Abstract
8-Oxo-7,8-dihydroguanine (OG) is the most common base damage found in the cell where it resides in many structural contexts including the nucleotide pool, single-stranded DNA at transcription forks and replication bubbles, and in duplex DNA base paired with either A or C. OG is prone to further oxidation to the highly mutagenic hydantoin products, spiroiminodihydantoin (Sp) and 5-guanidinohydantoin (Gh) in a sharply pH-dependent fashion within nucleosides. In the present work, studies were conducted to determine how the structural context affects OG oxidation to the hydantoins. These studies revealed a trend in which the Sp yield was greatest in unencumbered contexts, such as nucleosides, while the Gh yield increased in oligodeoxynucleotide (ODN) contexts or at reduced pH. Oxidation of oligomers containing hydrogen bond modulators (2,6-diaminopurine, N4-ethylcytidine) or alteration of the reaction conditions (pH, temperature, and salt) identify base stacking, electrostatics and base pairing as the drivers of the key intermediate 5-hydroxy-8-oxo-7,8-dihydroguanine (5-HO-OG) partitioning along the two hydantoin pathways, allowing us to propose a mechanism for the observed base pairing effects. Moreover, these structural effects cause an increase in the effective pKa of 5-HO-OG following an increasing trend from 5.7 in nucleosides to 7.7 in a duplex bearing an OG•C base pair, which supports the context-dependent product yields. The high yield of Gh in ODNs underscores the importance of further study on this lesion. The structural context of OG also determined its relative reactivity toward oxidation for which the OG•A base pair is ~2.5-fold more reactive than an OG•C base pair, and with the weak one-electron oxidant ferricyanide, the OG nucleoside reactivity is >6000-fold greater than that of OG•C in a duplex, leading to the conclusion that OG in the nucleoside pool should act as a protective agent for OG in the genome.
INTRODUCTION
Chemical modification of the genome occurs with many possible outcomes in which oxidation of DNA bases by the reactions associated with oxidative stress and inflammation is deleterious.1–3 Inflammation and oxidative stress have been linked to neurological disorders such as Alzheimer’s disease,4 amyotrophic lateral sclerosis,5 aging processes,6,7 and the initiation and propagation of some cancers.3,8 Oxidative stress can be initiated by electron-deficient species (HO•, O2•−) derived from the incomplete reduction of O2 to H2O in mitochondria, or from the further reactions that occur when NO is released during inflammation.9–11 These reactive oxygen species (ROS) and reactive nitrogen species (RNS) are initiators of oxidation and deamination of the nucleic acid bases, potentially leading to mutagenesis if not repaired.12–16
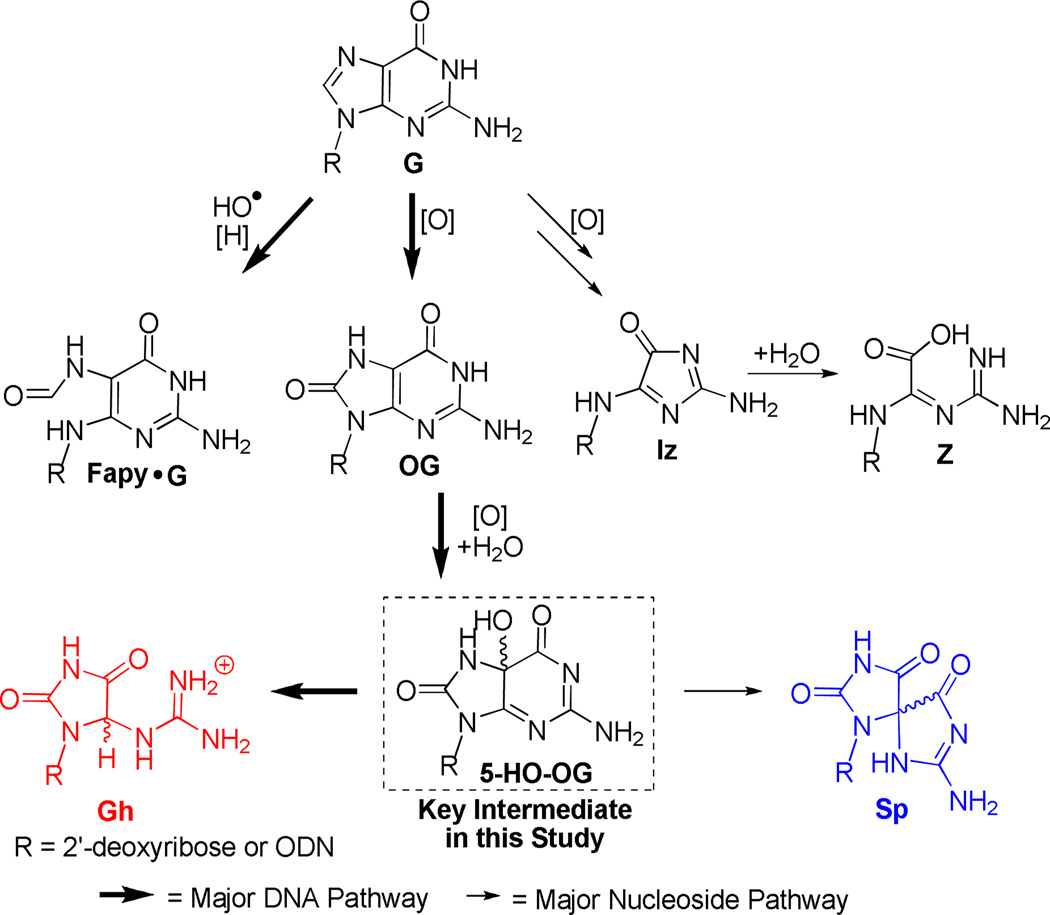
The nucleobase guanine (G) has the lowest one-electron redox potential and is the dominant site for oxidation within DNA.17 Studies concerning the oxidation of G in nucleoside and DNA contexts provide a wide spectrum of products that are oxidant and reaction context dependent.16,18–21 Earlier oxidation studies with G nucleoside, effected by ionizing radiation or Fenton chemistry (HO•), led to the identification of 2,5-diaminoimidazolone (Iz) and its hydrolysis product 2,2,4-triamino-2H-oxazol-5-one (Z) as the major nucleoside product ; 2,6-diamino-4-hydroxy-5- formamido - guanine (Fapy•G) was the major double-stranded oligodeoxynucleotide (dsODN) product observed under anaerobic reducing conditions; and 8-oxo-7,8-dihydroguanine (OG) was the major product observed under aerobic oxidizing conditions (Scheme 1).19,22–26 These experimental observations suggest that reaction context (i.e., nucleoside vs. dsODN), in addition to reaction conditions, alters the product distributions by affecting the stability of G oxidation intermediates. The one-electron oxidation of G yields a radical cation (G•+) with a pKa of ~3.9 (N1 proton), which at pH 7 in the nucleoside context, rapidly deprotonates to the neutral G•. Within dsODN, the base pairing of G to C effectively traps the acidic proton in the base pair, causing the intermediate radical to retain more cationic character.27 Therefore, nucleoside G oxidation products are dictated by the reactivity of G• leading to Iz/Z, whereas in dsODNs, G oxidation products may result from G•+, yielding Fapy•G and OG.19,28–33 Quantification of G oxidation products from genomic DNA consistently shows higher concentrations of OG than Fapy•G or Z in which the typical concentrations of OG in uncompromised cells is ~1 OG in 106 bases.34,35 These observations taken in their entirety suggest that G oxidation to OG is the major oxidation pathway for genomic DNA in vivo.35–40
Scheme 1.
Guanine oxidation pathways.
In duplex DNA, OG can form 3 hydrogen bonds with cytosine (C) or two hydrogen bonds with adenine (A) (Figure 1). The OG•A base pair is formed after rotation about the glycosidic bond to the syn conformation, thereby relieving the steric repulsion created in the OG•C base pair by the 8-oxo group with the C4’ oxygen of the same nucleotide, although the rotation introduces a minor distortion into the phosphate backbone.41–43 The relevance of OG within the genome is highlighted by the evolution of a repair system for removal of OG, which follows two different pathways depending on the base opposite OG to ensure accurate repair to the correct G•C base pair.44,45 The ability of OG to base pair with A ultimately leads to a transversion mutation (G→T) after a replication/transcription event occurs in the absence of repair, thus providing a chemical mechanism for the mutagenic properties of OG.46,47
Figure 1.
Base pairing schemes for the OG(anti)•C(anti) and OG(syn)•A(anti) base pairs.
The further oxidation of OG is more facile than that of G, due to its ~600 mV lower redox potential, making it a “hot spot” for additional oxidative damage leading to mutagenesis.48 OG nucleoside oxidation studies with ONOO−49 or HCO3•/CO3−s•, •NO2,50,51 riboflavin,52 HOCl,53 chromate,54 Na2IrCl6,55 K3Fe(CN)6,55 CoCl2/KHSO5,55 and Cu(II)/H2O2/reductant56,57 produce a high yield of spiroiminodihydantoin (Sp) at neutral pH. When the pH is decreased to ~6, or OG is oxidized in dsODNs, the major product observed was 5-guanidinohydantoin (Gh).49,52,58–61 The difference in Sp and Gh yield has been attributed to their common reaction intermediate 5-HO-OG, whose protonation state is proposed to determine partitioning along the two hydantoin pathways (Scheme 1).62,63 Specifically, the 5-HO-OG nucleoside has a measured pKa of ~5.8 for N1, such that under acidic reaction conditions, the protonated guanidinium group initiates the first step down the Gh pathway; whereas under neutral reaction conditions, N1 is not protonated, leading to a 1,2 acyl migration providing Sp.49,62
The high plasticity of the reaction surface with respect to pH, oxidant type and structural context underscores the need to understand the reactivity of OG in different reaction environments as a basis for predicting the molecular outcome of oxidative stress in the cell. Moreover, Sp and Gh are highly mutagenic leading to transversion mutations (G→T and G→C).64 These hydantoins also behave differently with respect to their repair by hNEIL1.65,66 Although, Sp has been the focus of many studies, Gh has received less attention despite initial evidence suggesting it is the principal product of OG oxidation in dsDNA. In this study, product distributions of OG oxidation were measured in nucleoside, nucleotide, and single- and double-stranded oligodeoxynucleotide (ssODN and dsODN) contexts with the oxidants K3Fe(CN)6, Na2IrCl6, and ONOO−/HCO3−. Within the dsODN context, product distributions were measured for OG base paired opposite C, A, and an abasic site. These studies provide insight into the products that would be anticipated when OG is oxidized in relevant contexts that could be the source of mutations. Also, the effect of structural context on the relative reactivity of OG toward oxidation was determined. The data reveal a high dependency of both overall reactivity and product outcome on the structural context. Additionally, a dramatic effect of apparent pH is observed in the microenvironment of N1 of OG in various base pairing contexts.
RESULTS
1. Experimental Design
Oxidation of 10.0 µM solutions of OG was initially compared in nucleoside, nucleotide and ss- or dsODN contexts. Within the dsODN context, OG oxidations for the base opposite C, A, or an abasic site analog (F, a stable THF analog) in the sequence contexts of flanking pyrimidines (5’-COT-3’) vs. flanking purines (5’-GOA-3’) were evaluated (Figure 2).
Figure 2.
18-mer ODN sequences utilized to study context effects on OG oxidation to hydantoin products. In these sequences O = OG and N = A, C, D, F, or 4EtC. The sequences within the gray boxes indicate the local contexts in which OG was oxidized.
Note that within the text, the dsODN duplexes are referred by the OG base pair of interest (e.g., OG•C denotes the dsODN bearing an OG•C base pair), and that in sequences, OG is denoted simply as “O”. All duplexes studied showed the expected native-gel shifts and thermal melting temperatures to justify the 37.0 °C reaction temperature utilized (Supporting Information S2 and S3).
Oxidation products were monitored to determine how reaction context, in particular base pairing, affects the partitioning of the common intermediate, 5-HO-OG, to Sp and Gh. Reaction product quantification in nucleoside, nucleotide, and ssODN contexts was achieved by HPLC analysis following previously established protocols in which nucleoside product peaks were integrated then normalized through their reported extinction coefficients for comparison.55,58,67 In the dsODN reactions, a method was developed that required dsODNs with strands of different length; this allowed purification of one strand from the other using denaturing-HPLC conditions, and subsequent analysis of the purified OG/Sp/Gh-containing single strands (18-mers) following a previous HPLC protocol (Supporting Information). In the ODN contexts, the 18-mer extinction coefficient was used to quantify peaks, with the assumption that OG-oxidation products do not significantly affect this value (Supporting Information S4–6). For all ODN reactions, piperidine cleavage controls were conducted and analyzed by polyacrylamide-gel electrophoresis (PAGE), to ensure that the products observed were a result of OG oxidation (Supporting Information S7). We observed only oxidation at the OG site under the reported reaction conditions, which is consistent with previous reports from our laboratory.68 Oxidation studies were conducted following literature procedures in which ONOOCO2− was generated in situ (5.0 equiv of SIN-1 for ODN, and 2.0 equiv for nucleoside and nucleotide contexts, all in the presence of 25.0 mM NaHCO3), and the one-electron oxidant Na2IrCl6 was delivered in three aliquots once every 5 min into the reaction mixture (total oxidant delivered was 10.0 equiv for ODN studies, and 2.0 equiv for nucleoside and nucleotide studies).49,55,69 Buffers utilized in these studies included acetate, phosphate and borate, depending upon the pH desired. Contrary to Tris buffer, none of these materials form base adducts during the oxidation of OG.61
2.1. Products observed from each OG structural context
All reactions were conducted at 37.0 °C, in 20.0 mM buffer (pH 4.0–8.5) with 100 mM NaCl, and triplicate trials were conducted to obtain suitable error bars (error = one standard deviation). Upon oxidation of nucleoside OG with ONOOCO2− derived CO3•−/•NO2 or Na2IrCl6, Sp was formed in >95% yield (Figure 3), similar to previous reports.49,55 Next, oxidation of 8-oxo-7,8-dihydro-2’-deoxyguanosine triphosphate (OGTP) with Na2IrCl6 or ONOOCO2− furnished Sp and Gh in a ratio of 5.5:1 (Figure 3). When OG was oxidized in an ssODN (5’-COT-3’ sequence) context with either oxidant, the relative yields of Sp and Gh were in an approximately 1:1 ratio (Figure 3). However, oxidation of OG in dsODNs (5’-COT-3’ sequence) with various base pairing partners led to changes in the Sp:Gh product ratios in which greater amounts of Gh were observed for more stable duplexes. In the end, the OG•C base pair context, upon oxidation with either oxidant system, led to Gh as the only detectable product (Figure 3). To further confirm the HPLC data, representative reactions were analyzed by mass spectrometry, which gave the anticipated masses for OG, Sp, and Gh at ~75% conversion (10.0 equiv Na2IrCl6, Supporting Information S8). It must be noted that at 100% conversion (20.0 equiv of Na2IrCl6) oxidation of OG-containing ODNs gave additional products that are all best explained by over oxidation of Gh (Supporting Information S9).58,70 Similar product distributions were observed in each reaction context utilizing the transition metal oxidant K3Fe(CN)6 as well as photochemically generated SO4•− (Supporting Information S10).
Figure 3.
Context effects on OG oxidation product distributions observed with the oxidants Na2IrCl6 (Ir(IV)) and ONOOCO2− (SIN-1). Each reaction was conducted as described in the text to give ~75% for Na2IrCl6 reactions, and ~10% for ONOOCO2− reactions. Product yields were estimated by integration of peak areas that were normalized by their molar extinction coefficients, and triplicate trials were conducted to obtain suitable errors.
2.2. Product distribution dependence on pH
Because the protonation state of 5-HO-OG appears to determine its partitioning between the two hydantoins, studies of the effective pKa of 5-HO-OG in each structural context provides information about the microenvironment in which oxidation occurs. The pH dependency on Sp and Gh yields within each structural context was evaluated, and then fitted to the Henderson-Hasselbalch equation (Figure 4). Effective pKa values increased in the order OG < OGTP < ssOG ~ OG•F < OG•A < OG•C giving the values 5.7, 6.0, 7.0, 7.0, 7.2, and 7.7, respecttively. For clarity the OG•F data is not shown because it overlaps with ssOG (see Supporting Information S11). For the OGTP data, the experimental data showed a non-ideal fit to the model, which likely results from the fact that the triphosphate group is also titratable at ~pH 6.7.71 At higher pH values (8.5–9.0), the OG•A and OG•C duplexes showed an additional inflection point (not shown) that likely reflects the pKa of OG at ~8.6.72 In this pH range, one expects that deprotonated OG disrupts base pairing and base stacking, leading to the Sp oxidation product as observed for the nucleoside and ssDNA.
Figure 4.
pH Dependency in Sp and Gh yield within each structural context studied. The OG-structural context studied include nucleoside OG (black circle), OGTP (red square), ssOG (aqua triangle), OG•A (dark green square), and OG•C (blue diamond). Solid lines represent fitting to the Henderson-Hasselbalch equation. Error in the data ~5%.
2.3. Sequence effect on OG oxidation products
In genomic DNA, OG could exist in 16 different sequence contexts, and it is possible that neighboring bases could influence the product distributions. In the study above, OG was flanked by two pyrimidines (5’-COT-3’, Figure 2); therefore, to minimize the number of reactions to evaluate, we chose to study OG flanked by two purines (5’-GOA-3’, Figure 5). These two sequence contexts provide two possible extremes in base stacking. Na2IrCl6-derived product distributions did not show a strong sequence dependency in ssODNs or in the OG•F and OG•A dsODN sequence contexts (Figure 5). However, the OG•C base pair context did yield a minor sequence effect on the products. The 5’-COT-3’ context led to exclusive formation of Gh, while the 5’-GOA-3’ context yield included Gh and Sp in a 19:1 ratio, respectively (Figure 5). Nevertheless, we conclude that sequence plays a minor role in determining OG oxidation product distributions.
Figure 5.
The effect of sequence context on OG oxidation product distributions in each ODN context studied.
2.4. Temperature and charge effects on OG oxidation products in ODNs
OG oxidations initiated with Na2IrCl6 were conducted to gain insight into why Sp was the major nucleoside product and Gh was the major dsODN oxidation product. The transition-metal oxidant Na2IrCl6 was selected because of its water solubility, ease of use, and specificity for oxidation of OG in ODNs.68,73 Moreover, ONOOCO2− and Na2IrCl6 appear to yield similar product distributions suggesting a similar mechanism (Figure 3); therefore, the following data may also be indicative of products derived from ONOOCO2− oxidation of OG in the contexts studied.
Nucleoside OG oxidations yielded ~95% Sp, whereas, ssOG oxidations yield Sp and Gh in a roughly a 1:1 ratio (Figure 3). Possible explanations for the increased yield of Gh in ssODNs requires understanding how the ODN context affects the common hydantoin intermediate, 5-HO-OG. Context effects that are not observed in the nucleoside include: (1) Steric effects from adjacent bases (i.e., base stacking),74 or (2) electrostatics, due to the anionic sugar-phosphate backbone in ODNs.75 To study the base stacking influence on OG-oxidation products, reactions were conducted at 4.0 °C. The decreased temperature increases base stacking around OG in ssODNs,76 thus an increase in the amount of Gh should be observed, while having no effect on nucleoside reactions. Indeed reactions at 4.0 °C had no effect on nucleoside OG, while the ssODN context yields more Gh (75 ± 5%) at 4.0 °C than at 37.0 °C (Gh = 50 ± 5%, Supporting Information S12). These observations support the hypothesis that base stacking in ODNs has an influence on the partitioning of 5-HO-OG to the hydantoins in which the ODN context favors the less sterically demanding product, Gh.
In the proposed oxidation pathway of OG to the hydantoins, one intermediate has been spectroscopically observed, 5-HO-OG, in which the experimentally derived pKa on the N1 nitrogen of ~5.8 (previous study) and ~5.7 (current study; from this point forward we used the previous value).49,63 It was proposed that titration of the N1 proton explains the strong pH dependence in Sp and Gh yield in which low pH (<5.8) favors Gh, and higher reaction pH favors Sp (>5.8).49,62 We hypothesized that the anionic sugar-phosphate backbone in ODNs could influence the acid/base chemistry of 5-HO-OG, thus modulating the product distribution of Sp and Gh. A similar effect has been described for the oxidation of G in dsODNs with an anthraquinone photooxidant.75 In an attempt to eliminate the base stacking effect, while observing the electrostatic effect that the anionic sugar-phosphate backbone has on partitioning of 5-HO-OG, oxidation of OGTP was conducted. In the OGTP oxidation reaction, both Gh and Sp were observed in a 1:5.5 ratio (Figure 3). A higher yield of Gh was observed in the OGTP reaction (Gh yield = 15% ± 3%) than the nucleoside OG reaction (Gh yield <5%), supporting the hypothesis that the anionic sugar-phosphate backbone, through electrostatics, has an influence on the partitioning of 5-HO-OG to the hydantoins. These data allow the following conclusions to be made: (1) Base stacking in ODNs favors the less sterically demanding product, Gh. (2) The anionic sugar- phosphate backbone influences 5-HO-OG, through an electrostatic effect, allowing more 5-HO-OG to partition toward the Gh pathway. Indeed, it is tempting to propose that these effects combine to give the measured change in effective pKa of 5-HO-OG (Figure 4), thus causing the observed modulation in product distribution.
2.5. Base pairing effect on OG oxidation products
Comparison of OG oxidation in ssODNs vs. dsODNs opposite an abasic site (OG•F duplex) aids our understanding of how the duplex context influences the reaction distribution without added base pairing effects. Oxidation of the OG•F duplex led to an Sp to Gh ratio of 1:1.5, whereas in ssODN contexts, the Sp to Gh ratio was 1:1 (Figures 3 and 4). Thus, more Gh was observed in the dsODN context than the ssODN context. These data further support the conclusion that base stacking drives the common intermediate 5-HO-OG toward the Gh pathway, because the dsODN context is better stacked than the ssODN context.74,77 However, it cannot be ruled out that electrostatics also influenced the partitioning of 5-HOOG to Gh in dsODNs when compared to ssODN contexts. In the dsODN contexts, OG forms well defined base pairs with A and C, with OG having the opposite orientation, syn vs. anti, about the glycosidic bond between the two base pairs (Figure 1).41,42 Because structural data on the OG•F context has not been determined, it was assumed that this OG exists primarily in the syn conformation, due to the steric clash between the C8 oxo group and the C4’ oxygen of the same nucleotide.41,78,79 Comparison between the OG•F context and OG•A base pair (5’-COT-3’ context, syn OG) provides details about how hydrogen bonding effects the product distribution. Oxidation of an OG•A base pair yields an Sp to Gh ratio of approximately 1:3, which is more Gh (~15%) than observed with the OG•F context (Figure 3). This observation likely results from the increased stability, or less dynamic nature80,81 of an OG•A base-paired duplex (Tm = 53.6 °C) relative to the OG•F context (Tm = 46.0 °C, Supporting Information S3). In conclusion, the dsODN’s stability influences hydantoin products, favoring Gh in less dynamic base-stacked contexts, while the yield of Sp increases in less stable or more dynamic contexts.
Oxidation of an OG•C base pair in the sequence context 5’-COT-3’, gave predominantly the Gh product (Figure 3). This observation has previously been reported,60,61 but the interesting contrast was the observation that oxidation of an OG•A base pair, in the same sequence context, gave a different result in which the Sp and Gh yield was 1:3, respectively (Figure 3). Therefore, the question was raised: What are the critical factors between these two dominant OG base-pairing schemes that alter the oxidation product distribution? Differences between the OG•C and the OG•A base pair include: (1) Glycosidic bond orientation (OG(anti)•C(anti) vs. OG(syn)•A(anti), Figure 1) resulting in the N1 hydrogen placed within the base pair vs. exposed in the major groove,41–43 (2) the number of base pair hydrogen bonds (OG•C = 3, and OG•A = 2, Figure 1),41–43 and (3) the duplex stability resulting from the previous two parameters (OG•C Tm = 56.8 °C; OG•A Tm = 53.6 °C, Supporting Information S3).82 Next, reactions were conducted to determine if any, or all, of these base-pair differences were the causative factor(s) in the observed oxidation product distributions.
As a first step, the duplex containing an OG•A base pair was stabilized by decreasing the reaction temperature to 4.0 °C; however, this procedure did not result in a significant effect on the product distribution (Supporting Information S12). From this data, we conclude that the oxidation product, Sp, observed from an OG•A base pair, does not result from decreased duplex stability alone. To confirm the importance of the OG base pairing partner, temperature-dependent studies were conducted with both OG•A (Tm = 53.6 °C) and OG•C (Tm = 56.8 °C) base pair contexts at higher temperatures, 45.0 and 55.0 °C. For both base pair contexts, the amount of Sp increased with increasing temperature (Supporting Information S12), as expected.
In a second step, to evaluate hydrogen bond strength in each base pair was important in establishing the product distribution; studies were conducted with analogs of A and C that modulate the hydrogen bonding. Diaminopurine (D), an analog of A, was selected to increase the number of hydrogen bonds in an OG•A base pair from 2 to 3 (Figure 6), providing the OG•D base pair. N4-Ethyl-C, an analog of C, was selected to decrease the hydrogen bond strength in an OG•C base pair, providing the OG•4EtC base pair (Figure 6). Both hydrogen bonding modulators, D and 4EtC, have previously been shown to cause the anticipated hydrogen bond effects with their typical hydrogen bonding partners (T•D and G•4EtC, respectively).83,84 Tm studies were conducted to analyze the hydrogen bond effects, and it was found that the OG•D duplex Tm was ~4 °C higher than the analogous duplex containing OG•A, supporting the proposed increased hydrogen bonding. Also in accord with predictions, the OG•4EtC duplex had a Tm that was ~6 °C lower than the OG•C duplex, supporting decreased hydrogen bond strength (Supporting Information S3).
Figure 6.
Proposed base pairing schemes for the OG•4EtC and OG•D base pairs. These modified base pairs are compared to their native OG base pairs with respect to structure and relative Tm.
Oxidation of an OG•4EtC base pair in dsDNA yields both Sp and Gh in a 1:9 ratio, representing an increase in the amount of Sp as predicted based on the destabilization of the OG•C base pair through introduction of the N4-ethyl group (Figure 7). However, this amount of Sp was less than that observed from the more stable OG•A base pair. This observation suggests that base-pair stability via hydrogen bonding plays a role in driving the oxidation of an OG•C base pair to Gh, but it is not the only factor. For example, the N4-ethyl group on 4EtC effects the base pair by altering the local hydration state;84 because H2O is a reaction partner with OG leading to the hydantoins,62 this altered hydration state cannot be ruled out as the causative factor, instead of the decreased hydrogen bonding. When the 3 hydrogen bond OG•A base pair analog OG•D was oxidized, the yields of Sp and Gh were similar to those observed from the OG•A base pair (Figures 3 and 7). In addition, an interstrand crosslink (~15% yield) was observed in both the denaturing HPLC and denaturing PAGE (Supporting Information S13). Attempts to study this crosslink were unsuccessful, due to its instability, so further studies on this new product were not pursued. Also, it should be pointed out that there is not a known biological relevance for this base pair. These observations rule out hydrogen bonding as the exclusive reason why an OG•A base pair yields Sp and Gh, and the OG•C base pair yields only Gh.
Figure 7.
Hydrogen bonding effect on OG oxidation product distributions.
3. Structural context effect on the relative reactivity of OG toward oxidation
The relative reactivity of each OG context toward oxidation by the diffusible one-electron oxidants K3Fe(CN)6, K2IrBr6, and Na2IrCl6, or the SIN-1 generated ONOOCO2− that yields CO3•−/•NO2 oxidants were ranked by using a series of competition assays. Reaction progress was monitored by gel-based assays with dsODN contexts, and HPLC-based assays with ssODN and nucleoside contexts. These studies were conducted with the ODN sequences shown in Figure 8 in which each OG structural context is represented while maintaining the same sequence context around OG. Each dsODN provided the expected native-gel shift and had a Tm value much greater than the 37.0 °C reaction temperature (Supporting Information S14). ODN1 allowed the ranking of base-paired OG relative reactivities toward oxidation without the complication of charge transport to occur between the two OG sites, because the three-base CCC bulge has previously been shown to diminish electron transfer.85 ODN2 compared base-paired OG to the OG•F context. ODN3 allowed the ranking of base-paired OG relative to single-stranded OG; in this strand two pyrimidine residues were placed between the ssOG and the dsODN part of the strand to ensure no electron transfer occurred;86 and ODN4 provided a single-stranded OG context to compare against nucleoside OG. The uracil (U) base placed between each OG in these sequences was used to ensure that the products remained the same when oxidations were conducted in ODNs with two OGs vs. ODNs with one OG, as indeed was the case (Supporting Information S15).
Figure 8.
ODN sequences studied to evaluate the context effect on the relative reactivity of OG toward oxidation. The gray highlighted regions show the sequence context in which the OG resides.
The ODN studies were conducted by 32P-radiolabeling of the 5’-end of the OG-containing ODN; because this method of analysis was conducted following strand breaks by gel electrophoresis, care was taken to design each sequence such that the most reactive OG context was placed on the 3’ end so that its reactivity would not be overestimated. After conducting the reactions under single-hit conditions (<30% conversion) the extent of reaction was monitored by following hot piperidine-induced strand breaks at the hydantoins. To overcome the questionable piperidine lability of OG,73,87–89 while ensuring high strand scission at the hydantoin lesions,60,90 the cleavage reactions were conducted with 250 mM β-mercaptoethanol in 1.0 M piperidine for 2.0 h at 90.0 °C. These conditions quench any unwanted OG strand scission, while providing high cleavage at both Sp and Gh sites (Supporting Information S16).
Ranking of the relative reactivity between single-stranded and nucleoside OG contexts was achieved by direct HPLC analysis. The nucleoside oxidation was followed by reversed-phase HPLC, and the ssDNA reactivity was quantified by ion-exchange HPLC following the previously described method. Next, uric acid was competed against OG for oxidant, to make a comparison to a known redox-active purine present in vivo. These reactions were monitored by HPLC (Supporting Information S17–21).
Table 1 provides the relative reactivity data for OG toward oxidation with the diffusible one-electron oxidants Na2IrCl6, K2IrBr6, and K3Fe(CN)6, and ONOOCO2−. These data were ranked from the least reactive OG context, the OG•C base pair, to the most reactive context, nucleoside OG. The results for SIN-1/HCO3− (ONOOCO2−) oxidations provided similar reactivity rankings for the OG•A and OG•C base pairs. To control for piperidine cleavage errors, the bases opposite OG in the complementary strand were flipped so that the OG base pairs were on opposite sides of the duplex, and again the OG•A and OG•C base pairs gave similar reactivities. The OG•F context was 10-times more reactive than base-paired OG, followed by single-stranded OG being 60-times more reactive, nucleoside OG 300-times more reactive, and uric acid being 3900-times more reactive than an OG•C base pair (Table 1).
Table 1.
Relative reactivities for each OG context with various oxidants.
| Oxidanta | Uric Acid |
OG | ssOG | OG • F |
OG • A |
OG • A |
|---|---|---|---|---|---|---|
| ONOOCO2− | 3900 | 300 | 60 | 10 | 1 | 1 |
| Na2IrCl6 | 2800 | 400 | 13 | 11 | 2.3 | 1 |
| K2IrBr6 | 1.8×104 | 1500 | 30 | 23 | 2.2 | 1 |
| K3Fe(CN)6 | >1.9×105 | >6000 | 125 | 120 | 2.7 | 1 |
All competition reactions were conducted at a 10.0 µM OG concentration in 20.0 mM NaPi (pH 7.0) with 100 mM NaCl at 37.0 °C. For each competition, enough oxidant was added to effect ~20–30% conversion to product and triplicate trials were conducted to obtain errors of ~5% of each value, which are reported in the Supporting Information S17–21.
The diffusible transition metal one-electron oxidants Na2IrCl6 (0.9 V), K2IrBr6 (0.8 V), and K3Fe(CN)6 (0.4 V, all vs. NHE)68 provided reactivity data that are dependent on the one-electron redox potential of the oxidant in which stronger oxidants had lower reactivity differences than the weaker oxidants. The OG•C and OG•A base pairs show a difference in their reactivity toward oxidation in which the OG•A base pair is 2.2–2.7 times more reactive toward oxidation than the OG•C base pair (Table 1). The relative reactivity of an OG•F context was one to two orders of magnitude more reactive than an OG•C base pair, which was oxidant dependent. A similar increase in reactivity was also observed for single-stranded OG compared to an OG•C base pair. Next, nucleoside OG relative reactivity, compared to the OG•C base pair, was highly oxidant dependent with the Ir(IV) complexes, and showed greater than two orders of magnitude increased reactivity with nucleoside OG. Furthermore, the K3Fe(CN)6 reaction gave a difference that was too large to measure; therefore, a lower limit was reported using the K2IrBr6 as the limiting value. Finally, the relative reactivity of OG was compared to uric acid. In these studies, it was found that uric acid was almost one order of magnitude more reactive than nucleoside OG, and three to five orders of magnitude more reactive than an OG•C base pair (Table 1).
DISCUSSION
Many studies have provided insight into structural context effects on guanine oxidation products, particularly in the pattern between Iz/Z, Fapy•G and OG reaction channels for which Iz/Z are the major nucleoside products, and Fapy•G/OG are major dsODN products.24–26 Furthermore, OG is the major product observed under aerobic oxidizing conditions, and consequently, its concentration provides a major biomarker of oxidative stress in vivo.34,35 The work reported here addresses the question of OG reactivity and product outcome as a function of structural context, for which cellular OG can reside in the nucleotide pool, in single-stranded DNA (i.e., replication forks and transcription bubbles), and in double-stranded DNA, particularly base paired with C or A, but also transiently opposite an abasic site when an OG•A base pair is repaired by MUTYH.44 The oxidation chemistry of OG is orders of magnitude more facile than that observed with G due to its ~600 mV lower redox potential,48 leading to the two hydantoin products upon oxidation with myriad oxidants.49,51,54,55 Both Sp and Gh are known to display high mutagenic potential due to polymerase stops and misinsertion of G or A opposite the lesions, which occur to varying extents depending on the lesion and sequence context.64,67,91–93 The hydantoins are excellent substrates for the NEIL base excision repair enzymes,65,93–95 and again the efficiency of repair depends on the hydantoin structure, Sp1 vs. Sp2 vs. Gh, as well as on the surrounding sequence and structural context; e.g., Gh is a substrate for hNEIL1 in ssDNA and in bulge and bubble structures, while Sp is not.96 Basal levels of Gh and Sp were recently detected in mouse liver DNA where the hydantoin levels were roughly 100-fold lower than OG, and in accord with our findings, Gh levels (~5 in 108 nt) were found to be somewhat higher than Sp.8 In a mouse model of inflammation-induced colon cancer, cellular levels of Sp were modestly correlated with progression of the disease.8 These reports underscore the need to understand the chemical mechanism of Gh and Sp formation in genomic DNA.
In the present work, we investigated the influence of the structural context surrounding OG on both the reactivity and product outcome of oxidation by one-electron oxidants. The first step of the oxidation process involves interaction of the oxidant with the DNA strand, either directly at the site of the OG base, or removal of an electron at a distant site, followed by hole transfer to OG. The present work was not focused on hole transfer chemistry within the helix, but rather on the innate reactivity of OG in different settings—nucleoside, oligonucleotide, base-paired, etc. Three diffusible one-electron oxidants were chosen that each has a different redox potential, and these included K3Fe(CN)6 (0.4 V), K2IrBr6 (0.8 V) and Na2IrCl6 (0.9 V, all vs. NHE).68 The relative rates were determined by conducting a series of competition assays between different OG contexts in which the ranking of relative reactivity toward oxidation was: OG nucleoside >> ssOG > OG•F > OG•A > OG•C (Table 1). The only exception to this trend was oxidation by ONOOCO2− in which the OG•A and OG•C base pairs showed nearly equal reactivity, which results from the large difference in redox potential between OG and the CO3•− (1.69 V) and •NO2 (1.04 V)50,51; therefore, minimizing any difference in the observed reactivity between the two base-paired OG contexts. These competition studies found that as the redox potential of the oxidant decreased the magnitude of the difference in OG’s relative reactivity increased; that is to say, the biggest differences were found in the most reactive OG context, the nucleoside, compared to the least reactive context, OG•C base pair, and this was observed with the weakest oxidant, K3Fe(CN)6. During the review process it was pointed out that back-electron transfer between the reduced oxidant and OG•+ might occur. Interestingly, studies with titrated K4Fe(CN)6 (FeII) into nucleoside reactions did indeed quench the reaction, supporting back-electron transfer (Supporting Information S23). However, the concentration of FeII, from spent oxidant, is low. Furthermore, uric acid, an established redox-active purine,97 displayed the highest reactivity toward oxidation, which is supported by the lower redox potential of uric acid (0.59 V)97 compared to OG nucleoside (0.74 V).48
From these relative reactivity data, a few conclusions can be drawn. First, in the base-paired OG contexts, the OG•A base pair is ~2.5-times more reactive toward oxidation than an OG•C base pair (Table 1). This base pair difference was observed with the diffusible transition-metal oxidants, which initiate oxidation by coming into the vicinity of OG; therefore, oxidation at the base-paired OG allows a relative measure of solvent exposure between the OG•A and OG•C base pairs. These data suggest when OG is base paired to A it “breathes” into the major groove ~2.5-times more often than when C is the base pairing partner. Previously, Johnston, et al. came to a similar conclusion for oxidation of G in dsODNs in various base-pair contexts in which they observed a G•A mismatch being ~16-times more reactive than a G•C base pair, for which they pose base dynamics as the best explanation.98 Our observed difference between OG base pairs should be less than those observed with G base pairs, as indeed it is, because the ΔTm between an OG•C and an OG•A base pair (ΔTm = −1.7 °C) is smaller than the difference between G•C and G•A pairs (ΔTm = −7.8 °C).99 Furthermore, another refinement on the base-pair effect on relative reactivity may include an Eox difference for OG between the base pairs, which has not been measured for the OG•C or OG•A base pairs, but has been described for other OG base pairs.100 In line with the trend that solvent accessibility increases reactivity, the OG•F context is more reactive than either base-paired OG context, and the single-stranded context, being the most solvent exposed of the ODN substrates, was even more reactive toward oxidation (Table 1). These data parallel the findings of Lee et al. who showed that abasic sites adjacent guanine increase the reactivity of GG sequences toward oxidation.101 Finally, uric acid was ~3 to 5 orders of magnitude more reactive than an OG•C base pair, which is consistent with both solvent accessibility and a lower redox potential.
Electron transfer to the oxidant results in formation of OG+•, which subsequently undergoes loss of protons and another electron as well as addition of a water molecule to form the observed intermediate 5-OH-OG (Scheme 2). The timing of these events, all expected to be rapid, has been explored computationally.62 That H2O is the source of the hydroxyl group in 5-OH-OG was previously shown by isotopic labeling.55 In addition, the structure of 5-OH-OG was confirmed by a low-temperature NMR study.63
Scheme 2.
Proposed mechanism for pH-dependent of OG oxidation to yield hydantoins Sp and Gh.
Although loss of one electron from OG is thought to be the rate-limiting step, the reactions of 5-OH-OG represent the product-determining steps.62 Previous studies have shown that Sp is the preferred product observed in nucleosides at pH > 5.8, whereas Gh is preferred at pH <5.8 and in the OG•C base pair context.49,58,60,61 The intermediate 5-HO-OG is a weak acid with a pKa of ~5.8 for the N1 proton,49 which we reproduced in this study (pKa ~5.7, Figure 4). As a consequence, the protonation state of 5-HO-OG is proposed to partition this intermediate along the different two hydantoin pathways:62 protonation of N1 of 5-HO-OG facilitates an acid-catalyzed amide-bond hydrolysis reaction leading to Gh after decarboxylation and tautomerization, while the neutral 5-OH-OG follows a thermodynamically preferred pathway to Sp via a 1,2 acyl migration mechanism (Scheme 2).62
In this study, the partitioning of the intermediate to Sp and Gh products was evaluated as a function of structural context of the OG base. Surrounding sequence effects were found to be minor, although we previously showed that a 3’ G stacked next to OG in a duplex led to an ~2.5-fold higher reactivity toward one-electron oxidants, as is the case with 5’-GG-3’ sequences.68 In the present work, we focused on the effects of the base opposite, which could affect solvent accessibility to OG, as well as comparisons of monomeric (nucleosides/nucleotides) and oligomeric structures. With unencumbered contexts, such as nucleosides and nucleotides, the intermediate 5-HO-OG partitioned to Sp in a higher yield than Gh, as expected at neutral pH (Figure 3).49,52,58 In the ssODN context, 5-HO-OG partitioned to both Sp and Gh in a ratio of about 1:1 (Figures 3 and 4). We hypothesize that base stacking and electrostatics in ssODN contexts provide the physical drivers to increase the yield of Gh. Support for this hypothesis comes from the low temperature reactions conducted on single-stranded OG, which increases base stacking, thus favoring the less sterically demanding product, Gh, in ssODNs.
Furthermore, oxidation of OG in all dsODN contexts led to Gh as the major product. Electrostatics in ODNs is proposed to be a second major factor increasing the yield of Gh relative to Sp. Support for this hypothesis came from the observation that OGTP furnished more Gh than nucleoside OG (Figure 3), cases in which steric factors should not apply. The effective pKa of 5-HO-OG in nucleotide contexts was measured to be ~0.3 greater than nucleoside contexts (Figure 4). Consequently, the triphosphate presence increases the basicity of the N1 proton on 5-HO-OG and slightly increases the Gh yield supporting the role of electrostatics in hydantoin yields. Electrostatics has previously been evaluated by Barnett, et al. in the oxidation of G within dsODNs contexts;75 they found that a decrease in negative charge, through incorporation of a methylphosphonate backbone, alters the reactivity of G, which is in accord with our observation that charge plays a role in OG oxidation. Further, the electrostatics in ssODNs causes an additional increase in the effective pKa of 5-HO-OG by ~1.3 compared to nucleoside OG (Figure 4), which is reflected in the increased Gh yield within ssOG contexts.
Base pairing effects on OG oxidation included the observations that an OG•F context yields Sp and Gh in a 1:2 ratio; the OG•A base pair yields Sp and Gh in a 1:3 ratio, and Gh (>95%) was the major product observed from oxidation of an OG•C base pair (Figures 3 and 4). Previous studies had concluded that the OG•C base pair yields exclusively Gh upon oxidation,60,61 so that the OG•A result was initially surprising. Because no structural data exist for the OG•F duplex, the following discussion will focus on the OG•A and OG•C base pair, for which structural data is available.41–43 The key differences between the base pairs of OG with A and C include: (1) base orientation (Figure 1), (2) number of hydrogen bonds (Figure 1), and (3) thermal stability of the duplex. The studies conducted in this report have identified the conformation of OG (syn vs. anti) as a second contributing factor beyond base stacking in determining the product distribution. Support for this conclusion comes from the observation that the hydrogen bonding modulators, D and 4EtC, had little effect on product distribution (Figure 6). Also, increasing duplex stability on the OG•A base pair, by dropping the reaction temperature, had no effect on product distribution for this base pair (Supporting Information S12). However, reactions conducted at temperatures near the Tm increase the Sp yield, presumably because the oligomers were more dynamic and approached the structure of an ssDNA strand. All together, these observations suggest that the orientation of 5-HOOG and the presence of a base opposite may have an influence on the oxidation products, arguments for which are detailed below.
As previously stated, the intermediate 5-HO-OG is a weak acid, and its protonation state at the N1 nitrogen helps dictate the partitioning to either Sp or Gh. This study further highlights a base pair effect on the partitioning to either Sp or Gh that cannot be ascribed to base dynamics alone. Therefore, we propose the following mechanism to describe this base pair effect. In the OG•C base pair, OG is in the anti conformation, which has the N1 proton involved in Watson-Crick base pairing to C, whereas, syn OG in the OG•A base pair positions the N1 proton exposed to solvent in the major groove. Upon oxidation of these base pairs to the intermediate 5-HOOG, the base pair with C opposite traps the N1 proton in a base pair hydrogen bond, providing a 5-HO-OG(H+)•C base pair intermediate (Scheme 3). The trapped proton is set up to catalyze the amide bond hydrolysis step that initiates partitioning down the Gh pathway; therefore, Gh is the dominant product observed. Proton trapping within the hydrogen bonds of a base pair best describes product distributions from the oxidation of a G•C base pair as well, which upon oxidation under aerobic conditions was shown to yield OG.19,28–32 This observation nicely parallels what we observe with the oxidation of an OG•C base pair.
Scheme 3.
Proposed mechanism for modulated hydantoin formation when OG is oxidized in the OG•C vs. OG•A base pair contexts.
In contrast, when 5-HO-OG is base paired with A, this proton trapping cannot occur, because the N1 proton is directed into the major groove and can be immediately transferred to the solvent (Scheme 3); therefore, the product distribution includes Sp and Gh in a 1:3 ratio, respectively, in which the distribution is determined by base stacking and electrostatics in the duplex context, as previously described. Support for the importance of the N1 proton location in the OG base pair is highlighted in the effective pKa difference for these base pairs (Figure 4): effective pKas for 5-OH-OG in the OG•A and OG•C base pairs are 7.2 and 7.7, respectively. This difference highlights a chemical rationale for the differing hydantoin yields under similar reaction conditions. As seen in Figure 4, the microenvironment of N1 of 5-OH-OG can mimic as much as a 100-fold change in acidity.
Moreover, the nucleus and mitochondrial matrix are maintained at pH ~7.2 and 8, respectively,102 which suggests that more Gh should be observed from genomic DNA oxidations, while Sp would be observed from damage to the mitochondrial genome. Furthermore, tumors can display slightly lower intracellular pH, which would favor the product Gh. Coupling the observation of the pH dependence in hydantoin yield, and the variable pH of biological systems, one could predict the hydantoin responsible for the observed mutation profiles in various tissues.
In the present studies, the sequence dependence of product distribution was tested in two extremes in sequence context, 5’-COT-3’ and 5’-GOA-3’ (Figure 5). From these data, the sequence context had no effect on ssOG, OG•F and OG•A base pairs; however, the OG•C base pair provided a modest (<5%) amount of Sp formation in the 5’-GOA-3’ sequence context. This observation indicates that base pairing and base stacking are more influential on product distribution than the subtleties of base sequence. Interestingly, Lim, et al. have studied the sequence variation in the reactivity of OG toward oxidation, and their studies with riboflavin-mediated oxidation of OG show the 5’-AOG-3’ is more reactive than the 5’-COT-3’ sequence.103 This observation highlights additional features to the complex OG oxidation story, which parallels our observation with a minor sequence variation in OG•C oxidation products; however, we note they did not study the OG•A base pair, therefore, a direct comparison cannot be made to the present work.
The biological relevance of these studies is multifold. First, oxidation of OG in the cell will be focused on the most solvent exposed source of the purine, the nucleotide pool. Because of the greater concentration of OG in the nucleotide pool (including OG from RNA oxidation),34,104,105 and the much higher reactivity of the OG nucleoside/nucleotide, oxidation of the monomer may serve a protective function for G and OG in the genome, much as uric acid has been proposed as a cellular antioxidant.97,106 Secondly, the higher reactivity of the OG•A base pair compared to OG•C provides a pathway to mutagenesis. Oxidation of the OG•C base pair to Gh•C and subsequent excision of Gh by NEIL1 should lead to accurate repair because the correct C partner still resides in the strand opposite the lesion. On the other hand, excision of the lesion from a Gh•A or Sp•A duplex derived from oxidation of the more reactive OG•A base pair would ensure mutation since a T would be inserted by pol β opposite the remaining A during the repair process. This type of repair-induced mutation is avoided in the repair of OG because hOGG1 (or bacterial Fpg) does not accept OG•A duplexes as substrates and instead the offending A is first removed by MUTYH.44 The NEIL enzymes that excise Sp and Gh have a much more promiscuous activity with respect to the base opposite the lesion, and therefore action on a hydantoin lesion with A opposite leads invariably to mutation.65,96 There is no known analog of MUTYH that removes misinserted bases opposite hydantoin lesions.
CONCLUSIONS
The lower redox potential of the common guanine-derived DNA lesion OG renders it extremely susceptible to further oxidation leading to the hydantoin products Sp and Gh. These secondary lesions are at least an order of magnitude more mutagenic than OG, and thus a thorough understanding of how the hydantoins are formed is desirable. Although the generally higher reactivity of nucleoside OG compared to duplex DNA and the pH sensitivity of the product outcome were previously known, we report here a complete study of the influence of structural context of the OG base on both its reactivity and pathway to products. One-electron oxidants spanning a range of potentials were chosen for study, including ferricyanide, hexachloroiridate, and the cellularly-relevant peroxynitrosocarbonate. Our findings reveal that the reactivity of OG is strongly governed by solvent accessibility with 3 orders of magnitude difference between the nucleoside context and base-paired OG in duplex DNA, suggesting that OG in the nucleotide pool should act as an antioxidant with respect to OG in the genome. In duplex DNA, the OG•A mispair is about 2.5X more reactive than the OG•C base pair, implying a more facile route to mutations via secondary oxidation that could occur at OG after misinsertion of A opposite.
Structural context also plays a dramatic role in determining which hydantoin product is formed from OG oxidation. The unstable intermediate 5-HO-OG partitions to Sp or Gh in a pH-dependent manner in the nucleoside context in which Sp is the dominant product above pH 6 while Gh predominates in more acidic environments; in contrast, Gh represents >95% of the product formed from the OG•C base pair in duplex DNA at physiological pH.49,52,58,63 The more detailed investigation described herein suggests two major factors contributing to the Sp:Gh product ratio: (1) steric effects, in which the less sterically demanding Gh is formed in more rigid duplex environments, and (2) electrostatic effects, where the microenvironment of N1 of the purine base plays a major role in the product-determining step. Illustrative of the latter, stable base pairing of OG opposite C in duplex DNA mimics the effect of a 100-fold higher H+ concentration, i.e. an effective local pH that is 2 units lower than the surrounding buffered solution, conditions that favor formation of Gh. Because DNA processing enzymes (polymerases, base excision repair glycosylases and nucleases) have differing reactivities with the two hydantoins, it is important to elucidate the context in which these mutagenic lesions are formed. The present studies highlight the unique chemical environment of bases in duplex DNA and how it differs from that of monomeric nucleosides, as well as providing a profile of where hydantoin lesions are likely to be found in nucleosides, nucleotides and oligomers in the cell.
EXPERIMENTAL METHODS
Materials
All chemicals were obtained from commercially available sources and used without further purification. Oligodeoxynucleotides were synthesized by the DNA/Peptide synthesis core facility at the University of Utah following standard solid-phase synthetic protocols and commercially available phosphoramidites (Glen Research, Sterling, VA).
Oligodeoxynucleotide preparation
All ODN strands were cleaved, deprotected and purified following previously described protocols.99 Mass spectrometry and Tm analysis was conducted following previously described protocols with the full details of all procedures outlined in the Supporting Information.99 Formation of dsODNs was conducted in a 300-µL sample by mixing 10.0 µM OG-containing strand with 12.5 µM complementary strand. Annealing occurred by heating the sample to 90.0 °C in a water bath, then holding the temperature constant for 5 min, followed by slowly cooling the water bath to room temperature over 3 h.
Oxidation reactions
All reactions were conducted in a 50-µL reaction volume that contained 10.0 µM OG, 20.0 mM buffer (pH 4.0–8.0), and 100.0 mM NaCl. The reactions were thermally equilibrated for 30 min at the desired temperature (4.0, 37.0, 45.0, or 55.0 °C) before the reaction was initiated. The oxidant concentrations and conditions were as follows. Na2IrCl6 and K2IrBr6 were titrated into the reaction in three aliquots delivered every 5 min to a final concentration of 100.0 µM, then after 30.0 min the reaction was quenched with 500 µM EDTA (pH 8.0). K3Fe(CN)6 was also titrated in three aliquots delivered every 5.0 min to a final concentration of 1–10 mM; then after 30.0 min, the reaction was quenched with 1–10 mM ascorbate. ONOOCO2− was generated in situ by the thermal decomposition (37.0 °C) of 100.0 µM SIN-1 over 3.0 h with 25.0 mM NaHCO3 (pH 7.0) present. Details of the gel-based and HPLC-based methods for conducting the analyses have previously been described, with the full details outlined in the Supporting Information.56,67
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH (CA090689) and ACD was funded by an NSF-REU grant (0649039). The authors would like to gratefully acknowledge Dr. Xiaoyun Xu for thoughtful discussions.
Abbreviations
- 4EtC
N4-ethylcytosine
- 5-OH-OG
5-hydroxy-8-oxo-7-8-dihydroguanine
- D
2,6-diaminopurine
- dsODN
double-stranded oligodeoxynucleotide
- F
tetrahydrofuran abasicsite analog
- Fapy•G
2,6-diamino-4-hydroxy-5-formamidoguanine
- Gh
5-guanidinohydantoin
- Iz
2,5-diaminoimidazolone
- O and OG
8-oxo-7-8-dihydroguanine
- dOGTP
8-oxo-7-8-dihydro-2’-deoxyguanosine 5’-triphosphate
- PAGE
polyacrylamide gel electrophoresis
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SIN-1
3-morpholinosydnonimine
- Sp
spiroiminodihydantoin
- ssODN
single-stranded oligodeoxynucleotide
- Tm
melting temperature
- Z
2,2,4-triamino-2H-oxazol-5-one.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Complete experimental details, HPLC, MS, Tm, piperidine cleavage studies, and effect of salt, pH and temperature on product distributions, Sp diastereomer ratios and FeII quenching reaction. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Holliday R. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 2.Son J, Pang B, McFaline JL, Taghizadeh K, Dedon PC. Mol. Bio. Syst. 2008;4:902–908. doi: 10.1039/b719411k. [DOI] [PubMed] [Google Scholar]
- 3.Lonkar P, Dedon PC. Int. J. Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangialasche F, Polidori MC, Monastero R, Ercolani S, Camarda C, Cecchetti R, Mecocci P. Ageing Res. Rev. 2009;8:285–305. doi: 10.1016/j.arr.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Aguirre N, Beal MF, Matson WR, Bogdanov MB. Free Rad. Res. 2005;39:383–388. doi: 10.1080/10715760400027979. [DOI] [PubMed] [Google Scholar]
- 6.Finkel T, Holbrook NJ. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 7.Ansari MA, Scheff SW. J. Neuropathol. Exp. Neurology. 2010;69:155–167. doi: 10.1097/NEN.0b013e3181cb5af4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Proc. Natl. Acad. Sci. U.S.A. 2012 doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. J. Biol. Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 10.Kussmaul L, Hirst J. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7607–7612. doi: 10.1073/pnas.0510977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckman JS, Koppenol WH. Am. J. of Physiol. Cell Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 12.Niles JC, Burney S, Singh SP, Wishnok JS, Tannenbaum SR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11729–11734. doi: 10.1073/pnas.96.21.11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mancardi D, Ridnour LA, Thomas DD, Katori T, Tocchetti CG, Espey MG, Miranda KM, Paolocci N, Wink DA. Curr. Mol. Med. 2004;4:723–740. doi: 10.2174/1566524043359854. [DOI] [PubMed] [Google Scholar]
- 14.Dong M, Wang C, Deen WM, Dedon PC. Chem. Res. Toxicol. 2003;16:1044–1055. doi: 10.1021/tx034046s. [DOI] [PubMed] [Google Scholar]
- 15.Dedon PC, Tannenbaum SR. Arch. Biochem. Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 16.Cadet J, Douki T, Ravanat J-L. Mut. Res. 2011;711:3–12. doi: 10.1016/j.mrfmmm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Steenken S, Jovanovic SV. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 18.Neeley WL, Essigmann JM. Chem. Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 19.Burrows CJ, Muller JG. Chem. Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 20.Gimisis T, Cismas C. Eur. J. Org. Chem. 2006;2006:1351–1378. [Google Scholar]
- 21.Cadet J, Douki T, Ravanat J-L. Free Radical Biol. Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 22.Cadet J, Berger M, Buchko GW, Joshi PC, Raoul S, Ravanat J-L. J. Am. Chem. Soc. 1994;116:7403–7404. [Google Scholar]
- 23.Candeias LP, Steenken S. Chem.--Eur. J. 2000;6:475–484. doi: 10.1002/(sici)1521-3765(20000204)6:3<475::aid-chem475>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Angelov D, Spassky A, Berger M, Cadet J. J. Am. Chem. Soc. 1997;119:11373–11380. [Google Scholar]
- 25.Ito K, Inoue S, Yamamoto K, Kawanishi S. J. Biol. Chem. 1993;268:13221–13227. [PubMed] [Google Scholar]
- 26.Kasai H, Yamaizumi Z, Berger M, Cadet J. J. Am. Chem. Soc. 1992;114:9692–9694. [Google Scholar]
- 27.Candeias LP, Steenken S. J. Am. Chem. Soc. 1989;111:1094–1099. [Google Scholar]
- 28.Steenken S. Chem. Rev. 1989;89:503–520. [Google Scholar]
- 29.Li X, Cai Z, Sevilla MD. J. Phys. Chem. B. 2001;105:10115–10123. [Google Scholar]
- 30.Adhikary A, Khanduri D, Sevilla MD. J. Am. Chem. Soc. 2009;131:8614–8619. doi: 10.1021/ja9014869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynisson J, Steenken S. Phys. Chem. Chem. Phys. 2002;4:5346–5352. [Google Scholar]
- 32.Reynisson J, Steenken S. Phys. Chem. Chem. Phys. 2002;4:527–532. [Google Scholar]
- 33.Pratviel G, Meunier B. Chem. Eur. J. 2006;12:6018–6030. doi: 10.1002/chem.200600539. [DOI] [PubMed] [Google Scholar]
- 34.Gedik CM, Collins A. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 35.Matter B, Malejka-Giganti D, Csallany AS, Tretyakova N. Nucleic Acids Res. 2006;34:5449–5460. doi: 10.1093/nar/gkl596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangal D, Vudathala D, Park J-H, Lee SH, Penning TM, Blair IA. Chem. Res. Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hailer MK, Slade PG, Martin BD, Sugden KD. Chem. Res. Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pouget JP, Douki T, Richard MJ, Cadet J. Chem. Res. Toxicol. 2000;13:541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
- 39.Collins A. Free Radical Biol. Med. 2003;34:1089–1099. doi: 10.1016/s0891-5849(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 40.Malins DC, Haimanot R. Cancer Res. 1991;51:5430–5432. [PubMed] [Google Scholar]
- 41.McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 42.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. Proc. Natl. Acad. Sci. U.S.A. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gannett PM, Sura TP. Chem. Res. Toxicol. 1993;6:690–700. doi: 10.1021/tx00035a015. [DOI] [PubMed] [Google Scholar]
- 44.David SS, O'Shea VL, Kundu S. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.David SS, Williams SD. Chem. Rev. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 46.Shibutani S, Takeshita M, Grollman AP. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 47.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steenken S, Jovanovic SV, Bietti M, Bernhard K. J. Am. Chem. Soc. 2000;122:2373–2374. [Google Scholar]
- 49.Niles JC, Wishnok JS, Tannenbaum SR. Chem. Res. Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- 50.Shafirovich V, Cadet J, Gasparutto D, Dourandin A, Geacintov NE. Chem. Res. Toxicol. 2001;14:233–241. doi: 10.1021/tx000204t. [DOI] [PubMed] [Google Scholar]
- 51.Crean C, Geacintov NE, Shafirovich V. Angew. Chem. Int. Ed. 2005;44:5057–5060. doi: 10.1002/anie.200500991. [DOI] [PubMed] [Google Scholar]
- 52.Luo W, Muller JG, Burrows CJ. Org. Lett. 2001;3:2801–2804. doi: 10.1021/ol0161763. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki T, Masuda M, Friesen MD, Ohshima H. Chem. Res. Toxicol. 2001;14:1163–1169. doi: 10.1021/tx010024z. [DOI] [PubMed] [Google Scholar]
- 54.Sugden KD, Campo CK, Martin BD. Chem. Res. Toxicol. 2001;14:1315–1322. doi: 10.1021/tx010088+. [DOI] [PubMed] [Google Scholar]
- 55.Luo W, Muller JG, Rachlin EM, Burrows CJ. Org. Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 56.Fleming AM, Muller JG, Ji I, Burrows CJ. Org. Biomol. Chem. 2011;9:3338–3348. doi: 10.1039/c1ob05112a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White B, Tarun MC, Gathergood N, Rusling JF, Smyth MR. Mol. Biosyst. 2005;1:373–381. doi: 10.1039/b511756a. [DOI] [PubMed] [Google Scholar]
- 58.Luo W, Muller JG, Rachlin EM, Burrows CJ. Chem. Res. Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 59.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. J. Am. Chem. Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 60.Duarte V, Muller J, Burrows C. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gremaud JN, Martin BD, Sugden KD. Chem. Res. Toxicol. 2010;23:379–385. doi: 10.1021/tx900362r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munk BH, Burrows CJ, Schlegel HB. J. Am. Chem. Soc. 2008;130:5245–5256. doi: 10.1021/ja7104448. [DOI] [PubMed] [Google Scholar]
- 63.McCallum JEB, Kuniyoshi CY, Foote CS. J. Am. Chem. Soc. 2004;126:16777–16782. doi: 10.1021/ja030678p. [DOI] [PubMed] [Google Scholar]
- 64.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 65.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kornyushyna O, Burrows CJ. Biochemistry. 2003;42:13008–13018. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
- 68.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. J. Am. Chem. Soc. 1999;121:9423–9428. [Google Scholar]
- 69.Delaney S, Delaney JC, Essigmann JM. Chem. Res. Toxicol. 2007;20:1718–1729. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henderson PT, Neeley WL, Delaney JC, Gu F, Niles JC, Hah SS, Tannenbaum SR, Essigmann JM. Chem. Res. Toxicol. 2005;18:12–18. doi: 10.1021/tx049757k. [DOI] [PubMed] [Google Scholar]
- 71.Jaffe EK, Cohn M. Biochemistry. 1978;17:652–657. doi: 10.1021/bi00597a014. [DOI] [PubMed] [Google Scholar]
- 72.Cho BP. Magn. Reson. Chem. 1993;31:1048–1053. [Google Scholar]
- 73.Muller JG, Duarte V, Hickerson RP, Burrows CJ. Nucleic Acids Res. 1998;26:2247–2249. doi: 10.1093/nar/26.9.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson AT, Wiest O. J. Phys. Chem. B. 2007;111:14398–14404. doi: 10.1021/jp076371k. [DOI] [PubMed] [Google Scholar]
- 75.Barnett RN, Bongiorno A, Cleveland CL, Joy A, Landman U, Schuster GB. J. Am. Chem. Soc. 2006;128:10795–10800. doi: 10.1021/ja061795y. [DOI] [PubMed] [Google Scholar]
- 76.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benevides JM, Stow PL, Ilas LL, Iacardona NL, Thomas GJ., Jr Biochemistry. 1991;30:4855–4863. doi: 10.1021/bi00234a004. [DOI] [PubMed] [Google Scholar]
- 78.Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, Patel DJ. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- 79.Uesugi S, Ikehara M. J. Am. Chem. Soc. 1977;99:3250–3253. doi: 10.1021/ja00452a008. [DOI] [PubMed] [Google Scholar]
- 80.Gelfand CA, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 81.Chen J, Dupradeau F-Y, Case DA, Turner CJ, Stubbe J. Nucleic Acids Res. 2008;36:253–262. doi: 10.1093/nar/gkm622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biochemistry. 2002;34:16148–16160. doi: 10.1021/bi00049a030. [DOI] [PubMed] [Google Scholar]
- 83.Cheong C, Tinoco I, Jr, Chollet A. Nucleic Acids Res. 1988;16:5115–5122. doi: 10.1093/nar/16.11.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nguyen HK, Auffray P, Asseline V, Dupret D, Thuong NT. Nucleic Acids Res. 1997;25:3059–3065. doi: 10.1093/nar/25.15.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hall DB, Barton JK. J. Am. Chem. Soc. 1997;119:5045–5046. [Google Scholar]
- 86.Bhattacharya PK, Barton JK. J. Am. Chem. Soc. 2001;123:8649–8656. doi: 10.1021/ja010996t. [DOI] [PubMed] [Google Scholar]
- 87.Chung M-H, Kiyosawa H, Ohtsuka E, Nishimura S, Kasai H. Biochem. Biophys. Res. Commun. 1992;188:1–7. doi: 10.1016/0006-291x(92)92341-t. [DOI] [PubMed] [Google Scholar]
- 88.Cullis PM, Malone ME, Merson-Davies LA. J. Am. Chem. Soc. 1996;118:2775–2781. [Google Scholar]
- 89.Gasparutto D, Ravanat J-L, Gerot O, Cadet J. J. Am. Chem. Soc. 1998;120:10283–10286. [Google Scholar]
- 90.Buchko GW, Cadet J, Berger M, Revanat J-L. Nucleic Acids Res. 1992;20:4847–4851. doi: 10.1093/nar/20.18.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 92.Henderson PT, Delaney JC, Gu F, Tannenbaum SR, Essigmann JM. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- 93.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hailer MK, Slade PG, Martin BK, Rosenquist TA, Sugden KD. DNA Repair. 2004;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 95.Sejersted Y, Hildrestrand GA, Kunke D, Rolseth Vy, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Asegg M, Fleming AM, Saugstad OD, Burrows CJ, Luna L, Bjoras M. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18802–18807. doi: 10.1073/pnas.1106880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Biochemistry. 2010 [Google Scholar]
- 97.Simic MG, Jovanovic SV. J. Am. Chem. Soc. 1989;111:5778–5782. [Google Scholar]
- 98.Johnston DH, Glasgow KC, Thorp HH. J. Am. Chem. Soc. 2002;117:8933–8938. [Google Scholar]
- 99.Schibel AEP, Fleming AM, Jin Q, An N, Liu J, Blakemore CP, White HS, Burrows CJ. J. Am. Chem. Soc. 2011;133:14778–14784. doi: 10.1021/ja205653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kawai K, Wata Y, Hara M, Tojo S, Majima T. J. Am. Chem. Soc. 2002;124:3586–3590. doi: 10.1021/ja016530s. [DOI] [PubMed] [Google Scholar]
- 101.Lee Y-A, Liu Z, Dedon PC, Geacintov NE, Shafirovich V. ChemBioChem. 2011;12:1731–1739. doi: 10.1002/cbic.201100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Casey JR, Grinstein S, Orlowski J. Nature Rev. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 103.Lim KS, Taghizadeh K, Wishnok JS, Babu IR, Shafirovich V, Geacintov NE, Dedon PC. Chem. Res. Toxicol. 2012;25:366–373. doi: 10.1021/tx200422g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haghdoost S, Sjolander L, Czene S, Harms-Ringdahl M. Free Radical Biol. Med. 2006;41:620–626. doi: 10.1016/j.freeradbiomed.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 105.Arnett SD, Osbourn DM, Moore KD, Vandaveer SS, Lunte CE. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2005;827:16–25. doi: 10.1016/j.jchromb.2005.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ames BN, Cathcart R, Schwiers E, Hochstein P. Proc. Natl. Acad. Sci. U.S.A. 1981;78:6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.