., 2011). Kinetic models of auxin-mediated signal transduction have resulted in the identification of genes involved in auxin/ethylene/cytokinin cross talk (Liu et al., 2010) and the effect of auxin/cytokinin cross talk in cell fate determination (Muraro et al., 2011). In the case of BR signaling, mathematical modeling is applied to evaluate auxin/BR cross talk (Ibañes et al., 2009; Sankar et al., 2011). Boolean logics have been used to analyze auxin and BR gene networks, which led to new insights in the role of the BRX gene in BR and auxin signaling (Sankar et al., 2011). In shoots, radial patterning of vascular bundles is controlled by auxin polar transport and BR signaling. Ibañes et al. (2009) used mathematical modeling combined with quantitative biological data. Their model uses the appearance of DR5::GUS expression as a readout for auxin maxima, whereas detailed studies of the vascular bundle patterning on various auxin transporter and bri1 mutants was used to evaluate the effect on radial patterning. However, to our knowledge, none of the modeling studies in Arabidopsis so far completed have employed the biochemical activity of a key component as a starting point. Therefore, the mathematical description of BRI1-mediated BR signaling in root growth as described here represents a novel approach in plant modeling.
At physiological concentrations, ligand availability rather than receptor concentration apparently determines BRI1-mediated BR signaling activity in roots. In addition, less than 1% of the total number of BRI1 receptors are occupied by ligand under these conditions. This does not appear to be unusual, as it is reported for many animal systems as well. Epidermal growth factor (EGF) binding to its receptor (EGFR) triggers a full cellular response when less than 1% of the receptors are occupied (Wiley et al., 1989; Uyemura et al., 2005; Teramura et al., 2006). In EGFR-mediated signaling, ligand-independent signal propagation is important (Verveer et al., 2000) and thought to be occurring at increased receptor density (Sawano et al., 2002). Clustering of receptors is important in autocrine signaling (DeWitt et al., 2001), while the ratio between high- and low-affinity EGF receptors in the plasma membrane also assumes a large number of non-ligand-binding receptors (Kuszynski et al., 1993). It has been proposed that EGFR affinity for EGF is reduced due to negative cooperativity (Macdonald and Pike, 2008; Alvarado et al., 2010), resulting in more single-occupied homodimers at low ligand concentration and double-occupied homodimers at high ligand concentration (Macdonald and Pike, 2008). EGFR is capable of regulating distinct biological processes, depending on the ligand and its concentration (Krall et al., 2011). In this model, the presence of a large number of unoccupied receptors could function as a mechanism to differentiate between downstream pathways seen in the hourglass model of EGF (Citri and Yarden, 2006; Warren and Landgraf, 2006). In the case of BR signaling in Arabidopsis root cells, it is not clear whether unoccupied BRI1 receptors contribute to a physiological response. Therefore, it was of interest to find that the switch between activation and inhibition of root growth depends on the BRI1 occupancy level. This implies that, like EGFR, BRI1 might function as a core regulator capable of activating multiple target proteins resulting in the activation of different genes dependent on the activated target.
Endocrine signaling pathways in animals often employ receptor availability in target cells as a means to regulate responses. Given our previous observation that BRI1 receptor density is kept constant throughout the root meristem (van Esse et al., 2011), it is unlikely that differential receptor density is an important mechanism in BR signaling. Our modeling suggests that at least a 10- to 30-fold reduction in BRI1 concentration is required to markedly change the BR signaling output. Taken together, ligand availability appears to be a more restrictive factor. In this respect, it is of interest that BRs appear to be largely immobile. This is based on the observation that cell type-specific expression in epidermal cells of BR synthesis can rescue the stature phenotype, while expression in vascular cells does not (Savaldi-Goldstein et al., 2007). A similar scenario was developed for the root meristem, where again the importance of the epidermis was noted as an important source of BR signaling activity (Hacham et al., 2011). Because all of the cells in shoots and in roots have BRI1 receptors, it was suggested that other diffusible BR pathway-related signals exist (Savaldi-Goldstein et al., 2007).
Unexpectedly, simulation experiments revealed that the properties of BR signaling are altered in commonly used BRI1-GFP reporter lines. The extra copy of the BRI1 gene results in a higher ligand sensitivity of the roots than predicted. To explain this, we propose that a negative regulator is out-titrated, rendering the roots almost constitutively sensitive to BRs. Alternatively, the GFP tag on the C-terminal end of BRI1 may interfere with the interaction with downstream targets, similar to what has been demonstrated for BAK1 (SERK3), one of the BRI1 coreceptors (Ntoukakis et al., 2011). In modeling terms, both mechanisms would result in a change in half-maximum response values. Interestingly, this is corroborated by the mathematical model that demonstrates that these values are indeed altered in the BRI1-GFP reporter lines. The bri1-301 mutant has a reduced sensitivity toward BRs (Xu et al., 2008) and is unable to transphosphorylate BAK1 (Kang et al., 2010). This indicates that the bri1-301 protein is hampered in its interaction with a positive regulator of the BRI1 signaling pathway, as predicted by our model. Recently, it has been reported that the early events in BRI1 signaling almost completely depend on the activity of the SERK coreceptor family (Gou et al., 2012). The model of BRI1-mediated BR signaling presented here is a starting point for more extended mathematical models to fit existing and new components of the BRI1 signaling pathway in a precise cellular context.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants (ecotype Columbia [Col-0]) were used in all growth assays for calibrating the mathematical model. The bri1 null mutants bri1-116 (Li and Chory, 1997) and bri1-201 (Bouquin et al., 2001), the weak allele bri1-301 (Xu et al., 2008), and two transgenic BRI1-GFP lines (Friedrichsen et al., 2000; Geldner et al., 2007) were used to verify the mathematical model. For more details on the plant lines used and genotyping information, see Supplemental File S4. Freshly harvested seeds were surface sterilized for 10 min in 10% (v/v) bleach in ethanol, after which the seeds were washed three times with ethanol and dried before plating. The seedlings were grown vertically under fluorescent light with 16-h-light/8-h-dark photoperiods on one-half-strength Murashige and Skoog medium (Duchefa) supplemented with 1% Suc (Sigma-Aldrich), 0.1% MES (Sigma-Aldrich), and 0.8% agar. To equalize germination, the plates were kept in the dark at 4°C for 2 d before they were placed in the light. For the root growth assays, the germinating medium was supplemented with 1 or 5 µm BRZ (Tokyo Chemical Industry [TCI]) and various concentrations of BL (Sigma-Aldrich) or HBL (Sigma-Aldrich). For wild-type lines, no significant difference between 1 and 5 µm BRZ was observed (Supplemental Fig. S11). For the hypocotyl assays, the germinating medium was supplemented with 1 µm BRZ and supplemented with various concentrations of BL. Hypocotyl lengths were measured after 5 d of growth in the dark. For all assays, at least three independent replicates were performed, measuring five to 10 roots or hypocotyls per assay (n ≥ 15 roots or hypocotyls in total). All values are shown ± se, and error bars indicate se.
Software and Modeling
The model was programmed in Matlab, version 7.8 (MathWorks).
The model parameters RL(0,0,t), Emax(t), k1, k2, and k3 were calibrated on wild-type root length of seedlings at 4, 6, and 8 DAG under varying BL concentrations. Emax and R(0,0,t) are two functions of time. Per time point t, they are regarded as free parameters. More details on the parameter estimation method can be found in Supplemental File S1. A Kd value of 7.4 nm (Wang et al., 2001) was used for all modeling experiments. To obtain an optimal fit between the mathematical model and the biological data set, several models were explored. More details about the different models tested and the criteria for model selection can be found in Supplemental Files S1 and S2. Estimates of endogenous BRI1 and BL levels are discussed in Supplemental File S4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic representation of BRI-mediated BR signaling.
Supplemental Figure S2. Root length assay on Col-0 lines in the absence of BRZ.
Supplemental Figure S3. BRI1 receptor occupancy levels at physiological ligand concentration.
Supplemental Figure S4. Model fit and predictions models 1 and 2.
Supplemental Figure S5. Model fit and predictions models 3 and 4.
Supplemental Figure S6. Modeling stimulation as well as inhibition are essential for a good fit and prediction of root length.
Supplemental Figure S7. Fit between model 1 and Col-0 after BL stimulation in the absence of BRZ.
Supplemental Figure S8. Root length assays on heterozygous and homozygous bri1-201 null mutants.
Supplemental Figure S9. Root length of bri1-301 at 4, 6, and 8 DAG.
Supplemental Figure S10. Model predicts root growth in response to less potent ligand.
Supplemental Figure S11. Root growth assays on BRI1-GFP reporter lines in the presence of 5 µm BRZ.
Supplemental Figure S12. Hypocotyl length assay using Col-0 when there is no BRZ in the medium.
Supplemental Figure S13. Model fit and predictions on hypocotyl length.
Supplemental Table S1. Comparison between different models using root length as readout.
Supplemental Table S2. Comparison between controlled random search and Matlab genetic algorithm combined with lsqnonlin.
Supplemental Table S3. Comparison between different model structures using hypocotyl length as readout.
Supplemental File S1. Model structures.
Supplemental File S2. Parameter estimation.
Supplemental File S3. The k values are altered in BRI1-GFP reporter lines.
Supplemental File S4. Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Joanne Chory for providing BRI-GFP line 2 and the bri1-116 mutant, Niko Geldner for providing BRI1-GFP line 1, John Mundy for providing the bri1-201 line, and Yonghong Wang for providing the bri1-301 line.
Glossary
- BR
brassinosteroid
- BL
24-epibrassinolide
- HBL
22-homobrassinolide
- BRZ
brassinazole
- AIC
Akaike information criteria
- BIC
Bayesian information criteria
- DAG
days after germination
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- Col-0
ecotype Columbia
References
- Alvarado D, Klein DE, Lemmon MA. (2010) Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell 142: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S. (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancoş S, Nomura T, Sato T, Molnár G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M. (2002) Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J. (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127: 450–458 [PMC free article] [PubMed] [Google Scholar]
- Brent R. (2009) Cell signaling: what is the signal and what information does it carry? FEBS Lett 583: 4019–4024 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng JC, Nam KH, Li J, Chory J. (2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131: 5341–5351 [DOI] [PubMed] [Google Scholar]
- Citri A, Yarden Y. (2006) EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 7: 505–516 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (1996) Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 10: 1–8 [DOI] [PubMed] [Google Scholar]
- DeWitt AE, Dong JY, Wiley HS, Lauffenburger DA. (2001) Quantitative analysis of the EGF receptor autocrine system reveals cryptic regulation of cell response by ligand capture. J Cell Sci 114: 2301–2313 [DOI] [PubMed] [Google Scholar]
- Fàbregas N, Ibañes M, Caño-Delgado AI. (2010) A systems biology approach to dissect the contribution of brassinosteroid and auxin hormones to vascular patterning in the shoot of Arabidopsis thaliana. Plant Signal Behav 5: 903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, et al. (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H. (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI. (2011) Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J. (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Wang Z-Y, Li J, Zhu Q, Lamb C, Ronald P, Chory J. (2000) Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288: 2360–2363 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañes M, Fàbregas N, Chory J, Caño-Delgado AI. (2009) Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci USA 106: 13630–13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J. (2011a) Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA 108: 8503–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J. (2011b) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev 25: 232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B, Wang H, Nam KH, Li J, Li J. (2010) Activation-tagged suppressors of a weak brassinosteroid receptor mutant. Mol Plant 3: 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY. (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp E, Liebermeister W, Wierling C, Kowald A, Lehrach H, Herwig H. (2009). Systems Biology: A Textbook. Wiley-Blackwell, Weinheim, Germany, pp 181–184. [Google Scholar]
- Krall JA, Beyer EM, MacBeath G. (2011) High- and low-affinity epidermal growth factor receptor-ligand interactions activate distinct signaling pathways. PLoS ONE 6: e15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuszynski CA, Miller KA, Rizzino A. (1993) Influence of cell density and receptor number on the binding and distribution of cell surface epidermal growth factor receptors. In Vitro Cell Dev Biol Anim 29A: 708–713 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, Scheres B. (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: 2721–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liu J, Mehdi S, Topping J, Tarkowski P, Lindsey K. (2010) Modelling and experimental analysis of hormonal crosstalk in Arabidopsis. Mol Syst Biol 6: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald JL, Pike LJ. (2008) Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc Natl Acad Sci USA 105: 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro D, Byrne H, King J, Voss U, Kieber J, Bennett M. (2011) The influence of cytokinin-auxin cross-regulation on cell-fate determination in Arabidopsis thaliana root development. J Theor Biol 283: 152–167 [DOI] [PubMed] [Google Scholar]
- Müssig C, Shin GH, Altmann T. (2003) Brassinosteroids promote root growth in Arabidopsis. Plant Physiol 133: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Min YK, Nakano T, Asami T, Yoshida S. (2000) Treatment of dark-grown Arabidopsis thaliana with a brassinosteroid-biosynthesis inhibitor, brassinazole, induces some characteristics of light-grown plants. Planta 211: 781–790 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Ntoukakis V, Schwessinger B, Segonzac C, Zipfel C. (2011) Cautionary notes on the use of C-terminal BAK1 fusion proteins for functional studies. Plant Cell 23: 3871–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price W. (1976) A controlled random search procedure for global optimisation. Comput J 20: 367–370 [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC. (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar M, Osmont KS, Rolcik J, Gujas B, Tarkowska D, Strnad M, Xenarios I, Hardtke CS. (2011) A qualitative continuous model of cellular auxin and brassinosteroid signaling and their crosstalk. Bioinformatics 27: 1404–1412 [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J. (2007) The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Sawano A, Takayama S, Matsuda M, Miyawaki A. (2002) Lateral propagation of EGF signaling after local stimulation is dependent on receptor density. Dev Cell 3: 245–257 [DOI] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S. (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramura Y, Ichinose J, Takagi H, Nishida K, Yanagida T, Sako Y. (2006) Single-molecule analysis of epidermal growth factor binding on the surface of living cells. EMBO J 25: 4215–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyemura T, Takagi H, Yanagida T, Sako Y. (2005) Single-molecule analysis of epidermal growth factor signaling that leads to ultrasensitive calcium response. Biophys J 88: 3720–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse GW, Westphal AH, Surendran RP, Albrecht C, van Veen B, Borst JW, de Vries SC. (2011) Quantification of the brassinosteroid insensitive1 receptor in planta. Plant Physiol 156: 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verveer PJ, Wouters FS, Reynolds AR, Bastiaens PI. (2000) Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science 290: 1567–1570 [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J. (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Seto H, Fujioka S, Yoshida S, Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Wang Q, Chong K, Wang F, Wang L, Bai M, Jia C. (2006) The brassinosteroid signal transduction pathway. Cell Res 16: 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CM, Landgraf R. (2006) Signaling through ERBB receptors: multiple layers of diversity and control. Cell Signal 18: 923–933 [DOI] [PubMed] [Google Scholar]
- Wiley HS, Walsh BJ, Lund KA. (1989) Global modulation of the epidermal growth factor receptor is triggered by occupancy of only a few receptors: evidence for a binary regulatory system in normal human fibroblasts. J Biol Chem 264: 18912–18920 [PubMed] [Google Scholar]
- Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J. (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Huang J, Li B, Li J, Wang Y. (2008) Is kinase activity essential for biological functions of BRI1? Cell Res 18: 472–478 [DOI] [PubMed] [Google Scholar]
- Ye H, Li L, Yin Y. (2011) Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J Integr Plant Biol 53: 455–468 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang Z-Y, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.