Abstract
The PHYTOCHROME AND FLOWERING TIME1 gene encoding the MEDIATOR25 (MED25) subunit of the eukaryotic Mediator complex is a positive regulator of jasmonate (JA)-responsive gene expression in Arabidopsis (Arabidopsis thaliana). Based on the function of the Mediator complex as a bridge between DNA-bound transcriptional activators and the RNA polymerase II complex, MED25 has been hypothesized to function in association with transcriptional regulators of the JA pathway. However, it is currently not known mechanistically how MED25 functions to regulate JA-responsive gene expression. In this study, we show that MED25 physically interacts with several key transcriptional regulators of the JA signaling pathway, including the APETALA2 (AP2)/ETHYLENE RESPONSE FACTOR (ERF) transcription factors OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 and ERF1 as well as the master regulator MYC2. Physical interaction detected between MED25 and four group IX AP2/ERF transcription factors was shown to require the activator interaction domain of MED25 as well as the recently discovered Conserved Motif IX-1/EDLL transcription activation motif of MED25-interacting AP2/ERFs. Using transcriptional activation experiments, we also show that OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59- and ERF1-dependent activation of PLANT DEFENSIN1.2 as well as MYC2-dependent activation of VEGETATIVE STORAGE PROTEIN1 requires a functional MED25. In addition, MED25 is required for MYC2-dependent repression of pathogen defense genes. These results suggest an important role for MED25 as an integrative hub within the Mediator complex during the regulation of JA-associated gene expression.
The plant hormone jasmonic acid and its conjugate derivatives, collectively known as jasmonates (JAs), are known to play an important role in the response to plant pathogens (Penninckx et al., 1998; Thomma et al., 1998), wounding, and herbivores (McConn et al., 1997; Zhang and Turner, 2008), as well as regulating plant developmental processes such as stamen development (Feys et al., 1994) and senescence (He et al., 2002). The involvement of JAs in this diverse range of processes suggests a mechanism for fine-tuning the activation of specific response pathways while controlling other nonessential response pathways. The activation of herbivory-associated JA genes and the suppression of pathogen defense genes by the basic helix-loop-helix (bHLH) transcription factor (TF) MYC2 is one example of this fine control of gene expression (Lorenzo et al., 2004; Dombrecht et al., 2007). In addition, JA signaling can act either synergistically or antagonistically with the signaling pathways of other plant hormones and understanding the cross talk between plant hormone pathways has been the focus of numerous research efforts (Penninckx et al., 1998; Schenk et al., 2000; Spoel et al., 2003; Anderson et al., 2004; Navarro et al., 2006, 2008).
The F-box protein CORONATINE INSENSITIVE1 (COI1) is important in JA signaling as it is required for almost all JA-responsive phenotypes (Feys et al., 1994; Devoto et al., 2005). Together with S-PHASE KINASE-ASSOCIATED PROTEIN1 (SKP1) and CULLIN proteins, COI1 forms an SKP/CULLIN/F-box complex, which attaches ubiquitin to target proteins to signal their degradation by the 26S proteasome (for review, see Howe, 2010; Pauwels and Goossens, 2011). The importance of COI1 in JA perception was revealed with the discovery of the JA ZIM domain (JAZ) proteins, which act as repressors of JA-associated transcription (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Genetic, biochemical, and structural studies have confirmed the COI1-JAZ complex as the coreceptor for the biologically active JA-Ile ligand and these findings provide insights into the regulation of the JA signaling pathway through release of transcriptional repression (Howe, 2010; Sheard et al., 2010).
Recently, a number of TFs have been identified as binding partners of the JAZ proteins with a variety of different roles in regulating JA-associated gene expression (for review, see Kazan and Manners, 2012). MYC2 was the first TF to be identified as a JAZ-binding partner (Chini et al., 2007), and recently, two MYC2-related bHLH TFs (MYC3 and MYC4) also have been shown to interact with JAZ proteins to regulate both pathogen and herbivore defense pathways (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). In addition, three other bHLH TFs (TRANSPARENT TESTA8, GLABRA3 [GL3], and ENHANCER of GL3) and two MYB domain TFs (MYB75 and GL1) interact with the JAZ proteins, and are known to have roles in both anthocyanin and trichome production (Qi et al., 2011). Additionally, JAZ proteins have been found to interact with two MYB domain TFs (MYB21 and MYB24) that control stamen development (Song et al., 2011), and also two ethylene (ET)-associated TFs (EIN3 and EIL1) that regulate pathogen defense and root growth through ET signaling (Zhu et al., 2011).
The removal of JAZ-mediated repression would allow TF proteins to recruit the general TFs and RNA polymerase II machinery to initiate transcription. In eukaryotes, the multiprotein complex that connects TFs with the core transcriptional machinery is called the Mediator complex and was recently purified in Arabidopsis (Arabidopsis thaliana; Bäckström et al., 2007). Approximately 30 Mediator subunits were isolated from Arabidopsis and functional roles in important plant processes such as development, flowering, and abiotic and biotic stress responses have been identified for some Mediator subunits (Autran et al., 2002; Boyce et al., 2003; Dhawan et al., 2009; Kidd et al., 2009; Gillmor et al., 2010; Wathugala et al., 2012). One subunit that exemplifies the diverse roles that a Mediator subunit can possess in different plant processes is the MEDIATOR25 (MED25) subunit. MED25 (previously known as PHYTOCHROME AND FLOWERING TIME1 [PFT1]) has been shown to play a role in the control of flowering time (Cerdán and Chory, 2003; Wollenberg et al., 2008; Iñigo et al., 2011), cell proliferation (Xu and Li, 2011), as well as abiotic and biotic stress responses (Kidd et al., 2009; Elfving et al., 2011). The role of MED25 in biotic stress responses was discovered with med25/pft1 mutants (hereafter referred to as med25) displaying increased susceptibility to leaf-infecting necrotrophic pathogens Alternaria brassicicola and Botrytis cinerea, yet increased resistance to the root-infecting hemibiotroph Fusarium oxysporum (Kidd et al., 2009). The med25 mutant also showed attenuated expression of both herbivore- and pathogen-associated JA genes as well as decreased production of anthocyanin and a reduced sensitivity to JA-mediated root inhibition (Kidd et al., 2009). These findings suggested an important role for MED25 in JA signaling, and thus potentially implicate MED25 as an interacting partner for JA-associated TFs in the Mediator complex.
In this study we aimed to identify MED25-interacting TFs that have roles in the MED25-mediated control of JA-associated gene expression. Through the use of yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) screening, we identified several MED25-interacting TFs known to function in the control of JA-associated gene expression. These included the defense-associated APETALA2 (AP2)/ETHYLENE RESPONSE FACTORs (ERFs); ERF1, OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59), and TDR1/ERF98 (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003; Gutterson and Reuber, 2004; Pré et al., 2008), as well as the defense- and herbivore-associated bHLH TFs; MYC2, MYC3, and MYC4 (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). We demonstrated the functional requirement for MED25 in ERF1- and ORA59-dependent activation of the PLANT DEFENSIN1.2 (PDF1.2) gene and MYC2-dependent activation of the VEGETATIVE STORAGE PROTEIN1 (VSP1) insect defense gene. Importantly, we provide evidence for the role of the EDLL motif, the recently discovered transcription activation domain from MED25-interacting AP2/ERFs, as an important domain for MED25 interaction. Together, our results suggest that MED25 acts an integrative hub for the regulation of JA-dependent gene expression.
RESULTS
Identification of MED25-Interacting TFs
To identify interacting partners of MED25, we carried out Y2H screening using two different bait constructs that corresponded to the N-terminal von Willebrand Factor A (vWF-A) domain responsible for Mediator binding and the C-terminal activator interaction domain (ACID) of the MED25 protein (Fig. 1A). We found that the first construct (MED25ΔACID/Q-rich) that consisted of amino acids 1 to 558 but lacked the ACID domain, as well as the Q-rich region at the C terminus of the MED25 protein, did not yield any interactions. However, when the second MED25 construct was expressed, which contained the ACID domain but lacked the N-terminal vWF-A domain and the C-terminal Q-rich domain (MED25ΔvWF-A/Q-rich, 228–681 amino acids), 12 interacting TFs were identified (Fig. 1B). Retesting these TFs with the MED25ΔACID/Q-rich construct did not show any interaction, suggesting that the interaction with the TFs requires the ACID domain of the MED25 protein. Among the TFs identified were a number of JA- and plant-defense-associated TFs. For instance, four members of group IX of the AP2/ERF TF family, ERF1, ORA59, TDR1/ERF98, and ERF15 were identified, as well as MYC2 and the related bHLH TFs, MYC3 and MYC4 (Fig. 1B). All of these TFs, with the exception of ERF15, have been previously implicated in the regulation of plant defense signaling (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003, 2004; Gutterson and Reuber, 2004; McGrath et al., 2005; Dombrecht et al., 2007; Pré et al., 2008; Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). While not currently having a published role in regulating defense signaling, ERF15 is the closest homolog of ORA59 based on its nucleotide sequence and forms a separate clade with ORA59 and ERF1 based on their similarity at the conserved AP2 domain (McGrath et al., 2005; Nakano et al., 2006). The Y2H screen also identified additional interacting TFs such as the drought and cold response regulator DREB2A, which belongs to group IV of the AP2/ERF TF family, as well as WRKY10, MYB104, and two uncharacterized TFs encoding a B-box zinc-finger (At4g39070) and a bZIP (At2g31370) TF. Four of the 12 TFs that we identified, DREB2A, ERF1, TDR1/ERF98, and the B-box zinc finger (At4g39070), have recently been reported as interacting partners of MED25 (Elfving et al., 2011; Ou et al., 2011). The independent discovery of these TFs in our interaction screen indicates the reliability of our Y2H system in detecting MED25-interacting TFs.
Figure 1.
The ACID domain of MED25 interacts with a number of Arabidopsis TFs in Y2H analyses. Representative schematics of the MED25 fragments used to screen the Y2H library along with the full-length MED25 protein (A). Only the fragment containing the ACID domain, MED25ΔvWF-A/Q-rich resulted in positive interactions in the Y2H screen and 12 positive interactions were identified (B). No interactions were seen for the binding domain (BD) only or when an empty activation domain (AD) was used. Y2Hs were performed with SD-Leu-Trp as well as SD-Leu-Trp-His supplemented with 3-AT. [See online article for color version of this figure.]
To verify the interactions between MED25 and the TF proteins identified through Y2H analysis, we carried out a single in vitro pull-down experiment using the MED25ΔvWF-A/Q-rich protein fused to a poly-His tag together with individual glutathione-S-transferase (GST)-tagged TF proteins. No interaction was observed with the control GST-GFP fusion protein, and with the exception of ORA59, 11 of the identified TFs interacted with the MED25ΔvWF-A/Q-rich protein, confirming the Y2H data (Fig. 2, A–C). We repeated the pull-down experiment with MYC2, ERF1, and ORA59 three times; however, only MYC2 and ERF1 proteins could be successfully pulled down. We also carried out a pull-down experiment with the His-GFP protein as an additional control; however, none of the GST-tagged TF proteins showed interaction with this protein (Supplemental Fig. S1). Overall, we conclude that MED25 can potentially interact with a number of Arabidopsis TFs, including those with known roles in JA and stress signaling.
Figure 2.
MED25 is able to bind to the interacting TF proteins in pull-down experiments. An in vitro His-pull-down experiment confirmed the Y2H interactions for 11 of the 12 interacting TFs. The GST-tagged TFs that were used as the input to the pull-down experiment are shown in an SDS polyacrylamide gel (A). After extensive washing, bound proteins were eluted and analyzed by IB assays with the indicated antibodies (B and C). Expected sizes of corresponding fusion proteins are marked by asterisks.
As the ORA59 protein was unable to be pulled down with the MED25 protein, we decided to investigate the interaction of ORA59 and MED25 further. We determined the MED25-interaction domain of ORA59 by performing Y2H experiments with three overlapping derivatives of the ORA59 protein (amino acids 1–139, 80–214, and 139–244). We observed an interaction only with the fragment containing the amino acids 139 to 244, suggesting that the interaction with MED25 requires the last 30 amino acids at the C terminus of ORA59 (Supplemental Fig. S2). Furthermore, the full length as well as overlapping fragments of ORA59 were fused to the N-terminal fragment of the yellow fluorescent protein (YFPN) and coexpressed with MED25 fragments fused to the C-terminal fragment of the yellow fluorescent protein (YFPC) for bimolecular fluorescent complementation (BiFC) experiments. These experiments confirmed the results of the Y2H analysis with the YFPC-fused MED25ΔvWF-A/Q-rich protein producing fluorescence only when expressed with the full-length ORA59 or the 139 to 244 amino acid fragment fused to YFPN and not with the other ORA59 fragments (Supplemental Fig. S3). We also expressed the full-length ORA59 and 139 to 244 amino acid fragment with the MED25ΔACID/Q-rich protein; however these combinations did not result in detectable fluorescence (Supplemental Fig. S3). These results suggest that the MED25 ACID domain is required for interaction with ORA59 and that a specific domain in the C terminus of ORA59 is required for MED25 interaction.
The Conserved Motif IX-1/EDLL Transcription Activation Motif of ERF1, ERF15, and TDR1/ERF98 Is Required for MED25 Binding
The ACID domain of the human MED25 protein has previously been shown to interact with the transcriptional activation domain (TAD) of VP16, the potent activator from the herpes simplex virus (Mittler et al., 2003; Yang et al., 2004; Milbradt et al., 2011; Vojnic et al., 2011). It is therefore likely that the interaction we observed between AP2/ERFs and MED25 also requires the ACID domain of MED25 and the TAD domain of the AP2/ERFs. The Conserved Motif IX-1 (CMIX-1) motif has been identified in the carboxy termini of eight AP2/ERF proteins belonging to group IXc (Nakano et al., 2006) although until recently the functional importance of this motif has been unknown. Interestingly, six of the eight proteins belonging to group IXc (ERF1, ERF15, ORA59, ESE1, TDR1/ERF98, and ERF91) have been found to interact with MED25 (Ou et al., 2011; this study). Recently, the CMIX-1 motif from TDR1/ERF98 has been shown to act as a strong TAD (Tiwari et al., 2012). The CMIX-1 motif was renamed as the EDLL motif based on the conserved amino acids between Arabidopsis TDR1/ERF98 and TDR1-like proteins from other plant species (Tiwari et al., 2012). It was shown that removal of the CMIX-1/EDLL motif or mutation of the conserved Leu residues abolished transcriptional activation ability of TDR1/ERF98 (Tiwari et al., 2012). Interestingly, while the specific Glu and Asp residues of the EDLL motif in TDR1/ERF98 are not conserved among the other CMIX-1 motif-containing proteins, the six MED25-interacting AP2/ERFs share a number of conserved Leu and Glu residues (Fig. 3A). As the Leu residues of TDR1/ERF98 were shown to be required for transcriptional activation, we mutated the same Leu residues in three selected TFs namely ERF1, ERF15, and TDR1/ERF98 to Val to determine whether this mutation would affect the interaction of these TFs with MED25 (Fig. 3B). Y2H experiments with full-length ERF1, ERF15, and TDR1/ERF98 proteins containing either mutated or native CMIX-1/EDLL motifs revealed that the conserved Leu in ERF1, ERF15, and TDR1/ERF98 are essential for MED25 interaction (Fig. 3C; Supplemental Fig. S4). These results together with the previous Y2H results with ORA59 suggest that the C-terminal CMIX-1/EDLL motif previously implicated in transcriptional activation is also important for interaction of ERF1, ERF15, TDR1/ERF98, and ORA59 with MED25.
Figure 3.
The EDLL motif found in the carboxy-terminal region of ERF1, ERF15, and TDR1/ERF98 is important for interaction with MED25. The sequence of the CMIX-1 domain from the MED25-interacting group IX AP2/ERFs (A). The conserved EDLL amino acid residues from TDR1/ERF98-like proteins (Tiwari et al., 2012) are highlighted in red (A). The mutated Leu residues of ERF1, ERF15, and TDR1/ERF98 are shown in red (B). Y2H analyses between MED25ΔvWF-A/Q-rich and full-length or mutated ERF1, ERF15, and TDR1/ERF98 show that the Leu residues in the EDLL motif are required for interaction with MED25 (C). Y2Hs were performed with SD-Leu-Trp as well as SD-Leu-Trp-His supplemented with 3-AT. [See online article for color version of this figure.]
MED25 Is Required for the Transcription Activation Ability of MYC2, ERF1, and ORA59
Of the MED25-interacting TFs, ERF1, ORA59, and MYC2 are better characterized for their roles as important regulators of JA-associated pathogen and herbivore defense genes (Lorenzo et al., 2003, 2004; Dombrecht et al., 2007; Pré et al., 2008). The interaction between these TFs and MED25 could potentially explain why JA-associated gene expression is reduced in the med25 mutant (Kidd et al., 2009). Therefore we decided to investigate these three TFs further to determine whether MED25 acts downstream to activate JA-associated transcription. To test whether MYC2, ERF1, or ORA59 proteins require MED25 for transcriptional activation, we performed in vivo transactivation experiments with MYC2, ERF1, or ORA59 using wild-type Arabidopsis plants and the previously characterized med25 mutant (Kidd et al., 2009). In these experiments, we cobombarded a reporter gene construct containing the GAL4 upstream activation sequence (pGAL4UAS) linked to the GUS gene (pGAL4UAS-GUS), together with cauliflower mosaic virus 35S expression constructs of MYC2, ERF1, and ORA59 fused to the GAL4 DNA-binding domain (GAL4BD) or GAL4BD alone (Fig. 4A). In addition, an expression construct of the firefly (Photinus pyralis) LUCIFERASE (LUC) gene was cobombarded as a normalization control.
Figure 4.
MED25 is required for proper transcriptional activation by ERF1, ORA59, and MYC2 in transient activation assays in Arabidopsis leaves. Schematic diagrams of the effector, reporter, and normalization control constructs are shown in insets (A–C). The activity of the reporter gene GUS was normalized to the activity of the firefly LUC gene cobombarded as a reference. Relative GUS/luciferase activity ratios after cobombardment of the reporter, effector, and control constructs are shown in A to C. MED25 is required by ERF1, ORA59, and MYC2 for both general transcriptional activation using the GAL4 binding domain (BD) and upstream activation sequence (GAL4-UAS; A), as well as for specific activation of the PDF1.2 promoter by ERF1 and ORA59 (B) and for activation of the VSP1 promoter by MYC2 (C). Error bars indicate sd. Data are from three biological replicates of two bombarded leaves. Asterisks represent significant differences between wild-type and med25 plants (two-way ANOVA, Tukey’s honestly significant difference [HSD] test; * = P < 0.05; ** = P < 0.01; and *** = P < 0.001). [See online article for color version of this figure.]
As expected, the addition of the vector constructs expressing either MYC2-, ERF1-, or ORA59-GAL4BD produced significantly higher transcription activity of the GUS reporter gene compared with the control effector plasmid (GAL4BD only) in wild-type Arabidopsis plants (Fig. 4A). No significant difference was observed in GUS reporter gene activity between the wild type and med25 with the control effector plasmid (Fig. 4A). However, transcription activation abilities of the MYC2-, ERF1-, and ORA59-GAL4BD fusion proteins were significantly reduced in med25 leaves compared with the wild-type leaves in independent experiments (Fig. 4A). As an additional control experiment, we cotransformed wild-type and med25 plants with a 35S:GUS and 35S:LUC construct without the effector plasmid. We did not observe any significant difference between GUS/LUC ratios, suggesting that the differences observed in transcription activation were not due to different transformation efficiencies (Supplemental Fig. S5). These results suggest that a functional MED25 is required for transcriptional activation by ERF1, ORA59, and MYC2 TFs.
MED25 Is Required for ORA59- and ERF1-Dependent Activation of PDF1.2
ERF1 and ORA59 have both been shown to activate expression of the PDF1.2 gene, with the expression of ORA59 being essential for PDF1.2 activation in Arabidopsis (Lorenzo et al., 2003; Pré et al., 2008). The med25 mutant has reduced expression of the PDF1.2 gene (Kidd et al., 2009), raising the hypothesis that both ERF1 and/or ORA59 may require a functional MED25 protein to activate PDF1.2 expression. To test this, we repeated the transactivation experiments as above but with the 1,187-bp fragment of the PDF1.2 promoter (pPDF1.2) linked to the GUS reporter gene (Fig. 4B). This promoter fragment has been used previously to test the activation of PDF1.2 in response to pathogens, hormone treatment, or the addition of TFs (Manners et al., 1998; Brown et al., 2003; Pré et al., 2008; Zarei et al., 2011). In agreement with previous studies, the expression of either ERF1 or ORA59 significantly induced the expression of pPDF1.2-GUS in wild-type leaves, and as previously reported (Pré et al., 2008), ORA59 activated pPDF1.2-GUS expression to higher levels than ERF1 (Fig. 4B). Interestingly, when either ORA59 or ERF1 was expressed in the med25 mutant plants, only partial activation of pPDF1.2-GUS expression was achieved (Fig. 4B). These experiments indicate that both ERF1- and ORA59-dependent activation of PDF1.2 are dependent on MED25.
MED25 Is Required for MYC2-Dependent Activation of VSP1
In contrast to ERF1 and ORA59, MYC2 acts as a negative regulator of JA-inducible defense genes such as PDF1.2 and a positive regulator of insect and herbivore response genes such as VSP1 (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007). Therefore, we tested the ability of MYC2 to activate VSP1 gene expression in the med25 mutant by using the VSP1 promoter in transactivation experiments. Expression of MYC2 in wild-type plants resulted in induction of VSP1 expression. However, in the med25 mutant line, expression of MYC2 resulted in significantly reduced expression of VSP1 (Fig. 4C). These results demonstrate that MED25 is required for MYC2-dependent activation of VSP1.
ERF1-, ORA59-, and MYC2-Regulated Genes Are Attenuated in med25
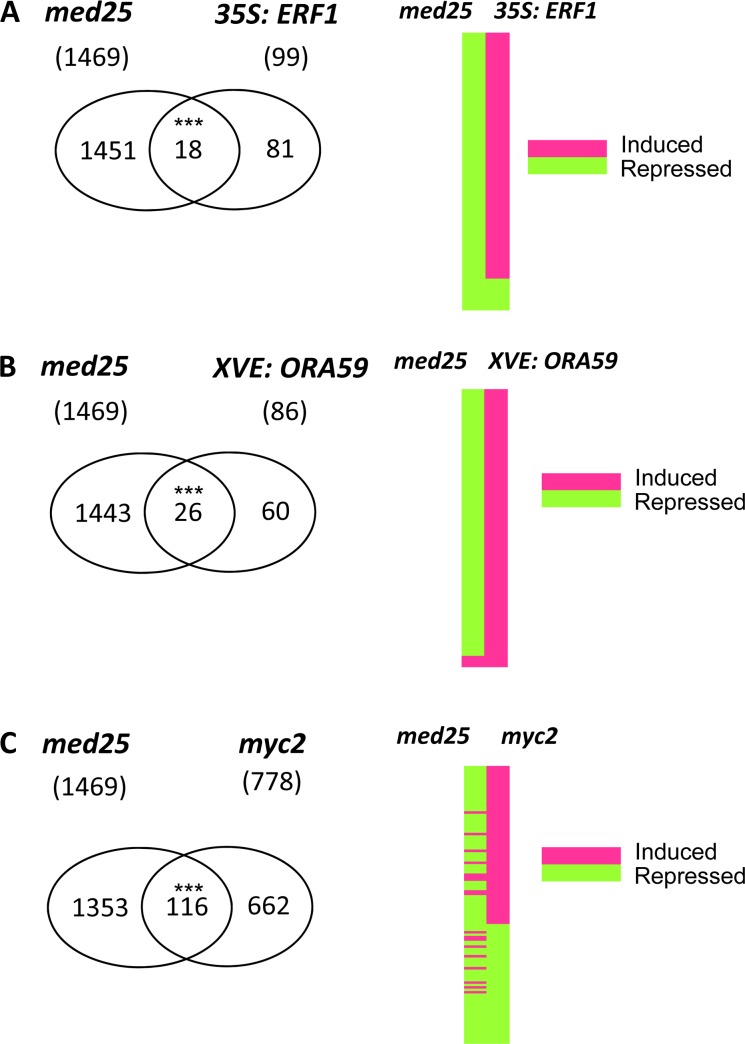
To further investigate the requirement of MED25 for ERF1-, ORA59-, or MYC2-regulated gene expression, we compared published microarray data from ERF1 and ORA59 overexpression lines (Lorenzo et al., 2003 Pré et al., 2008) and the myc2 mutant (Dombrecht et al., 2007) with microarray data obtained from the med25 mutant (Kidd et al., 2009; Fig. 5; Supplemental Tables S1–S4). The ERF1-regulated gene list (e.g. genes that are either activated or repressed in 35S:ERF1 relative to the wild type) used in this comparison (Lorenzo et al., 2003) consisted of 99 genes and 18 of these genes were found to be in common with the med25-regulated genes (Fig. 5A; Supplemental Table S2). The majority (16 out of 18) of the common genes were ERF1-activated genes, and all 16 of these genes showed reduced expression in the med25 microarray relative to the wild type (Fig. 5A; Supplemental Table S2). The gene list obtained from the ORA59 microarray experiment contained 86 ORA59-activated genes (Pré et al., 2008), and of the 26 genes that were common to the ORA59- and med25-regulated gene lists, almost all (25 out of 26) had reduced expression in med25 (Fig. 5B; Supplemental Table S3). Finally, 116 genes were common between the med25- and myc2-regulated gene lists (Dombrecht et al., 2007; Fig. 5C; Supplemental Table S4). MYC2-activated genes were found to be mostly repressed (41 out of 50) in med25, suggesting that MED25 is required for their activation (Supplemental Table S4). In addition, genes that are repressed by MYC2 had also predominantly reduced expression in med25 (57 out of 66; Fig. 5C; Supplemental Table S4). Therefore, both MYC2-activated and MYC2-repressed genes are attenuated in med25. Statistical tests using the hypergeometric distribution (P < 0.001) showed that the number of MED25-regulated genes found to be in common with the TF arrays was significantly higher than expected, given the total number of common genes present between each microarray experiment. In addition, as the majority of the common genes showed reduced expression in med25, this suggests that a functional MED25 may be required for activation of ERF1-, ORA59-, and MYC2-regulated genes.
Figure 5.
ERF1-, ORA59-, and MYC2-responsive genes have reduced expression in med25. Genes differentially expressed by ERF1, ORA59, and MYC2 were obtained from published gene lists; 35S:ERF1 (Lorenzo et al., 2003); XVE:ORA59 (Pré et al., 2008); and myc2 (Dombrecht et al., 2007). The total number of genes in each list is shown in brackets under each genotype. Common genes with the med25 microarray were identified and presented as Venn diagrams (A–C). The genes that were common between med25 and the TF lists (med25-35S:ERF1 = 18; med25-XVE:ORA59 = 26; med25-myc2 = 116) were then used to construct heat maps and their expression marked depending on whether they were induced or repressed compared with the wild type in the med25 array and the 35S:ERF1, XVE:ORA59, or myc2 arrays (A–C). Genes were then ranked according to their expression in the TF array from highest to lowest to produce the heat maps. Asterisks represent higher than expected common genes using the hypergeometric distribution. * = P < 0.001. [See online article for color version of this figure.]
MED25 Is Required for Repression of JA-Associated Defense Genes by MYC2
In the microarray comparisons, MED25 appeared to be required for both MYC2-activated genes such as VSP1, and MYC2-repressed genes such as PDF1.2, as expression of both genes were reduced in the med25 mutant (Supplemental Table S4). To further investigate whether MED25 is required for the repression of defense genes by MYC2, we created a med25 myc2 double mutant and analyzed gene expression in the double and single mutants with and without JA treatment, using real-time quantitative PCR (RT-qPCR; Fig. 6, A–E; Supplemental Table S5). In support of previous results, the expression of defense genes, PDF1.2, HEVEIN-LIKE (HEL), and BASIC CHITINASE (CHI-B), were significantly reduced in the med25 single mutant after JA treatment (Kidd et al., 2009; Fig. 6, A–C). In the myc2 single mutant these genes were expressed at a significantly higher level compared with the wild type after JA treatment, confirming the negative effect of MYC2 on JA-regulated defense gene expression (Lorenzo et al., 2004; Dombrecht et al., 2007). In the med25 myc2 double mutant, these genes were significantly attenuated and showed an expression level similar to that of the med25 single mutant (Fig. 6, A–C). These results suggest that MED25 is essential for the activation of PDF1.2, CHI-B, and HEL expression and that MYC-mediated repression of these genes acts on a pathway that is dependent on MED25.
Figure 6.
MED25 is required for the repression of defense-associated genes by MYC2. RT-qPCR analysis shows the med25 mutant has decreased expression of PDF1.2, HEL, CHI-B, and ORA59 relative to the wild type after JA treatment (A–C). The expression of PDF1.2, HEL, and CHI-B is increased in myc2 after JA treatment, however remained attenuated in the med25 myc2 double mutant (A–C). The expression of ORA59 and ERF1 in the med25 myc2 double mutant was partially attenuated relative to the myc2 single mutant after JA treatment and was expressed similar to the wild type (D and E). Data represents the relative transcript abundance to the housekeeping genes (β-ACTIN2, β-ACTIN7, and β-ACTIN8). Error bars are se of three biological replicates of 15 plants per replicate. a, b, c, and d represent significance (P < 0.05) using a 2-way ANOVA with the Bonferroni method applied as a post hoc test. [See online article for color version of this figure.]
To investigate how MYC2-mediated repression of JA-responsive defense genes may be occurring, we analyzed the expression of ORA59 and ERF1 in the single and double mutants. After JA treatment, ORA59 and ERF1 expression was induced higher than the wild type in the myc2 single mutant, confirming previously published results (Fig. 6, D and E; Dombrecht et al., 2007; Zander et al., 2010). Interestingly both ORA59 and ERF1 showed attenuated expression in the med25 myc2 double mutant, relative to the myc2 single mutant. However, ORA59 and ERF1 expression levels in the double mutant were not as strongly reduced as the defense genes (e.g. PDF1.2) and appeared similar to the wild type. These results suggest that MYC2 suppresses JA-associated defense genes through repression of the defense-associated TFs, ERF1 and ORA59, and that this repression by MYC2 is dependent on a functional MED25.
ORA59 Expression Is Reduced in the med25 Mutant
The reduced expression of ERF1-, ORA59-, and MYC2-dependent genes in the med25 microarray suggests that the interaction with MED25 is important for ERF1-, ORA59-, and MYC2 gene activation. However, an alternative reason for reduced expression of ERF1-, ORA59-, and MYC2-dependent genes could be reduced expression of the TFs themselves in the med25 mutant. We therefore examined the expression of ERF1, ORA59, and MYC2 in the med25 microarray experiment. The med25 microarray experiment involved treatment with the hemibiotrophic pathogen F. oxysporum, which is known to induce JA-associated defense gene expression (Kidd et al., 2009; Thatcher et al., 2009).
Interestingly, ORA59 expression was found to be induced in the wild type by F. oxysporum infection but was attenuated in med25 under both mock and infected conditions (Supplemental Fig. S6A). In contrast, ERF1 expression was unresponsive to F. oxysporum infection and no significant difference in ERF1 expression could be found between wild-type and med25 plants (Supplemental Fig. S6B). Therefore, down-regulation of JA-associated defense genes in the med25 microarray could also be due to a reduction in ORA59 expression but not ERF1. The expression of MYC2 in the med25 mutant appeared similar to the wild type in the microarray experiment (Supplemental Fig. S6C). This suggests that down-regulation of MYC2 target genes in the microarray is not due to reduced expression of the MYC2 gene but may be a result of the reduced transcriptional activation ability of MYC2 protein in med25 mutant plants. We also looked at the expression of MYC3 and MYC4 in the microarray. These genes also showed no significant change in expression in the med25 mutant (Supplemental Fig. S6, D and E). Therefore, alterations in MYC3 or MYC4 expression cannot explain the alterations in MYC2 target genes in med25. These results suggest MED25 is required for the expression of ORA59 but not ERF1 or the bHLH TFs MYC2, MYC3, or MYC4.
ORA59 and Other MED25-Interacting AP2/ERFs Bind to the ORA59 Promoter
The attenuation of ORA59 in the med25 mutant as well as the results from the transcriptional activation experiments suggests that MED25 is required both upstream for the expression of ORA59 as well as downstream for the activation of the JA/ET defense pathway by the ORA59 protein. We performed yeast one-hybrid (Y1H) experiments with the TF library using the promoter region of ORA59 as bait to determine which TFs were able to interact with the ORA59 promoter and therefore may be able to regulate ORA59 expression. Two overlapping fragments of the ORA59 promoter region were cloned and used to screen the TF library (Fig. 7A). Thirteen AP2/ERF TFs were identified that bind to the ORA59 promoter including ORA59 itself (Supplemental Table S6). With the exception of At2g47520 and At1g03800, all of the AP2/ERFs identified belonged to group IX of the AP2/ERF family and five of them were found to interact with MED25 (Supplemental Table S6). All 13 TFs were identified with promoter fragment 2, while only five of these TFs showed interaction with promoter fragment 1 (Supplemental Table S6). The ORA59 promoter contains a GCC box located between nucleotides −369 and −363 upstream of the translation start site (Fig. 7A) and it is likely that the identified TFs bind to this element. However, the small number of TFs identified with promoter fragment 1 in the initial screen could be due to the distance of this element from the translation start site present in the reporter vector that was used for the Y1H experiment as it has been shown that transcription activation diminishes with distance in yeast (Dobi and Winston, 2007). Supporting this, retesting of these TFs with the promoter fragment 1 in the presence of 3-amino-1,2,4-triazole (3-AT) abolished their transcription activation abilities, suggesting that the observed interaction is rather weak (data not shown). We therefore used promoter fragment 2 for further analyses. We retested the four group IX AP2/ERF TFs that interacted with MED25 as well as control TF proteins ERF4 and GFP using selective media supplemented with 3-AT to further assess the strength of the TF-promoter interaction using promoter fragment 2. In nonselective media (SD-Leu-Trp), we observed growth with the TFs ERF1, ORA59, and ERF15 as well as EFR4 and GFP. The TFs, ERF1, ORA59, and ERF15 showed strong interaction with promoter fragment 2 on selective media (SD-Leu-Trp-His) in the presence of 3-AT, while growth of the yeast cells with the TF TDR1/ERF98 was weak. No growth was observed with ERF4 and GFP on selective media with 3-AT (Fig. 7B). To test if the AP2/ERF TFs that were identified are binding to the GCC box located on the ORA59 promoter, we mutated the GCC box (GCCGCC to TCCTCA) in fragment 2 and retested the same TFs with ERF4 and GFP as controls. We found that the TFs were no longer able to bind to the mutated promoter with the addition of 25 mm 3-AT (Fig. 7B). These results suggest that the GCC box is an important element for TF binding in the ORA59 promoter. Overall these results show that a number of MED25-interacting AP2/ERFs, including ORA59 itself, are able to bind to the ORA59 promoter and therefore provides evidence for a positive feedback loop to further enhance ET/JA defense.
Figure 7.
MED25-interacting AP2/ERFs can bind to the ORA59 promoter in yeast. Schematics of the ORA59 promoter, with the GCC box motif (located on −369 to −363) and the promoter fragments used for the Y1H screen (A). mFragment2 contains point mutations in GCC box and the mutated nucleotides are indicated in lowercase. TSS, Translation start site. Binding analysis of the TFs to the promoter fragments Fragment2 and mFragment2 tested in Y1H using SD-Leu-Trp as well as SD-Leu-Trp-His supplemented with 25 mm 3-AT (B). [See online article for color version of this figure.]
DISCUSSION
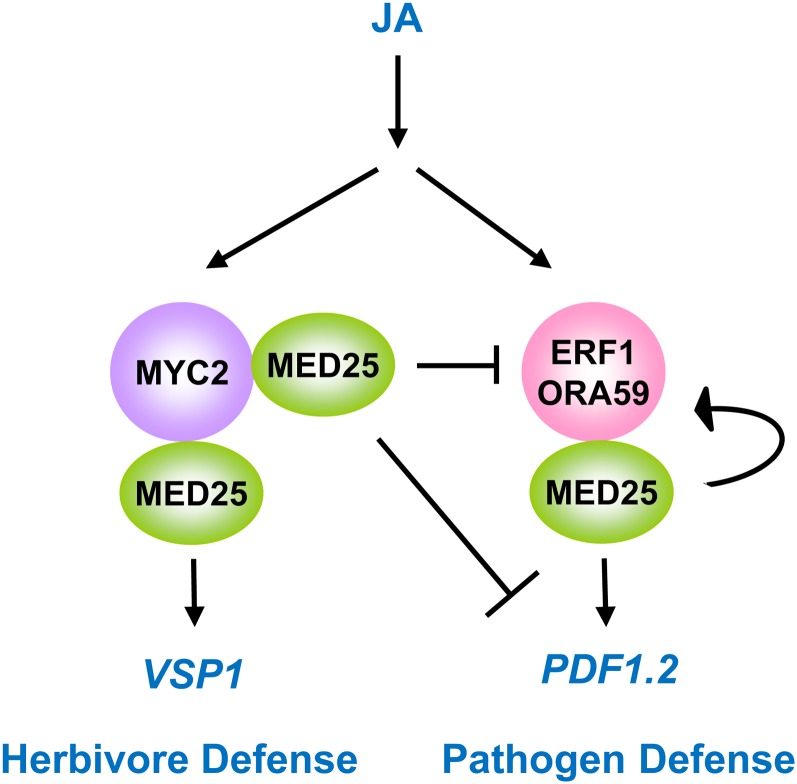
The Mediator complex, which functions as a universal adaptor for TFs, was recently identified in Arabidopsis (Bäckström et al., 2007) and is expected to have roles in integrating multiple plant signaling pathways (Kidd et al., 2011; Mathur et al., 2011). In accordance with this, the MED25 subunit of the Mediator complex has been shown to be a regulator of a number of developmental and physiological processes such as flowering time, cell size, abiotic and biotic stress responses, as well as regulation of the JA signaling pathway (Cerdán and Chory, 2003; Kidd et al., 2009; Elfving et al., 2011; Iñigo et al., 2011; Xu and Li, 2011). Here we have identified 12 TFs that interact with MED25. Some of the identified TFs (e.g. MYC2, MYC3, MYC4, and the group IX AP2/ERF TFs) have previously been shown to be important for correct regulation of JA-associated herbivore and pathogen defense genes. In this report, we explored the ability of three of these TFs, ERF1, ORA59, and MYC2, to activate JA-dependent gene expression through MED25. Using transcription activation assays, we found MED25 to be required for ERF1- and ORA59-dependent activation of the PDF1.2 gene as well as MYC2-dependent activation of the VSP1 gene. These results suggest that MED25 acts downstream of ERF1, ORA59, and MYC2 for the activation of PDF1.2 and VSP1 expression (Fig. 8).
Figure 8.
A proposed model of the role of MED25 in coordinating JA-associated gene expression. ERF1 and ORA59 interact with MED25 and MED25 is required for activation of the PDF1.2 gene by these TFs. ERF1 and ORA59 bind to the ORA59 promoter, providing evidence of a positive feedback loop (shown by a looped arrow) to enhance PDF1.2 expression and other ET- and JA-associated defense genes. MED25 interacts with MYC2 and is required for the correct activation of the VSP1 gene by MYC2. MED25 is also required for the suppression of ORA59, ERF1, and defense genes such as PDF1.2 by MYC2. [See online article for color version of this figure.]
To see whether MED25 may be required for other ERF1-, ORA59-, and MYC2-regulated genes, we compared published microarray data from ERF1 and ORA59 overexpressing lines as well as myc2 and med25 mutant lines. Despite the limitations of comparing across different microarray experiments, a higher than expected number of common genes were identified between the microarray comparisons. In addition, of the ERF1-, ORA59-, and MYC2-induced genes that were found to be common with the med25 array, the majority showed reduced expression in med25. While further experiments are needed to determine whether MED25 interacts directly with ERF1, ORA59, and MYC2 at the promoter regions of these genes, these results suggest that MED25 may be required for the regulation of ERF1-, ORA59-, and MYC2-activated genes. In addition to the MYC2-activated genes, the genes repressed by MYC2 were also reduced in med25. MYC2 is known to repress ERF1- and ORA59-regulated defense genes such as PDF1.2 (Lorenzo et al., 2004). To determine whether the repression of these genes by MYC2 was dependent on MED25, we created a med25 myc2 double mutant and examined the expression of defense genes (PDF1.2, HEL, and CHI-B), as well as the TFs ERF1 and ORA59. As previously reported, the myc2 mutant was found to have increased expression of PDF1.2, HEL, CHI-B, ERF1, and ORA59 relative to the wild type after JA treatment, suggesting that MYC2-mediated repression of JA-associated pathogen defense genes may potentially occur through repression of ERF1 and ORA59 expression (Lorenzo et al., 2004; Dombrecht et al., 2007; Zander et al., 2010). In the med25 myc2 double mutant, the expression of PDF1.2, HEL, CHI-B, ERF1, and ORA59 was significantly attenuated relative to the myc2 single mutant, suggesting that MYC2-mediated repression of JA-associated pathogen defense genes is at least partially dependent on a functional MED25 (Fig. 8).
Lastly, as the ORA59 gene has reduced expression in the med25 mutant, we decided to identify the TFs that bind to the ORA59 promoter to see whether they also interact with MED25. Using Y1H experiments we found that ORA59 as well as other MED25-interacting AP2/ERFs can bind to the ORA59 promoter. This suggests a positive feedback loop for activation of JA responses through ORA59 expression that may be attenuated in the med25 mutant (Fig. 8).
Integrating these conclusions, we propose a model for the role of MED25 in regulating the expression of JA-associated defense genes such as PDF1.2 through ERF1 and ORA59 as well as JA-associated herbivore genes such as VSP1 through MYC2 (Fig. 8). MED25 is also required for the suppression of defense genes by MYC2 and is therefore important in the regulatory cross talk between these two pathways. Finally, MED25 is also important in the amplification of the JA defense pathway through ORA59 expression (Fig. 8). Therefore MED25 plays a critical role in the regulation of the JA pathway for both pathogen and wound and herbivore responses.
The novel finding reported herein that ORA59 interacts with MED25 is particularly noteworthy, given the primary integrative role proposed for this gene in JA and ET signaling (Pré et al., 2008). The expression of ORA59 has been shown to be essential for activation of PDF1.2, as silencing of ORA59 leads to attenuated PDF1.2 expression (Pré et al., 2008). Our data supports this finding with reduced expression of both ORA59 and PDF1.2 expression in the med25 line despite wild-type-like activation of ERF1 expression. As ORA59 but not ERF1 expression was reduced in med25, this suggests that ORA59 is regulated by a different transcriptional pathway that is dependent on MED25. The Y1H experiments aimed to identify the TFs that can bind to the ORA59 promoter and revealed a number of MED25-interacting TFs. However, further work is required to determine whether the MED25-dependent regulation of ORA59 has some connection with its essential role in PDF1.2 expression. Nevertheless, our experiments suggest that the attenuation of PDF1.2 expression in med25 is compounded by at least two factors. First, ORA59 expression levels are attenuated in the med25 mutant, leading to reduced PDF1.2 expression and second, we showed that both ORA59 and ERF1 proteins have a reduced ability to activate PDF1.2 expression in med25 that may further contribute to the reduced level of PDF1.2 expression. Therefore our results reveal an important role for MED25 downstream of ORA59 and ERF1 for the activation of defense gene expression.
As well as the defense-associated AP2/ERFs, we identified the related bHLH TFs, MYC2, MYC3, and MYC4 as MED25-interacting proteins and despite having wild-type levels of MYC2, MYC3, and MYC4 expression in the microarray experiment, genes such as VSP1 that are positively regulated by MYC2, were reduced in med25. The reductions in VSP1 and other MYC2-regulated genes could be due to the reduced ability of MYC2 and potentially also MYC3 and MYC4 proteins, to activate transcription in the med25 mutant (Fig. 8B). However, further experiments need to be performed to ascertain the relative contribution of MYC2, MYC3, and MYC4 in controlling activation as well as repression through MED25.
Currently, 18 TFs have been identified to interact with MED25 and all of the identified TFs have been shown to interact with the MED25 fragment containing the ACID yet lacking the N-terminal vWF-A domain. Based on the results presented here as well as others (Elfving et al., 2011; Ou et al., 2011), the Arabidopsis MED25 ACID domain appears to be an effective interaction domain for a number of TF proteins. The TFs we identified belong to the AP2/ERF, bHLH, MYB, WRKY, bZIP, and zinc-finger TF families, demonstrating that the ACID domain of Arabidopsis MED25 is able to interact with a diverse range of plant TFs and is therefore an important component in MED25’s regulatory function in a range of environmental, biotic, and developmental processes.
Half of the MED25-interacting TFs that have been identified belong to the AP2/ERF family of TFs and two-thirds of the identified AP2/ERFs belong to the group IXc with the remaining three AP2/ERFs belonging to groups IV, VII, and X (Nakano et al., 2006). The identification of group IXc AP2/ERFs as opposed to other AP2/ERF family members suggests that this subgroup may preferentially bind to MED25. One explanation for the identification of group IXc AP2/ERFs as MED25-interacting proteins came with the recent report of the EDLL motif of TDR1/ERF98 (Tiwari et al., 2012). The EDLL motif, otherwise known as CMIX-1, is a conserved motif in the C terminus of group IXc AP2/ERFs (Nakano et al., 2006). Tiwari et al. (2012) recently reported that mutation of the conserved Leu or deletion of the CMIX-1 motif in TDR1/ERF98 removed the transcriptional activation ability of this TF. We demonstrate that deletion of the CMIX-1 motif in ORA59 as well as mutation of the conserved Leu in ERF1, ERF15, and TDR1/ERF98 abolished the interaction with MED25. This suggests a common domain among the group IXc AP2/ERFs that interact with MED25 to recruit the Mediator complex to activate transcription. Interestingly,Tiwari et al. (2012) suggested that the CMIX-1 motif of TDR1/ERF98 possessed a similar pattern of acidic amino acids as the TAD from the potent viral activator VP16. VP16 is able to interact with human MED25 and the NMR analysis of MED25 bound to the VP16 TAD has recently been published (Bontems et al., 2011; Eletsky et al., 2011; Milbradt et al., 2011; Vojnic et al., 2011). While detailed structural studies on the Arabidopsis MED25 have not been performed, further investigation may reveal a common acidic amino acid motif that can interact with MED25 to activate transcription.
Lastly, the interaction of MED25 with DREB2A has recently been characterized (Blomberg et al., 2012). As the CMIX-1 motif is not present in the DREB2A protein, it is likely that a different domain may be utilized for MED25 interaction with DREB2A. Further investigation of the CMIX-1 motif and the identification of activation domains in the other identified TFs may help uncover the ability of MED25 to interact with a diverse range of TF proteins. In addition, further investigation of the other subunits of the Mediator complex and their interaction partners could provide the potential for discovering new links into a range of plant signaling pathways.
CONCLUSION
In conclusion, the results presented in this article reveal a new role for MED25 as an integrative hub that coordinates JA signaling through interaction with multiple TFs. We demonstrate a mechanistic role for MED25 acting downstream of key JA-associated TFs and identify a common interaction domain in a subset of AP2/ERF TFs. Further investigation of the other subunits of the Mediator complex and their interaction partners could provide the potential for discovering new links into a range of plant signaling pathways. In addition future research into the Mediator complex should reveal new insights into the recruitment of the RNA polymerase II transcription apparatus to promoter regions and the transcription of protein-coding genes in eukaryotes.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used as the wild type and the med25-2/pft1.2 (SALK_129555) and the myc2 mutant (SALK_017005) was described previously (Anderson et al., 2004; Kidd et al., 2009). The med25 myc2 mutant was created by pollinating an emasculated myc2 mutant with med25 pollen. Double-mutant plants were confirmed by PCR using the primers obtained using the iSct primers tool (http://signal.salk.edu/cgi-bin/tdnaexpress) for the med25 and myc2 single transferred DNA lines (SALK_129555 and SALK_017005). Arabidopsis plants were grown at 20°C with a 16-h-light/8-h-dark photoperiod and a light intensity of 140 μm photons m−2 s−1. Nicotiana benthamiana plants were grown at 25°C under a 16-h-light/8-h-dark photoperiod. The methyl jasmonate treatments for the RT-qPCR experiment were performed according to the method published by Schenk et al. (2000).
TF Y2H and Y1H Library Screening
The TF library (REGIA + REGULATORS; RR Library) used in this study has been described previously (Castrillo et al., 2011) and is a kind gift from the authors. The TFs in this library had been fused to the GAL4 activation domain (GAL4AD) into pDEST22 (Invitrogen). Two derivatives of MED25 were fused to DNA-binding domain (GAL4BD) in pDEST32 (Invitrogen). pDEST32-MED25ΔACID/Q-rich and pDEST32-MED25ΔvWFA/Q-rich constructs were generated by fusing 1 to 558 amino acids and 228 to 681 amino acids of MED25 protein with GAL4BD, respectively. To carry out Y2H screening of the TF library with two different bait constructs, we first bulked the TF clones in pDEST22 in each 96-well plate. Both bait and prey constructs were then cotransformed into the yeast (Saccharomyces cerevisiae) strain MaV203 (Invitrogen) following manufacturer’s instructions, and the selection of the positive clones were carried out on SD-Leu-Trp-His plates with 12.5 mm 3-AT to prevent autoactivation by MED25. The positive protein-protein interactions were further investigated as described by the ProQuest Y2H system (Invitrogen). For the identification of MED25-interacting domain of ORA59, three fragments of ORA59 were amplified with the primers pGWORA-F1 (5′-AAAAAGCAGGCTTCATGGAATATCAAACTAACTTC-3′) and pGWORA-R1 (5′-CAAGAAAGCTGGGTCTAGGGGAAATTGAGTACTGCG-3′) for ORA591-139; pGWORA-F2 (5′-AAAAAGCAGGCTTCTCATACAGAGGAGTGAGGAAAC-3′) and pGWORA-R2 (5′-CAAGAAAGCTGGGTCTAACTCTGTTTTCTACTTCTTGACG-3′) for ORA5980-214; and pGWORA-F3 (5′-AAAAAGCAGGCTTCGCGGATGTCGTTGAAGAATCTC-3′) and pGWORA-R3 (5′-CAAGAAAGCTGGGTTCAAGAACATGATCTCATAAG-3′) for ORA59139-244 followed by second amplification with pAttB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and pAttB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′), and cloned into the pDONRZeo plasmid (Invitrogen). The entry clones were then recombined with the pDEST22 plasmid. Y2H experiments were carried out as above using pDest32-MED25ΔvWFA/Q-rich construct as bait.
To determine if the conserved Leu residues within ERF1, ERF15, and TDR1/ERF98 proteins are crucial for MED25 interaction, we mutated Leu to Val using QuickChange II site-directed mutagenesis kit (Agilent Technologies) following manufacturer’s recommendations. Y2H experiments expressing either the mutated or native proteins were carried out as above.
For Y1H screen, the TF clones in pDEST22 were individually transformed into the yeast strain AH109 (MATa) and pooled 12 clones to a well in two 96-well plates, in two alternative arrangements giving a total of four 96-well plates. To generate pHISLEU2, the LEU auxotrophy marker was amplified from the pDEST32 vector (Invitrogen) using primers SABR68 (5′-ggggggatcctccggaCAACTGTGGGAATACTCAGGTATCG-3′ contains a 5′ BamHI site followed by a Kpn2I site shown in lowercase) and SABR69 (5′-ccccggatcccctgc aggtcgactctaCTACCCTATGAACATATTCC-3′ includes a 5′ BamHI site followed by a SbfI site shown in lowercase) and the fragment was then ligated into the pHIS2.1 vector (Clontech) after SbfI and Kpn2I digestion replacing the TRP marker. Gateway conversion (Invitrogen) of the resulting vector was then carried out. Two promoter fragments of ORA59 (fragment 1: from −42 to −478; fragment 2: from −348 to −836) were amplified from Arabidopsis (Columbia-0) genomic DNA in a two-step PCR approach using the Gateway compatible specific primers pOra59F1-F (5′-AAAAAAGCAGGCTTC GTGCAATTGATCACTATATTAGTTGAACTG-3′) and pOra59F1-R (5′-CAAGAAAGCTGGGTCGTGTCTAAGTGGCACTAAGTTTGGG-3′) for fragment 1 or pOra59F2-F (5′-AAAAAAGCAGGCTTCCCGCCTTAGTTTCTGACAGAGTTTCGACTC-3′) and pOra59F2-R (5′-CAAGAAAGCTGGGTCGAGTGTATGACGTACGGCGGCGTATTCCCG-3′) for fragment 2, as well as the generic primers pAttB1 and pAttB2, and cloned into the pDONRZeo plasmid (Invitrogen). The mutated fragment2 (mFragment2), which contains mutated nucleotides in the GCC box, was generated using QuickChange II site-directed mutagenesis kit (Agilent Technologies) following manufacturer’s recommendations. The entry clones were then recombined with the pHISLEU2 plasmid. pHISLEU2 vectors with the promoter fragments were then transformed into the yeast strain Y187 (MATα). The pooled library and the bait clones were grown in SD-Trp or SD-Leu media. The pooled library clones were mixed with the bait clones and 3 μL of bait and prey were spotted and overlaid onto yeast peptone dextrose adenine plates. After 1 d of growth at 30°C, diploid cells were replica plated on screening plates (SD-Leu-Trp and SD-Leu-Trp-His ± 3-AT). Following a further overnight incubation at 30°C, plates were replica cleaned to thin out the yeast and grown for a further 3 d before scoring.
In Vitro Protein-Protein Interactions
His-tagged (6× His) MED25ΔVWA/Q-rich (228–681 amino acids) and GFP were cloned into pDEST17 (Invitrogen) via the Gateway cloning system. GST-tagged TFs as well as GFP were generated using the destination vector pDEST15. The constructs were then transformed into Escherichia coli Rosetta II cells (Merc4Biosciences) and proteins expressed at 30°C in the presence of 0.5 mm isopropyl b-d-thiogalactoside. For His pull-down experiments, nickel-nitrilotriacetic acid agarose (Ni-NTA) magnetic beads (Qiagen) were used following the manufacturer’s instructions. E. coli lysate containing His-MED25ΔVWA/Q-rich or His-GFP was incubated with Ni-NTA magnetic beads for 1 h at 4°C. Following one wash with the extraction/interaction buffer, the poly-His proteins immobilized with Ni-NTA magnetic beads were incubated with E. coli lysate containing GST-tagged GFP or TFs for 3 h at 4°C. Following four washes, proteins retained on the magnetic beads were eluted. The eluted proteins and input fractions separated by SDS-PAGE were analyzed by immunoblotting with anti-His (Invitrogen) or anti-GST (Sigma-Aldrich) antibodies.
Transcriptional Activation Assays
Full-length MYC2, ORA59, and ERF1 were used to construct effector plasmids by fusing with the yeast GAL4 DNA-binding domain (GAL4DB)-coding region under the control of the cauliflower mosaic virus 35S promoter into the Gateway (GWC_RfB; Invitrogen) converted pJG vector (pGAL4DBGW; Gonzalez et al., 2007; a kind gift from S. Conlan). pGAL4UAS:GUS was used as the reporter plasmid. To normalize reporter gene activity, we used p2X35S:fLUC as control. The effector, reporter, and reference plasmids (3, 3, and 2 μg, respectively) were delivered into rosette leaves of 5- to 6-week-old Arabidopsis plants by particle bombardment using the Bio-Rad PDS-1000/He machine (Bio-Rad). Cobombardment assays were performed as described previously (Rehmany et al., 2005). Three replicates (two to three leaves for each replicate) were used for each construct/genotype transformation experiment and each experiment was repeated at least twice. After bombardment, the samples were incubated on one-quarter-strength Murashige and Skoog medium at 22°C for 24 h, and then quantified for luciferase and GUS activities. Luciferase assay system (Promega) was used to quantify luciferase activity according to the manufacturer’s instructions. Fluorometric assays of GUS activity were performed as described previously (Jefferson et al., 1987). Luminescence and fluorescence were determined with GENios microplate reader (Tecan).
For transient expression assays involving the PDF1.2 and VSP1 promoter fragments, full-length MYC2, ORA59, and ERF1 were cloned into the plant expression vector pEarleyGate100 (Earley et al., 2006) using Gateway recombination system (Invitrogen). The 1,187-bp promoter fragment of the PDF1.2 gene (Zarei et al., 2011) and 1,860-bp VSP1 promoter fragment were amplified with Gateway compatible primers and cloned into pDONRZeo (Invitrogen) and then recombined with the binary vector pBGWFS7 (Karimi et al., 2002). The transient expression assays by particle bombardment were essentially carried out same as above except that the amount of the reporter plasmid used was 5 μg.
BiFC Experiments
For the BiFC experiments, YFPC:Med25ΔQ-rich and YFPC:Med25ΔACID/Q-rich constructs were generated by fusing 1 to 681 amino acids and 1 to 558 amino acids of MED25 protein with the binary vector pBiFP-3 (Azimzadeh et al., 2008), respectively. YFPN:Ora59, YFPN:ORA591-139, YFPN:ORA5980-214, and YFPN:ORA59139-244 were generated by fusing the full length or 1 to 139, 80 to 214, and 139 to 244 amino acids of ORA59 with the binary plasmid pBiFP-2 (Azimzadeh et al., 2008). We also coexpressed red fluorescent protein (mRFP), which accumulates in the cytoplasm and nucleus, and provides a positive transformation control. For this, we cloned mRFP into the binary plasmid pEarleyGate 100 (Earley et al., 2006) to generate 35S:mRFP. The constructs were then transformed into Agrobacterium tumefaciens strain GV3101 (pMP90). A. tumefaciens-mediated transient expression experiments were performed on 4- to 5-week-old N. benthamiana plants. A. tumefaciens strains grown in Luria broth were harvested by centrifugation and resuspended in infiltration medium (10 mm MES pH 5.6; 10 mm MgCl2; 150 μm acetosyringone) to a final OD600 of 0.5 and incubated at room temperature for 3 h. For all transient coexpression assays, A. tumefaciens strains carrying the constructs were mixed in a 1:1:1 ratio in infiltration medium to a final OD600 of 0.2 and infiltrated into N. benthamiana leaves using a needleless syringe. Two days after infiltration, the images were obtained using an LSM 710 laser scanning microscope (Carl Zeiss). Images were processed with Zen software (Carl Zeiss) and ImageJ (Abramoff et al., 2004).
Microarray Comparisons
The med25 array was analyzed as previously reported (Kidd et al., 2009). The genes differentially regulated by med25 were determined using a two-way ANOVA for either genotype or treatment using GeneSpring (Agilent Technologies) and the gene list obtained based on the effect of genotype was used for comparisons. To make the array more comparable to the myc2 gene list the Benjamini and Hochberg false discovery rate was removed and only genes that passed Student’s t test (P < 0.05) were compiled and compared with those genes found to be significantly different in the 35S:ERF1 (Lorenzo et al., 2003), XVE:ORA59 (Pré et al., 2008), and myc2 (Dombrecht et al., 2007) arrays using their published gene lists. Common genes between med25 and the TF gene lists were identified using either FiRe (Garcion et al., 2006) or manually using the VLOOKUP function in Microsoft Excel 2007 (Microsoft Corporation). The common genes were then sorted based on their fold-change expression in the TF array and color coded depending on whether they were induced or repressed. The statistical analysis on the common genes was performed using the hypergeometric distribution in R (R Development Core Team, 2011). This analysis calculates the likelihood of observing the number of common genes by chance given the size of the two gene lists and the total number of common genes from which the gene lists were derived.
RT-qPCR Analyses
RT-qPCR experiments were done as described previously using the 7900HT real-time PCR system (Applied Biosystems) in conjunction with SYBR Green fluorescence to detect transcript levels (McGrath et al., 2005; Kidd et al., 2009). Absolute gene expression levels relative to the previously validated reference genes β-ACTIN2, β-ACTIN7, and β-ACTIN8 were used for each complementary DNA sample using the equation: relative abundance = [Egene^(−Ct gene)]/[EACTIN^(−Ct ACTIN)]. The mean expression range of the reference gene was found to be within ±1 Ct across all experiments. The sequences of the primer pairs have been previously published (Anderson et al., 2004; McGrath et al., 2005).
Sequence data from this article can be found in the Arabidopsis Genome Initiative data libraries under the following accession numbers: MED25 (AT1G25540); MYC2 (AT1G32640); ERF1 (AT3G23240); ERF4 (AT3G15210); WRKY10 (AT1G55600); MYB104 (AT2G26950); ERF15 (AT2G31230); bZIP (AT2G31370); TDR1 (AT3G23230); MYC3 (AT5G46760); MYC4 (AT4G17880); B-Box (AT4G39070); DREB2A (AT5G05410); PDF1.2 (AT5G44420); and VSP1 (AT5G24780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Pull-down experiment using GST-tagged TF proteins with His-GFP protein as an additional control.
Supplemental Figure S2. Identification of MED25-interacting domain of ORA59 in yeast.
Supplemental Figure S3. Detection of MED25-ORA59 interactions via transient coexpression of YFPC:MED25ΔQ-rich and YFPN:ORA59, YFPN:ORA591-139, YFPN:ORA5980-214, or YFPN:ORA59139-244 constructs as well as YFPC:MED25ΔACID/Q-rich with YFPN:ORA59 or YFPN:ORA59139-244 constructs.
Supplemental Figure S4. Quantitative assays of β-galactosidase activity reveal a reduction in LacZ activity in the interactions of MED25ΔvWF-A/Q-rich fragment with mutant forms of TDR1, ERF1, and ERF15.
Supplemental Figure S5. Cotransformation of wild-type and med25 plants with 35S:GUS and 35S:LUC constructs using particle bombardment method shows no significant difference between wild-type and med25 plants for transformation efficiencies.
Supplemental Figure S6. The expression of ORA59, ERF1, MYC2, MYC3, and MYC4 is unchanged in med25 relative to the wild type.
Supplemental Table S1. The med25 gene list used for the TF microarray comparisons.
Supplemental Table S2. The genes that were common between the 35S:ERF1 microarray gene list (Lorenzo et al., 2003) and the med25 microarray gene list (Kidd et al., 2009; Supplemental Table S1).
Supplemental Table S3. The genes that were common between the XVE:ORA59 microarray gene list (Pré et al., 2008) and the med25 microarray gene list (Kidd et al., 2009; Supplemental Table S1).
Supplemental Table S4. The genes that were common between the myc2 microarray gene list (Dombrecht et al., 2007) and the med25 microarray gene list (Kidd et al., 2009; Supplemental Table S1).
Supplemental Table S5. The average relative expression of defense-associated genes in the med25, myc2, and med25 myc2 mutants with or without JA treatment using RT-qPCR.
Supplemental Table S6. The list of AP2/ERFs that were identified using two different ORA59 promoter sequences in Y1H analyses along with their groups according to Nakano et al. (2006).
Supplementary Material
Acknowledgments
We wish to thank Franziska Turck (Max Planck Institute, Cologne) for providing the TF library.
Glossary
- JA
jasmonate
- TFs
transcription factors
- ET
ethylene
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescent complementation
- TAD
transcriptional activation domain
- RT-qPCR
real-time quantitative PCR
- Y1H
yeast-one-hybrid
- 3-AT
3-amino-1,2,4-triazole
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- bHLH
basic helix-loop-helix
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophot Internat 11: 36–42 [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J. (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Nacry P, Christodoulidou A, Drevensek S, Camilleri C, Amiour N, Parcy F, Pastuglia M, Bouchez D. (2008) Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20: 2146–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Blomberg J, Aguilar X, Brännström K, Rautio L, Olofsson A, Wittung-Stafshede P, Björklund S. (2012) Interactions between DNA, transcriptional regulator Dreb2a and the Med25 mediator subunit from Arabidopsis thaliana involve conformational changes. Nucleic Acids Res 40: 5938–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontems F, Verger A, Dewitte F, Lens Z, Baert JL, Ferreira E, de Launoit Y, Sizun C, Guittet E, Villeret V, et al. (2011) NMR structure of the human Mediator MED25 ACID domain. J Struct Biol 174: 245–251 [DOI] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S. (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce JM, Knight H, Deyholos M, Openshaw MR, Galbraith DW, Warren G, Knight MR. (2003) The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J 34: 395–406 [DOI] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. (2003) A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 132: 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, Coupland G, Paz-Ares J, Oñate-Sánchez L. (2011) Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS ONE 6: e21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. (2011) The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG. (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58: 497–513 [DOI] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T. (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobi KC, Winston F. (2007) Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol Cell Biol 27: 5575–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Eletsky A, Ruyechan WT, Xiao R, Acton TB, Montelione GT, Szyperski T. (2011) Solution NMR structure of MED25(391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25. J Struct Funct Genomics 12: 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N, Davoine C, Benlloch R, Blomberg J, Brännström K, Müller D, Nilsson A, Ulfstedt M, Ronne H, Wingsle G, et al. (2011) The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc Natl Acad Sci USA 108: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion C, Baltensperger R, Fournier T, Pasquier J, Schnetzer MA, Gabriel JP, Métraux JP. (2006) FiRe and microarrays: a fast answer to burning questions. Trends Plant Sci 11: 320–322 [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Park MY, Smith MR, Pepitone R, Kerstetter RA, Poethig RS. (2010) The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. (2007) The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol 27: 5306–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7: 465–471 [DOI] [PubMed] [Google Scholar]
- He YH, Fukushige H, Hildebrand DF, Gan SS. (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA. (2010) Ubiquitin ligase-coupled receptors extend their reach to jasmonate. Plant Physiol 154: 471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iñigo S, Alvarez MJ, Strasser B, Califano A, Cerdan PD. (2011) PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J 69: 601–612 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17: 22–31 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Cahill DM, Manners JM, Schenk PM, Kazan K. (2011) Diverse roles of the Mediator complex in plants. Semin Cell Dev Biol 22: 741–748 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners JM, Penninckx IA, Vermaere K, Kazan K, Brown RL, Morgan A, Maclean DJ, Curtis MD, Cammue BPA, Broekaert WF. (1998) The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol 38: 1071–1080 [DOI] [PubMed] [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK. (2011) The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol 157: 1609–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbradt AG, Kulkarni M, Yi T, Takeuchi K, Sun ZY, Luna RE, Selenko P, Näär AM, Wagner G. (2011) Structure of the VP16 transactivator target in the Mediator. Nat Struct Mol Biol 18: 410–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G, Stühler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. (2003) A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J 22: 6494–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. (2011) Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou B, Yin KQ, Liu SN, Yang Y, Gu T, Wing Hui JM, Zhang L, Miao J, Kondou Y, Matsui M, et al. (2011) A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol Plant 4: 546–555 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Goossens A. (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF. (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. (2011) The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
- Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, Whisson SC, Kamoun S, Tyler BM, Birch PR, Beynon JL. (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17: 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. (2011) The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Manners JM, Kazan K. (2009) Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J 58: 927–939 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu GH, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Belachew A, Ma SF, Young M, Ade J, Shen Y, Marion CM, Holtan HE, Bailey A, Stone JK, et al. (2012) The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J 70: 855–865 [DOI] [PubMed] [Google Scholar]
- Vojnic E, Mourão A, Seizl M, Simon B, Wenzeck L, Larivière L, Baumli S, Baumgart K, Meisterernst M, Sattler M, et al. (2011) Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat Struct Mol Biol 18: 404–409 [DOI] [PubMed] [Google Scholar]
- Wathugala DL, Hemsley PA, Moffat CS, Cremelie P, Knight MR, Knight H. (2012) The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol 195: 217–230 [DOI] [PubMed] [Google Scholar]
- Wollenberg AC, Strasser B, Cerdán PD, Amasino RM. (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol 148: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Li Y. (2011) Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 4545–4554 [DOI] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, DeBeaumont R, Zhou S, Näär AM. (2004) The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci USA 101: 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M, La Camera S, Lamotte O, Métraux JP, Gatz C. (2010) Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J 61: 200–210 [DOI] [PubMed] [Google Scholar]
- Zarei A, Körbes AP, Younessi P, Montiel G, Champion A, Memelink J. (2011) Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol 75: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim JM, To TK, Li W, et al. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.