Abstract
Background
Cystic fibrosis is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Over 1,800 CFTR mutations have been reported, and about 12% of mutations are believed to impair pre-mRNA splicing. Given that several synthetic, non-splice-junction synonymous substitutions have been reported to alter splicing in CFTR, we predicted that naturally occurring synonymous substitutions may be erroneously classified as functionally neutral.
Methods
Computational tools were used to predict the effect of synonymous substitutions on CFTR pre-mRNA splicing. The functional consequences of selected substitutions were evaluated using a minigene splicing assay.
Results
Two synonymous mutations were shown to have a dramatic effect on CFTR pre-mRNA splicing, and consequently could alter protein integrity and phenotypic outcome. Conclusions: Traditional methods of mutation analysis overlook splicing defects that occur at internal positions in coding exons, especially synonymous substitutions. We show that bioinformatics tools and minigene splicing assays are a potent combination to prioritize and identify mutations that cause aberrant CFTR pre-mRNA splicing.
Introduction
Cystic fibrosis (CF) is an autosomal recessive disease caused by mutations in the cystic fibrosis transmembrane conductance regulator gene (CFTR), which encodes a cAMP-regulated chloride channel protein. Clinical manifestations of the disorder include lung disease, pancreatic insufficiency, male infertility, and elevated concentrations of chloride in sweat (1). Severity of the disease is highly variable and some patients demonstrate only a subset of the classic CF symptoms (2). Despite the phenotypic variability, CF is most often caused by the ΔF508 mutation, which accounts for about 70% of mutant alleles among CF patients. This mutation, which is located in the NBD1 domain, impairs processing of the CFTR protein in the endoplasmic reticulum (3). In total, 1,877 mutations appear in the Cystic Fibrosis Mutation Database (4), and 226 (12%) of these mutations are implicated in pre-mRNA splicing.
Mutations that interfere with accurate pre-mRNA splicing occur at acceptor and donor splice sites (SSs), the branch point sequence, and the polypyrimidine tract. Additionally, mutations can readily disrupt regulated splicing by interfering with exonic splicing regulators (ESRs) (5, 6). Such regulatory elements function as exonic or intronic splicing enhancers (ESEs and ISEs, respectively) or as exonic or intronic splicing silencers (ESSs and ISSs, respectively). Regardless of their type (nonsense, missense, or synonymous), substitutions can interfere with these internal splicing regulatory elements, causing a variety of outcomes, which include exon skipping, inclusion of intron sequences, or activation of cryptic SSs (7). These events could ultimately produce unstable transcripts or defective protein isoforms, as illustrated by splicing mutations in CF (8–11), breast cancer (12), and ataxia telangiectasia (13).
The Cystic Fibrosis Mutation Database lists 226 mutations implicated in CFTR splicing. Among these, five are exonic synonymous substitutions. All five changes involve a G>A substitution at the last nucleotide of the exon and disrupt splicing by interfering with the donor SS. All other synonymous substitutions in the CFTR Mutation Database are classified as polymorphisms, a fact that illustrates the common practice of overlooking synonymous substitutions in clinical diagnostics, because they are assumed to be functionally neutral. However, about 25% of synonymous substitutions introduced into human CFTR exons 10 and 13 have a deleterious effect on splicing (previously known as exons 9 and 12 (9,14)), confirming that pre-mRNA splicing depends on the regulatory information provided at the nucleotide level. Nevertheless, most patient-derived synonymous variants have not been systematically studied.
We searched for all reported variants located at synonymous codon positions in CFTR and used two computational tools, the Skippy web server (15) and SplicePort (16), to predict their effect on CFTR pre-mRNA splicing. Using a minigene splicing assay, which has been validated as a method to study CFTR splicing mutations (17), we identified two synonymous mutations that cause aberrant CFTR splicing through activation of cryptic exonic SSs for CFTR exons 8 and 22.
Methods
Exon numbering system
Exon numbering is taken from the 27-exon designation used in the Cystic Fibrosis Mutation Database (4).
Variant data
Synonymous substitutions in the CFTR gene were identified from dbSNP (v130) and the CF Mutation Database. For the former, we used the SNP mapping to the UCSC hg18 assembly of the human genome (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/snp130.txt.gz), and found 43 SNPs corresponding to synonymous changes in the CFTR gene. Additionally, we searched the CF Mutations database (http://www.genet.sickkids.on.ca/) for exonic mutations (a total of 1,444 entries were retrieved), from which we selected point mutations (1,065 entries) that corresponded to synonymous changes (a total of 94 entries). In total, we found 99 unique synonymous variants from the two databases (Table S1), three of which (c.1036C>T, c.2245C>T, c.3472C>A) occur at first codon positions.
Computational analysis of SNP impact on CFTR splicing
All synonymous substitutions were analyzed using the Skippy web server (15). Given mutation types and positions within exons, this tool evaluates several aspects of exonic splicing regulatory features that could cause aberrant splicing, such as loss of exonic splicing enhancers (ESEs) and gain of exonic splicing silencers (ESSs). Skippy also evaluates the possibility of cryptic exonic SS activation using MaxEnt scores (18), which evaluate SS strength. We compared the score difference for “AG” and “GT” dinucleotides within three and four nucleotides, respectively, of each variant. The SplicePort tool (16) evaluates the strength of SS based on sequence features located within 80 bps of each side of consensus “AG” and “GT” dinucleotides. We evaluated the impact of each variant on the strength of consensus dinucleotides by calculating the difference in SplicePort scores corresponding to reference and variant alleles. This approach has the advantage of linking variants located deep within exons to the performance of the donor or acceptor sites individually, or simultaneously when exons are shorter than 160 bps. Additionally, we checked whether the mutations created novel consensus “AG” and/or “GT” dinucleotide, and evaluated their strength using SplicePort.
Selection of variants for experimental testing
Based on our Skippy analysis of the 99 synonymous CFTR variants, we chose five for experimental testing. Three variants (rs1800083, rs1800084, rs1800122; variants 2, 3 and 5, respectively) have log-odds ratio (LOR) scores higher than the threshold value of 1.20 (Table 1), which indicates that they are statistically more similar to known splice-affecting variants (SAVs) than to neutral human polymorphisms that do not affect splicing (hSNPs). The threshold represents the minimal score for predicting variant involvement in exon skipping (15). We also chose to test rs1800105 (variant 4) because it scored well for activating a cryptic acceptor SSs in a Skippy analysis. In this case, the change in the MaxEnt 3′ SS score between the mutant and reference alleles (Δ3′SS) is higher than the suggested threshold of 1.0 (Table 2) (15). Rs35033453 (variant 1; Table 1) served as a test of a negative prediction, since it did not meet the threshold LOR score despite having a large loss of ESEs in the Skippy analysis (Table 2).
Table 1.
Summary information for the variants chosen for experimental testing with the minigene splicing assay.
| Variant Number | rsSNP ID | cDNA Name | Position on Chromosome 7 | CFTR Exon # | Exon Size (bps) | Position within Exon |
|---|---|---|---|---|---|---|
| 1 | rs35033453 | c.738G>A* | 116,962,696 | 6 | 164 | 159 |
| 2 | rs1800083 | c.915C>T | 116,967,435 | 8 | 247 | 46 |
| 3 | rs1800084 | c.927C>G | 116,967,447 | 8 | 247 | 58 |
| 4 | rs1800105 | c.2604A>G | 117,022,333 | 15 | 129 | 114 |
| 5 | rs1800122 | c.3594G>T | 117,054,937 | 22 | 249 | 126 |
Notes:
This mutation (rs35033453) is not currently included in the CF Mutation Database, but its corresponding cDNA name was determined based on its position. The cDNA names for all mutations include the reference and variant alleles observed at the corresponding locus. Genomic positions and variant information of all mutations are relative to the reference strand of the hg18 UCSC assembly of the human genome.
Table 2.
Skippy output for the five variants chosen for experimental testing.
| Variant Number | Number of ESE Losses | Number of ESE Gains | Number of ESS Gains | LOR Score | Exonic Environment* | Intronic ESS Density* | Enhancement of Cryptic 3′ Splice Signal (Δ3′SS) | Score of Cryptic 3′ Splice Signal (MaxEnt) | Score of Exon 3′ SS (MaxEnt) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| ESE Density | ESS Density | Upstream | Downstream | ||||||||
| 1 | 3 | 0 | 0 | 0.749 | >= 1 | 0 | >= 1 | 0 | NA | NA | 10.26 |
| 2 | 2 | 0 | 3 | 3.152 | 0 | 0 | 0 | 0 | NA | NA | 8.16 |
| 3 | 1 | 0 | 2 | 2.006 | 0 | 0 | 0 | 0 | 0.45999 | 8.92 | 8.16 |
| 4 | 0 | 0 | 0 | −1.284 | 0 | >= 2 | >= 1 | >= 2 | 1.16 | 4.62 | 6.71 |
| 5 | 4 | 0 | 0 | 1.36 | >= 1 | >= 2 | >= 1 | 0 | NA | NA | 9.83 |
Notes:
Skippy reports densities of exonic and intronic ESEs and ESSs as standard deviations from mean values observed for variants that do not affect splicing (hSNPs). Δ3′SS scores represent the difference between the original splice site score and the cryptic spice site score.
Limited information is available for these variants. For example, frequency information for variant 1 comes from dbSNP, which reports it in one out of 28 chromosomes sampled from a 15-individual multi-ethnic CFTR panel. Variant 2 was reported in one individual (19), but no further information is currently available (Edkins, pers. comm. 2011). Variant 3 was found in one out of 150 chromosomes sampled, in a relative of a CF patient (20), as well as in a control patient of a more recent survey (21). Variant 4 was found once among 60 chromosomes sampled, in a CF patient with two other debilitating mutations (22). Variant 5 is a rare allele found in one out of 274 chromosomes sampled from CF patients from Southern France (23).
Minigene constructs
Reference and variant allele sequences were synthesized by GenScript (Piscataway, NJ) and inserted into a PUC19 plasmid. Constructs included the exon of interest with at least 51 bps of flanking intronic sequences (Table 3). These sequences were transferred into the Invitrogen TOPO-TA cloning plasmid, pCR®8, after PCR amplification of the insert region using 30 second annealing and extension cycles and 55°C annealing temperature (primer sequences provided in Table 3). The completed constructs were verified by digestion with EcoRI and DNA sequencing. The CFTR regions were subsequently transferred into the pDESTsplice minigene splicing vector (24) using the second step of the Gateway cloning (known as an “LR” reaction) according to manufacturer’s instructions (Invitrogen). In the pDESTsplice vector, target exons lie between constitutively expressed exons from the rat insulin 2 gene. Final constructs were verified by restriction digestion with BsrGI followed by DNA sequencing.
Table 3.
Summary information for the source of genomic sequences (exons and their flanks) used in the minigene splicing assay.
| Variant | Exon Size (bps) | 5′ Flank (bps) | 3′ Flank (bps) | Total Size (bps) | Genomic Coordinates (hg18) | Primers | |
|---|---|---|---|---|---|---|---|
| Start | End | ||||||
| 1 | 164 | 61 | 57 | 282 | 116,962,478 | 116,962,757 | 5′-TTAGTTTCTAGGGGTGGAAGATACA-3′ 5′-GGGCTTTTTGAAAACATAATTTTTAA-3′ |
| 2 | 247 | 61 | 83 | 391 | 116,967,329 | 116,967,719 | 5′-GATCCCTGATATTTGAAAAATAAAAT-3′ 5′-ACATTTTTGCAAAGTTCATTAGAACT-3′ |
| 3 | 247 | 61 | 83 | 391 | 116,967,329 | 116,967,719 | 5′-GATCCCTGATATTTGAAAAATAAAAT-3′ 5′-ACATTTTTGCAAAGTTCATTAGAACT-3′ |
| 4 | 129 | 71 | 66 | 266 | 117,022,149 | 117,022,414 | 5′-TAAAAATAAAACCACAATGGTGGC-3′ 5′-ATACATCCCCAAACTATCTTAATTTAA-3′ |
| 5 | 249 | 51 | 51 | 351 | 117,054,761 | 117,055,111 | 5′-TGAAATTGTCTGCCATTCTTAAAA-3′ 5′-GATTCACTTACTGAACACAGTCTAACA-3′ |
In vitro transfection
Using a standard protocol for transfection, 4×105 K562 cells were transfected with 0.4 ug of plasmid DNA carrying either the reference allele or variant allele of interest. Three replicate transfections were performed for each plasmid by electroporation (using the Lonza 96-well Nucleofector II). Individual transfection reactions were plated in 1 mL media in a 12 well plate. After 24 hours the three replicates were combined for RNA harvest using the RNeasy® mini kit (Qiagen). Reverse transcription was performed using the iScript™ cDNA synthesis kit (Bio-Rad Laboratories). To generate cDNA, 900 ng of the RNA was used in a 20 uL iScript reaction mix and incubated in a PCR machine under the following conditions: 5 minutes at 25°C, 30 minutes at 42°C and 5 minutes at 85°C. Variants 2 and 5 were also transfected into IB3-1 cells. A total of 6×105 cells were transfected with 2 ug of DNA (in triplicate) by Lipofectamine 2000 (Invitrogen, cat. no. 11668-500) and plated into 2 mL of medium in a 6 well plate. After 24 hours, samples were handled the same way as indicated for K562 cells.
Analysis of minigene expression
The cDNA containing the spliced products of the minigene vector was amplified using PCR primers specific for the rat insulin 2 exons (5′-CCTGCTCATCCTCTGGGAGC-3′, 5′-AGGTCTGAAGGTCACGGGCC-3′). Conditions were set at 95°C denaturation for 5 min, followed by 35 cycles of 95°C for 1 min, 62°C for 1 min, and 72°C for 1 min, with a final 72°C hold for 5 min. Products were visualized by gel electrophoresis on a 2% agarose gel. Quantitation of the cDNA products was performed in parallel using the same forward PCR primer, with an additional tag at the 5′ end with 6-carboxyfluorescein to enable measurement of differences in the size and quantity of the reference and variant minigene products. Quantitation samples were run on the Applied Biosystems (ABI) Prism 3100 Genetic Analyzer, which included the use of a 36-cm capillary array and POP4 polymer. All samples were run with the ABI internal lane size standard ROX 500, which accurately sizes 35–500 bp fragments. Quantitation data was analyzed using ABI GeneScan (Version 3.7 II) and Genotyper (Version 2.5) software.
PCR products of the cDNA were subcloned using the pCR®8 TA cloning vector (Invitrogen) and sequenced to determine the precise boundaries generated during abnormal splicing. The sequences were verified by confirming that the spliced sequences were flanked by rat insulin 2 exon sequences, and were mapped to the CFTR genomic sequence using BLAT (25). We examined the effects of splicing mutations on the open reading frame (ORF) of the CFTR protein using Artemis Genome Browser and Annotation Tool (26). Genomic nucleotide sequences (accession numbers AC000111 and AC000061.1) were imported directly from EMBL-EBI using Artemis Dbfetch tool.
Results
Computational evaluation of variants
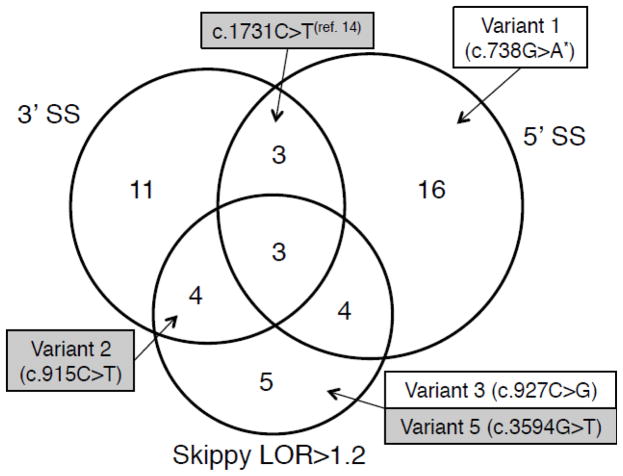
We first evaluated the impact of the synonymous substitutions using the NHGRI Skippy web server. The underlying model used by Skippy examines internal exons only, and therefore six mutations located in the first and last exons were not evaluated. Among the 93 variants evaluated by Skippy, 16 produced LOR scores larger than 1.2 (Figure 1, Tables S1, S2), suggesting that they could severely affect the exonic regulatory landscape.
Figure 1.
Summary of computational predictions for synonymous substitutions identified in CFTR. The Venn diagram depicts counts of mutations determined computationally to affect acceptor and donor SSs through SplicePort scoring (3′SS and 5′SS, respectively), or to affect the splicing regulatory landscape by Skippy (LOR>1.2). Mutations chosen for experimental testing represent their corresponding categories. Mutations found to affect splicing are shown with a shaded background. Variant 4 (c.2604A>G) is not included here since it does not have a high LOR score, and does not lower scores of SSs in exon 15. Mutation c.1731C>T was not evaluated in this study, but was found to affect skipping by Pagani et al. (14). *The name for this mutation was determined from its position, but is not currently found in the CF Mutation Database.
In addition to the comprehensive LOR metric, Skippy also indicates whether variants are likely to create de novo cryptic splice signals, which would alter the length of the original exons. Alternative acceptor splice signals were detected in 13 cases, with five meeting the Skippy threshold score of a Δ 3′SS greater than 1.0 (Tables S1, S2). Alternative cryptic donor splice signals with Δ 5′SS scores greater than 1.0 were identified in two additional variants (Tables S1, S2). We also verified the activation of cryptic SSs using the SplicePort scoring scheme. Only two, and six variants increased the score of alternative acceptor and donor SSs, respectively, beyond the scores of constitutive SSs (Table S1).
We also used SplicePort to evaluate the impact of variants on the strength of the constitutive SSs and found several variants that decreased their scores (Tables S1, S3). Specifically, 21 variants affected acceptor SSs (Figure S1, Table S1) and 26 affected donor SSs (Figure S2, Table S1). As anticipated, the mutations with the largest effect on SS scores were those located at the exon boundaries. We confirmed 5 known mutations at donor SSs (Figure S2, Table S1) and found two additional examples (c.1209G>A and c.1584G>A) (27,28) not reported in the CFTR database.
Overall, we concluded that 46 variants could have a detrimental effect on splicing (Figure 1). Of these, 14 appear to interfere with splicing by a dual mechanism (e.g. either affecting both acceptor and donor SSs or by affecting one SSs along with internal ESRs). Mutations chosen for experimental testing represent these mechanisms for interference with CFTR splicing. For example, variants 2, 3 and 5 alter the ESR environment, and variant 4 scores well for activating a cryptic acceptor splice signal while also affecting the ESR environment (see Methods, Table 1).
Minigene splicing assay results
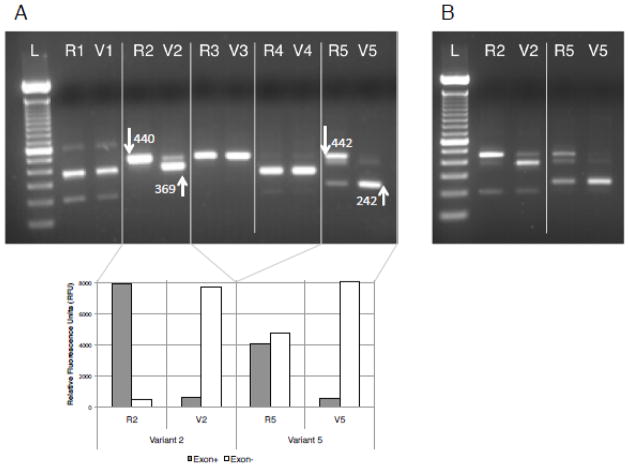
Functional consequences of the five selected variants (Table 1) were tested using a minigene splicing assay. Reference and variant alleles were cloned into the pDESTsplice vector and transiently transfected into K562 cells (see Methods). Evidence of altered splicing was identified in two of the five variants (variants 2 and 5; Figure 2A), and results were replicated in IB3-1 cells (a mutant CFTR lung epithelial cell line with the CFTR genotype of D508/W1282X) (Figure 2B). In the case of variant 2 the strongest band resulted from use of a cryptic splice site that shortened the 440-bp product by 71 bp (Figure 2). Additionally, full exon skipping occurred 2-times more frequently than for the reference allele (Figure S3). Similarly, variant 5 reduced the expected 442-bp reference allele to a product 200 bps smaller (Figure 2).
Figure 2.
Experimental assessment of splicing interference by five synonymous substitutions. A) Gel with splicing products collected following transient transfection into K562 cells. For each variant (1 through 5), constructs with the reference (R) and variant (V) alleles were analyzed separately. White arrows point to bands that indicate differential splicing; numbers correspond to band sizes. Results of quantitation with fluorescent labeled PCR primers are shown for variants 2 and 5, revealing different splicing patterns between reference and variant alleles. Exon+ and Exon− correspond to normal splicing (higher band) and cryptic splicing (lower band), respectively. B) Results for variants 2 and 5 were replicated in IB3-1 cells, a cell line more relevant for the phenotype of CF patients.
The remaining three variants showed virtually unaltered splicing patterns between reference and variant alleles, including full length exon skipping. Moreover, variant 1 showed substantial exon inclusion, which is consistent with having an LOR score lower than the threshold of 1.2 (Figure 2). The fluorescent quantitation also indicated that exon inclusion occurred at a slightly higher rate for the variant than for the reference allele (RFUs 7039 and 4984, respectively).
Effect of mutations on reading frame
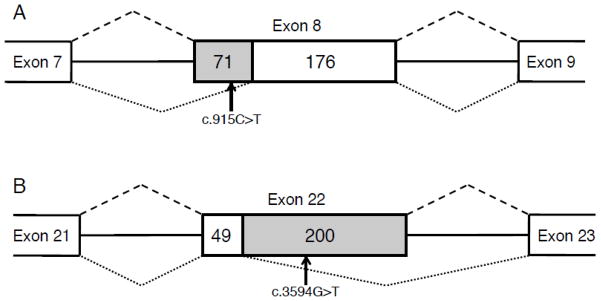
The splicing boundaries of each exon were determined through sequencing the PCR product corresponding to the band size of interest and identifying the vector-derived sequence that originated from the rat insulin 2 gene. In the case of variant 2, 71 bps were eliminated from the 5′ end of the CFTR exon 8 (Figure 3A). This would cause a shift in the ORF, which in turn would cause truncation of the protein through a premature STOP codon further downstream in exon 9 (TAA at chr7:116,969,329–116,969,331). In the case of variant 5, 200 bps were eliminated from the 3′ end of exon 22 (Figure 3B). Similarly to the case of variant 2, the shift in ORF predicted by this deletion would introduce an early STOP codon (TGA at chr7:117,069,799–117,069,801), which would cause truncation of the protein.”
Figure 3.
Schematic representation of splicing aberrations introduced by variants 2 and 5. A) Exon 8, in which variant 2 is located, is fully spliced with the reference allele (splicing pattern shown above by dashed lines). When the variant “T” allele is present at exon position 46, a total of 71 bps are skipped from the 5′ end of the exon (splicing pattern shown below by dotted lines). B) In the case of variant 5 (c.3594G>T), 200 bps are skipped from the 3′ end of exon 22. Flanking exons and introns are not shown to scale, whereas the middle exon is shown to scale. Numbers indicate size in bps of each section. The skipped region is shown with shaded background.
Conclusions
Our analyses demonstrate the dramatic effects of two seemingly innocuous SNPs on splicing in the CFTR gene. We have shown that synonymous substitutions in CFTR exons 8 (variant 2, c.915C>T) and 22 (variant 5, c.3594G>T) are capable of altering normal splicing patterns through ectopic SS activation in vitro and thus would represent mutations, not neutral polymorphisms, as listed in the dbSNP repository (29). Note that although all variants, except variant 1, are included in the CF Mutation Database (Table 1), they are described as neutral sequence variations, which may inaccurately describe variants 2 and 5. While the use of an in vitro minigene assay strongly suggests that these variants alter splicing, only in vivo assessment can validate the splicing outcome. The presence of other unknown variants in those genetic haplotypes could also influence the splicing outcome positively or negatively. RNA samples carrying these variants were not available for further testing.
Variant 2 enabled use of an ectopic SS, although some normal product was also produced. Notably, variant 2 coincided with the creation of three ESSs, a feature which we have previously observed at sites of de novo ectopic splicing (15). Moreover, SplicePort reported a reduced score of the acceptor SS due to the variant. Our manual examination of the sequence indicates that the mutation creates a new consensus branch point sequence YTNAY (where Y = pyrimidine, N = any nucleotide) (30), for which the C>T substitution introduces a T residue at the second position of the consensus sequence. Consistent with observed branch point locations in humans (30), the cryptic branch point ‘A’ residue is located 24 bps upstream from the ectopic acceptor SS. These observations suggest that the de novo branch point sequence can outcompete the original branch point sequence. This conclusion is supported by use of the alternative acceptor SS (at exon position 72), whose SplicePort score increases from −0.5 to −0.29 in the presence of variant 2, while remaining lower than the constitutive acceptor SS (that decreases from 0.4 to 0.03) and another alternative acceptor SS (at exon position 120), whose SplicePort score increases from −0.21 to −0.05).
Variant 5 also demonstrated activation of a cryptic SS, though it only eliminated ESEs without creating new ESSs. This example highlights the importance of complementary analyses by multiple computational tools, such as Skippy and SplicePort. Because the mutation happened in the middle of this 249-bp exon, SplicePort could not evaluate the effect on the reference SSs, being more than 80 bps away from either junction, while Skippy could assess the loss of ESEs and gain of ESSs. However, SplicePort additionally revealed that the alternative donor signal had the highest score (−0.28) among any SS signals affected by the variant. The score of the alternative signal was higher than that of the original reference SS (−0.64), indicating that the loss of four ESEs was the crucial event in the shift of SS usage. This mutation is located in the M11 region of the MSD2 transmembrane domain of the CFTR protein. Any product carrying the deletion would lack the C-terminal nuclear binding domain. Unfortunately, the effect of the mutation in vivo could not be tested due to death of the patient in infancy (23) (des Georges, pers. comm. 2011).
In this analysis we tested variant 1 despite the fact that it fell below the threshold LOR score recommended for Skippy assessment because the loss of three ESE sites provided a reasonable hypothesis for further examination. Nevertheless, the in vitro splicing of the variant allele was consistent with our predictions of no exon skipping. Moreover, our negative result from variant 4 might be explained by results from our previous work, which showed that ectopic SSs are predominantly located in the half of the exon closest to the natural SS they replace (15). In the case of variant 4, the predicted ectopic acceptor SS occurs in the unfavorable 3′ half of the exon. Furthermore, in agreement with the experimental result, the SplicePort score for the alternative splice signal remained much lower than the reference SS. Given that the Skippy and Spliceport tools utilize different models of SSs (i.e., MaxEnt scores at the SS core, or 160 bp around the SS, respectively) examination by more than one tool may be important for prioritization purposes.
All variants characterized in this study are likely to be overlooked by classical methods since their status as synonymous substitutions does not cause a change in the amino acid sequence. Additionally, the occurrence of these synonymous variants in the third position of the codon further supports an a priori conclusion of neutral substitutions. Nevertheless, we examined sequence conservation, which often indicates regulatory information (31) in intronic (32) and exonic splicing elements. At the single nucleotide level, reference alleles for variants 2 and 5 did not demonstrate a strong correlation between sequence conservation and functional impact. Although variant 1 was conserved, its effect of slightly increasing the splicing efficiency contradicts the predicted consequence of losing a conserved base. The variant 1 substitution is used in several species, including M. domestica, G. gallus, and X. tropicalis, providing evidence that it is not detrimental in vivo. Variant 3, which did not show splicing interference, also shows conservation across mammals. These results imply that conservation at single base resolution is not necessarily indicative of constraints applied to splicing regulation, and that variants replacing conserved positions will not a priori have a negative effect. This conclusion is supported by the example of c.1731C>T, which causes exon skipping (14). Despite this, the “T” variant, when present in rodents, is associated with 100% exon inclusion. This example underlines the fact that splicing regulation is done through a complex environment (e.g. rodents have additional sequence differences from humans which may compensate in distinct ways). Therefore the role of single nucleotides should only be considered in their specific environments. We also looked at negative selection measured over the neighborhood of a variant using the regulatory constraint (RC) model (15) implemented in Skippy. However we found no data supporting regulatory constraint at variants 2 and 5. Thus sequence conservation, while often used to implicate splicing elements, does not provide predictive strength in these cases.
The findings in this study indicate that synonymous substitutions can have a dramatic effect on splicing, and thus should be evaluated in functional studies. Because synonymous and non-synonymous substitutions can have consequences extending beyond the amino acid sequence, mutation analysis should routinely include the use of bioinformatics tools to evaluate the effects of sequence variations on splicing. It should be noted that the tools are useful for prioritization of variants; however due to the complexity of splicing events, they are prone to false positive and false negative predictions and necessitate experimental testing. Results obtained from mini-gene splicing assays often recapitulate the in vivo situation and provide justification for further experimental examination of patient samples, when available. Our findings provide evidence that pre-mRNA splicing ought to be evaluated in CF genomes where a second mutation eludes detection. In clinical diagnostics, synonymous substitutions should not be regarded as functionally neutral and may be significant contributors in compound heterozygous diseases. Proper classification of such mutations will be helpful in future assessment of CF, in therapeutic targeting, and to specifically address molecular mechanisms of disease.
Supplementary Material
Acknowledgments
We thank Ursula Harper of the NHGRI Genomics Core for performing the fluorescent quantitation. This work was supported by the Intramural program of the National Human Genome Research Institute, National Institutes of Health. Edward Edkins and Marie des Georges provided historical information on variants 2 and 5.
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cutting G, Accurso F, Ramsey B, Welsh M. Cystic fibrosis. In: Scriver C, et al., editors. Metabolic and molecular basis of inherited disease. 8. New York: McGraw-Hill; 2001. [Google Scholar]
- 2.Kerem E, Corey M, Kerem BS, Rommens J, Markiewicz D, Levison H, et al. The relation between genotype and phenotype in cystic-fibrosis - analysis of the most common mutation (delta-f508) New England Journal of Medicine. 1990;323:1517–22. doi: 10.1056/NEJM199011293232203. [DOI] [PubMed] [Google Scholar]
- 3.Lukacs GL, Mohamed A, Kartner N, Chang XB, Riordan JR, Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (delta-f508) occurs in the endoplasmic-reticulum and requires atp. Embo Journal. 1994;13:6076–86. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cystic Fibrosis Mutation Database. 2010 Http://www.Genet.Sickkids.On.Ca/cftr/app.
- 5.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–98. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 6.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mrna secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–4. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 8.Pagani F, Stuani C, Tzetis M, Kanavakis E, Efthymiadou A, Doudounakis S, et al. New type of disease causing mutations: The example of the composite exonic regulatory elements of splicing in cftr exon 12. Hum Mol Genet. 2003;12:1111–20. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 9.Pagani F, Buratti E, Stuani C, Baralle FE. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J Biol Chem. 2003;278:26580–8. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 10.Aznarez I, Chan EM, Zielenski J, Blencowe BJ, Tsui LC. Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum Mol Genet. 2003;12:2031–40. doi: 10.1093/hmg/ddg215. [DOI] [PubMed] [Google Scholar]
- 11.Faa V, Coiana A, Incani F, Costantino L, Cao A, Rosatelli MC. A synonymous mutation in the CFTR gene causes aberrant splicing in an italian patient affected by a mild form of cystic fibrosis. J Mol Diagn. 2010;12:380–3. doi: 10.2353/jmoldx.2010.090126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in brca1 and other genes. Nat Genet. 2001;27:55–8. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 13.Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, et al. Splicing defects in the ataxia-telangiectasia gene, atm: Underlying mutations and consequences. Am J Hum Genet. 1999;64:1617–31. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci U S A. 2005;102:6368–72. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolfe A, Mullikin JC, Elnitski L. Genomic features defining exonic variants that modulate splicing. Genome Biol. 2010;11:R20. doi: 10.1186/gb-2010-11-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dogan RI, Getoor L, Wilbur WJ, Mount SM. Spliceport - an interactive splice-site analysis tool. Nucleic Acids Research. 2007;35:W285–W91. doi: 10.1093/nar/gkm407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dujardin G, Commandeur D, Le Jossic-Corcos C, Ferec C, Corcos L. Splicing defects in the cftr gene: Minigene analysis of two mutations, 1811+1g>c and 1898+3a>g. J Cyst Fibros. 2011;10:212–6. doi: 10.1016/j.jcf.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to rna splicing signals. J Comput Biol. 2004;11:377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 19.NL#54 r. Http://www.Genet.Sickkids.On.Ca/resource/nl/CFnewslet.54.html.
- 20.NL#45 r. Http://www.Genet.Sickkids.On.Ca/resource/nl/CFnewslet.45.html.
- 21.Le Marechal C, Audrezet MP, Quere I, Raguenes O, Langonne S, Ferec C. Complete and rapid scanning of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by denaturing high-performance liquid chromatography (d-hplc): Major implications for genetic counselling. Hum Genet. 2001;108:290–8. doi: 10.1007/s004390100490. [DOI] [PubMed] [Google Scholar]
- 22.NL#40 r. Http://www.Genet.Sickkids.On.Ca/resource/nl/CFnewslet.40.html.
- 23.Claustres M, Laussel M, Desgeorges M, Giansily M, Culard JF, Razakatsara G, et al. Analysis of the 27 exons and flanking regions of the cystic fibrosis gene: 40 different mutations account for 91.2% of the mutant alleles in southern france. Hum Mol Genet. 1993;2:1209–13. doi: 10.1093/hmg/2.8.1209. [DOI] [PubMed] [Google Scholar]
- 24.Kishore S, Khanna A, Stamm S. Rapid generation of splicing reporters with pspliceexpress. Gene. 2008;427:104–10. doi: 10.1016/j.gene.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ. Blat - the blast-like alignment tool. Genome Research. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–5. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 27.Telleria JJ, Alonso MJ, Calvo C, Alonso M, Blanco A. Spectrum of CFTR mutations in the middle north of spain and identification of a novel mutation (1341g-->a). Mutation in brief no. 252. Online. Hum Mutat. 1999;14:89. doi: 10.1002/(SICI)1098-1004(1999)14:1<89::AID-HUMU18>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Dork T, Dworniczak B, Aulehla-Scholz C, Wieczorek D, Bohm I, Mayerova A, et al. Distinct spectrum of CFTR gene mutations in congenital absence of vas deferens. Hum Genet. 1997;100:365–77. doi: 10.1007/s004390050518. [DOI] [PubMed] [Google Scholar]
- 29.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. Dbsnp: The NCBI database of genetic variation. Nucleic Acids Research. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao KP, Masuda A, Matsuura T, Ohno K. Human branch point consensus sequence is yunay. Nucleic Acids Research. 2008;36:2257–67. doi: 10.1093/nar/gkn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci U S A. 2004;101:15700–5. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Research. 2003;13:1631–7. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.