Abstract
Soils in boreal forests contain large stocks of carbon. Plants are the main source of this carbon through tissue residues and root exudates. A major part of the exudates are allocated to symbiotic ectomycorrhizal fungi. In return, the plant receives nutrients, in particular nitrogen from the mycorrhizal fungi. To capture the nitrogen, the fungi must at least partly disrupt the recalcitrant organic matter–protein complexes within which the nitrogen is embedded. This disruption process is poorly characterized. We used spectroscopic analyses and transcriptome profiling to examine the mechanism by which the ectomycorrhizal fungus Paxillus involutus degrades organic matter when acquiring nitrogen from plant litter. The fungus partially degraded polysaccharides and modified the structure of polyphenols. The observed chemical changes were consistent with a hydroxyl radical attack, involving Fenton chemistry similar to that of brown-rot fungi. The set of enzymes expressed by Pa. involutus during the degradation of the organic matter was similar to the set of enzymes involved in the oxidative degradation of wood by brown-rot fungi. However, Pa. involutus lacked transcripts encoding extracellular enzymes needed for metabolizing the released carbon. The saprotrophic activity has been reduced to a radical-based biodegradation system that can efficiently disrupt the organic matter–protein complexes and thereby mobilize the entrapped nutrients. We suggest that the released carbon then becomes available for further degradation and assimilation by commensal microbes, and that these activities have been lost in ectomycorrhizal fungi as an adaptation to symbiotic growth on host photosynthate. The interdependence of ectomycorrhizal symbionts and saprophytic microbes would provide a key link in the turnover of nutrients and carbon in forest ecosystems.
Introduction
The total soil organic matter (SOM) corresponds to more than three times as much carbon (C) as that contained in the atmosphere or within terrestrial vegetation (Schmidt et al., 2011). A major part of this carbon occurs in forests (Falkowski et al., 2000). It was earlier believed that the C turnover of SOM is mainly controlled by the input of above-ground plant material and the decomposing activity of saprophytic organisms. However, an increasing amount of evidence now suggests that plant roots and their associated microbial communities play an important role in SOM dynamics (Read et al., 2004; Högberg and Read, 2006; Schmidt et al., 2011). The organisms that provide the largest sink for this C in boreal and temperate forests are the fungal root symbionts that form ectomycorrhizae (ECM). Estimates suggest that 10–50% of the C fixed by photosynthesis is allocated belowground to the ECM fungi (Simard et al., 2002). Other experiments show that up to one third of the soil microbial biomass and half of the dissolved organic carbon produced in forest soils originate from ECM symbionts (Högberg and Högberg, 2002).
The ECM mycelia prospect the soil for essential nutrients such as nitrogen (N) and transfer these to the host plant. A large portion of the soil N is present in an organic form including in particular proteins and amino acids (Nannipieri and Eldor, 2009). These N compounds are associated with polyphenols, polysaccharides and other degradation products of microbial and plant biopolymers that are present in SOM (Piccolo, 2001; Chengrong and Zhihong, 2008). The facts that polyphenols, such as tannins, humic acids and fulvic acids, can bind to, and form recalcitrant complexes with proteins limits their accessibility for the fungi (Bending and Read, 1996), suggesting that the saprotrophic activities of ECM fungi should be directed not only to organic nutrient compounds in soil, but also to other C-containing soil polymers. Several studies have demonstrated that at least some species of ECM fungi can decompose components of the major classes of organic compounds like proteins, pectins, cellulose, hemicelluloses and polyphenols commonly found in soils (Norkrans, 1950; Trojanowski et al., 1984; Haselwandter et al., 1990). Studies in soil microcosms have also shown that patches containing SOM are actively colonized by ECM fungi and depleted of their nutrients (Bending and Read, 1995a, b). More recently, it has been proposed by Talbot and colleagues (2008) that ECM fungi may live as facultative saprotrophs, i.e. they can degrade and metabolize soil C compounds as an alternative C source when the supplies of photosynthate from the host plants are low. Considering the large biomass of the ECM mycelia, this type of saprotrophic activity might represent a significant, but as yet largely unaccounted for, pathway of C loss in forest ecosystems.
However, the evidences that ECM fungi can act as decomposers have been questioned and the mechanisms by which ECM fungi may decompose organic compounds are poorly characterized. Analyses of the genomes of ECM fungi have shown that in contrast to saprophytic fungi ECM fungi have a reduced set of genes encoding plant cell wall-degrading enzymes (Martin et al., 2008; Nagendran et al., 2009). Furthermore, results from experiments examining the saprotrophic activity of ECM fungi in soil microcosms and field settings have been contradictory (Bending and Read, 1995a, b; Colpaert and Van Laere, 1996). It has also been argued that the assays commonly employed to measure the saprophytic activity of ECM fungi are unspecific and do not properly capture the decomposing activities (Baldrian, 2009). Other authors have suggested that because ECM fungi are confined to deeper soil horizons (Lindahl et al., 2007), which contain more decomposed litter and humus material of low energetic value, they are unlikely to be able to grow as facultative saprophytes on this material (Baldrian, 2009).
In the present study, we investigated the mechanisms by which the ECM fungus Paxillus involutus degrade complex organic matter extracted from plant litter material. Pa. involutus (Batsch) Fr. (Basidiomycetes; Boletales) is widely distributed in the Northern hemisphere, and is one of the best-studied ECM fungi, especially with respect to its ecology and physiology (Wallander and Söderström, 1999). Chemical modifications of the organic matter were analysed by spectroscopic methods. We find that during the assimilation of organic N, Pa. involutus modifies the major components of the organic matter by producing hydroxyl radicals through a Fenton system similar to that of saprophytic, wood decomposing brown-rot fungi (Green III and Highley, 1997). Transcriptome analyses showed that Pa. involutus expresses an extracellular enzyme system that mediates radical-based biodegradation of polymers contained in the organic matter, including polyphenols. However, the transcriptome lacked transcripts encoding enzymes needed for metabolizing the C liberated by depolymerization of soluble polysaccharides. We suggest that the liberated C is acquired by commensal microbes (such as soil-living, saprophytic bacteria and fungi) and that the ability to metabolize this C has been lost in ECM fungi as an adaptation to symbiotic growth on host photosynthate.
Results
Growth conditions and experiments
Three organic extracts were used: forest litter extracted with hot water (FH), and a maize compost extracted with cold (MC) or hot (MH) water. The extracts varied considerably in their content of C, N and other nutrients. During the 7 days of incubation, Pa. involutus assimilated between 33% and 53% of the total N present in the organic matter extracts. The added glucose was not detected at the end of the incubations (Table S1). At the end of the incubations, the pH of the FH medium decreased (from pH 4.0 to 3.7), while it increased in the MH medium (pH 5.5 at the end of incubation) and the MC medium (pH 5.4).
Chemical conversion of organic matter
Comparison of the FTIR spectra of the FH, MH and MC extracts before and after incubation showed that the chemical composition of the organic extracts changed during growth of the Pa. involutus mycelium. Pronounced differences were observed in four spectral regions characteristic of specific vibration modes: sugar modes (970–1200 cm−1), nitrate stretching modes (1350–1450 cm−1), C–O/C=O stretching modes (1500–1800 cm−1) and aliphatic C–H stretching modes (2850–3000 cm−1) (Fig. 1). In all inoculated extracts, several prominent peaks of the sugar region substantially decreased in intensity, which indicates changes in the relative abundance of sugar molecules including polysaccharides (Fig. S1). However, due to overlapping bands and spectral similarities, detailed assignment of the sugar composition based on infrared spectra is difficult and not very precise. The changes of the FITR spectra at 2850–3000 cm−1 showed that relative abundance of molecules containing aliphatic C–H groups were modified, which is consistent with the changes discussed in the sugar region. The spectral region containing C–O/C=O stretching modes differed among the organic matter extracts; still the effect of fungal incubation was evident for all three samples in this region as well. The spectral changes suggest that the relative concentrations of carboxyl, ketone, aldehyde and amide groups were altered.
Fig. 1.
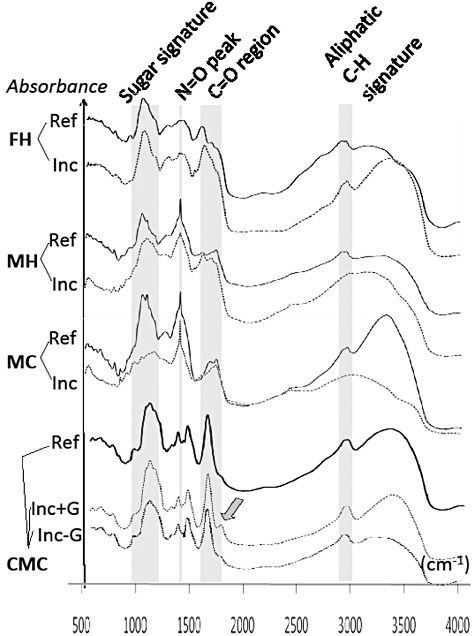
FTIR spectra of the three organic matter extracts and carboxy methyl cellulose (CMC) before (Ref, reference) and after 7 days of incubation (Inc, Inoculated) with Pa. involutus. FH, forest litter extracted with hot water; MH, maize compost extracted with hot water; MC, maize compost extracted with cold water. In the CMC spectra, the arrow indicates the appearance of a new peak located in the carbonyl region which is indicative of Fenton induced, oxidative modification of cellulose; +G and −G indicate CMC medium with or without supplement of glucose. Three replicates were analysed for the Inoculated and five for the Reference samples. The variation of the spectra between replicates was very low (Fig. S1).
The synchronous fluorescence spectra of the FH, MH and MC extracts contained three prominent peaks: Peak 1, located around 280–300 nm, could be associated with monoaromatic rings; Peak 2 (330–360 nm), with more complex molecules with two condensed aromatic rings; and Peak 3 (360–400 nm) with more complex aromatic ring systems bearing carbonyl or carboxyl groups (Senesi et al., 1991) (Fig. 2; Table S2). In the FH extracts, the fluorescence intensity of all peaks decreased during incubation, which suggests that both simple and more complex aromatic compounds were at least partly degraded (decrease in concentration) or transformed (modification of the degree of polycondensation, amount of conjugated chromophores, or of the degree of electron-donating substitution) by Pa. involutus. In the MH and MC extracts, it was mainly the more complex aromatic compounds (Peak 3) that were degraded or transformed during the incubation.
Fig. 2.
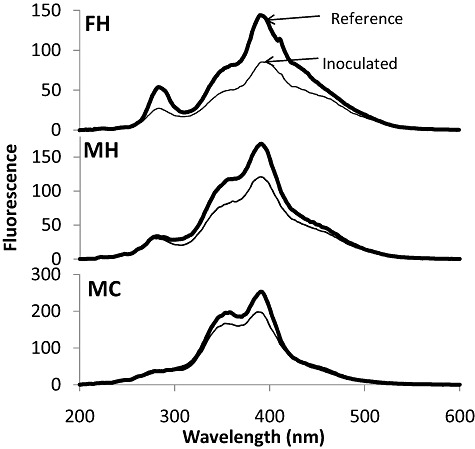
Synchronous fluorescence spectroscopy of the three organic matter extracts before (Reference, thick line) and after 7 days of incubation (Inoculated, thin line) with Pa. involutus. FH, forest litter extracted with hot water; MH, maize compost extracted with hot water; MC, maize compost extracted with cold water. Replicates (n = 3) of the samples were pooled before being analysed.
Analysis of the FH litter extract using Py-GC/MS confirmed that Pa. involutus degraded and converted the major classes of organic compounds present in dissolved organic matter (DOM) extracts (Fig. 3A; Table S3). The relative amount of pyrolysates related to aromatics, polyaromatics and N compounds decreased at least twice during the incubation. Relative amounts of pyrolysates issued from lignin subunits, present in the humic acids as residuals of the degradation process (Piccolo, 2001), and polysaccharides were less affected. However, detailed analysis of the pyrolysates related to lignin residuals showed that its two major subunits guaiacyl and syringyl were modified during growth of the fungus (Fig. 3B). The concentration of propenylguaiacol relative to guaiacol increased, indicating depletion of inter-unit ether linkages. The decrease in syringol/guaiacol ratio and the increase in 3-methoxycatechol pyroylsates relative to syringyl units showed that the syringyl units were both demethoxylated and demethylated.
Fig. 3.
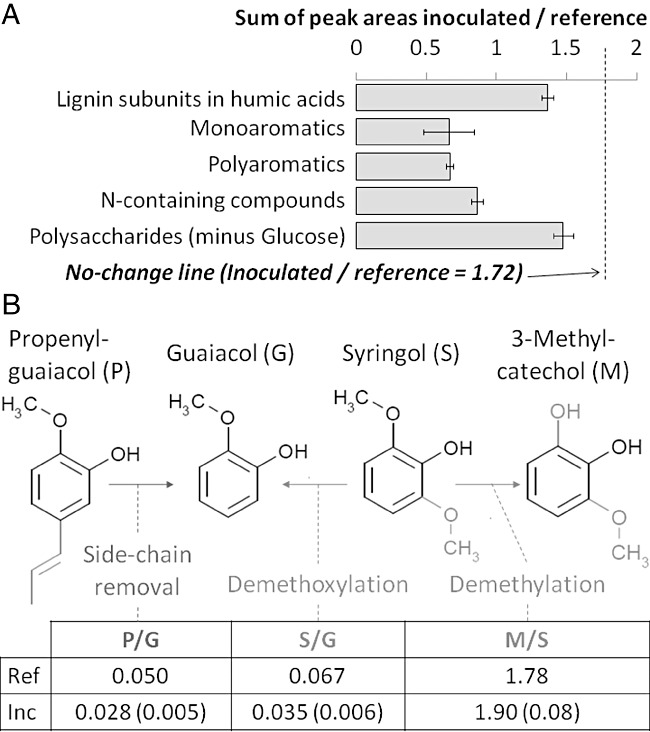
Pyrolysis GC/MS analysis of the organic matter extracted from forest litter using hot water (FH) after 7 days of incubation (Inoculated) and before incubation (Reference). A. Relative amounts of the major groups of organic compounds. A ratio below the ‘No change line’ indicates that this particular class of pyrolysis products was depleted in the Inoculated as compared with the Reference samples. ‘Lignin’ does not refer to genuine plant lignin but rather parts of the lignin molecule that are present in the humic acids as residuals of the degradation process. B. Chemical modification of lignin residuals subunits. Numbers indicate the relative peak area of the different lignin subunits in the reference sample against the average relative peak area in the incubated samples (n = 5, error bars denote standard error).
Size-exclusion chromatography showed that a majority of the soluble aromatic compounds and carbohydrate containing molecules present in the three organic matter extracts were in the size range of ∼ 35–70 kDa (Fig. S2). We did not observe an alteration of the size distribution of the aromatic compounds in the incubated extracts; these polymers had the typical size (Piccolo, 2001) and synchronous fluorescence spectrum of humic acids (Ferrari and Mingazzini, 1995). In contrast, the size of the main peak of polysaccharides decreased during the incubation, which suggests that they were at least partially depolymerized during fungal growth.
We also examined whether Pa. involutus was able to chemically modify a cellulose substrate (carboxy methyl cellulose, CMC) and if this conversion was consistent with a mechanism involving free radical oxidation. The FTIR spectra of the substrate before and after 7 days of incubation were almost identical apart from the appearance of a new peak (λ∼ 1730 cm−1) located in the C=O stretching region (Fig. 1). Notably, such distinct increase in the intensity of carbonyl groups did only occur if the medium was supplemented with glucose (Fig. 1).
Extracellular enzyme and iron reducing activities
When comparing the activity of extracellular enzymes in the organic matter media (FH, MH and MC) before and after 7 days of incubation, a significant increase in the activities of laccase and other oxidases was detected in two out of the three media (Fig. S3). In contrast, the activities of lignin peroxidase and cellulase were very low and most likely Pa. involutus did not secrete such enzyme activities during the degradation of DOM. Among hemicellulases, the activity of xylanase, but not β-glucuronidase, increased during the incubation.
A key requirement for the Fenton mechanism is a system for reduction of Fe3+ to Fe2+, which might be accomplished by extracellular fungal metabolites or reductive enzymes (Green III and Highley, 1997; Baldrian and Valaskova, 2008). Analysis of the iron-reducing activity in the organic matter extracts showed that it was significantly increased during decomposition (Fig. S4). Accordingly, iron-reducing compound(s) was formed during organic matter degradation.
Secretome analysis
A cDNA library constructed from mycelia grown on the FH, MH and MC substrates was screened for transcripts encoding secreted or membrane bound proteins (Grell et al., 2011). In total, 18 contigs were identified that encode secreted enzymes with a possible role in organic matter degradation (Table S4). Five cDNA sequences were homologous to plant cell wall polysaccharide degrading enzymes. One sequence was predicted to encode an expansin family protein (TAST-1). Consistent with the enzymatic activity measurements, we did not identify any enzyme from the canonical crystalline cellulose decomposition system (GH6, 7 and 45) (Lynd et al., 2002). Instead, we found one endo-beta-1,4-glucanase of the GH9 family (TAST-2). However, the gene model did not bear a cellulose-binding module, and was therefore unlikely to bind and efficiently degrade crystalline cellulose (Kostylev et al., 2012). We identified three GH61 genes (TAST-5, 6, 7).
A number of oxidoreductases has been suggested to support Fenton chemistry, among them glyoxal oxidases that produces H2O2 (Baldrian and Valaskova, 2008). We identified a putative galactose oxidase/glyoxal oxidase among the TAST clones. Accumulation and regulation of oxalic acid secreted by the fungus is thought to be also involved in Fenton chemistry (Baldrian and Valaskova, 2008). TAST-13 is a predicted oxalate decarboxylase, the key enzyme in one of the two oxalate removal systems described in white-rot fungi (Mäkeläet al., 2010). Three TAST sequences were encoding enzymes potentially involved in decomposition of lignin residues: an aromatic peroxygenase (TAST-9), a cytochrome p450 oxidoreductase (TAST-10) and a laccase (TAST-11).
Transcriptome profiling
Microarray analysis showed that the transcriptome profiles of the mycelium grown on the three organic extracts were similar but distinctly different from the ones grown on the MMN medium (Fig. S5). In total, 73 isotigs (transcripts) that encoded putative organic matter degrading enzymes and proteins were significantly upregulated more than twofold in at least one of the pairwise comparisons FH/MMN, MH/MMN and MC/MMN (Fig. 4A). The 73 isotigs belonged to 60 isogroups (genes). The regulations of the isotigs from the same isogroup were almost identical.
Fig. 4.
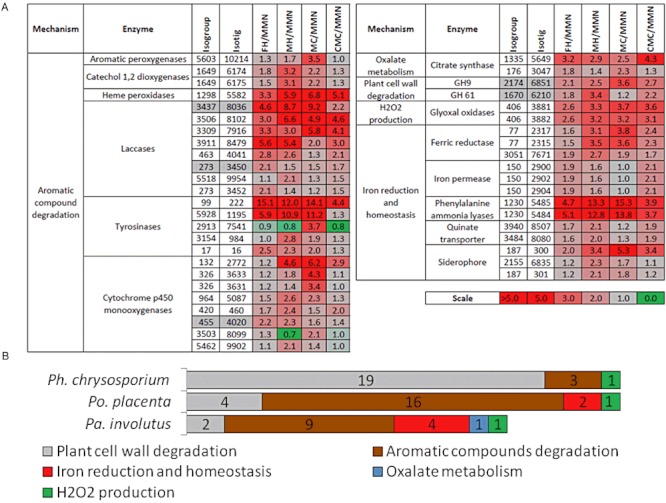
Regulation of genes potentially involved in organic matter degradation by Pa. involutus. A. Expression profile of 44 genes that were manually annotated as potentially involved in organic matter degradation, and were upregulated more than twice (false discovery rate q < 0.01) in at least one of pairwise comparisons in media containing extracts of complex organic material versus mineral nutrient medium (MMN). The data presented are average ratio of expression (n = 3). Four different types of organic substrates were used: forest litter extracted with hot water (FH), maize compost extracted with hot water (MH), maize compost extracted with cold water (MC) and carboxy methyl cellulose (CMC). Isotigs and isogroups refer to transcripts and genes respectively. In grey boxes are 5 isotigs that were also identified in the TAST screening (Table S4). B. Comparison of the transcriptional response of Po. placenta, Ph. chrysosporium and Pa. involutus when growing on a cellulose medium as compared with a medium containing glucose as the carbon source. The number of genes that were upregulated at least twofold (in average of three replicates) and those with annotations consistent with a potential role in organic matter degradation are shown. Microarray data for Ph. chrysosporium and Po. placenta growing on microcrystalline cellulose (AVICEL) and glucose media were downloaded from the GEO database (accession numbers GSE14736 and GSE12540 respectively).
Twenty-seven of the 60 upregulated genes were predicted to encode Carbohydrate-Active enzymes (CAZymes). However, only two of them, a GH9 (endoglucanase) and a GH61, were expected to be involved in plant cell wall degradation. The remaining CAZymes most likely have a role either in the synthesis and turnover of the fungal cell wall, the degradation of chitin-containing compounds or in glycoprotein modification (not shown in Fig. 4A). Twenty-two genes were induced encoding enzymes possibly involved in lignin degradation, such as multicopper oxidases, p450 oxidases, tyrosinases and catechol dioxygenase. Nine of the upregulated genes were found to encode for iron metabolism-related proteins and were therefore potentially involved in Fenton reaction. We also found two genes coding for proteins involved in oxalate/citrate metabolism (citrate synthase) and one coding for a glyoxal oxidase.
The transcriptional response of the fungus when grown on cellulose (CMC) was similar to the response when grown on the FH, MH and MC organic matter extracts (Fig. 4A; Fig. S5). In all media, at least one transcript was significantly upregulated among the glyoxal oxidases, phenylalanine ammonia lyases, haem peroxidases, laccases, tyrosinases, cytochrome p450 oxidoreductases, citrate synthases and endoglucanases (GH9) respectively. However, the expression levels of specific transcripts within these families differed depending on the substrate.
Comparative transcriptome analysis
Finally, we compared the set of genes that were significantly upregulated during conversion of cellulose in the ECM fungus Pa. involutus, the white-rot fungus Phanerochaete chrysosporium and the brown-rot fungus Postia placenta (Fig. 4B). The cohort of overexpressed genes of Ph. chrysosporium was dominated by plant cell-wall degrading CAZymes. Moreover, Ph. chrysosporium lacked transcripts encoding proteins involved in pigment production, oxalic acid metabolism and iron metabolism. Such transcripts were together with oxidases involved in lignin degradation, prevalent in the upregulated transcriptomes of both Po. placenta and Pa. involutus. Six plant cell-wall degrading CAZymes were upregulated in Po. placenta including two GH1 (β-glucosidases), two GH3 (one β-glucosidase and one β-xylosidase), one GH 10 (endoxylanase/hemicellulase) and one GH28 (polygalacturonase). None of these CAZymes were upregulated in Pa. involutus. In this fungus two plant cell-wall degrading CAZymes were induced namely a GH9 and a GH61 (Fig. S6).
Discussion
During the assimilation of organic N from the soluble organic material, Pa. involutus degraded at least part of the polysaccharides, while the polyphenols were not digested, even though some of their aromatic constituents were chemically modified. Such a selective depolymerization of polysaccharides is very similar to a brown-rot decay mechanism where cellulose and hemicelluloses (polysaccharides) are depolymerized and lignin (aromatic polymer) is not, but is still chemically modified (Green III and Highley, 1997). The pyrolysis analysis showed that the lignin subunits present in the polyphenols were modified by Pa. involutus through a significant reduction in the number of side-chains. Depletion of original C3 side-chains indicates oxidation of these groups during incubation, and is a good indicator of the degradation of lignin residues (Nierop et al., 2005). Demethylation of methoxyl groups linked to syringyl subunits, similar to our findings, has recently been detected in in situ analyses of brown-rotted wood by 2D NMR (Martinez et al., 2011) and has also been found to be consistent with an oxidative degradation of the lignin through Fenton reaction by the brown-rot fungus Po. placenta (Yelle et al., 2011). Likewise, the introduction of a sharp carbonyl-groups signal in the FTIR spectrum of cellulose, as observed when Pa. involutus was grown on CMC, has been associated with a Fenton reaction mechanism (Green III and Highley, 1997).
The Fenton reaction requires the reduction of Fe3+ to Fe2+ and iron reducing compounds were produced during the degradation of plant litter by Pa. involutus. Three mechanisms have been proposed for ferric reduction in basidiomycetes: (i) iron-reducing enzymes (ferric reductases, cellobiose oxidases, cellobiose dehydrogenase) (Baldrian and Valaskova, 2008), (ii) low-molecular-weight (LMW) glycopeptides (Vanden Wymelenberg et al., 2010), and (iii) redox cycling by small-molecular mass compounds like dimethoxyhydro- and benzoquinones: DMHQ, DMBQ (Newcombe et al., 2002). In support for the first mechanism, we identified two genes encoding putative ferric reductases that were significantly upregulated during the degradation of organic matter. However, we did not recognize any transcripts displaying sequence similarity to cellobiose dehydrogenase. This enzyme is also absent in the brown-rot fungus Po. placenta (Martinez et al., 2009). We identified three transcripts encoding proteins with sequence similarity to LMW iron reducing glycoproteins in Ph. chrysosporium (Tanaka et al., 2007). However, none of them were upregulated during organic matter degradation. The induction of genes involved in pigment synthesis (phenylalanine ammonia lyases and tyrosinases) and quinate transport suggests the involvement of a quinone redox-cycling iron reduction mechanism (Martinez et al., 2009; Vanden Wymelenberg et al., 2010). Pa. involutus is known to produce the pigment involutin (Feling et al., 2001): this molecule bear a catechol group that could undergo oxidation in the corresponding quinone, which could then be reduced again intracellularly after transport through the quinate transporter (see Fig. S7). However, the lack of electron-donating methoxyl groups (that are present in characterized hydroquinones that contribute to Fenton reaction, see Suzuki et al., 2006) makes this molecule more of a free radical scavenger rather than an iron reducing compound. We therefore hypothesize that the pigment is secreted by the fungus as a protection against its own oxidative machinery.
When degrading organic matter, Pa. involutus overexpressed a number of transcripts of oxidases like laccases, catechol dioxygenase, haem peroxidase, tyrosinases and cytochrome p450 monooxygenases that have also been showed to be produced by the brown-rot fungus Po. placenta when grown on wood or cellulose media (Martinez et al., 2009; Vanden Wymelenberg et al., 2010). No transcripts encoding class II peroxidases (Mn and lignin peroxidases) that are signatures for a white-rot mechanism were detected in Pa. involutus. Considering the expression of CAZymes involved in the degradation of plant cell walls, there were large differences between Pa. involutus and Po. placenta (see Fig. S6). Although, none of them expressed genes coding for enzymes of the canonical crystalline cellulose decomposition system (Lynd et al., 2002), the transcript profile of Po. placenta employs an array of CAZymes like endoglucanases, β-glucosidases and hemicellulase when it is grown on cellulose or aspen (Martinez et al., 2009; Vanden Wymelenberg et al., 2010). Except for one endoglucanase (GH9), no such glycosyl hydrolases were induced in Pa. involutus during growth on cellulose or plant litter. In addition to the GH9, a member of the GH61 family was significantly upregulated in Pa. involutus. GH61 is the most abundant CAZyme family acting on plant cell walls in the genome of L. bicolor (Martin et al., 2008). Recently, it has been reported that GH61 can depolymerize cellulose oxidatively in cooperation with cellobiose dehydrogenase or LMW reducing agents (Langston et al., 2011; Quinlan et al., 2011). Hence, GH61 could be an important component of the radical-based cellulose degrading mechanism of ECM fungi.
In saprophytic brown-rot fungi, the radical-based and enzymatic parts of the lignocellulose degrading system act synergistically (Baldrian and Valaskova, 2008; Yelle et al., 2011). Most likely, the capacity of Pa. involutus to express the enzymatic part has been lost during the evolutionary transition from a saprophytic brown-rot precursor (Binder and Hibbett, 2006) to a symbiotic ECM fungus relying on the plant host for carbohydrate provision. The genomic mechanisms that could account for this loss are not known. However, considering previous genome studies of ECM fungi showing a large reduction in the number of plant cell wall degrading enzymes (Martin et al., 2008; Nagendran et al., 2009), gene loss and deletions have likely played an important role. Moreover, in contrast to the situation in brown-rot fungi (Martinez et al., 2009), the polysaccharides present in the organic matter, even though depolymerized, were not assimilated. This was also consistent with the absence of gene models coding for cellobiohydrolase, the enzyme cleaving cellobiose into two glucoses, acting in the last step of cellulose degradation. Moreover, in contrast to the situation in brown-rot fungi (Martinez et al., 2009), oxidative degradation of cellulose did not occur in Pa. involutus without adding glucose to the medium. This observation suggests that mutations affecting transcriptional regulation have also contributed to the symbiotic adaptation of the brown-rot decay system.
The decomposing activities of ECM fungi in soils are commonly analysed using enzyme assays that include plant cell wall degrading enzymes (Courty et al., 2005). Considering the fact that such enzymes were not expressed during the degradation of plant litter material by Pa. involutus, and that very little enzymatic activities were measured in the organic matter extracts, measuring the activity of those enzymes might give misleading results. Laccases are also used as enzyme marker for saprophytic ECM activity (Courty et al., 2009). Although being upregulated by Pa. involutus during the decay of litter material, laccases are found in multigene families having members with complex patterns of expression. Hence, further studies are needed to develop molecular markers that can correctly capture the saprotrophic activity of ECM fungi in the field.
That ECM fungi do not assimilate C from decomposed plant litter has been indicated in field experiments using 14C-labelled litter material (Treseder et al., 2006). Several studies have also shown that the ECM mycelia are surrounded by distinct communities of saprophytic bacteria and fungi (Boer et al., 2005; Izumi and Finlay, 2011). Most likely, these communities contain commensals that grow on the C resources that become available during the radical-based degradation by the ECM fungi. Some of these microbes may strive for the same nutrient resources as the ECM fungi. Thus, it can be expected that ECM fungi have evolved mechanisms including the secretion of toxic metabolites that could control the activity of saprophytic microorganisms (Boer et al., 2005). Indeed, studies in soil microcosms have shown that the ECM mycelium of Pa. involutus can reduce the activity of saprophytic bacteria (Olsson et al., 1996). The combined metabolic activity of symbiotic fungi and saprophytic microbes may have a significant impact on the turnover of carbon and nutrients in forest soils. Moreover, being supplied by energy from the plant, this pathway could operate at deeper soil horizons that are energetically unavailable for traditional saprophytes (Lindahl et al., 2007).
A caveat of the present study is that the experiments were done using organic litter material in solution, i.e. the DOM fraction. Dissolved organic matter represents only a small proportion of the total SOM and lacks insoluble components like lignin, cellulose and chitin as well as aggregates formed between organic compounds and minerals (Bolan et al., 2011). It is well known that mineral particles could stabilize organic compounds against degradation by microbial enzymes (Allison, 2006). It remains to be determined to what extent the radical-based biodegradation system of ECM fungi can act on intact SOM material.
Experimental procedures
Fungal strain and culture conditions
Cultures of Pa. involutus (Batsch) Fr. (strain ATCC 200175) were maintained aseptically on 1.5% agar plates containing minimum Melin-Norkrans medium (MMN) (composition: 2.5 g l−1 glucose, 500 mg l−1 KH2PO4, 200 mg l−1 NH4Cl, 150 mg l−1 MgSO4·7H2O, 25 mg l−1 NaCl, 50 mg l−1 CaCl2, 12 mg l−1 FeCl3·6H2O, and 1 mg l−1 Thiamine-HCl; pH 4.0). The fungus was grown in Petri dishes on a layer of glass beads immersed in liquid medium (van Schöll et al., 1996). A monolayer of autoclaved 4 mm diameter glass beads was poured into the bottom of a 9 cm Petri dish and 10 ml of the MMN medium was added. A mycelial plug (c. 5 mm in diameter) was cut from the margin of an actively growing mycelium (MMN agar medium) and transferred to the centre of the glass bead plate. After 9 days of incubation (18°C, dark) when the diameter of the colony reached a size of approximately 4 cm in diameter and a biomass of 10 mg (dry weight), the MMN medium was removed. The glass beads and the mycelium were washed with 10 ml of sterile MilliQ (MQ) water, and 10 ml of MMN medium without N was added to induce a N-deprived mycelium. After 24 h, the mycelium was washed with MQ water and extracts of organic matter (10 ml) were added. In addition, the fungus was grown in CMC (10 g l−1). The organic matter extracts and the CMC medium were supplemented with glucose (final concentration 2.5 g l−1) before inoculation, to avoid a situation of carbon limitation. The cultures were incubated for 7 days at 18°C in the dark, just before the mycelium started being space-limited in the Petri dish.
Preparation of organic matter extracts
Forest litter material was collected from the upper 10 cm soil layer in a 61-year-old pure spruce stand growing in N-poor site in central Sweden (soil pH = 5.0). The maize compost was produced by cutting maize leaves into small pieces and composting them in an isolated plastic compost bin for 12 months. The litter and compost material was extracted with either cold or hot water (Davidson et al., 1987). Three organic extracts were generated: forest litter extracted with hot water (FH), and a maize compost extracted with cold (MC) or hot (MH) water. Particles were removed by sequentially filtering (0.2 µm) and LMW compounds by ultra-filtration (cut-off 10 kDa). Further details are given in Supporting Information (Appendix S1).
Chemical analysis
Samples for Fourier transform infrared (FTIR) spectroscopy were prepared by drying (vacuum over night at 4°C) 5 ml of the organic matter extracts. The FTIR spectrum was recorded using a Bruker IFS66 v/s spectrometer. Data were collected in diffuse reflectance mode using a praying mantis diffuse reflectance attachment (Harrick Sci.). Each spectrum was the result of 1000 consecutive scans at a resolution of 4 cm−1. Synchronous fluorescence spectra were obtained using a Perkin-Elmer LS50B fluorescence spectrophotometer. Samples (750 µl) were kept at room temperature (20°C) and processed at a 10 nm bandwidth and 25 nm offset (Δλ = 25 nm) between excitation and emission. Pyrolysis gas chromatography-mass spectrometry (Py-GC/MS) was performed using a Perkin Elmer TurboMass/ Autosystem XL with Frontier Lab double Shot pyrolyser. Size-exclusion chromatography was performed using a HiLoad 16/60 Superdex200 column (GE Healthcare). Further details are given in Supporting Information (Appendix S1).
Enzyme activity measurements and ferrozine assay
To remove interfering compounds, the extracts were treated with PVPP (PolyVinyl Poly Pyrrolidone) (Pierpoint, 1996), followed by acetone precipitation. Laccase activity was measured using syringaldazine as a substrate (Leonowicz and Grzywnowicz, 1981), lignin peroxidase using veratryl alcohol (Tien and Kirk, 1988), overall oxidase activity using ABTS (Palmieri et al., 1997), cellobiohydrolase activity using methylumbelliferyl-β-d-cellobioside (Courty et al., 2005), glucuronidase activity using methylumbelliferyl-β-d-glucuronide hydrate (Courty et al., 2005), and xylanase activity using RBB-Xylan as substrate (Biely et al., 1985). The capacity of Pa. involutus to produce iron-reducing compounds was examined using a ferrozine assay (Goodell et al., 2006). Further details are given in Supporting Information (Appendix S1).
Construction and screening of TAST cDNA library
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen). A cDNA library was generated from RNA isolated of mycelia grown on the FH, MH and MC substrates. Poly(A) RNA isolation, cDNA synthesis, and transposon assisted signal trapping (TAST) cDNA library construction were essentially carried out as described previously (Grell et al., 2011). A collection of 576 TAST clones were selected for sequencing. The sequences were assembled into 348 contigs which were manually annotated. Further details are given in Supporting Information (Appendix S1).
Transcriptome sequencing and microarray
The transcriptomes expressed by the fungus during growth on the FH, MH and MC extracts and the MMN medium, respectively, was sequenced using the 454 technology. In total, the sequencing yielded 2 029 605 reads that were assembled into a set of 12 873 isotigs. These isotigs represent various splice variants and they were mapped to 8620 genes or isogroups. Sequences may be accessed from http://mbio-serv2.mbioekol.lu.se/Paxillus/Hybrid/ (add ‘paxillus_’ to the given isotig and isogroup numbers). EST sequences are also available at GenBank SRA046093. Based on manual annotations, we identified 269 transcripts among the 12 873 isotigs that encodes for enzymes and proteins with a possible role in the degradation of organic matter (polysaccharide modifications, lignin degradation, iron reduction and homeostasis, oxalate metabolism and H2O2 production). Roche NimbleGen arrays were designed to assess expression of 12 214 isotigs. The data deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE34402 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34402). Further details are given in Supporting Information (Appendix S1).
Acknowledgments
The work was supported by grants from the Swedish Research Council (VR), the strategic research program Biodiversity and Ecosystem Services in a Changing Climate (BECC), the Danish Agency for Science and Technology, and the Research Foundation – Flanders (FWO). 454 sequencing was conducted by the US Department of Energy Joint Genome Institute, supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH1123.1. We thank Charles G. Kurland for help with the manuscript, and Jan Czech and professor Carleer for support with the Py-GC/MS. Novozymes is acknowledged for a royalty-free license to use the TAST technology for research purposes.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. FTIR spectra of the sugar region.
Fig. S2. Size-fractionation of the organic matter extracts.
Fig. S3. Extracellular enzyme activities secreted by Pa. involutus.
Fig. S4. Iron-reducing capabilities in the organic extracts degraded by Pa. involutus.
Fig. S5. Principal component analysis of transcriptome data.
Fig. S6. Comparison of CAZYme repertoires ofPa. involutus, Po. placenta and Ph. chrysosporium when growing on cellulose.
Fig. S7. Proposed contribution of involutin in a hydroquinone-mediated Fenton reaction.
Table S1. Changes in C, N and nutrients during the conversion of soil organic matter.
Table S2. Data used for interpreting peaks in the synchronous fluorescence (SF) spectra.
Table S3. Putative identification of the pyrolysis compounds.
Table S4. Identification of secreted proteins using the TAST method.
Appendix S1. Supplementary experimental procedures.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Allison SD. Soil minerals and humic acids alter enzyme stability: implications for ecosystem processes. Biogeochemistry. 2006;81:361–373. [Google Scholar]
- Baldrian P. Ectomycorrhizal fungi and their enzymes in soils: is there enough evidence for their role as facultative soil saprotrophs? Oecologia. 2009;161:657–660. doi: 10.1007/s00442-009-1433-7. [DOI] [PubMed] [Google Scholar]
- Baldrian P, Valaskova V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev. 2008;32:501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- Bending GD, Read DJ. The structure and function of the vegetative mycelium of ectomycorrhizal plants V. Foraging behaviour and translocation of nutrients from exploited litter. New Phytol. 1995a;130:401–409. [Google Scholar]
- Bending GD, Read DJ. The structure and the function of the vegetative mycelium of ectomycorrhizal plants. VI. Activities of nutrient mobilizing enzymes in birch litter colonized by Paxillus involutus (Fr.) Fr. New Phytol. 1995b;130:411–417. [Google Scholar]
- Bending GD, Read DJ. Nitrogen mobilization from protein-polyphenol complex by ericoid and ectomycorrhizal fungi. Soil Biol Biochem. 1996;28:1603–1612. [Google Scholar]
- Biely P, Mislovicova D, Toman R. Soluble chromogenic substrates for the assay of endo-1,4-beta-xylanases and endo-1,4-beta-glucanases. Anal Biochem. 1985;144:142–146. doi: 10.1016/0003-2697(85)90095-8. [DOI] [PubMed] [Google Scholar]
- Binder M, Hibbett DS. Molecular systematics and biological diversification of Boletales. Mycologia. 2006;98:971–981. doi: 10.3852/mycologia.98.6.971. [DOI] [PubMed] [Google Scholar]
- Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Bolan NS, Adrianao DC, Kunhikrishnan A, James T, McDowell R, Senesi N. Dissolved organic matter: biogeochemistry, dynamics, and environemntal significance in soils. Adv Agron. 2011;100:1–75. [Google Scholar]
- Chengrong RC, Zhihong HX. Analysis and behaviour of soluble organic nitrogen in forest soils. J Soils Sediments. 2008;8:363–378. [Google Scholar]
- Colpaert JV, Van Laere A. A comparison of the extracellular enzyme activities of two ectomycorrhizal and a leaf-saprotrophic basidiomycete colonizing beech leaf litter. New Phytol. 1996;134:133–141. [Google Scholar]
- Courty P-E, Pritsch K, Schloter M, Hartmann A, Garbaye J. Activity profiling of ectomycorrhiza communities in two forest soils using multiple enzymatic tests. New Phytol. 2005;167:309–319. doi: 10.1111/j.1469-8137.2005.01401.x. [DOI] [PubMed] [Google Scholar]
- Courty P-E, Hoegger PJ, Kilaru S, Kohler A, Buée M, Garbaye J, et al. Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol. 2009;182:736–750. doi: 10.1111/j.1469-8137.2009.02774.x. [DOI] [PubMed] [Google Scholar]
- Davidson EA, Galloway LF, Strand MK. Assessing available carbon: comparison of techniques across selected forest soils. Commun Soil Sci Plant Analysis. 1987;18:45–65. [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski P, Scholes RJ, Boyle E, Canadell J, Canfield D, Elser J, et al. The global carbon cycle: a test of our knowledge of earth as a system. Science. 2000;290:291–296. doi: 10.1126/science.290.5490.291. [DOI] [PubMed] [Google Scholar]
- Feling R, Steglich W, Muhlbacher J, Bringmann G. The absolute configuration of the mushroom metabolite involutin and chamonixin. Tetrahedon. 2001;57:7857–7863. [Google Scholar]
- Ferrari GM, Mingazzini M. Synchronous fluorescence spectra of dissolved organic matter (DOM) of algal origin in marine coastal waters. Mar Ecol Prog Ser. 1995;125:305–315. [Google Scholar]
- Goodell B, Daniel G, Jellison J, Qian Y. Iron-reducing capacity of low-molecular weight compounds produced in wood by fungi. Holzforschung. 2006;60:630–636. [Google Scholar]
- Green F, III, Highley TL. Mechanism of brown-rot decay: paradigm or paradox. Int Biodeterior Biodegradation. 1997;39:113–124. [Google Scholar]
- Grell MN, Jensen AB, Olsen PB, Eilenberg J, Lange L. Secretome of fungus-infected aphids documents high pathogen activity and weak host response. Fungal Genet Biol. 2011;48:343–352. doi: 10.1016/j.fgb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Haselwandter K, Bobleter O, Read DJ. Degradation of 14C-labelled lignin and dehydropolymer of coniferyl alcohol by ericoid and ectomycorrhizal fungi. Arch Microbiol. 1990;153:352–354. [Google Scholar]
- Högberg MN, Högberg P. Extramatrical ectomycorrhizal mycelium contributes one-third of microbial biomass and produces, together with associated roots, half of the dissolved organic carbon in a forest soil. New Phytol. 2002;154:791–795. doi: 10.1046/j.1469-8137.2002.00417.x. [DOI] [PubMed] [Google Scholar]
- Högberg P, Read DJ. Towards a more plant physiological perspective on soil ecology. Trends Ecol Evol. 2006;21:548–554. doi: 10.1016/j.tree.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Izumi H, Finlay RD. Ectomycorrhizal roots select distinctive bacterial and ascomycete communities in Swedish subarctic forests. Environ Microbiol. 2011;13:819–830. doi: 10.1111/j.1462-2920.2010.02393.x. [DOI] [PubMed] [Google Scholar]
- Kostylev M, Moran-Mirabal JM, Walker LP, Wilson DB. Determination of the molecular states of the processive endocellulase Thermobifida fusca Cel9A during crystalline celulose depolymerization. Biotechnol Bioeng. 2012;109:295–299. doi: 10.1002/bit.23299. [DOI] [PubMed] [Google Scholar]
- Langston JA, Shaghasi T, Abbate E, Xu F, Vlasenko E, Sweeney MD. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol. 2011;77:7007–7015. doi: 10.1128/AEM.05815-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonowicz A, Grzywnowicz K. Quantitative estimation of laccase forms in some white-rot fungi using syringaldazine as a substrate. Enzyme Microb Technol. 1981;3:55–58. [Google Scholar]
- Lindahl BD, Ihrmark K, Boberg J, Trumbore SE, Hogberg P, Stenlid J, Finlay RD. Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol. 2007;173:611–620. doi: 10.1111/j.1469-8137.2006.01936.x. [DOI] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä MR, Hilden K, Lundell TK. Oxalate decarboxylase: biotechnological update and prevalence of the enzyme in filamentous fungi. Appl Microbiol Biotechnol. 2010;87:801–814. doi: 10.1007/s00253-010-2650-z. [DOI] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Martinez AT, Rencoret J, Nieto L, Jimenez-Barbero J, Gutierrez A, Rio JC. Selective lignin and polysaccharide removal in natural fungal decay of wood as evidenced by in situ structural analyses. Environ Microbiol. 2011;13:96–107. doi: 10.1111/j.1462-2920.2010.02312.x. [DOI] [PubMed] [Google Scholar]
- Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA. 2009;106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagendran S, Hallen-Adams HE, Paper JM, Aslam N, Walton JD. Reduced genomic potential for secreted plant cell-wall-degrading enzymes in the ectomycorrhizal fungus Amanita bisporigera, based on the secretome of Trichoderma reesei. Fungal Genet Biol. 2009;46:427–435. doi: 10.1016/j.fgb.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Nannipieri P, Eldor P. The chemical and functional characterization of soil N and its biotic components. Soil Biol Biochem. 2009;41:2357–2369. [Google Scholar]
- Newcombe D, Paszczynski A, Gajewska W, Kröger M, Feis G, Crawford R. Production of small molecular weight catalysts and the mechanism of trinitrotoluene degradation by several Gloeophyllum species. Enzyme Microb Technol. 2002;30:506–517. [Google Scholar]
- Nierop KGJ, van Bergen PF, Buurman P, van Lagen B. NaOH and Na4P2O7 extractable organic matter in two allophanic volcanic ash soils of the Azores Islands-a pyrolysis GC/MS study. Geoderma. 2005;127:36–51. [Google Scholar]
- Norkrans B. Studies in growth and cellulolytic enzymes of Tricholoma. Symbolae Botanicae Upsaliensis. 1950;11:1–126. [Google Scholar]
- Olsson PA, Chalot M, Bååth E, Finlay RD, Söderström B. Ectomycorrhizal mycelia reduce bacterial activity in a sandy soil. FEMS Microbiol Ecol. 1996;21:77–86. [Google Scholar]
- Palmieri G, Giardina P, Bianco C, Scaloni A, Capasso A, Sannia G. A novel white laccase from Pleurotus ostreatus. J Biol Chem. 1997;272:31301–31307. doi: 10.1074/jbc.272.50.31301. [DOI] [PubMed] [Google Scholar]
- Piccolo A. The supramolecular structure of humic substances. Soil Sci. 2001;166:810–832. [Google Scholar]
- Pierpoint WS. The extraction of enzymes from plant tissues rich in phenolic compounds. Methods Mol Biol. 1996;59:69–80. doi: 10.1385/0-89603-336-8:69. [DOI] [PubMed] [Google Scholar]
- Quinlan RJ, Sweeney MD, Lo LL, Otten H, Poulsen JC, Johansen KS, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read DJ, Leake JR, Perez-Moreno J. Mycorrhizal fungi as drivers of ecosystem processes in heathland and boreal forest biomes. Can J Bot. 2004;82:1243–1263. [Google Scholar]
- Schmidt MW, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA, et al. Persistence of soil organic matter as an ecosystem property. Nature. 2011;478:49–56. doi: 10.1038/nature10386. [DOI] [PubMed] [Google Scholar]
- van Schöll L, Hoffland E, van Bremen N. Organic anion exudation by ectomycorrhizal fungi and Pinus sylvestris in response to nutrient deficiencies. New Phytol. 1996;170:153–163. doi: 10.1111/j.1469-8137.2006.01649.x. [DOI] [PubMed] [Google Scholar]
- Senesi N, Miano TM, Provenzano MR, Brunetti G. Characterization, differentiation, and classification of humic substances by fluorescence spectroscopy. Soil Science. 1991;152:259–271. [Google Scholar]
- Simard SW, Jones MD, Durall DM. Carbon and nutrient fluxes in within and between mycorrhizal plants. In: van der Heijden MGA, Sanders IR, editors. Mycorrhizal Ecology. Berlin, Germany: Springer; 2002. pp. 33–74. [Google Scholar]
- Suzuki MR, Hunt CG, Houtman CJ, Dalebroux ZD, Hammel KE. Fungal hydroquinones contribute to brown rot of wood. Environ Microbiol. 2006;8:2214–2223. doi: 10.1111/j.1462-2920.2006.01160.x. [DOI] [PubMed] [Google Scholar]
- Talbot JM, Allison SD, Treseder KK. Decomposer in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Funct Ecol. 2008;22:955–963. [Google Scholar]
- Tanaka H, Yoshida G, Baba Y, Matsumura K, Wasada H, Murata J, et al. Characterization of a hydroxyl-radical-producing glycoprotein and its presumptive genes from the white-rot basidiomycete Phanerochaete chrysosporium. J Biotechnol. 2007;128:500–511. doi: 10.1016/j.jbiotec.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Tien M, Kirk TK. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 1988;161:238–249. [Google Scholar]
- Treseder KK, Torn MS, Masiello CA. An ecosystem-scale radiocarbon tracer to test use of litter carbon by ectomycorrhizal fungi. Soil Biol Biochem. 2006;38:1077–1082. [Google Scholar]
- Trojanowski J, Haider K, Huttermann A. Decomposition of 14C labelled lignin, holocelllulose and lignocellulose by mycorrhizal fungi. Arch Microbiol. 1984;139:202–206. [Google Scholar]
- Vanden Wymelenberg A, Gaskell J, Mozuch M, Sabat G, Ralph J, Skyba O, et al. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl Environ Microbiol. 2010;76:3599–3610. doi: 10.1128/AEM.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallander H, Söderström B. Paxillus. In: Cairney JWG, Chambers SM, editors. Ectomycorrhizal Fungi: Key Genera in Profile. Berlin, Germany: Springer-Verlag; 1999. pp. 231–252. [Google Scholar]
- Yelle DJ, Wei D, Ralph J, Hammel KE. Multidimensional NMR analysis reveals truncated lignin structures in wood decayed by the brown rot basidiomycete Postia placenta. Environ Microbiol. 2011;13:1091–1100. doi: 10.1111/j.1462-2920.2010.02417.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


