Ubiquitin ligation to other proteins modulates the activity, longevity, and/or localization of the target proteins in eukaryotic systems. As components of the ubiquitin pathway, plant hormone receptors determine the abundance of key transcriptional regulators of auxin, GA, and jasmonate response pathways (for review, see Kelley and Estelle, 2012). Ubiquitin has also been shown to alter the abundance, function, and localization of membrane- and endoplasmic reticulum (ER)-resident proteins. Other hormone signaling cascades, including the ethylene and brassinosteroid pathways, depend on the regulated proteolysis of membrane-associated receptor-like protein kinases. Significantly, the consequences of ubiquitin modification in some cases are independent of the proteasome. An appropriate general term, inclusive of all processes involving ubiquitin, is the ubiquitin modification system (UMS). This Update, in addition to reviewing general aspects of the UMS, specifically focuses on the roles of ubiquitin in the plant endomembrane system.
UBIQUITINATION: A VERSATILE POSTTRANSLATIONAL MODIFICATION
Ubiquitin is a highly conserved 76-amino acid protein whose posttranslational covalent ligation to other proteins, including itself, serves myriad physiological functions. It was shown to be an essential protein in Saccharomyces cerevisiae, as deletion of all ubiquitin-encoding genes resulted in complete loss of viability (Finley et al., 1994). The breadth and depth of ubiquitin’s known physiological roles suggest that it is also essential in plants, although this has not been directly demonstrated. Initially, ubiquitin was identified in two independent areas of investigation: as a single ubiquitin covalently attached to Lys-119 of histone 2A in mammals (Böhm et al., 1980), and as a required component for the in vitro degradation of model proteins in a reticulocyte extract (for review, see Wilkinson, 2005; Ciechanover, 2009). Our knowledge of the in vivo functions of ubiquitin have greatly expanded subsequently to include the regulated proteolysis of transcription factors; internalization of plasma membrane (PM)-resident transporters and receptors; clearance of misfolded ER-localized proteins via endoplasmic reticulum-associated degradation (ERAD); and cell cycle progression, among others (Smalle and Vierstra, 2004; Deshaies and Joazeiro, 2009).
THE UMS
Ubiquitin alters the longevity, localization, and/or activity of proteins via one or more covalent bonds between the C-terminal carboxyl group of ubiquitin and a nucleophilic moiety on the substrate protein (referred to here as the “target”). Canonically, ligation occurs at ɛ-amino groups of target lysyl residues or at the target’s N terminus. More recently, Ser, Thr, and Cys esters at the C terminus of ubiquitin have been reported in yeast (Saccharomyces cerevisiae) and mammalian cells (for review, see Wang et al., 2012), although the in vivo abundance of these linkages is still unclear. For some known ubiquitination targets, substitutions of Lys residues with nonconjugatable Arg residues result in reduced degradation, implicating Lys residues as the primary modification sites. On the other hand, N-terminal or Ser/Thr ubiquitination is made more likely when substitutions of all Lys residues have a minimal effect.
Ubiquitin can also be polymerized, as its Lys residues serve as acceptor sites for additional ubiquitin molecules. A global proteomic study using mammalian cells found that while Lys-48 ubiquitin-ubiquitin chains are the most abundant, all seven ubiquitin Lys linkages were present (Kim et al., 2011). Similar global analyses in Arabidopsis (Arabidopsis thaliana) yield the same relative ubiquitin chain abundance pattern: Lys-48 chains are most abundant, followed by Lys-63, Lys-11, and then Lys-33 and Lys-29 in much lower abundance (Saracco et al., 2009). Linear polyubiquitin called M1 chains (in which the C terminus of one ubiquitin is attached to the N terminus of the next) are also possible, and in mammals, M1 chains play a role in the activation of Nuclear factor kappa-light-chain-enhancer of activated B cells, a transcription factor activated in response to a number of signals including extracellular cytokines (Emmerich et al., 2011). Site-specific substitution of ubiquitin Lys residues uncovered the role of Lys-63 chains in DNA repair and kinase activation in mammalian cells (Peng et al., 2003). Lys-63 chains are also implicated in vacuolar targeting (for review, see Lauwers et al., 2010). In vivo roles of these linkages remain to be defined in plants, as there are only a few intriguing results: mutations in RING Domain Ligase1-2 (RGLG1-2), which forms Lys-63 chains, suppress apical dominance in Arabidopsis (Yin et al., 2007), and recently, Lys-63 chains were found to play a role in PIN-FORMED2 (PIN2) degradation (Leitner et al., 2012).
Attachment of ubiquitin to target proteins, including ubiquitin itself to form chains, typically requires a cascade of three enzymes: E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme (UBC); and E3, the ubiquitin ligase. The cascade can be thought of as a protein pyramid. For example, in Arabidopsis, there are two highly related E1s, approximately 37 predicted E2s, and more than 1,000 predicted E3s (Smalle and Vierstra, 2004). While plant-specific gene duplication events surely contribute to this diversity, the pyramidal structure is present in yeast and mammals as well. However, it should be borne in mind that, to date, only a subset of predicted plant E2s and E3s have been demonstrated to have the predicted activity.
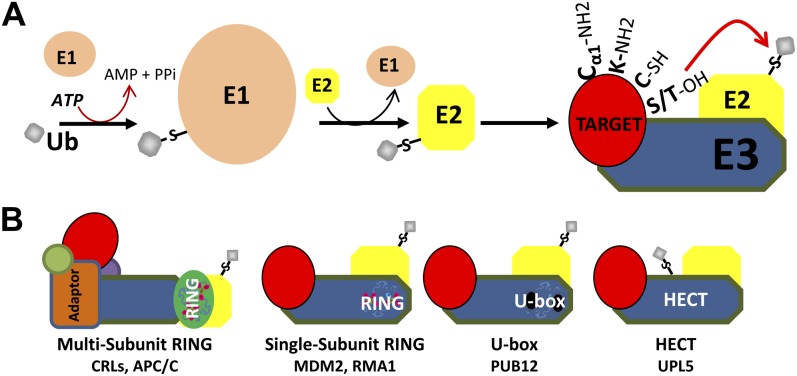
E1 and E2 function sequentially, with E1 catalyzing activation of the C-terminal carboxyl group via adenylylation and subsequent thioester formation (Fig. 1) and then transferring the ubiquityl group to an E2 Cys, denoted as Ub∼E2. A subset of E3s called the HECT type (for homology to E6-AP C terminus) also carry thioester-linked ubiquitin prior to transfer to target proteins, but the majority of E3s catalyze ubiquitin transfer by serving as scaffolds for the target and the Ub∼E2, and allosterically activating ubiquitin transfer. Named for the E2-interacting domain shared among their respective family members, these scaffold ligases fall into two very general classes: U-box (in plants, termed plant U-box) and RING (for really interesting new gene) E3s (Yee and Goring, 2009). A hybrid of HECT and RING types has been reported in the RING-between-RING proteins. RING-between-RING proteins contain a functional RING domain, which interacts with the Ub∼E2, and additionally resemble the HECT-type E3s, with a Cys acceptor for activated ubiquitin prior to transfer to the target (Wenzel and Klevit, 2012). RING proteins are also found as subunits in multisubunit E3s, such as the well-characterized SCF-type complex (Hua and Vierstra, 2011; for review, see Kelley and Estelle, 2012) and the anaphase-promoting complex (Peters, 2006). Examples exist of targets with multiple E3s (Popov et al., 2010) and E3s with relaxed target specificities (Kostova et al., 2007). Adding another layer of complexity, some E3s (in conjunction with particular E2s) even specify different linkage types to certain targets (for review, see Ikeda and Dikic, 2008). The organization of E3s, particularly the discernment of accessory proteins, remains an active area of investigation.
Figure 1.
The ubiquitination enzyme cascade. A, The modification of eukaryotic intracellular proteins with the small protein ubiquitin (Ub; gray sphere) depends on a three-enzyme cascade. E1 (tan oval) activates ubiquitin at the expense of two ATP equivalents by forming a high-energy thioester bond between the E1 active-site Cys and the C terminus of ubiquitin. Ubiquitin is then passed to an active-site Cys on E2 (yellow octagon). Lastly, the Ub∼E3 thioester is brought into proximity with the desired protein target (red oval) by a scaffold protein called E3 (blue). Ubiquitin can be added to a variety of nucleophilic groups on the target protein, including the N terminus (Cα1-NH2), Lys residues (K-NH2), Ser and Thr residues (S/T-OH), and Cys residues (C-SH). B, Types of E3s. Multisubunit RING E3s, such as CULLIN RING Ligases (CRLs), recognize substrates with adaptor proteins (i.e. F-box proteins). Single-subunit RINGs like MDM2 consist of one polypeptide with binding sites for the E2 (the RING domain) and the target protein. The U-box is a modified RING domain lacking zinc-binding sites. HECT-type E3s form a Ub∼E3 thioester prior to ubiquitin being transferred to the target. APC, Anaphase-promoting complex.
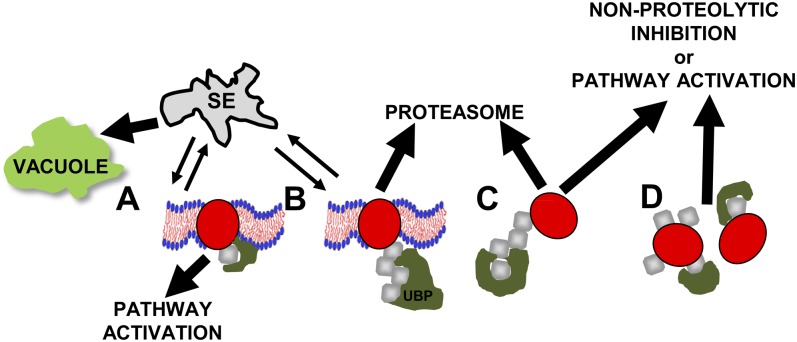
In addition to families of E2 and E3 enzymes, eukaryotes express proteins with ubiquitin-deconjugating and ubiquitin-binding abilities. The former serve to remove ubiquitin (D’Andrea and Pellman, 1998), while the latter interact with different types of ubiquitinated proteins (Andersen et al., 2005). Ubiquitin-binding proteins can mediate interaction between ubiquitin-modified targets and the proteasome (Fu et al., 2010), shuttle ubiquitinated proteins from one compartment to another, or serve to modulate the activities of others in the same complex (Hicke and Dunn, 2003). Alternative fates of ubiquitinated targets include proteolysis in the lytic compartment of the vacuole or the central cavity of the proteasome, nonproteolytic inhibition, and even protein activation (Fig. 2).
Figure 2.
Fates of ubiquitinated proteins. The fate of a ubiquitinated target protein, typically mediated by ubiquitin-binding proteins (UBP; dark green), depends on the number of ubiquitin moieties attached to the substrate, the type of ubiquitin-ubiquitin linkage, and whether the substrate protein is membrane associated. A, Monoubiquitinated integral membrane proteins are internalized, leading to activation or vacuolar degradation upon exit from the sorting endosome (SE). B, Polyubiquitinated integral membrane proteins can be degraded either by vacuoles or, via ERAD, the proteasome. C, Polyubiquitinated soluble proteins, in which ubiquitin moieties are linked via Lys-48, are proteasomally degraded; in contrast, Lys-63-linked substrates tend not to be proteolyzed but have been shown to play roles in DNA repair. D, Monoubiquitination and multiubiquitination of soluble proteins can bring about activation or inhibition of a protein’s activity.
The complexity of the UMS presents a challenge to studying the consequences of ubiquitination. In the simple case of a cytosolic protein with a short half-life, the compounds MG132 (a proteasome inhibitor with some activity against Cys proteases [Woffenden et al., 1998]) and cycloheximide (CHX; an inhibitor of protein synthesis) will lead to the hyperaccumulation and rapid disappearance of the target, respectively. This simple reciprocal response will not be observed if, for instance, the target’s E3 turns over faster than the target, if the treatment is prolonged (proteasome inhibition artificially depletes the free ubiquitin pool, which in turn affects proteolysis and endocytosis), or if the target’s degradation depends on new protein synthesis.
What do we know about the sequence specificity of ubiquitin attachment? Proteomic surveys in yeast and cultured mammalian cells have failed to detect a specific sequence around the ubiquitin Lys attachment site, although there is a slight enrichment of basic amino acids, Ala, and Gly one to six residues N terminal to the given Lys. Overall, accessibility of the lysyl amino group seems to be a key factor in determining whether ubiquitination will take place (Xu et al., 2010).
The presence of membrane-localized E2 and E3 enzymes suggests that ubiquitination occurs near membrane surfaces. Following a general description of the biogenesis of membrane proteins, we discuss the role of ubiquitin in ER protein quality control and in regulating membrane protein abundance.
MEMBRANE-RESIDENT SOLUBLE AND INTEGRAL PROTEINS ARE COTRANSLATIONALLY PROCESSED IN THE ER
In eukaryotes, the nuclear envelope, ER, Golgi, PM, and lysosomes form a continuum of lipid bilayer-enclosed compartments known collectively as the endomembrane system. Lumenal and/or integral membrane proteins follow a discrete pathway from the ER to their final destinations. Prior to entering the ER, a stretch of approximately 20 to 30 amino acids at the N terminus of the nascent protein is cotranslationally recognized by the signal recognition particle, which relocates the translating ribosome to the Secretion61 translocon on the ER, thus funneling the new protein into the ER lumen (for review, see von Heijne, 2007). Once inside the ER, secreted and membrane proteins are subjected to chaperone-assisted folding, protein disulfide isomerase-mediated disulfide synthesis, and N-glycosylation (for review, see Kleizen and Braakman, 2004). In yeast, nascent polypeptides are modified via N-glycosylation and subsequent glycan trimming, followed by association with the chaperone calnexin (Cnx), if soluble, or calreticulin (Crt), if membrane associated. Cnx-Crt recruits protein disulfide isomerase and binding immunoglobulin protein, a chaperone that aids in the folding of a nascent protein’s nonglycosylated domains. Properly folded proteins are then exported to the Golgi. Improperly folded proteins are recognized by UDP-Glc:glycoprotein glucosyltransferase, which reglucosylates them, thereby enabling additional rounds of Cnx-Crt-driven folding (for review, see Hong and Li, 2012). If a protein is properly folded, it is exported to the cis-Golgi, where it is further processed via modifications such as O-glycosylation, phosphorylation, and assembly into multisubunit complexes before being packaged into exocytic vesicles that merge with the PM (for review, see Reynders et al., 2011).
ERAD: WHEN MATTERS UNFOLD
As described above, the ER is equipped with multiple mechanisms to ensure that proteins reach their native conformations. However, a stochastic fraction of proteins fail to achieve this state, and these become substrates for ERAD. Codiscovered in yeast and mammalian cells, the ERAD pathway is a ubiquitin-mediated process by which misfolded or damaged transmembrane and lumenal ER proteins are retrotranslocated into the cytosol and degraded by the proteasome (for review, see Meusser et al., 2005). There are two multipass transmembrane E3 ligases with cytosolic RING domains known to participate in ERAD: HMG-CoA reductase degradation1 (Hrd1), which primarily recognizes transmembrane and intraluminal target domains, and the Degradation of α2-10 (Doa10) complex, which recognizes target cytosolic domains (Kostova et al., 2007). Hrd1 requires an ensemble of accessory proteins to aid in target recruitment and recognition (notably the membrane-spanning Hrd1 adaptor Hrd3 and the soluble ER-resident lectin yeast osteosarcoma9) and preferentially interacts with the membrane E2s Ubc7 and Ubc1 (Hirsch et al., 2009). Doa10 relies on the E2s Ubc6 and Ubc7 as well as the ATPase Cell division control48 (Cdc48) and the Ubc7 cofactor Cue1 (Ravid et al., 2006). Defectively folded proteins interact with these E3s via hydrophobically exposed regions and glycan moieties that are added prior to E3-target binding (Hong and Li, 2012).
While the ERAD machinery is stimulated by stresses that induce an unfolded protein response (Tsai and Weissman, 2010), it has been shown to regulate basal physiological processes as well. When sterol concentrations are high, ER-localized HMG-CoA reductase, which catalyzes the rate-limiting step in sterol biosynthesis, is proteasomally degraded following ubiquitination by the E3 Hrd1 (Bays et al., 2001; Sever et al., 2003).
There is also some precedent for protein quality control pathways that occur at the Golgi and the PM (for review, see Arvan et al., 2002). Okiyoneda et al. (2010) demonstrated that a pathological mutant form of the cystic fibrosis transmembrane conductance regulator (CFTR; ΔF508CFTR), which is normally degraded by ERAD, could localize at the PM at a permissive temperature. At a nonpermissive temperature, PM-localized ΔF508CFTR was rapidly internalized and lysosomally proteolyzed in a ubiquitin-dependent manner, indicating that a quality control mechanism operates at the PM.
ERAD IN PLANTS
In silico methods have uncovered putative plant homologs of yeast and mammalian ERAD components, including the E3s Hrd1 and Doa10, the E2s Ubc6 and Ubc7, and accessory proteins such as the ATPase Cdc48 and the chaperone Hrd3 (Müller et al., 2005). Insightful functional data on plant ERAD came from studies of missense mutations in the receptor-like kinase BRASSINOSTEROID INSENSITIVE1 (BRI1), the brassinosteroid (BR) receptor (Hong et al., 2008). The altered BRI1 proteins, bri1-5 and bri1-9, have increased ER retention and degradation via interaction with plant ERAD pathway components (Hong et al., 2008), and the BR-insensitive bri1-5 and bri1-9 mutant phenotypes were partially rescued in bri1-5 hrd3a-1 and bri1-9 hrd3a-1 double mutants (Liu et al., 2011). Simultaneous knockout of both Arabidopsis Hrd1 orthologs in the bri1-9 background also partially suppressed the BR-insensitive phenotype of soil-grown plants (Su et al., 2011), suggesting that degradation of BRI1 mutants depends on a HRD1/HRD3A system in plants. In another investigation, a T-DNA knockout of the Arabidopsis E2 UBC32 (most closely related to yeast Ubc6) was found to partially suppress several aspects of the bri1-9 phenotype (Cui et al., 2012). Considering that the UBC32 promoter is highly induced by reactive oxygen species-producing abiotic stresses, it appears that UBC32, like Ubc6 in yeast, plays a role in the ERAD response.
Additional functional evidence for plant ERAD comes through salinization stress experiments. Liu et al. (2007) found that Arabidopsis T-DNA homozygotes for a putative homolog of Site1 Protease (S1P), a Golgi-localized Ser protease required for mounting a robust unfolded protein response in mammals, were hypersensitive to various monovalent salts. Yellow fluorescent protein-S1P localized to Golgi bodies and facilitated the cleavage of an ER-localized bZIP transcription factor with homology to a mammalian ERAD transit amplifier. T-DNA knockout of an ER-localized zinc transporter led to salt hypersensitivity and repressed the expression of salt-inducible UPR marker genes (Wang et al., 2010). Recently, Liu et al. (2011) showed that only HRD3A (not the homolog HRD3B) was required in Arabidopsis for basal tolerance to the ER stress inducer tunicamycin, which disrupts ER protein glycosylation. Furthermore, hrd3a mutants were NaCl hypersensitive, exhibiting higher induction of ER chaperones and diminished greening on agar plates.
THE ER-LOCALIZED ETHYLENE RECEPTOR IS REGULATED BY UBIQUITINATION
Recent progress has been made in understanding the ubiquitin-dependent regulation of ethylene receptors in plants. The hormone ethylene plays diverse roles in plant biology, including seed dormancy breaking and fruit maturation (for review, see Klee, 2004). In Arabidopsis, ethylene is perceived by a receptor kinase family (Moussatche and Klee, 2004). Focusing on a single paralog, Chen et al. (2007) determined that the ER-resident ethylene receptor ETR2 is degraded in the presence of high concentrations of ethylene, whereas low to moderate ethylene concentrations lead to enhanced protein accumulation. Ethylene-induced degradation could be blocked with MG132 and the protease inhibitor ALLN. In contrast, ETR2 was proteolyzed in the presence of CHX, making it reasonable to postulate that the ethylene-induced reduction in ETR2 protein levels occurs via the proteasome through the ERAD pathway.
NONPROTEASOMAL FATES FOR UBIQUITINATED MEMBRANE PROTEINS
While polyubiquitination via Lys-48 ubiquitin chains typically leads to proteasome-dependent degradation of cytosolic proteins and ER-retained proteins are degraded in a proteasome-dependent fashion via ERAD, PM-resident receptors and transporters tend to be vacuolar protease substrates following internalization and sorting into multivesicular bodies (MVBs). MVB assembly and maturation depend on the ESCRT (for endosomal sorting complex required for transport) assemblies (Katzmann et al., 2001; Babst et al., 2002), which are complexes that recognize proteins destined for the vacuole by binding ubiquitin (Shields et al., 2009). The emerging picture in yeast and mammals is that while monoubiquitination is usually sufficient for internalization, multiubiquitination and Lys-63 polyubiquitination enhance endocytic rates, and several PM-resident transmembrane proteins have been shown to require Lys-63 polyubiquitination for ESCRT-dependent sorting into MVBs (for review, see Lauwers et al., 2010). Evidence for PIN2 regulation in plants is consistent with this model (see below), but whether this general principle applies in plants has yet to be determined.
The yeast Sterile2 G-protein-coupled receptor, which activates a pheromone-sensing pathway, is ubiquitinated and degraded in the vacuole upon ligand binding (Hicke and Riezman, 1996). In mammals, the ubiquitin ligase Casitas B-Lineage ubiquitinates the epidermal growth factor receptor ERBB1 (Levkowitz et al., 1999), mediating its endocytosis and lysosomal degradation (Soubeyran et al., 2002). A recent report by Herberth et al. (2012) suggests that ubiquitin may be a signal for both endocytosis and MVB-to-vacuole sorting in plants: C-terminal fusions of ubiquitin to several PM and MVB-resident proteins led to their vacuolar localization. Mammalian Mahogunin Ring Finger1 down-regulates the physiological output of the melanocortin-1 receptor via competition with the effector G-protein Gαs (Pérez-Oliva et al., 2009). This is a particularly interesting example of E3 target protein inhibition via nonproteolytic means.
Leitner and colleagues (2012) very recently determined that the auxin efflux protein PIN2 is spatially regulated by Lys-63 polyubiquitination in vivo. Substitution of any one Lys to Arg in a conserved hydrophilic loop of the PIN2 cytosolic domain had no discernible effects, but mutation of a 12-Lys cluster caused PIN2 to become equally distributed in the basolateral and apical domains of root PMs, and PIN212KR failed to rescue gravitropism defects of the eir1-4 loss-of-function allele of PIN2. Intriguingly, the in-frame fusion of ubiquitin to the PIN2 cytosolic loop caused constitutive internalization and diminished steady-state levels of PIN2, effects that could be blocked by MG132. While the authors did not determine how PIN2 accumulates exclusively at the apical PM, its distribution and abundance are clearly modulated by ubiquitination.
DOWN-REGULATION OF RECEPTOR-LIKE KINASES BY UBIQUITINATION
Like mammals, higher plants are equipped with pattern recognition receptors that allow an infected host to mount a defense against pathogens. FLAGELLIN SENSING2 (FLS2), a PM-localized receptor-like kinase conserved in plants, recognizes the bacterial filamentous protein flagellin and, together with BRI1-ASSOCIATED KINASE1 (BAK1), initiates an innate immune response upon flagellin binding (Gómez-Gómez and Boller, 2000; Boller and Felix, 2009). Since acquired pathogen resistance often involves programmed cell death, immune responses are attenuated to preserve the life of the host. Accordingly, FLS2 is internalized upon exposure to flagellin (Robatzek et al., 2006). Performing a yeast two-hybrid screen with the BAK1 kinase domain, Lu et al. (2011) uncovered PLANT U-BOX13 (PUB13). BAK1 phosphorylation of PUB13 enables it to associate with FLS2 in the presence of flagellin, and PUB13 ubiquitinates FLS2 (but not BAK1), suggesting that the PUB13-BAK1 relationship is not ligase to target. Immune response genes are hyperinduced in pub13 loss-of-function backgrounds when exposed to pathogenic bacteria, indicating that PUB13 dampens FLS2-dependent innate immune responses by counteracting FLS2 accumulation. Thus, the FLS2-BAK1-PUB13 axis is an example of a membrane receptor system regulated by endocytosis, ubiquitination, and uniquely E3 (rather than target) phosphorylation.
Flagellin-mediated FLS2 is not the only example of a bacterial pattern recognition receptor regulated by ubiquitination. The abundance of SYMBIOSIS RECEPTOR-LIKE KINASE (SYMRK), a receptor-like kinase necessary for fungal and bacterial symbioses in legumes and nonlegumes, is controlled by the E3 ligase SEVEN IN ABSENTIA4 (SINA4; Den Herder et al., 2012). SINA4 presumably maintains SYMRK above the threshold to sense microbial metabolites but low enough to inhibit excessive bacterial colonization.
PM MICRONUTRIENT TRANSPORTERS ARE REGULATED BY UBIQUITINATION
Boron is required by plants for cell wall synthesis, although excess boron is, for reasons that are unclear at present, toxic (Cervilla et al., 2007). In Arabidopsis, the PM-localized BORON TRANSPORTER1 (BOR1) has been shown to facilitate boron uptake in roots (Takano et al., 2010). BOR1 is internalized and degraded by a vacuolar pathway in high-boron conditions, whereas a mutant lacking a conserved Lys (BOR1K590A) remains stable (Kasai et al., 2011). Monoubiquitin- and diubiquitin-modified transporters were detected in BOR1 immunoprecipitates from wild-type (but not BOR1K590A) plants. Intriguingly, while both wild-type and mutant BOR1 were internalized upon treatment with CHX and brefeldin A (a compound that inhibits recycling to the PM after endocytosis), only the wild-type transporter was trafficked to the lytic vacuole. In other words, while BOR1K590A could be internalized, loss of ubiquitination blocked its vacuolar proteolysis. This is a reasonable conclusion, since ESCRT-dependent MVB maturation relies on substrate ubiquitination in animals and yeast.
Like boron, iron uptake is maintained at levels between deficiency and toxicity. Root-localized IRON-REGULATED TRANSPORTER1 (IRT1) is more abundant in Arabidopsis roots than shoots, localizes to early endosomes and the PM, and is constitutively degraded in the vacuole (Vert et al., 2002). IRT1 has also been shown to be regulated by ubiquitination (Barberon et al., 2011). Indeed, while wild-type IRT1 overexpression expectedly led to a moderate accumulation of iron and other metals, overexpression of an IRT1 harboring mutations of Lys residues in the cytosolic domain (likely candidates for ubiquitin modification) caused phenotypes consistent with prodigious metal accumulation (Kerkeb et al., 2008; Barberon et al., 2011). Furthermore, IRT1 immunoprecipitates were antiubiquitin immunoreactive (but not anti-polyubiquitin immunoreactive). This observation makes sense, considering that vacuolar down-regulation of many PM proteins is facilitated by monoubiquitination.
While E3s are the specificity factors of the ubiquitin pathway, dedicated roles for a deubiquitinating enzyme and an E2 in phosphate homeostasis have also been described. UBIQUITIN-SPECIFIC PROTEASE14 (UBP14) was recently found necessary for root hair elongation, an adaptation to phosphate starvation in Arabidopsis (Li et al., 2010). Diminished UBP14 transcript accumulation correlated with poorer adaptation to low-phosphate growth conditions, while introduction of a UBP14 transgene in a ubp14 loss-of-function background reversed these effects. Strikingly, phosphate levels did not strongly differ between wild-type and mutant Arabidopsis, and supplementation with structurally analogous phosphite partially suppressed the phenotype, indicating that UBP14 is probably required for phosphate sensing rather than phosphate transport per se.
PHOSPHATE2 (PHO2), an E2 alternatively known as UBC24, seems to curb phosphate import, since Arabidopsis homozygous for the early termination pho2 mutation prodigiously accumulate phosphate in shoots (Delhaize and Randall, 1995; Aung et al., 2006; Bari et al., 2006). PHO2 is posttranscriptionally down-regulated by miR399 microRNA, as miR399 and PHO2 abundance vary reciprocally. Recent work suggests that PHO1, a membrane protein important for phosphate loading into the vascular system, may be a substrate for PHO2-dependent ubiquitination and subsequent vacuolar proteolysis (Liu et al., 2012).
RECENTLY CHARACTERIZED MEMBRANE-ASSOCIATED RING PROTEINS PLAY DISTINCT ROLES IN PLANT BIOLOGY
A typical response to pathogens by plants is to initiate programmed cell death in the vicinity of the infection. Lin et al. (2008) described the RING gene RING1, whose microRNA-mediated knockdown correlated with reduced programmed cell death when the fungal toxin fumonisin B1 (FB1) was applied. The half-life of RING1 was lengthened in the presence of FB1, indicating that RING1 probably autoubiquitinates in the absence of infection. The RING protein KEEP ON GOING (KEG), which primarily localizes to endosomes, is also implicated in programmed cell death (Gu and Innes, 2011). The keg1-4 missense allele suppresses the enhanced programmed cell death seen in mutants of the enhanced disease resistance1 (edr1) kinase. EDR1 interacted with KEG in vivo, and KEG has been shown to target EDR1 to membranes (Gu and Innes, 2011), but the nature of their relationship is not clearly established. To further complicate the picture, KEG appears to regulate the abundance of ABI5, which is a nucleus-localized transcription factor (Stone et al., 2006). The reported dual localization of KEG is not a singular case either. Cheng et al. (2012) recently described RGLG2, a ubiquitin ligase implicated in the down-regulation of the abiotic stress tolerance-promoting transcription factor ERF53. While GFP-tagged RGLG2 ordinarily exhibits apparent PM localization, salt stress leads to its accumulation in the nucleus and its interaction with ERF53.
Although the hormone abscisic acid (ABA) facilitates drought stress adaptation, there are also ABA-independent pathways by which plants cope with water deficit. Water homeostasis is maintained in part by channels called aquaporins, integral PM proteins found in all three domains of life (Törnroth-Horsefield et al., 2006). Overexpression of hot pepper (Capsicum annuum) RING MEMBRANE ANCHOR1-H1 (Rma1H1) conferred enhanced drought tolerance to Arabidopsis, and Rma1 (the Arabidopsis ortholog of Rma1H1) ubiquitinates the aquaporin PLASMA MEMBRANE INTRINSIC PROTEIN2;1 (PiP2;1) in vivo (Lee et al., 2009). Rma1H1 is transcriptionally induced by drought and cold but not ABA. Given that PiP2;1 increases with MG132 treatment and that overexpression of Rma1H1 reduces PiP2;1 levels while causing the remaining PiP2;1 to accumulate in the ER, Rma1 appears to affect drought tolerance by targeting a water transport facilitator for ubiquitination and down-regulation. MG132 inhibition suggests proteasomal degradation; however, more direct evidence is needed to define its proteolytic pathway.
RMA1 is not unique in this regard. SALT- AND DROUGHT-INDUCED RING FINGER1 (SDIR1) is a predicted transmembrane RING protein transcriptionally up-regulated by salt and drought but not by ABA (Zhang et al., 2007). Promoter-GUS fusions indicate that it is expressed in guard cells, and biochemical fractionation experiments place it in the endomembrane system. Loss-of-function and overexpression lines exhibited ABA hyposensitivity and hypersensitivity, respectively, findings that were mirrored by sdir1 hypersensitivity and 35S:SDIR1 hyposensitivity to drought stress.
Many E3s ubiquitinate PM-resident transporters or transporter adaptor proteins, thereby curbing the influx of nutrients to within a physiologically desirable range. Falling outside this paradigm, LOG2 (for LOSS OF GLUTAMINE DUMPER2) is a membrane-localized RING E3 ligase that appears to positively regulate the activity of an amino acid export system (Pratelli et al., 2012). In Arabidopsis, overexpression of GLUTAMINE DUMPER (GDU) genes (particularly GDU1) increases amino acid efflux, curtails biomass production, and enhances viability on growth media supplemented with certain amino acids: the Gdu1D phenotype. The molecular mechanism of amino acid export is not clear, although the RING protein LOG2 is necessary for its manifestation (Pratelli et al., 2012). LOG2 colocalizes with GDU1 in the PM fraction of transgenic Arabidopsis and ubiquitinates GDU1 in vitro, but whether GDU1 is an in vivo substrate of LOG2 is not established. Paradoxically, the Gdu1D phenotype is suppressed when GDU1 is overexpressed in log2-2 T-DNA or LOG2-directed artificial microRNA backgrounds. This suggests that the amino acid efflux response requires physical interaction between LOG2 and GDU1. Supporting this hypothesis, a mutant form of GDU1 that fails to cause the Gdu1D phenotype (GDU1G100R; Pratelli and Pilot, 2006) is also unable to interact with LOG2 (Pratelli et al., 2012). It is conceivable that GDU1 is an adaptor protein that recruits LOG2 to an amino acid transporter(s).
In addition to RING-type E3s, the physiological roles for a number of PUB-encoding genes (such as PUB13; see above) have been demonstrated. SENESCENCE-ASSOCIATED UBIQUITIN LIGASE1 (SAUL1) is a PUB that localizes to the PM via its ARMADILLO repeats (Drechsel et al., 2011). Association with the PM was found to be important for in vivo function, as a wild-type SAUL1 transgene (but not a SAUL1 transgene lacking ARMADILLO repeats) was able to rescue the senescence phenotype of plants challenged with low light (Salt et al., 2011). In the legume Medicago truncatula, PUB1 was demonstrated to modulate nodulation by down-regulating the nodulation factor RECEPTOR-LIKE KINASE3 (Mbengue et al., 2010).
MEMBRANE-ANCHORED UBIQUITIN-FOLD PROTEINS BIND A SUBSET OF E2S
In all eukaryotes, proteins with structural similarity to ubiquitin have been described, constituting a ubiquitin-fold family. While others have been demonstrated to attach to targets in an analogous manner to ubiquitin, such as RELATED TO UBIQUITIN (Nedd8 in animals), AUTOPHAGY8 (ATG8) and ATG12, and SMALL UBIQUITIN-LIKE MODIFIER, membrane-anchored ubiquitin-fold proteins (MUBs) are distinct, since prenylation precludes C-terminal covalent attachment to proteins (Downes et al., 2006). Instead, this modification localizes MUBs to the PM. MUBs bind noncovalently to a subfamily of E2s (Dowil et al., 2011), presumably to increase local E2 concentration and possibly to orient Ub∼E2 complexes for specific chain formation.
PERSPECTIVE
Our understanding of the role of ubiquitin in plant biology has expanded greatly in the last several years to comprise ER protein quality control, nonproteasomal down-regulation of PM-resident receptors and transporters, abiotic stress adaptation, and amino acid homeostasis. Major challenges include identifying in vivo targets for recently characterized E3 ligases, assessing the in vivo relevance of specific E2-E3 combinations, and determining what functions E3-associated adaptor and ubiquitin-binding proteins play in target recognition and in determining the ultimate consequence of ubiquitination. New technologies, such as proteasome inhibitors that do not concomitantly cause depletion of the cellular free ubiquitin pool, will be invaluable assets in these endeavors.
Glossary
- ER
endoplasmic reticulum
- UMS
ubiquitin modification system
- ERAD
endoplasmic reticulum-associated degradation
- PM
plasma membrane
- CHX
cycloheximide
- Cnx
calnexin
- Crt
calreticulin
- BR
brassinosteroid
- MVB
multivesicular body
- ABA
abscisic acid
- MUBs
membrane-anchored ubiquitin-fold proteins
References
- Andersen KM, Hofmann K, Hartmann-Petersen R. (2005) Ubiquitin-binding proteins: similar, but different. Essays Biochem 41: 49–67 [DOI] [PubMed] [Google Scholar]
- Arvan P, Zhao X, Ramos-Castaneda J, Chang A. (2002) Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic 3: 771–780 [DOI] [PubMed] [Google Scholar]
- Aung K, Lin S-I, Wu C-C, Huang Y-T, Su CL, Chiou T-J. (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. (2002) Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell 3: 271–282 [DOI] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G. (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible W-R. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. (2001) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol 3: 24–29 [DOI] [PubMed] [Google Scholar]
- Böhm L, Crane-Robinson C, Sautière P. (1980) Proteolytic digestion studies of chromatin core-histone structure: identification of a limit peptide of histone H2A. Eur J Biochem 106: 525–530 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Cervilla LM, Blasco B, Ríos JJ, Romero L, Ruiz JM. (2007) Oxidative stress and antioxidants in tomato (Solanum lycopersicum) plants subjected to boron toxicity. Ann Bot (Lond) 100: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-F, Shakeel SN, Bowers J, Zhao X-C, Etheridge N, Schaller GE. (2007) Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J Biol Chem 282: 24752–24758 [DOI] [PubMed] [Google Scholar]
- Cheng MC, Hsieh EJ, Chen JH, Chen HY, Lin TP. (2012) Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol 158: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. (2009) Tracing the history of the ubiquitin proteolytic system: the pioneering article. Biochem Biophys Res Commun 387: 1–10 [DOI] [PubMed] [Google Scholar]
- Cui F, Liu L, Zhao Q, Zhang Z, Li Q, Lin B, Wu Y, Tang S, Xie Q. (2012) Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea A, Pellman D. (1998) Deubiquitinating enzymes: a new class of biological regulators. Crit Rev Biochem Mol Biol 33: 337–352 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Herder G, Yoshida S, Antolín-Llovera M, Ried MK, Parniske M. (2012) Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 24: 1691–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dowil RT, Lu X, Saracco SA, Vierstra RD, Downes BP. (2011) Arabidopsis membrane-anchored ubiquitin-fold (MUB) proteins localize a specific subset of ubiquitin-conjugating (E2) enzymes to the plasma membrane. J Biol Chem 286: 14913–14921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Saracco SA, Lee SS, Crowell DN, Vierstra RD. (2006) MUBs, a family of ubiquitin-fold proteins that are plasma membrane-anchored by prenylation. J Biol Chem 281: 27145–27157 [DOI] [PubMed] [Google Scholar]
- Drechsel G, Bergler J, Wippel K, Sauer N, Vogelmann K, Hoth S. (2011) C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. J Exp Bot 62: 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich CH, Schmukle AC, Walczak H. (2011) The emerging role of linear ubiquitination in cell signaling. Sci Signal 4: re5. [DOI] [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol 14: 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HY, Lin YL, Fatimababy AS. (2010) Proteasomal recognition of ubiquitylated substrates. Trends Plant Sci 15: 375–386 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gu Y, Innes RW. (2011) The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol 155: 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberth S, Shahriari M, Bruderek M, Hessner F, Müller B, Hülskamp M, Schellmann S. (2012) Artificial ubiquitylation is sufficient for sorting of a plasma membrane ATPase to the vacuolar lumen of Arabidopsis cells. Planta 236: 63–77 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R. (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H. (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84: 277–287 [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. (2009) The ubiquitylation machinery of the endoplasmic reticulum. Nature 458: 453–460 [DOI] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J. (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Li J. (2012) The protein quality control of plant receptor-like kinases in the endoplasmic reticulum. In F Tax, B Kemmerling, eds, Receptor-Like Kinases in Plants from Development to Defense, Vol 13. Springer, Berlin, pp 275–307
- Hua Z, Vierstra RD. (2011) The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol 62: 299–334 [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. EMBO Rep 9: 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai K, Takano J, Miwa K, Toyoda A, Fujiwara T. (2011) High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J Biol Chem 286: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Kelley DR, Estelle M. (2012) Ubiquitin-mediated control of plant hormone signaling. Plant Physiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkeb L, Mukherjee I, Chatterjee I, Lahner B, Salt DE, Connolly EL. (2008) Iron-induced turnover of the Arabidopsis IRON-REGULATED TRANSPORTER1 metal transporter requires lysine residues. Plant Physiol 146: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ. (2004) Ethylene signal transduction: moving beyond Arabidopsis. Plant Physiol 135: 660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B, Braakman I. (2004) Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol 16: 343–349 [DOI] [PubMed] [Google Scholar]
- Kostova Z, Tsai YC, Weissman AM. (2007) Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol 18: 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol 20: 196–204 [DOI] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT. (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J, Petrásek J, Tomanov K, Retzer K, Parezová M, Korbei B, Bachmair A, Zazímalová E, Luschnig C. (2012) Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci USA 109: 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. (1999) Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 4: 1029–1040 [DOI] [PubMed] [Google Scholar]
- Li W-F, Perry PJ, Prafulla NN, Schmidt W. (2010) Ubiquitin-specific protease 14 (UBP14) is involved in root responses to phosphate deficiency in Arabidopsis. Mol Plant 3: 212–223 [DOI] [PubMed] [Google Scholar]
- Lin SS, Martin R, Mongrand S, Vandenabeele S, Chen KC, Jang IC, Chua NH. (2008) RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J 56: 550–561 [DOI] [PubMed] [Google Scholar]
- Liu J-X, Srivastava R, Che P, Howell SH. (2007) Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 51: 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cui F, Li Q, Yin B, Zhang H, Lin B, Wu Y, Xia R, Tang S, Xie Q. (2011) The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res 21: 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-Y, Huang T-K, Tseng C-Y, Lai Y-S, Lin S-I, Lin W-Y, Chen J-W, Chiou T-J. (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L. (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Hervé C, et al. (2010) The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. (2005) ERAD: the long road to destruction. Nat Cell Biol 7: 766–772 [DOI] [PubMed] [Google Scholar]
- Moussatche P, Klee HJ. (2004) Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem 279: 48734–48741 [DOI] [PubMed] [Google Scholar]
- Müller J, Piffanelli P, Devoto A, Miklis M, Elliott C, Ortmann B, Schulze-Lefert P, Panstruga R. (2005) Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17: 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T, Barrière H, Bagdány M, Rabeh WM, Du K, Höhfeld J, Young JC, Lukacs GL. (2010) Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pérez-Oliva AB, Olivares C, Jiménez-Cervantes C, García-Borrón JC. (2009) Mahogunin ring finger-1 (MGRN1) E3 ubiquitin ligase inhibits signaling from melanocortin receptor by competition with Galphas. J Biol Chem 284: 31714–31725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J-M. (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Popov N, Schülein C, Jaenicke LA, Eilers M. (2010) Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol 12: 973–981 [DOI] [PubMed] [Google Scholar]
- Pratelli R, Guerra DD, Yu S, Wogulis M, Kraft E, Frommer WB, Callis J, Pilot G. (2012) The ubiquitin E3 ligase LOSS OF GDU2 is required for GLUTAMINE DUMPER1-induced amino acid secretion in Arabidopsis. Plant Physiol 158: 1628–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Pilot G. (2006) The plant-specific VIMAG domain of Glutamine Dumper1 is necessary for the function of the protein in Arabidopsis. FEBS Lett 580: 6961–6966 [DOI] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. (2006) Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J 25: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders E, Foulquier F, Annaert W, Matthijs G. (2011) How Golgi glycosylation meets and needs trafficking: the case of the COG complex. Glycobiology 21: 853–863 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt JN, Yoshioka K, Moeder W, Goring DR. (2011) Altered germination and subcellular localization patterns for PUB44/SAUL1 in response to stress and phytohormone treatments. PLoS ONE 6: e21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Hansson M, Scalf M, Walker JM, Smith LM, Vierstra RD. (2009) Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever N, Song B-L, Yabe D, Goldstein JL, Brown MS, DeBose-Boyd RA. (2003) Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J Biol Chem 278: 52479–52490 [DOI] [PubMed] [Google Scholar]
- Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, Piper R. (2009) ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J Cell Biol 185: 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. (2002) Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature 416: 183–187 [DOI] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Liu Y, Xia Y, Hong Z, Li J. (2011) Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc Natl Acad Sci USA 108: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Tanaka M, Toyoda A, Miwa K, Kasai K, Fuji K, Onouchi H, Naito S, Fujiwara T. (2010) Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törnroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P. (2006) Structural mechanism of plant aquaporin gating. Nature 439: 688–694 [DOI] [PubMed] [Google Scholar]
- Tsai YC, Weissman AM. (2010) The unfolded protein response, degradation from the endoplasmic reticulum, and cancer. Genes Cancer 1: 764–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (2007) The membrane protein universe: what’s out there and why bother? J Intern Med 261: 543–557 [DOI] [PubMed] [Google Scholar]
- Wang M, Xu Q, Yu J, Yuan M. (2010) The putative Arabidopsis zinc transporter ZTP29 is involved in the response to salt stress. Plant Mol Biol 73: 467–479 [DOI] [PubMed] [Google Scholar]
- Wang X, Herr RA, Hansen TH. (2012) Ubiquitination of substrates by esterification. Traffic 13: 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Klevit RE. (2012) Following Ariadne’s thread: a new perspective on RBR ubiquitin ligases. BMC Biol 10: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KD. (2005) The discovery of ubiquitin-dependent proteolysis. Proc Natl Acad Sci USA 102: 15280–15282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woffenden BJ, Freeman TB, Beers EP. (1998) Proteasome inhibitors prevent tracheary element differentiation in zinnia mesophyll cell cultures. Plant Physiol 118: 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR. (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol 28: 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR. (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Yin X-J, Volk S, Ljung K, Mehlmer N, Dolezal K, Ditengou F, Hanano S, Davis SJ, Schmelzer E, Sandberg G, et al. (2007) Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]