Abstract
MicroRNA are structural components of an epigenetic mechanism of post-transcriptional regulation of messenger RNA translation. Recently, there is significant interest in the application of microRNA as a blood-based biomarker of underlying physiologic conditions, and the therapeutic administration of microRNA inhibitors and mimics. The purpose of this review is to describe the current body of knowledge on microRNA regulation of genes involved in lipid metabolism, and to introduce the role of microRNA in development and progression of atherosclerosis.
Keywords: MicroRNA, Lipids, Epigenetics
Introduction
Dyslipidemia is a major risk factor for atherosclerosis, and reductions in levels of atherogenic lipoproteins substantially decrease risk. The current selection of behavioral and pharmacological interventions has variable and often inadequate effects on improving lipoprotein profiles. MicroRNA (miR) are a regulatory mechanism effecting mRNA translation, with downstream alterations in lipoprotein synthesis, reverse cholesterol transport and insulin signaling, making miR a potential biomarker of underlying pathology and therapeutic target and/or agent for regulating lipoprotein metabolism. Thus far, two miRs are proven to have a direct role in lipoprotein metabolism [1, 2], and a number of others have been implicated and warrant investigation. This paper will summarize the current knowledge of miR expression and function in lipid metabolism. Because the body of knowledge is limited, we will include miR findings from both in vitro and in vivo studies of animal models and humans.
MicroRNA Biogenesis
MicroRNA are nucleotide sequences 18–25 nucleotides in length that are composed of ribonucleic acid and possess a regulatory role in mRNA translation. The first observation of miR described binding of short antisense RNA to mRNA in the cellular cytoplasm of Caenorhabditis elegans. [3] Subsequently, numerous miR have been found in diverse species, including humans, the sequences of which are highly conserved across species, supporting a critical role of these molecules in post-transcriptional gene regulation. [4–6] The first description of miR in metabolic processes in animals was in Drosophila melanogaster. [7]
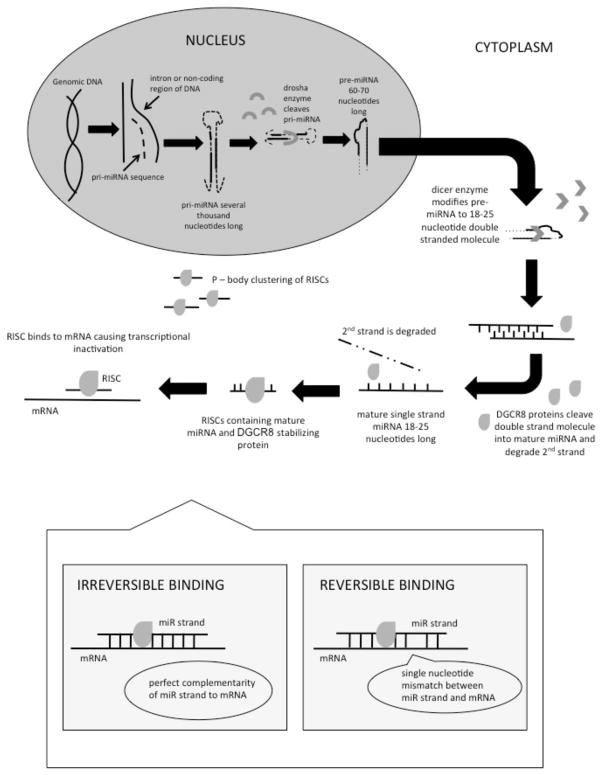
MiR sequences originate in the cell nucleus, where miR encoding genes are transcribed, generating miR precursors (Figure 1). Initially, a primary miR (pri-miR) transcript several thousand base pairs in length is formed, which adopts a double strand stem-loop structure for stability. [8] Pri-miR is processed into a 60–70 nucleotide molecule called pre-miR by a nuclear ribonuclease called Drosha. [9, 10] Pre-miR is then exported to the cytoplasm where it is further modified by the Dicer ribonuclease into a double stranded nucleotide sequence 18–25 nucleotides in length. [11] This double stranded molecule is cleaved into a functional strand called the “guide strand” and the complementary strand termed the “passenger strand,” which is subsequently degraded by the DiGeorge syndrome critical region gene 8 (DGCR8) protein, commonly called Pasha [12], yielding the mature single strand miR. [13, 14] In order to preserve these molecules from degradation in the cytoplasm, the functional miR is bound to DGCR8 protein to form a ribonucleoprotein called the ribonucleoprotein miR-induced silencing complex (RISC). RISCs accumulate in cytoplasmic foci called P-bodies. [15–17] In addition to storing miR molecules, RISCs contain enzymes necessary for mRNA degradation following cleavage caused by miR binding. [17] Ultimately, RISCs bind to the 3′ un-translated region of mRNA in order to inhibit initiation of translation, thus acting to modulate the quantity and rate of protein translation. [18] MiR that are perfectly complimentary to the mRNA sequences cause mRNA cleavage, whereas imperfect complementarity causes reversible inhibition of mRNA translation. [19]
Figure 1. MicroRNA processing pathways in the nucleus and cytoplasm.
Dotted lines: segments of RNA that are cleaved from precursor molecules during processing of mature microRNA
Solid lines: segments of RNA that are retained during processing and incorporated in mature microRNA
RISC: ribonucleoprotein miR induced silencing complex
DGCR8 protein: DiGeorge syndrome critical region gene 8, whish is an essential cofactor protein for primary miR processing
Individual MiR species are assigned a numeric name in sequential order by date of discovery and classification. [20] The full name of a miR species is preceded by a three-letter prefix designating the species (e.g., hsa-miR-101 was the 101st miR to be reported in Homo sapiens). [20] Orthologous, or identical miR sequences found in multiple species are assigned the same numeric value with a species-specific prefix. [20] For example, the mouse orthologue to the human example given above is mmu-miR-101 (Mus musculus miR-101). MiR species whose sequence differs at only one or two locations are called paralogues and are given a single letter suffix (e.g. hsa-miR-30a, hsa-miR30b, hsa-miR30c). [20]
MiR regulate numerous disease processes, including cell differentiation and proliferation in cardiac tissue during fetal cardiac development and cardiac hypertrophy, conduction physiology via potassium channels, angiogenesis in myocardium, glucose metabolism, lipid metabolism, and adipocyte differentiation. [21, 22] The first studies of miR and metabolism were performed in fruit flies and determined that dme-miR-14 regulates fat metabolism by down-regulating the production of triacylglycerol and diacylglycerol. [7] Further work demonstrated that flies lacking dme-miR-278 become insulin resistant with an increased insulin production accompanied by elevated circulating glucose. [23] MiR expression patterns can differ by tissue type, suggesting that this mechanism of gene expression regulation is dynamic and can be highly specific. [24] Because miR regulation of mRNA translation is a highly dynamic process, it is thought that miR activity may be triggered by external stimuli, mediating an organism’s response to changes in its environment. Thus there is potential for miRs to be biomarkers for disease progression and/or response to therapy, and ultimately targets of therapeutics for cardiovascular and metabolic disorders.
MiR-33
The miR showing the greatest therapeutic promise to date is miR-33. [25] The transcripts for the two hsa-miR-33 isoforms (a and b) are located in a non-coding region of Sterol Regulatory Binding Element Factor (SREBF) genes, which regulates cholesterol synthesis by several mechanisms, including transcriptional regulation of LDL-c receptors (Table 1, Figure 2). [2] MiR-33 is highly conserved across species, and can be found in numerous cell types, including macrophages, hepatocytes, and endothelial cells. [2] Microarray experiments found hsa-miR-33 to be differentially expressed in response to cholesterol enrichment or depletion in human macrophage cells in vitro. [2] Hsa-miR-33-a expression is directly correlated with levels of SREBF-2, and inversely correlated with expression of Adenosine Triphosphate Binding Cassette Transporter (ABCA1), a cholesterol efflux pump. [2] Similar findings were observed in mouse models of hypercholesterolemia (i.e., homozygous LDL-c receptor knockout and homozygous apolipoprotein E knockout). [2] Also, mmu-mir-33 was inversely associated with serum cholesterol, with decreased expression in homozygous LDL-c receptor knockout mice fed high fat diets compared to normal diet. Corresponding changes in SREBF-2 (positive) and ABCA1 (negative) were observed. Importantly, these findings prove that miR-33 is responsive to environmental changes (i.e. alterations in cholesterol levels secondary to diet or medication), making miR potential biomarkers of response to environmental stimuli and targets of therapeutic interventions.
Table 1.
Gene Targets of MicroRNA Regulating Lipid Metabolism
| miR species | Direction of Effect | Observed Targets | Target Gene Function | Citation |
|---|---|---|---|---|
| 33 | ↓ | Adenosine Triphosphate Binding Cassette Transporter (ABCA1) | Cholesterol efflux pump; initiation of HDL-c formation |
Rayner et. al., 2010 Rayner, Sheedy, et. al., 2011 Rayner, Esau, et. al., 2011 |
| 33 | ↓ | Neimann Pick Type C1 (NPC1) | Transports LDL-c to endosomes/lysosomes for hydrolysis to free cholesterol | Rayner et. al., 2010 |
| 33 | ↓ | Hydroxylacyl-CoA Dehydrogenase/3 Ketoacyl-CoA Thiolase, beta subunit (HADHB) | Mitochondrial oxidation of fatty acids |
Rayner, Esau, et. al., 2011 Dávalos et. al., 2011 |
| 33 | ↓ | Carnitine O-Octanoyltransferase (CROT) | Mitochondrial oxidation of fatty acids |
Rayner, Esau, et. al., 2011 Dávalos et. al., 2011 |
| 33 | ↓ | Carnitine Palmitoyltransferase 1A (CPT1A) | Mitochondrial oxidation of fatty acids |
Rayner, Esau, et. al., 2011 Dávalos et. al., 2011 |
| 33 | ↑ | Sterol Regulatory Element Binding Transcription Factor (SREBF) | Regulates transcription of LDL receptor, fatty acids, and cholesterol synthesis pathways | Rayner, Esau, et. al., 2011 |
| 33 | ↓ | AMP Kinase (AMPK) | Regulates ATP-consuming biopathways to prevent cellular ATP depletion |
Rayner, Esau, et. al., 2011 Dávalos et. al., 2011 |
| 33 | ↓ | Insulin Receptor Substrate 2 (IRS2) | Mediates effects of insulin |
Rayner, Esau, et. al., 2011 Dávalos et. al., 2011 |
| 122 | ↓ | Glycogen Synthase 1(GYS1) | Catalyzes the addition of glucose to glycogen | Esau et. al., 2006 |
| 122 | ↓ | Solute Carrier Family 7 (SLC7A1, CAT1) | Transport of cationic amino acids in hepatic cells |
Esau et. al., 2006 Cirera et. al., 2010 |
| 122 | ↓ | Misshapen/NIK-Related Kinase 1 (MINK1) | Activates JNK and p38 pathways, which are involved in stress response | Esau et. al., 2006 |
| 122 | ↓ | Aldoase A, Fructose-Biphosphate (ALDOA) | Catalyzes conversion of fructose to glyceraldehydes | Esau et. al., 2006 |
| 122 | ↓ | Cyclin G1 (CCNG1) | DNA repair | Esau et. al., 2006 |
| 122 | ↓ | Procollagen-Proline, 2- Oxoglutarate-4-Dioxygenase (P4HA1) | Post-translational modification of collagens | Esau et. al., 2006 |
| 122 | ↑ | Sterol Regulatory Element Binding Transcription Factor (SREBF) | Regulates transcription of LDL receptor, fatty acids, and cholesterol synthesis pathways | Iliopoulos et. al., 2010 |
| 122 | ↑ | Diacylglycerol O-Acyltransferase 2 (DGAT2) | Catalyzes synthesis of triglycerides | Iliopoulos et. al., 2010 |
| 122 | ↑ | Fatty Acid Synthase (FASN) | Synthesis of long chain fatty acids | Iliopoulos et. al., 2010 |
| 122 | ↑ | Nuclear Receptor Subfamily 1, group H, member 3/Liver X Receptor α (NR1H3) | Cholesterol homeostasis | Iliopoulos et. al., 2010 |
| 122 | ↑ | Acetyl-CoA Carboxylase α (ACACA) | Fatty acid synthesis | Iliopoulos et. al., 2010 |
| 122 | ↑ | Cholesterol 7α-hydroxylase (CYP7A1) | Bile acid production, absorption of cholesterol | Song et. al., 2010 |
| 370 | ↓ | Carnitine Palmitoyltransferase 1A (CPT1A) | Mitochondrial oxidation of fatty acids | Iliopoulos et. al., 2010 |
| 370 | ↑ | Sterol Regulatory Element Binding Transcription Factor 1c (SREBF1c) | Regulates transcription of LDL receptor, fatty acids, and cholesterol synthesis pathways | Iliopoulos et. al., 2010 |
| 370 | ↑ | Diacylglycerol O-Acyltransferase 2 (DGAT2) | Catalyzes synthesis of triglycerides | Iliopoulos et. al., 2010 |
| 370 | ↑ | Fatty Acid Synthase (FASN) | Synthesis of long chain fatty acids | Iliopoulos et. al., 2010 |
| 370 | ↑ | Nuclear Receptor Subfamily 1, group H, member 3/Liver X Receptor α (NR1H3) | Cholesterol homeostasis | Iliopoulos et. al., 2010 |
| 370 | ↑ | Acetyl-CoA Carboxylase α (ACACA) | Fatty acid synthesis | Iliopoulos et. al., 2010 |
| 758 | ↓ | Adenosine Triphosphate Binding Cassette Transporter (ABCA1) | Cholesterol efflux pump; initiation of HDL-c formation | Ramirez et. al., 2011 |
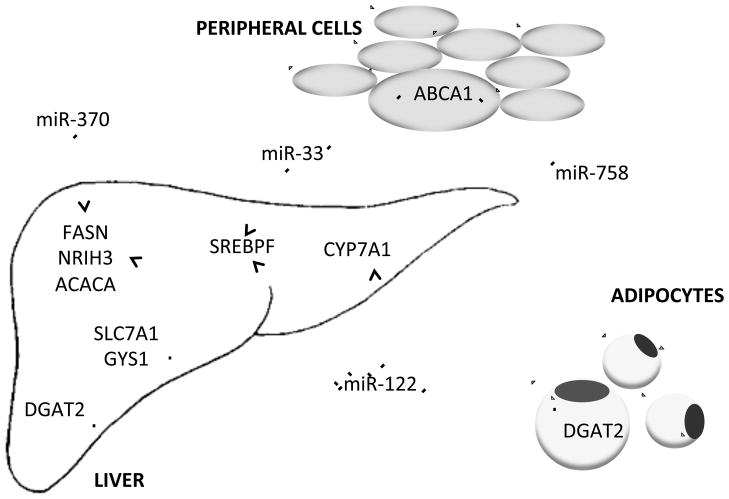
Figure 2. Messenger RNA targets of microRNA that are involved in regulation of lipoprotein metabolism.
Pointed tip arrows: up-regulation of target mRNA expression
Blunted tip arrows: down-regulation of target mRNA expression
ABCA1: Adenosine Triphosphate Binding Cassette Transporter
SREBPF: Sterol Regulatory Element Binding Transcription Factor
DGAT2: Diacylglycerol O-Acyltransferase 2
CYP7A1: Cholesterol 7α-hydroxylase
FASN: Fatty Acid Synthase
NRIH3: Nuclear Receptor Subfamily 1, group H, member 3/Liver X Receptor α
ACACA: Acetyl-CoA Carboxylase α
SLC7A1: Solute Carrier Family 7
GYS1: Glycogen Synthase 1
Additional putative binding sites for miR-33 occur on ATP-Binding Cassette, SubFamily G (ABCG1) and Neimann Pick type C1 (NPC1) mRNA. In vitro transfection studies demonstrated that miR-33 decreased ABCA1 and NPC1 mRNA, but not ABCG1 mRNA, in human macrophages. Finally, lentivirus transfection of mice with mmu-miR-33 substantially decreased ABCA1, resulting in 22% decrease in plasma high-density lipoprotein cholesterol (HDL-c), while transfection of mmu-miR-33 inhibitor resulted in increased expression ABCA1 and accompanying 25% increase in HDL-c. [2] Increased HDL-c in response to miR-33 inhibitors corresponds with several anti-atherogenic events, including changes in reverse cholesterol transport, decreased plaque size and lipid accumulation in foam cells, decreased number of macrophages, and increased plaque stability in the aortic root of mouse hearts. [26] An intriguing observation is that the transfected miR-33 inhibitors were found to actually enter atherosclerotic lesions, and increase expression of ABCA1 in macrophages residing in the plaque. [26] The same macrophages displayed changes in expression of inflammatory genes. [26]
MiR-33 also regulates mRNA associated with β–oxidation of fatty acids, including Hydroxylacyl-CoA Dehydrogenase/3 Ketoacyl-CoA Thiolase (HADHB), Carnitine O-Octanoyltransferase (CROT), and Carnitine Palmitoyltransferase 1A (CPT1A) (Table 1). [27] Overexpression of miR-33b in radioactively labeled hepatocytes (Huh7 and HepG2) decreased the rate of oxidation, while inhibition induced β–oxidation. [27] In Huh7 cells, transfection with miR-33b also led to accumulation of a greater number of triglycerides and larger lipid droplets. [27]
Until recently, studies of miR-33 function were stymied by the lack of the presence of both the miR-33b isoform and the SREBF1 gene in mice. A recent study of African green monkeys, who, like humans, have the SREBF1 gene and sae-miR-33b isoform, found substantial alterations in gene expression and lipoprotein levels after sae-miR-33-b inhibition. [28] Animals receiving sae-miR-33b inhibitor had decreased triglycerides (50%) and increased HDL-c (50%) after 12 weeks of treatment, with no apparent side effects. [28] Numerous corresponding changes in mRNA involved in regulation of lipoprotein levels with putative binding sites for sae-miR-33b, including ABCA1, CROT, HADHB, SREBF1, Protein Kinase AMP-Activated 1 (AMPK), and CPT1A were associated with changes in lipoprotein levels (Table 1, Figure 2). [28]
In both in vitro studies of Huh7 cells and in vivo studies of mice, miR-33 appears to inhibit translation of Insulin Receptor Substrate 2 (IRS2) mRNA, with accompanying downstream alterations in IRS2 targets (Table 1). [27] Similarly, the expression of IRS2 is increased in African Green Monkeys treated with sae-miR-33b inhibitor. [28] IRS2 is a mediator of insulin and insulin-like growth factor 1, and decreased levels are associated with development of insulin resistance. [29] Impaired insulin metabolism is widely known to be a risk factor for cardiovascular disease, and is associated with lipoprotein metabolism in that impaired insulin secretion resulting from insulin resistance also leads to decreased lipoprotein lipase response and accumulation of plasma fatty acids. [30] Correspondingly, in vitro inhibition of miR-33a and miR-33b up-regulates fatty acid oxidation and hepatocyte insulin response. [27] Similar to studies of miR-mediated lipoprotein metabolism, further studies of insulin metabolism in animal models are limited by the lack of miR-33b in traditional animal models of diabetes (i.e. mice, rats).
MiR-122
MiR-122 accounts for 70% of all miR in the adult mouse liver and is conserved in species ranging from fish to vertebrate mammals. [24, 31] Several experimental studies of miR-122 indicate a regulatory role in lipid metabolism. Transfection of an anti-sense oligonucleotide inhibitor of mmu-miR-122 into a mouse hepatocyte-derived cell line (AML12) caused an increase in six mRNA (Glycogen Synthase 1 (GYS1), Solute Carrier Family 7 (SLC7A1), Misshapen/NIK-Related Kinase 1 (MINK1), Aldoase A, Fructose-Biphosphate (ALDOA), Cyclin G1 (CCNG1), and Procollagen-Proline, 2-Oxoglutarate-4-Dioxygenase, Alpha Subunit, Isoform 1 (P4HA1)) [1], some of which may have indirect effects on lipid metabolism (Table 1, Figure 2). The same results were observed with mmu-miR-122 inhibition in mice, and in both experiments, an inhibitor dose-dependent response of mRNA expression was observed. [1] Importantly, the mmu-miR-122 inhibited mice had a significant decrease in plasma total cholesterol and plasma triglycerides compared to controls. [1]
Similarly, hsa-miR-122, along with hsa-miR-422, inhibits translation of the Cholesterol 7α-hydroxylase (CYP7A1) mRNA, and therefore bile acid synthesis. [32] In a human hepatocyte-derived cell line (HepG2), cells treated with hsa-miR-122 inhibitor displayed increased bile acid concentrations compared to untreated controls, implicating hsa-miR-122 regulation of CYP7A1 as a possible mechanism for decreasing serum cholesterol and triglycerides. [32] In addition, inhibition of hsa-miR-122 in Hep2G cells decreased the expression of SREBF-1c, a gene that regulates transcription of the low density lipoprotein cholesterol (LDL-c) receptor and cholesterol synthesis pathway, along with other enzymes known to regulate fatty acid and triglyceride accumulation in the liver. [33] The activity of hsa-miR-122 in this context appears to be regulated by hsa-miR-370, making the latter an additional potential therapeutic target. [33] In a porcine model of obesity, liver levels of ssc-miR-122 were down regulated in response to a high cholesterol diet compared to normal diet, and corresponding increases in body weight, total cholesterol, and HDL-c were observed. [34] However, no changes in SLC7A1, a target of miR-122 with a regulatory role in endothelial function, were observed.
MiR-370
As described above, miR-370 has an indirect effect on lipid metabolism via its up-regulation of miR-122 and its targets. Hsa-miR-370 has a direct inhibitory effect on CPT1A in Hep2G cells, which facilitates β-oxidation and is also a target of miR-122, as well as direct effects on SREBF-1c (Table 1, Figure 2). [33] Inhibition of three enzymes (diacylglycerol O-acyltransferase 2 (DGAT2), fatty acid synthase (FASN), and acetyl-CoA carboxylase 1 (ACACA)) is also seen in Hep2G cells in vitro, and it appears that changes in FASN and ACACA are mediated by the effects on SREBF and DGAT2 (Table 1, Figure 2). [33] Transfection of mmu-miR-370 in mice resulted in increased hepatic triglyceride and cholesterol levels, but corresponding changes in mRNA targets were not described. [33]
Emergent Candidate MiR and Lipoprotein Regulation
Several additional miR show preliminary evidence of a role in the regulation of lipoprotein metabolism. MiR-758 was a bioinformatically predicted regulatory agent for ABCA1, the protein enabling cellular cholesterol efflux by apolipoprotein AI containing particles (i.e., HDL-c). In vitro studies of mouse (J774 cells) and human (THP-1 cells) macrophages revealed miR-758 is inversely associated with ABCA1, with inhibition of miR-758 causing increased ABCA1 expression, and administration of a miR-758 mimic causing decreased expression. [35] Further study of an animal model found expression of mmu-miR-758 was decreased in peritoneal macrophages from mice fed a high fat diet, with accompanying increases in the expression of ABCA1. [35]
MiR-106b also appears to target ABCA1, impairing cellular cholesterol efflux in mouse neuroblastoma (Neuro2a) cells treated with radioactively labeled cholesterol. [36] This miR shows important neurologic implications as amyloid β peptide, a precursor to Alzheimer’s Disease that appears to be regulated by ABCA1, is substantially increased by transfection of miR-106b in vitro. [36]
Emergent Candidate MiR and Atherosclerosis
A primary consequence of dyslipidemia (i.e., elevated non-HDL-c) is the development of atherosclerosis. Thus miR-modulated regulation of lipid metabolism influences the development of atherosclerotic lesions. Several studies show that miR influence the development and progression of atherosclerotic lesions via a number of mechanisms, including inflammation of macrophages and foam cell development, smooth muscle cell proliferation and migration, and apoptosis. Here we provide a brief summary of miR involved in atherosclerosis. Additional review of this body of literature can be found elsewhere. [37]
The hypothesis that miR may have overlapping regulatory functions in lipid metabolism and atherosclerosis is supported by the observation that miR-21 is a prevalent miR in cardiomyocytes, regulating apoptosis and vascular smooth muscle cell proliferation. [38] MiR-21 also appears to have a role in liver function and cholesterol regulation. Hepatic expression of miR-21 is up-regulated by free unsaturated fatty acids, with increased expression in the livers of rats fed a high fat diet and human liver tissue of obese individuals. [39] The mechanism of action of miR-21 in this setting is down-regulation of expression of the Phosphatase and Tensin Homolog protein, a tumor suppressor involved in early hepatic insulin hypersensitivity. [39] These findings provide additional evidence for dynamic activity of miR in response to environmental stimuli, resulting in alterations of metabolic function. In humans, has-miR-21 is detectable in blood [40, 41], and therefore feasibly measured in a research context, making this an ideal target for testing whether variable expression is associated with risk reduction interventions.
Microarray measurement of miR in human monocytes in vitro exposed to oxidized LDL-c showed differential expression of several miR, with as much as 11-fold change in expression (hsa-miR-125-5p) in exposed cells versus controls. [42] Oxidized LDL-c is a pro-atherogenic species that has a high affinity for macrophages, facilitating in the development of foam cells and atherosclerotic lesions. Administration of hsa-miR-125-5p inhibitors in human monocytic leukemia (THP-1) cells results in significant increases in total cholesterol after exposure to oxidized LDL-c, as well as decreased oxidized LDL-c uptake in macrophages. [42] Inhibition studies also show alterations in inflammatory markers associated with atherosclerosis, including Interleukin-6, Interleukin-2, Transforming Growth Factor-β, and Tumor Necrosis Factor-α. [42] Ischemia-induced arterial injury is associated with changes in miR expression in endothelial cells in rats, including up-regulation of rno-miR-21 and down-regulation of rno-miR-125. [43] Numerous additional studies report that miR expression is associated with changes in vascular smooth muscle cell function in mammalian species, particularly mmu-miR-143, mmu-miR-145, rno-miR-145 [44–47], rno-miR-221, and rno-miR-222. [48]
Further research is needed to identify where atherosclerosis-associated differential expression of miR occurs in tissues associated with this disease (e.g. endothelial cells, smooth muscle cells, macrophages). Furthermore, understanding of all of the regulatory effects of miR on expression of genes associated with atherosclerosis mechanisms, including which mRNA are targeted, remains incomplete. And finally, the temporally and spatially (i.e., tissue type, cell type) specific expression represents a critical additional layer of complexity that warrants detailed study.
Conclusions
MiR are a class of small RNA molecules with reversible regulatory actions on gene expression. MiR regulate a wide variety of physiologic processes including dyslipidemia, a major risk factor for atherosclerosis. MiR-33 targets ABCA1, and has tremendous prospective therapeutic implications, given its direct role on HDL-c, non-HDL-c, and triglycerides. Importantly, exogenous miR-33 treatment has recently been studied in large primates, with dramatic improvements in lipoprotein levels and no apparent short-term side effects. In addition, inhibition of miR-33 appears to improve insulin sensitivity.
MiR-122 is the most prevalent miR in the livers of many species, and targets several genes that control cholesterol metabolism. Moreover, miR-122 appears to act in tandem with other miR (i.e. mir-370) to modify lipoprotein metabolism in the liver. The aggregate effects of miR-122 appear to be decreased total cholesterol and triglycerides and decreased production of the LDL-c receptor. There are several additional miR with early evidence suggesting functional roles in cholesterol metabolism, and the activity of miR appears to be responsive to changes in the environment. Thus, these miR may represent plausible anti-atherogenic therapeutic targets.
Clinical Implications
There are three potential implications for miR in clinical practice. The first is measurement of risk for significant clinical outcomes. Taking miR-33 as an example, differential expression is observed in genetically altered mice lacking LDL-c receptors compared to controls. Importantly, these differences were observed in hepatic tissue. However, in order for miR expression to be clinically practical in humans, differential expression must be observed in readily accessible tissue, such as blood. Measurement of miR in peripheral blood leukocytes for a wide array of conditions including cancer [49, 50], myocardial infarction and injury [51, 52], stroke [53], and sepsis [54], show distinct patterns of expression in disease states compared to controls. These findings suggest that use of differential miR expression patterns in peripheral blood of humans with abnormal lipid metabolism is a plausible clinical biomarker. To our knowledge, no studies of human miR expression in dyslipidemia have been reported (Table 2). There are three possible implications for measurement of miR profiles in peripheral blood: (1) indirect markers of underlying pathology, (2) direct markers of pathology, and (3) reflective makers of pathology. In order to study the latter, studies measuring simultaneous expression of miR in blood and organs are needed. No studies comparing miR expression levels in blood to organ specific (e.g. macrophages and liver) expression were found, however comparative studies in animals have been reported for other disease processes. [55]
Table 2.
Evidence for Clinical Applications of microRNA in in vitro Studies of Human-Derived Cell Lines and in vivo Studies of Humans and Related Primates
| miR species | Markers of response | Targeted interventions |
|---|---|---|
| 33 | miR-33 is down regulated by cholesterol enrichment in human THP-1 macrophages in vitro (Rayner et. al., 2010) | miR-33 inhibition decreased serum triglycerides and increased HDL-c in African Green Monkeys and upregulated fatty acid oxidation and hepatic insulin response in Huh2 hepatocytes in vitro (Rayner et. al., 2011); miR- 33 inhibition decreased expression of Insulin Receptor Substrate 2 mRNA in HepG2 hepatocytes in vitro (Davalos, et. al., 2011) |
| 122 | miR-122 inhibition decreased expression of Sterol Regulatory Element Binding Factor 1c mRNA (Iliopoulos, et. al., 2010) and increased bile acid concentrations in HepG2 hepatocytes in vitro (Song et. al., 2010) | |
| 370 | miR-370 transfection upregulated Sterol Regulatory Binding Factor 1c, Diacylglycerol O-Acyltransferase 2, Fatty Acid Synthase, and acyl-CoA carboxylase 1 in HepG2 hepatocytes in vitro (Iliopoulos et. al., 2010) | |
| 758 | Inhibition of miR-758 decreased Adenosine Triphosphate Binding Cassette Transporter mRNA in THP-1 macrophages in vitro (Ramirez et. al., 2011) | |
| 21 | miR-21 is upregulated by elevated free fatty acids secondary to obesity in human liver (Vinciguerra et. al., 2009) | |
| 125-5p | miR125-5p is upregulated in THP-1 monocytes exposed to oxidized LDL-c in vitro (Chen et. al., 2009) | Inhibition of mir-125-5p increased total cholesterol and oxidized LDL-c uptake, and altered expression of inflammatory markers in THP-1 monocytes in vitro (Chen et. al., 2009) |
microRNA (miR)
High-density lipoprotein cholesterol (HDL-c)
Messenger RNA (mRNA)
Low-density lipoprotein cholesterol (LDL-c)
Secondly, a translational application for blood-based miR measurement is to assess expression before and after risk reduction interventions (e.g., medication, diet, physical activity) to determine whether blood-based expression is differential, and whether miR are biomarkers for changes in clinical risk profile. Data suggest that miR are highly responsive to changes in the in vitro cellular environment and to an organisms environment. Keeping with the miR-33 example provided above, differential expression has been observed between mice fed high fat compared to normal diets. Studies report encouraging preliminary findings supporting environment-induced response in human-derived cell lines in vitro (Table 2). Future studies employing repeated measures designs for miR expression in humans are warranted in order to determine whether miR can serve as a meaningful measure of the effects of interventions and underlying physiologic changes.
Finally, there are currently two plausible mechanisms for miR as an intervention. The first is administration of antisense oligonucleotide inhibitors to specific miR (also termed antagomiRs), into the peripheral circulation. Inhibitors block the activity of the endogenously occurring miR, resulting in alteration of gene expression at a specific location. Animal studies have demonstrated successful inhibition of miR-33 activity following inhibitor administration, accompanied by substantial changes in lipoprotein profile. The second application is miR mimics, which are synthesized and chemically modified copies of miR species that can be administered into the peripheral circulation in order to enhance the overall effect of a specific miR. Both applications are effective in in vitro studies of human-derived cells (Table 2). A review of these methods is described in detail elsewhere. [56]
Strengths and Weaknesses
Significant strengths of miR research are the potential for an important complementary approach to assessing cardiovascular risk, measurement of response to behavioral and pharmacologic interventions, and as therapeutic targets for cardiovascular risk reduction. MiR are highly dynamic, and alterations in expression are likely to precede changes in current standard clinical biomarkers. A limitation to clinical translation of miR research to the clinical setting is that the majority of experiments on miR expression and lipoprotein metabolism were performed on tissues that are not feasible to access in routine clinical care (e.g. liver, endothelium). In order for a biomarker to be clinically useful, the tissue source must be easily obtained, and further research on feasibility and utility of blood-based detection in humans is needed.
Acknowledgments
Funding for Elena Flowers’ doctoral studies was provided by
NIH/NIGMSR25 GM56847
NIH/NCRR/OD UCSF-CTSI Grant TL1 RR024129
Funding for Bradley E. Aouizerat was provided by
NIH/NCRR/OD UCSF-CTSI Grant Number KL2 RR024130
Abbreviations
- miR
microRNA
- mRNA
messenger RNA
- RNA
ribosomal nucleic acid
- pri-miR
primary microRNA
- DGCR8
DiGeorge syndrome critical region gene 8
- RISC
ribonucleoprotein miR-induced silencing complex
- LDL-c
low density lipoprotein cholesterol
- HDL-c
high density lipoprotein cholesterol
- SREBF
Sterol Regulatory Binding Element Factor
- ABCA1
Adenosine Triphosphate Binding Cassette Transporter
Footnotes
Author Contributions:
Elena Flowers conducted literature search, synthesized data, composed manuscript, and created tables and figures.
Erika S. Froelicher advised first author on manuscript composition, editorial process, and provided critical review of content.
Bradley E. Aouizerat provided expert review and content checking, and advised first author on manuscript composition and scope.
Disclosures/conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Rayner KJ, Suarez Y, Davalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–3. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 7.Xu P, Vernooy SY, Guo M, et al. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–5. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Jeon K, Lee JT, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein E, Caudy AA, Hammond SM, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 12.Yeom KH, Lee Y, Han J, et al. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34(16):4622–9. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matranga C, Tomari Y, Shin C, et al. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Lee Y, Yeom KH, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 16.Hammond SM, Bernstein E, Beach D, et al. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Valencia-Sanchez MA, Hannon GJ, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nature cell biology. 2005;7(7):719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30(4):363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 19.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278(45):44312–9. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poy MN, Spranger M, Stoffel M. microRNAs and the regulation of glucose and lipid metabolism. Diabetes Obes Metab. 2007;9 (Suppl 2):67–73. doi: 10.1111/j.1463-1326.2007.00775.x. [DOI] [PubMed] [Google Scholar]
- 22.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91(3):209–17. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20(4):417–22. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 25.Rottiers V, Najafi-Shoushtari SH, Kristo F, et al. MicroRNAs in Metabolism and Metabolic Diseases. Cold Spring Harbor symposia on quantitative biology. 2011 doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayner KJ, Sheedy FJ, Esau CC, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121(7):2921–31. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davalos A, Goedeke L, Smibert P, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108(22):9232–7. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–7. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirola L, Johnston AM, Van Obberghen E. Modulation of insulin action. Diabetologia. 2004;47(2):170–84. doi: 10.1007/s00125-003-1313-3. [DOI] [PubMed] [Google Scholar]
- 30.Reaven GM. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes. 1988;37(12):1595–607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- 31.Chang J, Nicolas E, Marks D, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1(2):106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 32.Song KH, Li T, Owsley E, et al. A putative role of micro RNA in regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes. J Lipid Res. 2010;51(8):2223–33. doi: 10.1194/jlr.M004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iliopoulos D, Drosatos K, Hiyama Y, et al. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res. 2010;51(6):1513–23. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cirera S, Birck M, Busk PK, et al. Expression profiles of miRNA-122 and its target CAT1 in minipigs (Sus scrofa) fed a high-cholesterol diet. Comp Med. 2010;60(2):136–41. [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez CM, Davalos A, Goedeke L, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011;31(11):2707–14. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Yoon H, Ramirez CM, et al. miR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Experimental neurology. 2011 doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Najafi-Shoushtari SH. MicroRNAs in cardiometabolic disease. Current atherosclerosis reports. 2011;13(3):202–7. doi: 10.1007/s11883-011-0179-y. [DOI] [PubMed] [Google Scholar]
- 38.Cheng Y, Zhang C. MicroRNA-21 in cardiovascular disease. J Cardiovasc Transl Res. 2010;3(3):251–5. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinciguerra M, Sgroi A, Veyrat-Durebex C, et al. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 up-regulation in hepatocytes. Hepatology. 2009;49(4):1176–84. doi: 10.1002/hep.22737. [DOI] [PubMed] [Google Scholar]
- 40.Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaz C, Ahmad HM, Sharma P, et al. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics. 2010;11:288. doi: 10.1186/1471-2164-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen T, Huang Z, Wang L, et al. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83(1):131–9. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 43.Ji R, Cheng Y, Yue J, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100(11):1579–88. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Y, Liu X, Yang J, et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105(2):158–66. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elia L, Quintavalle M, Zhang J, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16(12):1590–8. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C. MicroRNA-145 in vascular smooth muscle cell biology: a new therapeutic target for vascular disease. Cell Cycle. 2009;8(21):3469–73. doi: 10.4161/cc.8.21.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Cheng Y, Zhang S, et al. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104(4):476–87. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keller A, Leidinger P, Borries A, et al. miRNAs in lung cancer - studying complex fingerprints in patient’s blood cells by microarray experiments. BMC Cancer. 2009;9:353. doi: 10.1186/1471-2407-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila Pa) 2009;2(9):807–13. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ai J, Zhang R, Li Y, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391(1):73–3. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Ji X, Takahashi R, Hiura Y, et al. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55(11):1944–9. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 53.Tan KS, Armugam A, Sepramaniam S, et al. Expression profile of MicroRNAs in young stroke patients. PLoS One. 2009;4(11):e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasilescu C, Rossi S, Shimizu M, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 2009;4(10):e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu DZ, Tian Y, Ander BP, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2009;30(1):92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103(9):919–28. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]