Abstract
Snake-venom α-bungarotoxin is a member of the α-neurotoxin family that binds with very high affinity to the nicotinic acetylcholine receptor (AChR) at the neuromuscular junction. The structure of the complex between α-bungarotoxin and a 13-mer peptide (WRYYESSLEPYPD) that binds the toxin with high affinity, thus inhibiting its interactions with AChR with an IC50 of 2 nM, has been solved by 1H-NMR spectroscopy. The bound peptide folds into a β-hairpin structure created by two antiparallel β-strands, which combine with the already existing triple-stranded β-sheet of the toxin to form a five-stranded intermolecular, antiparallel β-sheet. Peptide residues Y3P, E5P, and L8P have the highest intermolecular contact area, indicating their importance in the binding of α-bungarotoxin; W1P, R2P, and Y4P also contribute significantly to the binding. A large number of characteristic hydrogen bonds and electrostatic and hydrophobic interactions are observed in the complex. The high-affinity peptide exhibits inhibitory potency that is better than any known peptide derived from AChR, and is equal to that of the whole α-subunit of AChR. The high degree of sequence similarity between the peptide and various types of AChRs implies that the binding mode found within the complex might possibly mimic the receptor binding to the toxin. The design of the high-affinity peptide was based on our previous findings: (i) the detection of a lead peptide (MRYYESSLKSYPD) that binds α-bungarotoxin, using a phage-display peptide library, (ii) the information about the three-dimensional structure of α-bungarotoxin/lead-peptide complex, and (iii) the amino acid sequence analysis of different AChRs.
The nicotinic acetylcholine receptor (AChR) is a ligand-gated ion channel that is activated by binding of acetylcholine. α-Neurotoxins such as the 74-aa α-bungarotoxin (α-BTX) bind specifically to AChR and exhibit high toxicity because of inhibition of the AChR function at the neuromuscular junction. Synthetic peptides that bind α-BTX may yield competitive inhibitors of α-BTX binding to AChR, and could serve as potential lead compounds for effective antidotes against α-BTX poisoning. Moreover, understanding the nature of the very high binding affinity of α-BTX to AChR is obviously of considerable importance in the study of the ligand-binding site of AChR.
The tertiary structure of AChR has not yet been obtained. Its membrane-bound nature and its size have so far curbed all attempts to obtain suitable crystals for x-ray analysis or to carry out NMR studies. Some efforts to produce large quantities of the extracellular domain in a soluble form have been reported (ref. 1 and references therein). The acetylcholine-binding site is located mainly at the α-subunit of AChR, in the vicinity of Cys-192 and Cys-193 (2–4), and partially overlaps the binding domain for α-BTX (Torpedo AChR residues α185–196) (5). α-BTX binds to the pentameric muscle AChR with a Kd of 10−11 M (6), and to the α-subunit of Torpedo AChR with a Kd of 10−9 M (7). NMR studies of α-BTX complex with a 12-mer fragment of the Torpedo AChR, α185–196 (8), and with an 18-mer fragment, α181–198 (9), revealed that the bound peptides adopt an extended conformation, and interactions with α-BTX were formed only between a segment corresponding to residues α186–190 of the receptor. One approach to obtain a better understanding of the structural requirements for AChR binding would be to study longer and longer peptide analogs of the α-subunit of AChR. On the other hand, searching for a minimal synthetic peptide that efficiently inhibits the binding of AChR to α-BTX would greatly accelerate the search for effective antidotes against α-BTX poisoning, and would pinpoint those structural requirements for AChR/α-BTX binding.
Using a combinatorial phage-displayed peptide library, we previously identified a 13-residue peptide (MRYYESSLKSYPD) that binds α-BTX with an affinity 15-fold higher than that of AChR α185–196 (10). The library-peptide consensus motif, YYXSS, is similar to the receptor motif YYXCC (α189–193). We solved the three-dimensional solution structure of α-BTX complex with this library-lead peptide (designated LLPep) by 1H-NMR spectroscopy (11). According to the NMR structure, the bound peptide was found to adopt a globular conformation around a hydrophobic core created by the side chain of Y11P of the peptide, whereas the free peptide in solution was characterized by a rather extended conformation. The amino acid residues that bind tightly with α-BTX or whose side chains interact internally with other residues of the LLPep are R2P, Y3P, Y4P, E5P, L8P, and Y11P. However, none of the side-chain protons of M1P, K9P, or S10P interact directly with the toxin (11). In a recent study (12), we have used this NMR-derived structural information, together with a comparative sequence analysis of AChR of animal species ranking their resistance to α-BTX (13–15), to design and prepare peptides that interact with α-BTX with affinities higher than that of the above mentioned LLPep. Using this approach, peptides that inhibit α-BTX binding to AChR in the low nanomolar range were obtained. The inhibition potency of these high-affinity peptides is stronger by at least two orders of magnitude than that achieved by the original LLPep, and is stronger than that achieved by any known synthetic peptide that corresponds to α-BTX binding site of AChR (12). The high-affinity peptides have a protective effect on mice against α-BTX lethality (12).
In this paper, we report the 1H-NMR spectroscopy study of the solution structure of a 13-mer high-affinity peptide (WRYYESSLEPYPD, designated HAPep) in complex with α-BTX. It is pertinent to note that our NMR studies revealed the presence of two isoforms of α-BTX that correspond to α-BTXAla-31 and α-BTXVal-31. These isoforms were separated by chromatography, and the NMR studies were carried out with α-BTXAla-31. We studied the structure of the binding site of α-BTX as well as its interactions with the peptide, and identified key residues within the toxin and the peptide that contribute to the binding. This peptide differs from the LLPep by only three replacements: M1W, K9E, and S10P. The detailed reasoning for the 165-fold increase in the inhibition potency of this peptide (12), compared with the original LLPep (10), is expected to emerge from such structural studies. The structure of the complex indeed indicates the formation of a novel structural motif that combines a β-hairpin within the bound peptide with an intermolecular β-sheet. We expect that the information acquired will shed new light on the nature of the AChR residues that enable it to adopt a conformation that strengthens its binding to α-BTX and increases the specificity of the interaction.
Materials and Methods
Peptide Synthesis.
The design, selection, synthesis, and purification of the 13-mer HAPep (WRYYESSLEPYPD) were described (12). HPLC-purified peptide was >98% pure. Both mass spectrometry and NMR sequential assignment verified the amino acid composition of the purified peptide (12).
Separation and Characterization of the Two Isoforms of α-BTX.
α-BTX from the Bungarus multicinctus venom (Sigma) was separated into its two isoforms by reversed-phase HPLC using a prepacked Vydac (Hesperia, CA) Protein C-4 column (25 × 250 mm, 7-μm bead size), employing a binary gradient of 0.1% trifluoroacetic acid (TFA) in water (solution A) and 0.1% TFA in 75% acetonitrile in water (solution B) eluted at t = 0 min, B = 5%; t = 5 min, B = 5%; t = 60 min, B = 75% (flow rate 8.0 ml/min), using the same apparatus as described (12). Purity validation of the separated isoforms and of the crude α-BTX was performed by analytical reversed-phase HPLC using a prepacked Vydac Protein C-4 column (4 × 250 mm, 5-μm bead size) and a gradient of t = 0 min, B = 0%; t = 5 min, B = 0%; t = 50 min, B = 100% (flow rate 0.8 ml/min). The purity of α-BTXAla-31 was ≈90% (Rt 29.5 min), and that of α-BTXVal-31 was 98% (Rt 30.4 min). Samples of the two isoforms were subjected to amino acid analysis and to electrospray mass spectrometry. The biotinylation of HPLC-purified isoforms, α-BTXAla-31 and α-BTXVal-31, as well as the inhibition of their binding to Torpedo AChR by the HAPep and by nonpurified α-BTX, were carried out as described (12).
NMR Spectroscopy.
NMR samples of α-BTX in complex with HAPep contained 1 mM of complex (1:1 stochiometry) in 10 mM sodium phosphate buffer at pH 6. NMR samples were prepared as previously described (11), in either 90% H2O/10% 2H2O or 99.9% 2H2O and contained trace amounts of NaN3 as a preservative. 1H-NMR spectra were recorded at 30°C, 37°C, and 42°C on Bruker (Karlsruhe, Germany) DMX-500 and DRX-800 spectrometers. Spectra were acquired with 4096–8192 complex data points in t2 and 512–800 increments in t1, using a spectral width of 14–16 ppm. Two-dimensional (2D) nuclear Overhauser effect (NOE) spectra (16) of the complex were acquired by using mixing times of 150, 100, 70, and 45 ms, and 2D homonuclear Hartmann-Hahn spectra (17) were acquired by using mixing times of 140 and 70 ms. The double quantum-filtered correlation spectroscopy (18) was acquired by conventional procedures. To identify slowly exchanging amide protons, the complex was lyophilized from H2O and redissolved in 99.9% 2H2O, and a series of 2D homonuclear Hartmann-Hahn spectra were recorded within 1,440 min. All data were processed and analyzed by using the program xwinnmr v.2.6 and aurelia (Bruker).
Experimental Restraints.
The cross-peak intensities in 2D-NOE spectra of the complex, recorded with several mixing times, were compared to eliminate the possibility of spin diffusion effects. The NOE intensities from the spectrum, recorded with a mixing time of 150 ms, were used for structure calculation. NOE restraints were derived as described (11). Backbone hydrogen bonds within the β-sheet were identified from amide proton exchange data and interstrand NOEs. Additional restraints were included to define the five known disulfide bridges of the toxin (3/23, 16/44, 29/33, 48/59, and 60/65). The φ angle restraints were derived from 3JHN-Hα coupling constants measured from double quantum-filtered correlation spectra. The spectra were acquired with 8,192 data points in F2 dimension and subsequently zero-filled to 16,384 points. Stereospecific assignments were based on analysis of both JHα-Hβ (determined from double quantum-filtered correlation spectra measured in 2H2O), and HN-Hβ and Hα-Hβ NOEs (measured from 2D-NOE spectra recorded with a 45-ms mixing time; ref. 19).
Structure Calculations.
Three-dimensional structures of the complex were calculated on the basis of the NMR data using a combination of distance geometry and dynamic simulated annealing protocols, using the program cns (20) developed by Brünger, Clore, Nilges, and coworkers. The dynamic simulated annealing procedure was similar to that described by Nilges et al. (21). Distance restraints were used with a square-well potential, and the NOE force constant was set to 50 kcal⋅mol−1⋅Å−2.
Results
Identification and Characterization of the Two α-BTX Isotoxins.
While doing a sequence-specific resonance assignment (22) of the HAPep/α-BTX complex, it became apparent that more than one population of the toxin resonance signals was present in the NMR spectra. The possibility that the appearance of the two populations was due to the presence of bound and nonbound toxin was excluded, because stepwise addition of the peptide did not result in any change of the relative populations. FPLC gel-filtration of the complex of α-BTX with the peptide revealed a single peak, implying for the proximity in molecular weight. However, reversed-phase HPLC analysis of the toxin revealed the presence of a second species of α-BTX, accounting for ≈40% of the total. In a previous study, Basus and coworkers (23) identified an isoform of α-BTX (≈10%) in which Ala-31 was replaced by Val. We analyzed the HPLC-separated fractions by mass spectrometry (the first fraction had a molecular weight of 7,982.9 and the second of 8,011.3) and by amino acid analysis, which indicated that the two fractions indeed differ by the Ala/Val replacement. The separated isoforms, namely α-BTXAla-31 and α-BTXVal-31, were tested for their ability to bind Torpedo AChR, and were found to similarly bind the receptor, as described by Basus and coworkers (23). Furthermore, the HAPep was found to inhibit equally the binding of both biotinylated isoforms to Torpedo AChR (IC50 of 2 nM). [In our previous study of the complex of α-BTX with LLPep (11), α-BTX Val-31 accounted for only 10% of the total toxin.] For further NMR studies of the complex, α-BTXAla-31 was used.
Resonance Assignment.
The sequence-specific resonance assignment of the bound toxin and bound peptide was done according to the well-established procedure of Wüthrich and coworkers (22). Toxin residues located at fingers I and II underwent considerable changes in their chemical shift when complexed with the HAPep and with the LLPep (11). It is interesting to note that, although the HAPep differs in sequence from the lead-peptide only at positions 1, 9 and 10 (Table 1), a large change in the chemical shift of bound-peptide protons was also observed for residues R2P, Y3P, Y4P, Y11P, and D13P. This difference implies that the three replacements, yielding an increased affinity, induced changes in the conformation of other residues of the peptide as well.
Table 1.
Sequence and affinity of peptides that bind α-BTX
| Peptide | Amino acid sequence* | IC50,
M† |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Library lead (LLPep) | M | R | Y | Y | E | S | S | L | K | S | Y | P | D | 3.3 × 10−7 | |
| No. 39† | W | R | Y | Y | E | S | S | L | K | S | Y | P | D | 3.5 × 10−8 | |
| No. 29† | M | R | Y | Y | E | S | S | L | K | P | Y | P | D | 3.2 × 10−8 | |
| No. 48† | W | R | Y | Y | E | S | S | L | K | P | Y | P | D | 1.0 × 10−8 | |
| No. 56† | W | R | Y | Y | E | C | C | L | D | P | Y | P | D | 1.9 × 10−9 | |
| No. 50† (HAPep) | W | R | Y | Y | E | S | S | L | E | P | Y | P | D | 2.0 × 10−9 | |
| AChR numbering | 187 | ⋅ | ⋅ | 190 | ⋅ | ⋅ | ⋅ | ⋅ | 195 | ⋅ | ⋅ | ⋅ | ⋅ | 200 | |
| Muscle (Torpedo) α1 | W | V | Y | Y | T | C | C | P | D | T | P | Y | L | 10−6 | |
| Muscle (Torpedo) α1 | W | V | Y | Y | T | C | C | P | D | T | P | Y | L | D‡ | 2.6 × 10−8 |
| Neuronal (rat) α7 | E | K | F | Y | E | C | C | K | E | P | Y | P | D | 1.8 × 10−5 | |
Residues that were replaced in synthetic peptides deriving from the LLPep are bold.
Numbering of peptides, as well as the inhibition values of α-BTX-binding to Torpedo AChR, are taken from ref. 12.
Adding residues WKH to the N terminal of this peptide, to derive fragment α184–200 of muscle AChR, yielded IC50 = 10−6 M (unpublished results). Further extension of this peptide on either the C or N terminus did not result in better inhibition values (33).
Structure Calculations.
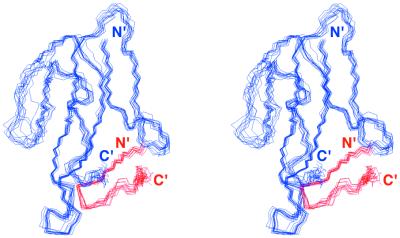
For the final structure refinements, a total of 1,228 distance restraints were used: 986 intramolecular interactions in the bound toxin (including 245 long-range interactions, >5 residues apart), 150 intrapeptide interactions (including 12 long-range interactions), and 92 intermolecular interactions between the toxin and the HAPep. Sixty-two φ angle restraints were used in the structure calculation: 55 angles for the toxin and 7 for the bound peptide. In addition, 42 χ1 angle restraints and 23 stereospecific assignments of β-methylene protons were introduced. Ten final structures that satisfy the experimental distance restraints with no violation larger than 0.4 Å and that show good covalent geometry (Table 2) are shown in Fig. 1. The atomic rms deviation (rmsd) between the individual structures and the energy-minimized average structure is 0.70 Å for the backbone atoms of α-BTX residues 1–46 and 56–72 and all 13 peptide residues, and 1.26 Å for the heavy atoms. The backbone rmsd for all 13 residues of the bound peptide is 0.57 Å, and for all 87 residues of the complex is 1.07 Å.
Table 2.
Structural statistics of the high-affinity peptide/α-BTX complex
| Structural statistics | 〈SA〉* |
|---|---|
| rmsd from experimental distance restraints, Å | 0.034 ± 0.002 |
| rmsd from experimental dihedral restraints, degree | 0.49 ± 0.06 |
| rmsd from idealized covalent geometry: | |
| Bonds, Å | 0.0033 ± 0.0002 |
| Angles, degree | 0.52 ± 0.02 |
| Impropers, degree | 0.45 ± 0.02 |
| CNS (20) potential energies, kcal⋅mol−1 | |
| Etotal | 310.2 ± 32.0 |
| Erepulsion† | 94.2 ± 12.0 |
| Eimproper | 21.4 ± 1.8 |
| Enoe | 74.7 ± 12.1 |
| Ecdihedral | 3.1 ± 0.90 |
〈SA〉 is the ensemble of 10 NMR structures of the complex.
The Crystallography & NMR System (CNS) Frepulsion function was used to simulate van der Waals interactions with atomic radii set to 0.75 times their CHARM force field values.
Figure 1.
Stereoview of the superposition of 10 NMR-derived structures of the HAPep/α-BTX complex. Backbone atoms of α-BTX (residues 1–73) are shown in blue; backbone atoms of the peptide are shown in red.
Structure of Bound α-BTX.
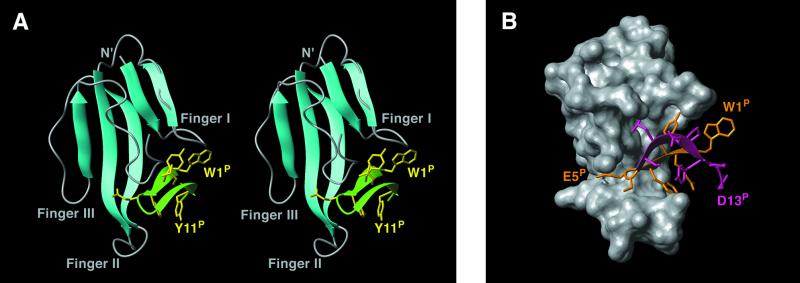
A schematic ribbon diagram of the NMR-derived structure of the bound toxin in its complex with the HAPep is shown in Fig. 2A. α-BTX is a member of the three-finger toxin family: three long fingers and a tail protrude from the tangle-like region of the molecule. Finger I is formed by two anti-parallel β-strands consisting of residues 2–5 and 12–15 connected by a loop, and finger II is formed by β-strands consisting of residues 22–30 and 37–45. A third strand comprising residues 56–60 of finger III combines with the two β-strands of finger II to form a triple-stranded antiparallel β-sheet, as previously found in the LLPep/α-BTX complex (11). It should be indicated that, in the present study, the two β-strands that form the second finger stretch further than previously found (11). As emerges from the NMR data, toxin residues 1–4, which make an antiparallel β-sheet with residues 12–15, are involved in an additional secondary structural element, a parallel β-sheet with the C terminus residues 63–66, not previously observed. At least three slow-exchanging amide protons and several NOE interactions support this conclusion.
Figure 2.
Solution structure of the HAPep/α-BTX complex. (A) Ribbon diagram of the complex. α-BTX backbone is shown in cyan (β-sheet regions) and in gray. Peptide backbone is shown in green; side chains of W1P, Y3P, Y4P, E5P, L8P, and Y11P are shown in yellow. (B) Surface view of the complex. α-BTX is shown in gray, peptide is shown in orange (residues 1–6) and pink (residues 7–13). The figures were prepared by using the program molmol (38).
Structure of the Bound High-Affinity Peptide.
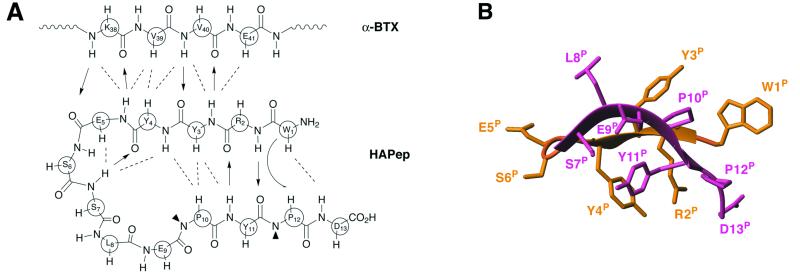
The bound peptide folds into a β-hairpin structure created by two antiparallel β-strands consisting of residues 2–4 and 7–11 of the peptide (Fig. 3). The two strands are connected by an elongated loop characterized by several short-range distances (Y4P/S6P, Y4P/S7P, and E5P/S7P) as revealed by NMR. These interactions are most likely responsible for the characteristic hairpin that facilitates the proximity between the two antiparallel peptide strands. The β-hairpin conformation is stabilized by four intrapeptide hydrogen bonds created by the amide protons of residues R2P, Y4P, S7P, and Y11P, interacting with residues Y11P, S7P, Y4P, and R2P, respectively. This conformation is further supported by several short-distance interactions between nonvicinal peptide residues that are brought close in space because of the folding of the bound peptide (W1P/P10P, W1P/P12P, W1P/D13P, Y3P/Y11P, Y3P/P10P, and Y4P/P10P). S7P, L8P, and E9P form a bulge that protrudes from the β-hairpin tongue: L8P points toward the toxin whereas E9P points outside, toward the solvent (Figs. 2B and 3B). In the present structure, the aromatic rings of Y4P and Y11P are in close proximity and form a hydrophobic patch that further stabilizes the β-hairpin (Figs. 2B and 3B). It should be indicated that the free peptide is characterized by a rather extended conformation.
Figure 3.
(A) Schematic representation of inter- and intramolecular β-sheet interactions between the HAPep and residues 38–41 of α-BTX. One-letter code of each residue is circled at the αC position. Dashed-lines indicate interstrand NOEs, arrows show hydrogen bonds, and curved arrow represents interactions in which side-chain atoms are involved. (B) Structure of the HAPep when bound to α-BTX. Residues 1–6 are shown in orange, residues 7–13 in pink.
Binding Interactions and the Contact Surface.
The NMR-derived intermolecular interactions in the HAPep/α-BTX complex are listed in Table 3. Fifteen residues of the bound toxin (T6, A7, T8, S9, and I11 of finger I; D30, R36, G37, K38, V39, V40, and E41 of finger II; and H68, K70, and Q71 of the C terminus) interact with nine peptide residues (W1P, R2P, Y3P, Y4P, E5P, S6P, S7P, L8P, and Y11P). Residues Y3P, E5P, and L8P have the highest contact area with α-BTX, indicating their importance for intermolecular interactions. W1P, R2P, and Y4P also contribute significantly (Table 3). In most of the intermolecular NOEs (≈70%), side-chain, rather than backbone protons of the peptide are involved, implying the specificity of the interactions.
Table 3.
Intermolecular NMR-observed interactions and contact areas of the high-affinity peptide and the library-lead peptide in their corresponding complexes with α-BTX
| High-affinity
peptide (HAPep)
|
Library-lead
peptide† (LLPep)
|
||||||
|---|---|---|---|---|---|---|---|
| Peptide residues | Interacting α-BTX residues | No. of interactions | Contact area, Å2 | Peptide residues | Interacting α-BTX residues | No. of interactions | Contact area, Å2 |
| W1 | A7*, T8*, S9* | 6 | 74 | M1 | — | — | — |
| R2 | D30*, V39* | 6 | 86 | R2 | V39* | 1 | 46 |
| Y3 | T6*, V39, V40, E41, H68* | 17 | 139 | Y3 | T6*, A7*, I11*, V39, H68* | 17 | 135 |
| Y4 | R36*, V39*, V40* | 19 | 72 | Y4 | D30*, R36*, V39*, V40 | 21 | 125 |
| E5 | K38*, V39, V40, H68 | 17 | 134 | E5 | K38*, V39, V40*, H68* | 8 | 104 |
| S6 | R36* | 1 | 43 | S6 | — | — | — |
| S7 | G37*, H68, K70 | 4 | 5 | S7 | H68, R72 | 3 | 32 |
| L8 | I11, H68*, K70*, Q71* | 20 | 102 | L8 | I11, H68*, K70*, Q71* | 14 | 97 |
| Y11 | R36* | 2 | 23 | Y11 | — | — | — |
Interactions in which side-chain atoms of the peptide are involved.
Values taken from ref. 11.
The two peptide strands forming the β-hairpin (Fig. 3B) actually combine with the triple-stranded antiparallel β-sheet of the toxin (residues 22–30, 37–45, and 56–60) to form a five-stranded intermolecular, antiparallel β-sheet (Fig. 2A). The connection between the two β-sheet elements within the two molecules is created via interactions between α-BTX residue 38–41 and peptide residues R2P-E5P (Fig. 3A). This conformation is revealed by four intermolecular hydrogen bonds between the amide protons of Y3P, E5P, K38, and V40 and the carbonyl atoms of V40, K38, E5P, and Y3P, respectively. Additional classical interstrand interactions typical of an antiparallel β-sheet further support the formation of this intermolecular structure: short-distance amide/amide interactions (Y3P/V40 and E5P/K38), a short distance alpha/alpha interaction (Y4P/V39), as well as interstrand interactions between amide and alpha protons (V40/Y4P, Y3P/E41, and E5P/V39) (Fig. 3A).
The total decrease in the accessible surface area of the peptide on complex formation is 1,228 Å2. The contact area of the peptide with the toxin is 710 Å2, whereas the rest (>500 Å2) represents contact area within the peptide. Y3P, E5P, and L8P each contribute about 20% of the total intermolecular contact area, whereas W1P, R2P, and Y4P make a lower contribution. The loss of the accessible surface of α-BTX on complex formation is 585 Å2. Most of the intermolecular contact area is contributed by toxin residues located on finger II (>50%), whereas residues from finger I and from the C terminus contribute relatively little to the intermolecular surface.
Discussion
In this study, the structure of the complex between α-BTX and a 13-mer HAPep, which inhibits α-BTX binding to AChR with an IC50 of 2 nM, was solved by NMR spectroscopy. The HAPep adopts a β-hairpin structure, and forms an intermolecular antiparallel β-sheet with finger II of α-BTX (Fig. 3A). The fact that a short peptide in its bound form has such a well organized and a well-defined structural element, which exists both intra- and intermolecularly, seems to be a unique phenomenon. Frequently, the conformation of a peptide bound to a protein consists of one or more β-turns (24, 25). In rare cases, the bound peptide adopts a β-hairpin-type conformation (26–28). However, the formation of a short stretch of antiparallel β-sheet, not only within the bound peptide but also between the peptide and the protein, represents a novel structural motif as previously reported (29). We thus assume that this rather unique structure accounts for most of the increase in the binding affinity of the HAPep to α-BTX. The overall stability of a β-hairpin structure is the outcome of different contributions, including main-chain hydrogen bonding, secondary structure propensities of the turn region, and side-chain to side-chain interactions (30). A study of Ma and Nussinov (ref. 31 and references therein) suggests that hydrophobic interactions provide the primary force for peptide folding and for its thermodynamic stability. In our study, the hydrophobic interaction between the side-chain atoms of Y4P and Y11P [not found in the LLPep/α-BTX structure (11)], as well as the intramolecular backbone hydrogen bonds followed by several short- and long-range intrabound-peptide interactions (Fig. 3 A and B), stabilize the β-hairpin within the peptide. Further stabilization results from the intermolecular backbone/backbone hydrogen bonding, the hydrophobic interactions between the side chains of R2P, Y3P, Y4P, and E5P with K38, V39, V40, and E41 (Table 3 and Fig. 3A), as well as the electrostatic interactions R2P/D30 and E5P/K38.
Comparison of the Newly Established NMR Structure with That of the LLPep/α-BTX Complex.
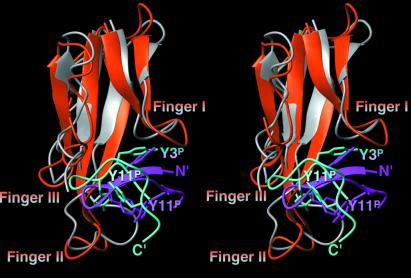
Although the NMR structure of the HAPep/α-BTX complex bears many similarities to the previously reported structure of the LLPep/α-BTX complex (11), it is clear that the present structure represents a much tighter interaction between the peptide and α-BTX. A superposition of the structures of the two complexes is shown in Fig. 4. In both structures, R2P, Y3P, Y4P, E5P, and L8P interact with the same regions of α-BTX (Table 3). However, the intermolecular contact areas between HAPep residues and the toxin are in most cases larger than the corresponding values obtained in the LLPep/α-BTX complex (ref. 11 and Table 3). In the LLPep/α-BTX complex, the intermolecular contact surfaces corresponding to the peptide and toxin were 590 and 530 Å2, respectively (11). In the HAPep/α-BTX complex, the corresponding values obtained were 710 Å2 for the peptide and 585 Å2 for the toxin. It is noteworthy that the reduction in the accessible surface area of the HAPep in coiling from its extended form into its bound conformation is higher by ≈310 Å2 than the corresponding loss in surface area of the bound LLPep. The larger number of intramolecular interactions within the bound HAPep contributes to this increase in area loss. In the LLPep/α-BTX complex (11), the peptide folds into an almost globular conformation around the side chain of Y11P. This residue is involved in several intrapeptide interactions, mainly with backbone atoms of peptide residue R2P and Y4P (11). In the HAPep/α-BTX complex, the side chain of Y11P is tilted from the center of the loop (Figs. 3B and 4) so as to enable the formation of the dense network of intrabound peptide interactions between the two strands of the elongated β-hairpin structure. Although the side chain of Y11P points outside the β-hairpin, it retains a large intrapeptide contact surface and forms a hydrophobic patch with the aromatic ring of Y4P (Fig. 3B). Intrapeptide interactions between the amino acid residues at the N- and C-termini appear in both structures. In the LLPep/α-BTX complex, they are formed by R2P/S10P, R2P/Y11P, and Y4P/Y11P. However, in the HAPep/α-BTX complex, the interactions comprise the very end residues: W1P with D13P, P12P, and with P10P (Fig. 3B).
Figure 4.
A ribbon diagram comparison of the solution structures of the HAPep/α-BTX complex (the toxin is shown in orange, the peptide in magenta) and LLPep/α-BTX complex (the toxin is shown in gray, the peptide in cyan; ref. 11). Side chains of residues Y3P, Y4P, and Y11P, common to both peptides, are shown as well. All three fingers of α-BTX, as well as the N and C terminus of the HAPep and the LLPep, respectively, are indicated.
As shown in Table 1, two single-residue replacements, M1W or S10P, resulted in peptides (nos. 39 and 29, respectively) each inhibiting the binding of α-BTX to AChR 10-fold stronger as compared with LLPep (12). It seems that a proline residue at position 10 changes the backbone conformation in a manner that stabilizes the β-hairpin. In the LLPep/α-BTX complex, S10P forms intrapeptide interactions through its backbone and beta protons. In the HAPep/α-BTX complex, P10P also forms side-chain/side-chain interactions with W1P (Fig. 3B). The latter interacts through its side-chain with finger I of the toxin (Table 3) and forms intramolecular interactions with C-terminal residues of the peptide. However, in the LLPep/α-BTX complex, M1P is not involved in any NMR-observed inter or intramolecular interactions (Table 3). As a result of the W1P intermolecular interaction, R2P is brought into closer proximity to the toxin (Fig. 2B) and is thus involved in a larger number of intermolecular interactions and a greater intermolecular contact area (Table 3).
Y3P and Y4P, found in the two peptide-sequences, interact with the same regions of the toxin (see Table 3 and Fig. 4). In the HAPep/α-BTX complex, Y3P interacts mainly with α-BTX residues from finger II that constitute the intermolecular β-sheet structure, and the contribution of finger I residues to its intermolecular contact is low (Table 3). In the HAPep/α-BTX complex, Y4P has fewer intermolecular interactions; however, it forms new and important intrapeptide interactions with Y11P (Fig. 3B).
α-BTX residues of fingers I and II undergo the largest change in chemical shift when binding with either HAPep or LLPep, which is in accordance with the differences in the intermolecular interactions formed within the two complexes (Table 3). The C terminus of the toxin bound to the HAPep has a high degree of freedom (Fig. 1) because of the absence of long-range or intermolecular interactions for toxin residues R72-G74 (Table 3).
Implication for Binding to AChR.
A careful analysis of the amino acid sequence of the HAPep, in comparison with muscle as well as neuronal AChR sequences, reveals similarities in all residues except for positions 6, 7, and 8 of the peptide (Table 1). W1P that corresponds to position 187 of the muscle AChR (Table 1) contributes, through its side chain, to the formation of both intermolecular contacts (hydrophobic interactions as well as hydrogen bonding with A7, T8, and S9 of α-BTX), as well as intramolecular interactions within the HAPep/α-BTX complex. Previous studies revealed the importance of this residue for AChR binding to α-BTX (5). The motif PYPD in the HAPep (residues 10–13) is homologous to positions 196–199 of neuronal α7 AChR (Table 1). The presence of P10P and P12P seems to represent the proline subsite of muscle AChR that was shown to be of critical importance in establishing the affinity of the receptor to the toxin (32). Replacement of S6P and S7P in the HAPep with two cysteine residues (peptide no. 56, Table 1), to mimic all AChR sequences at the corresponding positions (α192–193, Table 1; ref. 33), did not increase the binding affinity of the resulting peptide to α-BTX.
The HAPep has a leucine residue at position 8 that was shown by NMR to play a major role in the binding and to interact directly with several toxin residues (Table 3). Replacement of this leucine by either proline or lysine, which are present at the corresponding position (α194) in muscle and neuronal AChRs that bind α-BTX (Table 1), resulted in a significant decrease in the affinity (12). L8P protrudes from the β-hairpin tongue (Figs. 2B and 3B), raising the possibility that it may mimic a leucine residue of the receptor (34) outside its known binding region (α184–200), thus forming a conformational epitope.
The appearance of β-sheet structural elements in the HAPep/α-BTX complex may represent a similar motif in the AChR/α-BTX complex. Independent studies of tritium-hydrogen exchange kinetics of the AChR (35) as well as difference CD spectroscopy, in the presence and absence of α-BTX (36), indicated that significant exchange retardation and a net increase in the β-structure component occurred, on α-BTX binding to AChR segments. Furthermore, a recent study of the secondary structure of the major AChR α-subunit determinant interacting with α-BTX revealed a similar pattern of inter- and intramolecular β-sheet in the complex of AChR-derived peptide α182–202 with α-BTX (37).
The design of the HAPep was based on combinatorial methods that take into consideration both structural information of the LLPep/α-BTX complex, as well as sequence data of AChR from different animal species. We assumed that, to obtain a high-affinity peptide, the general mode of binding of a given lead peptide should be maintained. Consequently, we systematically replaced residues only at those positions shown by NMR to be not essential for binding. Indeed, all peptide residues that contributed to the binding of α-BTX and/or to the stabilization of the peptide conformation in the LLPep/α-BTX complex (11) were found to have similar functions in the HAPep/α-BTX complex. The increased affinity of HAPep to α-BTX, in comparison to the LLPep, can be explained by the structure of the complex. The bound peptide folds into a β-hairpin structure created by two antiparallel β-strands, which combine with the already existing triple-stranded β-sheet of the toxin to form a five-stranded intermolecular, antiparallel β-sheet. The HAPep approaches the toxin more closely than the LLPep does, and creates a larger number of hydrogen bonds and electrostatic and hydrophobic interactions. It might represent a conformational epitope of the receptor and thus could serve as an effective antidote against toxin poisoning. Sequence similarity of the high-affinity peptide and of different AChRs implies that the binding-mode found in the HAPep/α-BTX complex probably mimics the receptor binding to the toxin.
Abbreviations
- 2D
two-dimensional
- AChR
nicotinic acetylcholine receptor
- α-BTX
α-bungarotoxin
- NOE
nuclear Overhauser effect
- rmsd
rms deviation. α-BTX residues are designated with a one-letter code followed by a number identifying their position in the sequence (e.g., V14)
- a superscript P is used to mark a peptide residue (e.g.
Y3P). The high-affinity peptide, WRYYESSLEPYPD, is designated HAPep. The library-lead peptide, MRYYESSLKSYPD, is designated LLPep
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1haa and 1haj).
References
- 1.Alexeev T, Krivoshein A, Shevalier A, Kudelina I, Telyakova O, Vincent A, Utkin Y, Hucho F, Tsetlin V. Eur J Biochem. 1999;259:310–319. doi: 10.1046/j.1432-1327.1999.00041.x. [DOI] [PubMed] [Google Scholar]
- 2.Kao P N, Dwork A J, Kaldany R R, Silver M L, Wideman J, Stein S, Karlin A. J Biol Chem. 1984;259:11662–11665. [PubMed] [Google Scholar]
- 3.Neumann D, Gershoni J M, Fridkin M, Fuchs S. Proc Natl Acad Sci USA. 1985;82:3490–3493. doi: 10.1073/pnas.82.10.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraenkel Y, Shalev D E, Gershoni J M, Navon G. Crit Rev Biochem Mol Biol. 1996;31:273–301. doi: 10.3109/10409239609106586. [DOI] [PubMed] [Google Scholar]
- 5.Neumann D, Barchan D, Fridkin M, Fuchs S. Proc Natl Acad Sci USA. 1986;83:9250–9253. doi: 10.1073/pnas.83.23.9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stroud R M, McCarthy M P, Shuster M. Biochemistry. 1990;29:11009–11023. doi: 10.1021/bi00502a001. [DOI] [PubMed] [Google Scholar]
- 7.Tzartos S J, Changeux J P. J Biol Chem. 1984;259:11512–11519. [PubMed] [Google Scholar]
- 8.Basus V J, Song G, Hawrot E. Biochemistry. 1993;32:12290–12298. doi: 10.1021/bi00097a004. [DOI] [PubMed] [Google Scholar]
- 9.Gentile L N, Basus V J, Shi Q L, Hawrot E. Ann NY Acad Sci. 1995;757:222–237. doi: 10.1111/j.1749-6632.1995.tb17479.x. [DOI] [PubMed] [Google Scholar]
- 10.Balass M, Katchalski-Katzir E, Fuchs S. Proc Natl Acad Sci USA. 1997;94:6054–6058. doi: 10.1073/pnas.94.12.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scherf T, Balass M, Fuchs S, Katchalski-Katzir E, Anglister J. Proc Natl Acad Sci USA. 1997;94:6059–6064. doi: 10.1073/pnas.94.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasher R, Balass M, Scherf T, Fridkin M, Fuchs S, Katchalski-Katzir E. Chem Biol. 2001;8:147–155. doi: 10.1016/s1074-5521(00)90063-2. [DOI] [PubMed] [Google Scholar]
- 13.Neumann D, Barchan D, Horowitz M, Kochva E, Fuchs S. Proc Natl Acad Sci USA. 1989;86:7255–7259. doi: 10.1073/pnas.86.18.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barchan D, Kachalsky S, Neumann D, Vogel Z, Ovadia M, Kochva E, Fuchs S. Proc Natl Acad Sci USA. 1992;89:7717–7721. doi: 10.1073/pnas.89.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barchan D, Ovadia M, Kochva E, Fuchs S. Biochemistry. 1995;34:9172–9176. doi: 10.1021/bi00028a029. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A, Ernst R R, Wüthrich K. Biochem Biophys Res Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 17.Davis D G, Bax A. J Am Chem Soc. 1985;107:2820–2821. [Google Scholar]
- 18.Piantini U, Sorenson O, Ernst R R. J Am Chem Soc. 1982;104:6800–6801. [Google Scholar]
- 19.Wagner G, Braun W, Havel T F, Schaumann T, Go N, Wüthrich K. J Mol Biol. 1987;196:611–639. doi: 10.1016/0022-2836(87)90037-4. [DOI] [PubMed] [Google Scholar]
- 20.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Nilges M, Gronenborn A M, Brünger A T, Clore G M. Protein Eng. 1988;2:27–38. doi: 10.1093/protein/2.1.27. [DOI] [PubMed] [Google Scholar]
- 22.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 23.Kosen P A, Finer-Moore J, McCarthy M P, Basus V J. Biochemistry. 1988;27:2775–2781. doi: 10.1021/bi00408a018. [DOI] [PubMed] [Google Scholar]
- 24.Scherf T, Hiller R, Naider F, Levitt M, Anglister J. Biochemistry. 1992;31:6884–6897. doi: 10.1021/bi00145a004. [DOI] [PubMed] [Google Scholar]
- 25.Stanfield R L, Wilson I A. Curr Opin Struct Biol. 1995;5:103–113. doi: 10.1016/0959-440x(95)80015-s. [DOI] [PubMed] [Google Scholar]
- 26.Hrabal R, Komives E A, Ni F. Protein Sci. 1996;5:195–203. doi: 10.1002/pro.5560050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tugarinov V, Zvi A, Levy R, Hayek Y, Matsushita S, Anglister J. Struct Fold Des. 2000;8:385–395. doi: 10.1016/s0969-2126(00)00119-2. [DOI] [PubMed] [Google Scholar]
- 28.Derrick J P, Maiden M C, Feavers I M. J Mol Biol. 1999;293:81–91. doi: 10.1006/jmbi.1999.3144. [DOI] [PubMed] [Google Scholar]
- 29.Lescar J, Stouracova R, Riottot M M, Chitarra V, Brynda J, Fabry M, Horejsi M, Sedlacek J, Bentley G A. J Mol Biol. 1997;267:1207–1222. doi: 10.1006/jmbi.1997.0950. [DOI] [PubMed] [Google Scholar]
- 30.Blanco F, Ramirez-Alvarado M, Serrano L. Curr Opin Struct Biol. 1998;8:107–111. doi: 10.1016/s0959-440x(98)80017-1. [DOI] [PubMed] [Google Scholar]
- 31.Ma B, Nussinov R. J Mol Biol. 2000;296:1091–1104. doi: 10.1006/jmbi.2000.3518. [DOI] [PubMed] [Google Scholar]
- 32.Kachalsky S G, Jensen B S, Barchan D, Fuchs S. Proc Natl Acad Sci USA. 1995;92:10801–10805. doi: 10.1073/pnas.92.23.10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conti-Tronconi B M, McLane K E, Raftery M A, Grando S A, Protti M P. Crit Rev Biochem Mol Biol. 1994;29:69–123. doi: 10.3109/10409239409086798. [DOI] [PubMed] [Google Scholar]
- 34.Changeux J P, Edelstein S J. Neuron. 1998;21:959–980. doi: 10.1016/s0896-6273(00)80616-9. [DOI] [PubMed] [Google Scholar]
- 35.McCarthy M P, Stroud R M. Biochemistry. 1989;28:40–48. doi: 10.1021/bi00427a007. [DOI] [PubMed] [Google Scholar]
- 36.Conti-Tronconi B M, Diethelm B M, Wu X D, Tang F, Bertazzon T, Schroder B, Reinhardt-Maelicke S, Maelicke A. Biochemistry. 1991;30:2575–2584. doi: 10.1021/bi00224a003. [DOI] [PubMed] [Google Scholar]
- 37.Samson, A., Chill, J. H., Rodriguez, E., Scherf, T. & Anglister, J. (2001) Biochemistry, in press. [DOI] [PubMed]
- 38.Koradi R, Billeter M, Wüthrich K. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. , 29–32. [DOI] [PubMed] [Google Scholar]