Abstract
Rubisco is composed of eight small subunits coded for by the nuclear RBCS multigene family and eight large subunits coded for by the rbcL gene in the plastome. For synthesis of the Rubisco holoenzyme, both genes need to be expressed coordinately. To investigate this molecular mechanism, the protein synthesis of two subunits of Rubisco was characterized in transgenic rice (Oryza sativa) plants with overexpression or antisense suppression of the RBCS gene. Total RBCS and rbcL messenger RNA (mRNA) levels and RBCS and RbcL synthesis simultaneously increased in RBCS-sense plants, although the increase in total RBCS mRNA level was greater. In RBCS-antisense plants, the levels of these mRNAs and the synthesis of the corresponding proteins declined to a similar extent. The amount of RBCS synthesized was tightly correlated with rbcL mRNA level among genotypes but not associated with changes in mRNA levels of other major chloroplast-encoded photosynthetic genes. The level of rbcL mRNA, in turn, was tightly correlated with the amount of RbcL synthesized, the molar ratio of RBCS synthesis to RbcL synthesis being identical irrespective of genotype. Polysome loading of rbcL mRNA was not changed. These results demonstrate that the availability of RBCS protein up-regulates the gene expression of rbcL primarily at the transcript level in a quantitative manner for stoichiometric assembly of Rubisco holoenzyme.
A number of key photosynthetic components in chloroplasts are multisubunit protein complexes composed of both nucleus- and chloroplast-encoded subunits (Malkin and Niyogi, 2000). For their proper assembly and the formation of photosynthetic machinery, gene expression between these cellular compartments must be well coordinated, and photosynthetic organisms have signaling systems between the nucleus and chloroplasts for this purpose (Pesaresi et al., 2007; Woodson and Chory, 2008; Waters and Langdale, 2009; Stern et al., 2010). It is also understood that the availability of nucleus-encoded subunits modulates the gene expression of chloroplast-encoded subunits at translational levels.
The photosynthetic carbon-fixing enzyme Rubisco has been extensively used for the model system, as Rubisco is composed of only two kinds of subunits, a nuclear multigene family-encoded small subunit (RBCS) and a chloroplast-encoded large subunit (RbcL; Dean et al., 1989; Spreitzer, 2003). In higher plants and green algae (Chlamydomonas reinhardtii), Rubisco holoenzyme is a hexadecamer composed of eight RBCS and eight RbcL subunits. Rubisco is the most abundant leaf protein in C3 plants and catalyzes the first steps in photosynthesis and photorespiration (Ellis, 1979; Lorimer, 1981; Evans, 1989; Makino et al., 1992), these rates being determined by Rubisco activity under conditions of saturating light and current atmospheric CO2 and O2 levels (Evans, 1986; Makino et al., 1988).
When the gene expression of RBCS was suppressed in higher plants such as tobacco (Nicotiana tabacum; Rodermel et al., 1988; Hudson et al., 1992), the C4 plant Flaveria bidentis (Furbank et al., 1996), and rice (Oryza sativa; Makino et al., 1997), the amounts of RBCS and RbcL proteins declined in a coordinated manner. In RBCS-suppressed tobacco, polysome loading of rbcL mRNA was reduced without a change in rbcL mRNA level (Rodermel et al., 1996; Wostrikoff and Stern, 2007), suggesting repression of the translation of rbcL mRNA. It has been suggested that a repressor motif in unassembled RbcL protein, otherwise not accessible, interacts with rbcL mRNA for the repression of its translation (Wostrikoff and Stern, 2007). Translational suppression of rbcL has also been observed in Chlamydomonas spp. with suppression of RBCS gene by its deletion (Khrebtukova and Spreitzer, 1996). These results demonstrate that the gene expression of rbcL undergoes negative-feedback regulation at translational levels in response to the availability of RBCS protein. This mechanism is similar to that first described for the hierarchical synthesis of the cytochrome b6/f complex within a chloroplast in Chlamydomonas spp., namely, the control by epistasy of synthesis (Kuras and Wollman, 1994; Choquet et al., 1998; Choquet and Vallon, 2000; Boulouis et al., 2011).
A similar mechanism is operative for other photosynthetic components in Chlamydomonas spp. For example, in the synthesis of the F1 subunit of chloroplast ATP synthase, oligomers of chloroplast-encoded subunits α and β repressed the translation of subunit β when these subunits were not assembled with a nucleus-encoded subunit γ (Drapier et al., 2007). Synthesis of the chloroplast-encoded cytochrome f was repressed when a nucleus-encoded subunit Rieske iron-sulfer protein was deleted or its assembly with cytochrome f was disrupted by a point mutation (de Vitry et al., 2004). These studies imply that translational modulation of chloroplast-encoded genes plays a key role in the stoichiometric assembly of various chloroplast multimetric protein complexes.
On the other hand, when endogenous RBCS was overexpressed in rice, the rbcL mRNA level also increased concomitant with a drastic increase in the total RBCS mRNA level (Suzuki et al., 2007, 2009a, 2009b). Rubisco content increased to an extent similar to that of the rbcL mRNA level. In addition, the rbcL mRNA level declined to a level similar to that of total RBCS mRNA in RBCS-suppressed rice plants with the antisense technique or the RNA interference technique for individual RBCS genes (Suzuki et al., 2009a, 2009b; Ogawa et al., 2012). These results imply that the coordinated expression of two genes is not fully explained by the translational modulation of the rbcL gene.
To explore the molecular mechanism of Rubisco synthesis in detail, the coordinated gene expression of RBCS and rbcL was characterized in transgenic rice plants with an overexpression or suppression of RBCS. Young, expanding leaves were used as samples, because Rubisco synthesis is the most active and Rubisco degradation is inactive during the development of rice leaves (Mae et al., 1983; Makino et al., 1984; Suzuki et al., 2001). Syntheses of RBCS and RbcL proteins were separately determined, and their relationships with the mRNA levels of RBCS and rbcL were quantitatively analyzed. Polysome loading was also examined as an index for the translational status of these genes. We obtained evidence for the positive regulation of transcript levels of rbcL by the availability of RBCS protein in the synthesis of the Rubisco holoenzyme.
RESULTS AND DISCUSSION
The Synthesis of Rubisco Subunits Is Coordinated in a Quantitative Manner in RBCS-Transgenic Rice Plants
Rubisco activities, Rubisco and total nitrogen (N) contents, and Rubisco N/total N ratios were determined in RBCS-sense (lines 26-8 and 35-4; Suzuki et al., 2007), RBCS-antisense (line AS-71; Makino et al., 2000), and wild-type rice plants (Table I). Rubisco activities were 1.5- to 1.7-fold higher in RBCS-sense plants and 76% in RBCS-antisense plants in comparison with wild-type plants. Similar result was obtained for Rubisco contents. In order to examine whether Rubisco contents were specifically changed in the transgenic plants, total N contents and Rubisco N/total N ratios were measured. Total N contents were used as an index for organic N contents. It is shown that most of the organic N is used as protein N in leaf tissue (Peoples and Dalling, 1988). Total N did not greatly differ among the genotypes, although the values in transgenic plants were slightly higher. Rubisco N/total N ratios were 1.3- to 1.4-fold higher in RBCS-sense plants and declined to 65% in RBCS-antisense plants. These results indicated that the amount of Rubisco was specifically increased in RBCS-sense plants but declined in RBCS-antisense plants. Since expanding leaves were used as samples, the Rubisco N/total N ratios were different from our previous results obtained from the uppermost, fully expanded leaves of rice (Makino et al., 2000; Suzuki et al., 2007, 2009a). The Rubisco N/total N ratios in the uppermost, fully expanded leaves of the same plants were 25.9% ± 0.4%, 31.5% ± 0.6%, 30.8% ± 0.6%, and 11.0% ± 0.8% for wild-type plants, RBCS-sense plants (lines 26-8 and 35-4), and RBCS-antisense plants (AS-71), respectively. These values were equivalent to those in our previous studies.
Table I. Rubisco activities, Rubisco protein and total N contents, and Rubisco N/total N ratios.
These values were determined in young, expanding leaves of wild-type plants and RBCS-sense (26-8 and 35-4) and RBCS-antisense (AS-71) plants. Asterisks indicate statistically significant differences from the values of wild-type plants by Dunnett’s test (P < 0.05). Data are presented as means ± se (n = 3). FW, Fresh weight.
| Line | Rubisco Activity | Rubisco Protein Content | Total N Content | Rubisco N/Total N |
| mmol CO2 kg−1 FW s−1 | mmol active site kg−1 FW | mol kg−1 FW | % | |
| Wild type | 0.126 ± 0.008 | 0.092 ± 0.002 | 0.423 ± 0.003 | 16.8 ± 0.2 |
| 26-8 | 0.184 ± 0.006* | 0.127 ± 0.000* | 0.446 ± 0.004 | 22.1 ± 0.2* |
| 35-4 | 0.220 ± 0.024* | 0.152 ± 0.016* | 0.501 ± 0.017* | 23.5 ± 1.8* |
| AS-71 | 0.096 ± 0.002 | 0.065 ± 0.001 | 0.471 ± 0.016 | 10.8 ± 0.5* |
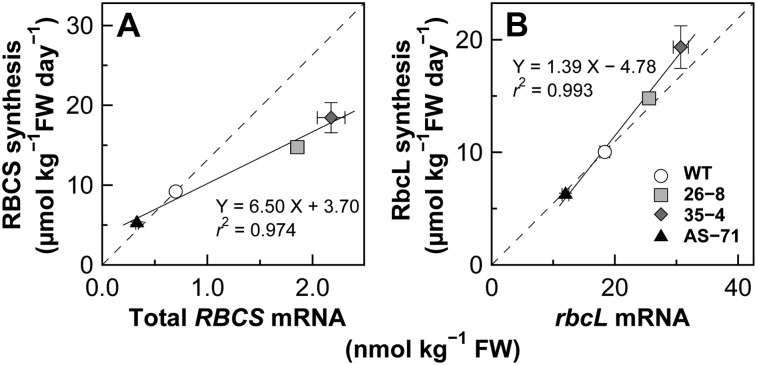
The amounts of RBCS and RbcL synthesized per unit of leaf fresh weight were determined by the incorporation of 15N label by these subunits. RBCS synthesis was 1.6- to 2.1-fold higher in RBCS-sense plants and about 55% in RBCS-antisense plants (Fig. 1A). The amounts of RbcL synthesis were almost identical to those of RBCS synthesis irrespective of genotype, being 1.5- to 1.9-fold higher in RBCS-sense plants and about 60% in RBCS-antisense plants (Fig. 1B). These results indicate that the synthesis of Rubisco subunits is coordinated in a quantitative manner in young, expanding leaves of RBCS-sense and antisense rice plants to maintain stoichiometry between the subunits. It has been suggested in Chlamydomonas spp. that degradation of RBCS plays a role in the maintenance of stoichiometry between Rubisco subunits when the level of RbcL was decreased by blocking chloroplast protein synthesis (Schmidt and Mishkind, 1983). However, it was unknown how the synthesis and/or degradation of RbcL and RBCS are coordinated when RBCS is overproduced either in Chlamydomonas spp. or in higher plants.
Figure 1.
Protein synthesis and the mRNA levels of Rubisco. Protein synthesis of RBCS (A) and RbcL (B) and transcript levels of RBCS (C) and rbcL (D) were determined in expanding young leaves of wild-type (WT), RBCS-sense (26-8 and 35-4), and RBCS-antisense (AS-71) plants. Values are expressed per unit of leaf fresh weight (FW). Data are presented as means ± se (n = 3). Asterisks indicate statistically significant differences from the values of wild-type plants by Dunnett’s test (P < 0.05). In C, white, light gray, dark gray, and black columns represent transcript levels of RBCS2, -3, -4, and -5, respectively.
The Transcript Level of rbcL Is Modulated to Synthesize RbcL Equivalent to RBCS
Four out of five members of the RBCS multigene family that are mainly expressed in leaf blades (Suzuki et al., 2007, 2009a, 2009b) were determined on a leaf fresh weight basis. In RBCS-sense plants, total RBCS mRNA levels were 2.7- to 3.1-fold higher than in wild-type plants, because of drastic increases in the mRNA level of the transgene RBCS2 without changes in the levels of other members (Fig. 1C). rbcL mRNA levels also increased 1.5- to 1.9-fold of the wild-type level, but the difference was smaller than that in total RBCS mRNA levels (Fig. 1D). The total RBCS mRNA level in RBCS-antisense plants was about 50% of the wild-type level, because the mRNA level of each member of RBCS declined (Fig. 1C). The rbcL mRNA level also declined to about 65% of the wild-type level (Fig. 1D). The 18S ribosomal RNA per total RNA was determined as an internal standard, and no statistically significant difference was found among the genotypes (data not shown).
To examine the relationships between the synthesis of Rubisco subunits and the corresponding mRNA levels, the relationships between the amounts of RBCS and RbcL syntheses and their corresponding mRNA levels were analyzed (Fig. 2). In each graph, a dashed line passing through the origin and the wild-type data are drawn to quantitatively predict the amounts of protein synthesis as a function of the mRNA levels on the assumption that this relationship remains constant in the wild-type plants. When the relationships between Rubisco contents and the mRNA levels of total RBCS and rbcL at their maxima were analyzed in leaves of rice plants grown under different N nutrition levels, data approximately fell on the line passing through the origin and the control N data (Suzuki et al., 2007). Although RBCS synthesis was positively correlated with the total RBCS mRNA levels irrespective of genotype, RBCS synthesis measured in RBCS-sense plants was 60% to 64% of that predicted from the wild-type data, whereas RBCS synthesis measured in RBCS-antisense plants was almost identical to the predicted value (Fig. 2A). RbcL synthesis and the rbcL mRNA level were also positively correlated with each other irrespective of genotype (Fig. 2B). Measured RbcL synthesis was very close to that predicted from the wild-type data (95%–115%).
Figure 2.
Relationships between protein synthesis and transcript levels of Rubisco. A, RBCS synthesis versus total RBCS mRNA level. B, RbcL synthesis versus rbcL mRNA level. Circles, squares, diamonds, and triangles represent the wild-type plant (WT), RBCS-sense line 26-8, RBCS-sense line 35-4, and RBCS-antisense line AS-71, respectively. Data are taken from Figure 1. A dotted line is drawn through the origin and data from wild-type plants. FW, Fresh weight.
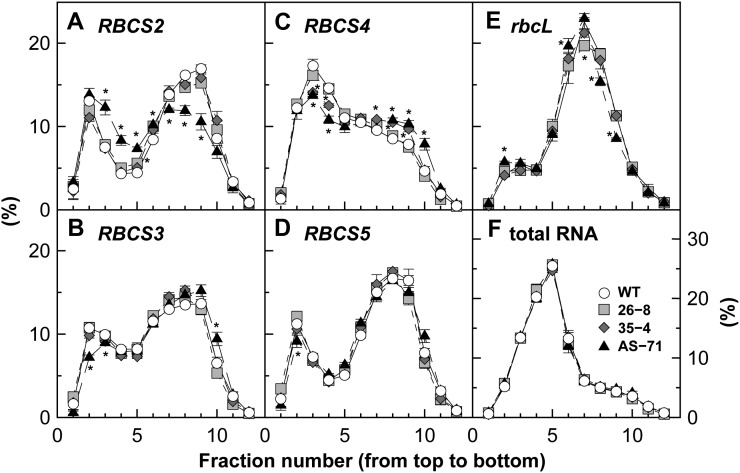
In addition, polysome loading of Rubisco transcripts was examined after fractionation with the Suc gradient as an index for their translational status. In wild-type plants, the pattern of mRNA distribution showed two peaks in RBCS2, -3, and -5 and a single peak at lighter fractions in RBCS4 (Fig. 3, A–D). In rbcL, a single peak with a small shoulder at lighter fractions was observed in the middle of the fractions (Fig. 3E). In RBCS-sense plants, substantial changes in the distribution pattern were not observed even in the transgene RBCS2. In RBCS-antisense plants, drastic changes were also not observed, although a slight shift to lighter fractions was observed in RBCS2 (Fig. 3A), which was used for antisense suppression. Although a similar tendency was found for rbcL (Fig. 3E), the extent was not as great as that observed by Rodermel et al. (1996) with RBCS-antisense tobacco, where the distribution of rbcL mRNA drastically shifted to lighter fractions. The amounts of total RNA were highest in fraction 5 irrespective of genotype (Fig. 3F). The distribution pattern was similar irrespective of genotype and was independent of the pattern of mRNA distribution.
Figure 3.
Polysome loading of Rubisco mRNAs. Transcript levels of each fraction were expressed as percentages of the sum of all fractions. A to E show the data of RBCS2, -3, -4, and -5 and rbcL, respectively. F shows the distribution of total RNA among the fractions. Symbols are the same as in Figure 2. Asterisks indicate statistically significant differences from the values of wild-type plants (WT) by Dunnett’s test (P < 0.05).
Since mRNA levels and protein synthesis were tightly correlated to each other without a change in polysome loading in rbcL, it is indicated that RbcL synthesis is primarily determined by rbcL mRNA level in RBCS-transgenic rice plants. Since the amounts of RBCS and RbcL synthesized were almost identical irrespective of genotype (Fig. 1, C and D), it is also indicated that rbcL mRNA level is adjusted to synthesize RbcL equivalent to RBCS. It is understood that translational modulation of chloroplast-encoded subunits plays a key role in the assembly of chloroplast multimetric proteins (Rodermel et al., 1996; de Vitry et al., 2004; Drapier et al., 2007; Wostrikoff and Stern, 2007). In the case of Rubisco, it has been suggested that gene expression of rbcL undergoes negative-feedback regulation by excessive RbcL protein in tobacco (Rodermel et al., 1996; Wostrikoff and Stern, 2007). The reason for the discrepancy between these and our studies is not clear. It may be that transcriptional and translational regulations differ in importance in terms of Rubisco synthesis in different plant species. It has previously been suggested that these regulations are not mutually exclusive (Rodermel, 1999). For example, we have found in Arabidopsis (Arabidopsis thaliana) plants that the suppression of one or two major RBCS genes led to decreases in rbcL and total RBCS mRNA levels, although the decline in rbcL mRNA level was relatively smaller (Izumi et al., 2012). This finding suggests that modulation of rbcL mRNA level is also operative in Arabidopsis but that the extent is weaker than in rice and the translational regulation contributes to a greater extent. In addition, it has been suggested that nuclear factors are involved in the assembly of a specific chloroplast multisubunit protein complex at a posttranslational level. For example, maize (Zea mays) transposon mutants lacked the cytochrome b6/f complex, although each subunit was normally synthesized (Voelker and Barkan, 1995). It is still unknown whether such a mechanism is operative for the assembly of Rubisco holoenzyme.
On the other hand, increases in the amounts of RBCS synthesized were much lower than those of the total RBCS mRNA levels in RBCS-sense plants (Fig. 2A), suggesting posttranscriptional down-regulation of RBCS synthesis. However, since polysome loading of RBCS mRNAs did not greatly differ among the genotypes (Fig. 3, A–D), it is unlikely, at least, that gene expression of RBCS is primarily modulated at translation initiation. Although the mechanism for the down-regulation of Rubisco synthesis in RBCS-sense plants is still unclear, it has been reported that the rate of translation was independent of polysome loading of the mRNA in some cases (Fütterer and Hohn, 1996). For example, protein synthesis of Rubisco rapidly declined when illuminated amaranth (Amaranthus hypochondriacus) seedlings were transferred to dark conditions (Berry et al., 1988). Light intensity did not affect polysome loading of the leaf catalase mRNA in rye (Secale cereale; Schmidt et al., 2002), although its protein turnover rate increased in a dose-dependent manner along with light intensity (Hertwig et al., 1992). In these cases, it has been suggested that translational initiation and elongation are modulated in a coordinated manner.
The Availability of RBCS Up-Regulates Transcript Levels of rbcL
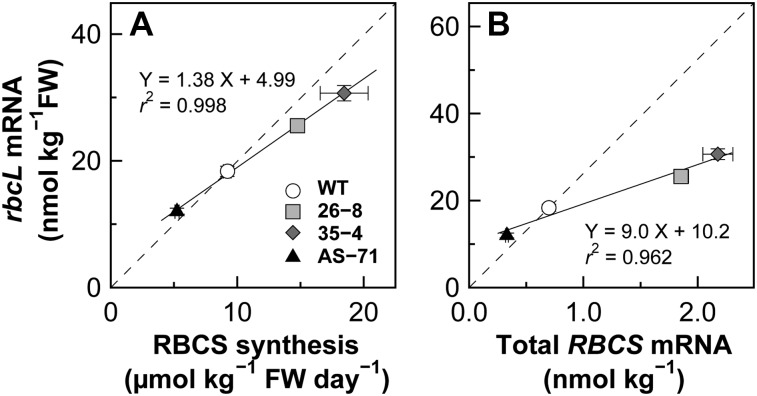
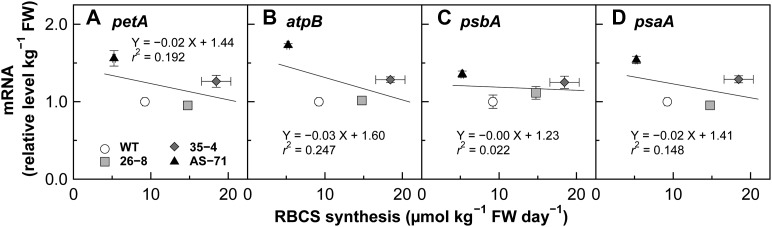
In order to examine the interaction of gene expression of RBCS with that of rbcL, relationships between rbcL mRNA levels and the amounts of RBCS synthesized and RBCS mRNA level were analyzed. A dashed line passing thorough the origin and the wild-type data are also drawn in each graph. The rbcL mRNA level and RBCS synthesis were positively correlated with each other irrespective of genotype (Fig. 4A). Measured rbcL mRNA levels were close to those predicted from the wild-type data (83%–115%). On the other hand, although rbcL and total RBCS mRNA levels were also positively correlated with each other, rbcL mRNA levels measured in RBCS-sense plants were about 50% of the predicted values (Fig. 4B). These results show that rbcL mRNA level is tightly coupled not with total RBCS mRNA level but with the amounts of RBCS synthesized. petA (cytochrome f apoprotein), atpB (β-subunit of chloroplast ATP synthase), psbA (reaction center protein of PSII), and psaA (reaction center protein of PSI) were selected as reference genes of chloroplast-encoded major photosynthetic components. The expression levels of these genes were not affected by changes in the amounts of RBCS synthesized (Fig. 5). Therefore, it is indicated that the availability of RBCS protein primarily up-regulates rbcL mRNA level in a specific, quantitative manner for stoichiometric assembly of Rubisco holoenzyme in rice. This means that a nucleus-encoded subunit can play a role as a positive regulator for gene expression of its chloroplast-encoded assembly partner in the synthesis of chloroplast multimetric protein complexes, as predicted previously by Ellis (1977).
Figure 4.
Relationships between RBCS and rbcL. A, rbcL mRNA level versus RBCS synthesis. B, rbcL mRNA level versus total RBCS mRNA level. Data are taken from Figure 1. Symbols and lines are the same as in Figure 2. FW, Fresh weight, WT, wild type.
Figure 5.
Relationship between mRNA levels of other chloroplast photosynthetic components and RBCS synthesis. A to D show the results of petA, atpB, psbA, and psaA, respectively. These transcript levels are expressed as relative values where the data from the wild-type plants (WT) are defined as 1. Symbols are the same as in Figure 2. Data are presented as means ± se (n = 3). FW, Fresh weight.
It is still an open question how the rbcL mRNA level is positively regulated by the RBCS protein. One possible explanation is enhancement of the transcription of rbcL. Chloroplast photosynthetic genes are transcribed by plastid-encoded RNA polymerase (PEP). PEP presumably requires nucleus-encoded σ factors, which determine the promoter specificity of the major PEP (Lysenko, 2007; Lerbs-Mache, 2011). Thus, the σ factors regulate the first step of chloroplast gene expression. However, the function of σ factors is likely to be redundant, and a specific one(s) for rbcL has not been reported. Protein factors such as CSP41 (Bollenbach et al., 2009) and NARA5 (Ogawa et al., 2009) have been reported to be involved in the transcription of rbcL via PEP in Arabidopsis, but they also affect the mRNA levels of genes for other chloroplast photosynthetic components. Thus, a pathway of specific transcriptional activation of rbcL has not yet been clarified. The stability of an mRNA also affects its steady-state level. In Chlamydomonas spp., nucleus-encoded factors that stabilize a specific chloroplast mRNA by preventing 5′-to-3′ exonucleolytic degradation has been found for genes encoding light-harvesting and electron transport components in the thylakoid membrane such as petA (Loiselay et al., 2008; Boulouis et al., 2011), psbD (Kuchka et al., 1989; Nickelsen et al., 1999), and psbB (Monod et al., 1992; Vaistij et al., 2000a, 2000b). An mRNA-stabilizing factor has also been found for rbcL, but its role in higher plants may be marginal because a defect in its ortholog in Arabidopsis only slightly affected the amount of Rubisco (Johnson et al., 2010). Thus, major factors that stabilize rbcL mRNA have not yet been identified in higher plants. Unknown factors may be involved in the process of the coordinated gene expression between RBCS and rbcL.
CONCLUSION
In summary, we demonstrated in young, expanding leaves of rice that the availability of RBCS protein primarily up-regulates rbcL mRNA level, which determines the amount of RbcL synthesized, and that, consequently, the stoichiometry between RBCS and RbcL is maintained. Which factor(s) mediates between RBCS protein and rbcL mRNA level and to what extent transcriptional and posttranscriptional processes contribute to adjustments of the expression of Rubisco genes are questions of interest to be studied. In addition, since the amount of Rubisco synthesized drastically changes during leaf development (Mae et al., 1983; Makino et al., 1984; Nikolau and Klessig, 1987; Bate et al., 1991; Suzuki et al., 2001, 2010; Imai et al., 2008), it is also of interest whether the rbcL mRNA level is regulated by RBCS protein in the leaves at different developmental stages.
MATERIALS AND METHODS
Plant Culture and 15N Labeling
Rice (Oryza sativa ‘Notohikari’) was transformed with RBCS2 complementary DNA in the sense orientation under the control of its own promoter (Suzuki et al., 2007). From these varieties, T3 progeny of lines Sr-26-8 and Sr-35-4 were used here. Rice transformed with RBCS2 complementary DNA in the antisense orientation (line AS-71 with about 40% of wild-type Rubisco in mature leaves; Makino et al., 2000) and nontransformed rice ‘Notohikari’ were also used. Plants were grown hydroponically in an isolated and temperature-controlled greenhouse (Suzuki et al., 2009a) with slight modification. The greenhouse was maintained with a 14-h photoperiod (5 am to 7 pm) under natural sunlight conditions supplemented with six 400-W metal halide lamps, day/night temperatures of 25°C/20°C, and 60% relative humidity. The final concentration of N in nutrient solution (Makino et al., 1988) was increased to 3 mm (1.5 mm NH4NO3). Plants were labeled with 15N for the measurements of RBCS and RbcL synthesis. When 11th leaves became one-third of their final length, the plants were fed with nutrient solution without N. A solution of (15NH4)2SO4 (30.5 atom % excess) was then added to the nutrient solution to feed 0.94 mmol of N per plant. The next day, the pH of the nutrient solution was adjusted to 5.5 to 6.0 and the plants were fed with the same amount of (15NH4)2SO4 again. The following day, the 11th leaves were collected, weighed, immediately frozen in liquid N2, and stored at −80°C until analysis. All samples were collected between 11 am and 1 pm.
Determination of Total N and Rubisco Protein and Activity
Frozen leaves were homogenized in sodium phosphate buffer, and their N contents were determined with Nessler’s reagent after Kjeldahl digestion as described by Suzuki et al. (2007). Rubisco content was determined by formamide extraction of Coomassie Brilliant Blue R-250-stained bands corresponding to the large and small subunits of Rubisco separated by SDS-PAGE using calibration curves made with purified rice Rubisco (Makino et al., 1985) or by image analysis using Multi Gauge version 3.1 (Fuji Film). Rubisco activity was measured spectrophotometrically by coupling 3-phosphoglyceric acid formation with NADH oxidation at 25°C as described by Nakano et al. (2000) with slight modifications. Prior to the assay, sample homogenate was treated with Na2SO4 at 4°C for 30 min and then desalted (Parry et al., 1997). A final concentration of 250 mm Na2SO4 was found to be best for the removal of sugar-phosphate inhibitors with respect to Rubisco activity (data not shown). For desalting, a Zeba Spin Desalting Column (Thermo Scientific) was used.
Measurement of Rubisco Synthesis
RBCS and RbcL were purified by preparative SDS-PAGE (Suzuki et al., 2010). The amounts of RBCS and RbcL were calculated from the amounts of Rubisco holoenzyme and the ratio of molecular mass between RBCS and RbcL, since these subunits in unassembled form did not accumulate highly, even when gene expression of the other was suppressed (Rodermel, 1999). 15N abundances of these proteins were measured by emission spectography (Yoneyama et al., 1975) using a 15N analyzer (N-151; JASCO), and the amounts of RBCS and RbcL synthesized were calculated as described by Mae et al. (1983).
RNA Analysis
Sample leaves were homogenized with a small amount of acid-washed quartz sand in the presence of liquid N2 using a mortar and pestle. An aliquot was used for the extraction of total RNA according to Suzuki et al. (2004) with slight modification (Suzuki et al., 2009a). Another aliquot was used for the analysis of polysome loading. Samples were prepared according to Sugimoto et al. (2004) with slight modifications. Heparin was removed from all the solutions used. A sample (0.8 mL) was layered onto 8 mL of Suc gradient (15%–55%) and centrifuged at 32,000g for 140 min at 4°C using an ultracentrifuge model 55P-72S equipped with a RPS40T rotor (Hitachi Koki). Twelve 0.69-mL fractions were collected by gentle pipetting from the top of the gradient and transferred into new tubes. After the addition of 69 μL of a solution of 5% (w/v) SDS, 0.2 m EDTA (pH 8), and 2 μL of nucleic acid carrier (Ethachinmate; Nippon Gene), RNA was extracted from each fraction from the step of chloroform-isoamyl alcohol extraction, all the steps being carried out at room temperature. The obtained RNA pellet was dissolved in nuclease-free water, isopropanol precipitated, washed with 75% (v/v) ethanol, and dissolved in 100 μL of nuclease-free water. RNA concentration was determined by A260. The mRNA levels of RBCS multigene family and rbcL were determined by real-time quantitative PCR after reverse transcription (Ogawa et al., 2012). In the case of polysome analysis, equal volumes of RNA samples (less than 300 ng of RNA) were reverse transcribed with the PrimeScript RT Reagent Kit with gDNA Eraser (Takara). The RBCS2 mRNA levels in RBCS-antisense plants were determined by amplification of the region including the open reading frame followed by agarose gel electrophoresis and SYBR Green detection (Suzuki et al., 2007). The primer pairs used for other than Rubisco genes were as follows: 5′-TGAATGTGGGTGCTGTTCTTATTT-3′ and 5′-TCGGGCGGCGCTAAT-3′ for petA; 5′-GGGAGCTGGAGTAGGTAAAACAGTA-3′ and 5′-CCCCCGTGAGCTTTAGCAA-3′ for atpB; 5′-GGCATACCATCAGAGAAACTTCCT-3′ and 5′-GTTGCAGCTGCTACTGCTGTTTT-3′ for psbA; and 5′-GAGGCTCATAAAGGCCCATTT-3′ and 5′-GAGCATGCCATGACGTTGTT-3′ for psaA.
Sequence data from this article can be found in GenBank/EMBL databases under the following accession numbers: RBCS2, Os12g0274700; RBCS3, Os12g0291100; RBCS4, Os12g0292400; RBCS5, Os12g0291400; rbcL, OrsajCp033; petA, OrsajCp041; atpB, OrsajCp032; psbA, OrsajCp002; and psaA, OrsajCp025.
Acknowledgments
We thank Dr. Kensuke Kusumi (Kyushu University) for his advice on polysome analysis. We also thank Dr. Louis Irving (Tuskuba University) for his critical comments on the manuscript.
Glossary
- N
nitrogen
- PEP
plastid-encoded RNA polymerase
References
- Bate NJ, Rothstein SJ, Thompson JE. (1991) Expression of nuclear and chloroplast photosynthetic-specific genes during leaf senescence. J Exp Bot 42: 801–811 [Google Scholar]
- Berry JO, Carr JP, Klessig DF. (1988) mRNAs encoding ribulose-1,5-bisphosphate carboxylase remain bound to polysomes but are not translated in amaranth seedlings transferred to darkness. Proc Natl Acad Sci USA 85: 4190–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Sharwood RE, Gutierrez R, Lerbs-Mache S, Stern DB. (2009) The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Mol Biol 69: 541–552 [DOI] [PubMed] [Google Scholar]
- Boulouis A, Raynaud C, Bujaldon S, Aznar A, Wollman FA, Choquet Y. (2011) The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Stern DB, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman FA. (1998) Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc Natl Acad Sci USA 95: 4380–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Vallon O. (2000) Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82: 615–634 [DOI] [PubMed] [Google Scholar]
- Dean C, Pichersky E, Dunsmuir P. (1989) Structure, evolution and regulation of rbcS genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol 40: 415–439 [Google Scholar]
- de Vitry C, Ouyang Y, Finazzi G, Wollman FA, Kallas T. (2004) The chloroplast Rieske iron-sulfur protein: at the crossroad of electron transport and signal transduction. J Biol Chem 279: 44621–44627 [DOI] [PubMed] [Google Scholar]
- Drapier D, Rimbault B, Vallon O, Wollman FA, Choquet Y. (2007) Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J 26: 3581–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. (1977) Protein synthesis by isolated chloroplasts. Biochim Biophys Acta 463: 185–215 [Google Scholar]
- Ellis RJ. (1979) The most abundant protein in the world. Trends Biochem Sci 4: 241–244 [Google Scholar]
- Evans JR. (1986) The relationship between CO2-limited photosynthetic rate and ribulose-1,5-bisphosphate-carboxylase content in two nuclear-cytoplasm substitution lines of wheat and coordination of ribulose-bisphosphate-carboxylation and electron-transport capacities. Planta 167: 351–358 [DOI] [PubMed] [Google Scholar]
- Evans JR. (1989) Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78: 9–19 [DOI] [PubMed] [Google Scholar]
- Furbank RT, Chitty JA, Von Caemmerer S, Jenkins C. (1996) Antisense RNA inhibition of rbcS gene expression reduces Rubisco level and photosynthesis in the C4 plant Flaveria bidentis. Plant Physiol 111: 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J, Hohn T. (1996) Translation in plants: rules and exceptions. Plant Mol Biol 32: 159–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig B, Streb P, Feierabend J. (1992) Light dependence of catalase synthesis and degradation in leaves and the influence of interfering stress conditions. Plant Physiol 100: 1547–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson GS, Evans JR, von Caemmerer S, Arvidsson YBC, Andrews TJ. (1992) Reduction of ribulose-1,5-bisphosphate carboxylase/oxygenase content by antisense RNA reduces photosynthesis in transgenic tobacco plants. Plant Physiol 98: 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Suzuki Y, Mae T, Makino A. (2008) Changes in the synthesis of Rubisco in rice leaves in relation to senescence and N influx. Ann Bot (Lond) 101: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Tsunoda H, Suzuki Y, Makino A, Ishida H. (2012) RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J Exp Bot 63: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. (2010) MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrebtukova I, Spreitzer RJ. (1996) Elimination of the Chlamydomonas gene family that encodes the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA 93: 13689–13693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka MR, Goldschmidt-Clermont M, van Dillewijn J, Rochaix JD. (1989) Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell 58: 869–876 [DOI] [PubMed] [Google Scholar]
- Kuras R, Wollman FA. (1994) The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J 13: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011) Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Loiselay C, Gumpel NJ, Girard-Bascou J, Watson AT, Purton S, Wollman FA, Choquet Y. (2008) Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol Cell Biol 28: 5529–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer GH. (1981) The carboxylation and oxygenation of ribulose 1,5-bisphosphate: the primary events in photosynthesis and photorespiration. Annu Rev Plant Physiol 32: 349–383 [Google Scholar]
- Lysenko EA. (2007) Plant sigma factors and their role in plastid transcription. Plant Cell Rep 26: 845–859 [DOI] [PubMed] [Google Scholar]
- Mae T, Makino A, Ohira K. (1983) Changes in the amounts of ribulose bisphosphate carboxylase synthesized and degraded during the life span of rice leaf (Oryza sativa L.). Plant Cell Physiol 24: 1079–1086 [Google Scholar]
- Makino A, Harada M, Kaneko K, Mae T, Shimada T, Yamamoto N. (2000) Whole-plant growth and N allocation in transgenic rice plants with decreased content of ribulose-1,5-bisphosphate carboxylase under different CO2 partial pressure. Aust J Plant Physiol 27: 1–12 [Google Scholar]
- Makino A, Mae T, Ohira K. (1984) Relation between nitrogen and ribulose-1,5-bisphosphate carboxylase in rice leaves from emergence through senescence. Plant Cell Physiol 25: 429–437 [Google Scholar]
- Makino A, Mae T, Ohira K. (1985) Enzymic properties of ribulose-1,5-bisphosphate carboxylase/oxygenase purified from rice leaves. Plant Physiol 79: 57–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Mae T, Ohira K. (1988) Differences between wheat and rice in the enzyme properties of ribulose-1,5-bisphosphate carboxylase/oxygenase and their relationship to photosynthetic gas exchange. Planta 174: 30–38 [DOI] [PubMed] [Google Scholar]
- Makino A, Sakashita H, Hidema J, Mae T, Ojima K, Osmond B. (1992) Distinctive responses of ribulose-1,5-bisphosphate carboxylase and carbonic anhydrase in wheat leaves to nitrogen nutrition and their possible relationships to CO2-transfer resistance. Plant Physiol 100: 1737–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Shimada T, Takumi S, Kaneko K, Matsuoka M, Shimamoto K, Nakano H, Miyao-Tokutomi M, Mae T, Yamamoto N. (1997) Does decrease in ribulose-1,5-bisphosphate carboxylase by antisense rbcS lead to a higher N-use efficiency of photosynthesis under conditions of saturating CO2 and light in rice plants? Plant Physiol 114: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin R, Niyogi K. (2000) Photosynthesis. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 568–628
- Monod C, Goldschmidt-Clermont M, Rochaix JD. (1992) Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol Gen Genet 231: 449–459 [DOI] [PubMed] [Google Scholar]
- Nakano H, Muramatsu S, Makino A, Mae T. (2000) Relationship between the suppression of photosynthesis and starch accumulation in the pod-removed bean. Aust J Plant Physiol 27: 167–173 [Google Scholar]
- Nickelsen J, Fleischmann M, Boudreau E, Rahire M, Rochaix JD. (1999) Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell 11: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolau BJ, Klessig DF. (1987) Coordinate, organ-specific and developmental regulation of ribulose-1,5-bisphosphate carboxylase gene expression in Amaranthus hypochondriacus. Plant Physiol 85: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Suzuki Y, Yoshizawa R, Kanno K, Makino A. (2012) Effect of individual suppression of RBCS multigene family on Rubisco contents in rice leaves. Plant Cell Environ 35: 546–553 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Nishimura K, Aoki T, Takase H, Tomizawa K, Ashida H, Yokota A. (2009) A phosphofructokinase B-type carbohydrate kinase family protein, NARA5, for massive expressions of plastid-encoded photosynthetic genes in Arabidopsis. Plant Physiol 151: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MAJ, Andralojc PJ, Parmar S, Keys AJ, Habash D, Paul MJ, Alred R, Quick WP, Servaites JC. (1997) Regulation of Rubisco by inhibitors in the light. Plant Cell Environ 20: 528–534 [Google Scholar]
- Peoples MB, Dalling MJ. (1988) The interplay between proteolysis and amino acid metabolism during senescence and nitrogen reallocation. In LD Noodén, AC Leopold, eds, Senescence and Aging in Plants. Academic Press, San Diego, pp 181–217
- Pesaresi P, Schneider A, Kleine T, Leister D. (2007) Interorganellar communication. Curr Opin Plant Biol 10: 600–606 [DOI] [PubMed] [Google Scholar]
- Rodermel S. (1999) Subunit control of Rubisco biosynthesis: a relic of an endosymbiotic past? Photosynth Res 59: 105–123 [Google Scholar]
- Rodermel S, Haley J, Jiang CZ, Tsai CH, Bogorad L. (1996) A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc Natl Acad Sci USA 93: 3881–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel SR, Abbott MS, Bogorad L. (1988) Nuclear-organelle interactions: nuclear antisense gene inhibits ribulose bisphosphate carboxylase enzyme levels in transformed tobacco plants. Cell 55: 673–681 [DOI] [PubMed] [Google Scholar]
- Schmidt GW, Mishkind ML. (1983) Rapid degradation of unassembled ribulose 1,5-bisphosphate carboxylase small subunits in chloroplasts. Proc Natl Acad Sci USA 80: 2632–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Dehne S, Feierabend J. (2002) Post-transcriptional mechanisms control catalase synthesis during its light-induced turnover in rye leaves through the availability of the hemin cofactor and reversible changes of the translation efficiency of mRNA. Plant J 31: 601–613 [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ. (2003) Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys 414: 141–149 [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K. (2004) The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol 45: 985–996 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kawazu T, Koyama H. (2004) RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37: 542–544, 544 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kihara-Doi T, Kawazu T, Miyake C, Makino A. (2010) Differences in Rubisco content and its synthesis in leaves at different positions in Eucalyptus globulus seedlings. Plant Cell Environ 33: 1314–1323 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Makino A, Mae T. (2001) Changes in the turnover of Rubisco and levels of mRNAs of rbcL and rbcS in rice leaves from emergence to senescence. Plant Cell Environ 24: 1353–1360 [Google Scholar]
- Suzuki Y, Miyamoto T, Yoshizawa R, Mae T, Makino A. (2009a) Rubisco content and photosynthesis of leaves at different positions in transgenic rice with an overexpression of RBCS. Plant Cell Environ 32: 417–427 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi K, Yoshizawa R, Mae T, Makino A. (2009b) Differences in expression of the RBCS multigene family and Rubisco protein content in various rice plant tissues at different growth stages. Plant Cell Physiol 50: 1851–1855 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ohkubo M, Hatakeyama H, Ohashi K, Yoshizawa R, Kojima S, Hayakawa T, Yamaya T, Mae T, Makino A. (2007) Increased Rubisco content in transgenic rice transformed with the ‘sense’ rbcS gene. Plant Cell Physiol 48: 626–637 [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Boudreau E, Lemaire SD, Goldschmidt-Clermont M, Rochaix JD. (2000b) Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 97: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Goldschmidt-Clermont M, Wostrikoff K, Rochaix JD. (2000a) Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J 21: 469–482 [DOI] [PubMed] [Google Scholar]
- Voelker R, Barkan A. (1995) Nuclear genes required for post-translational steps in the biogenesis of the chloroplast cytochrome b6f complex in maize. Mol Gen Genet 249: 507–514 [DOI] [PubMed] [Google Scholar]
- Waters MT, Langdale JA. (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J. (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Stern DB. (2007) Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc Natl Acad Sci USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama T, Arima K, Kumazawa K. (1975) Sample preparation from dilute ammonium solution for emission spectrographic analysis of heavy nitrogen. Jpn J Soil Sci Plant Nutr 46: 146–147 [Google Scholar]