Abstract
DNA damage checkpoints delay mitotic cell-cycle progression in response to DNA stress, stalling the cell cycle to allow time for repair. CDKB is a plant-specific cyclin-dependent kinase (CDK) that is required for the G2/M transition of the cell cycle. In Arabidopsis, DNA damage leads the degradation of CDKB2, and the subsequent G2 arrest gives cells time to repair damaged DNA. G2 arrest also triggers transition from the mitotic cycle to endoreduplication, leading to the presence of polyploid cells in many tissues. In contrast, in rice (Oryza sativa), polyploid cells are found only in the endosperm. It was unclear whether endoreduplication contributes to alleviating DNA damage in rice (Oryza sativa). Here, we show that DNA damage neither down-regulates Orysa;CDKB2;1 nor induces endoreduplication in rice. Furthermore, we found increased levels of Orysa;CDKB2;1 protein upon DNA damage. These results suggest that CDKB2 functions differently in Arabidopsis and rice in response to DNA damage. Arabidopsis may adopt endoreduplication as a survival strategy under genotoxic stress conditions, but rice may enhance DNA repair capacity upon genotoxic stress. In addition, polyploid cells due to endomitosis were present in CDKB2;1 knockdown rice, suggesting an important role for Orysa;CDKB2;1 during mitosis.
Keywords: CDKB2, DNA damage, rice, endomitosis, ploidy, cell cycle
Introduction
Repairing DNA damage in a timely fashion is essential to maintaining genome integrity. DNA damage triggers activation of the DNA repair machinery and delay or arrest of cell-cycle progression to allow sufficient time for damaged DNA to be repaired before proceeding to mitosis. DNA damage signals are transmitted via several proteins, suppressing the activity of cyclin-dependent kinase (CDK) to arrest the cell cycle (Sancar et al., 2004). Like animals, plants have multiple CDK-related protein kinases, which are classified into six types: CDKA–F (Joubès et al., 2000). A-type CDK (CDKA) is closely related to yeast Cdc2/Cdc28, and is expressed throughout the cell cycle (Colasanti et al., 1991; Ferreira et al., 1991; Hirt et al., 1991, 1993; Fobert et al., 1996). However, B-type CDKs (CDKBs) are plant-specific, and are further classified into two sub-types: CDKB1 and CDKB2. Expression of CDKBs is under strict cell-cycle control; CDKB1 is expressed from late S to M phase, while CDKB2 is expressed from G2 to M phase, as confirmed in experiments in alfalfa (Medicago sativa) (Magyar et al., 1997), rice (Oryza sativa) (Umeda et al., 1999b), tobacco (Nicotiana tabacum) (Porceddu et al., 2001) and Arabidopsis (Menges et al., 2005).
In Arabidopsis, DNA double-strand breaks (DSBs) have been reported to enhance endoreduplication (Ramirez-Parra and Gutierrez, 2007; Adachi et al., 2011). Cells undergoing endoreduplication replicate chromosomal DNA without intervening mitoses, and the resulting larger, higher-ploidy nucleus is often associated with an increase in cell size. A major component of the switch to endoreduplication is prevention of mitosis by reduction of mitotic CDK activity to a level that does not initiate mitosis but is able to drive replication of DNA. Consistent with this, reduced expression of CDK and cyclin B was seen during endoreduplication in trichome, epidermal and mesophyll cells of Arabidopsis leaves (Schnittger et al., 2003; Verkest et al., 2005). Endoreduplication often occurs in cell types that undergo specialized differentiation, such as hair cells and xylem cells, or in plant cell types that have high metabolic activity, such as endosperm and embryo suspensor cells. Plants use endoreduplication as a bypass pathway to avoid cell-cycle arrest or cell death by DNA damage, because endoreduplicated cells remain where they were generated, and thus contribute to organogenesis as a constituent of tissues.
In contrast to Arabidopsis, and other plants in which polyploid cells are found in many tissues, polyploid cells are found only in the endosperm in rice. Furthermore, the effect of DNA damage on cell cycle and ploidy in rice has not yet been explored. Thus, it is of great interest to determine how rice responds to damaged DNA in terms of the cell cycle. We are currently analyzing the effect of DNA damage on ploidy and CDKB2 expression in rice. Arabidopsis has two B2-type CDKs: CDKB2;1 and CDKB2;2. Both genes show a peak in expression during the G2/M phase transition of the cell cycle (Menges et al., 2003). DNA damage degrades Arath;CDKB2;1 (Adachi et al., 2011), and Arath;CDKB2;1 down-regulation is associated with endoreduplication (Andersen et al., 2008). In contrast, rice has a single gene (CDKB2;1) encoding CDKB2 (Umeda et al., 1999b). Transcription of Orysa;CDKB2;1 is abundant during progress from G2 to M phase (Umeda et al., 1999b).
Here, we reveal that DNA damage neither induces endoreduplication nor down-regulates Orysa;CDKB2;1 expression. Instead, Orysa;CDKB2;1 accumulates in response to DNA damage. As in Arabidopsis, an Orysa;CDKB2;1 knockdown mutant showed polyploidy in rice calli, but this was shown to be due to endomitosis rather than endoreduplication.
Results and Discussion
DSBs do not induce polyploidy in rice
In rice plants, mitotic cells are limited exclusively to the root, shoot and intercalary meristems, thus we used callus cells cultured on solid medium to investigate the effects of DNA damage on rice cell-cycle progression. We first monitored the extent of DNA DSBs in the genomic DNA of rice calli irradiated with various doses of X-rays using the comet assay (Menke et al., 2001). In this assay, the ‘tail moment’ represents the level of DNA damage.
Upon Xray exposure, DNA damage increased in a dose-dependent manner (Figure 1a). Beyond a certain threshold, when DNA damage is too severe, cell viability itself is also decreased, and it becomes difficult to evaluate the biological effects precisely. Thus, we used 5 Gy as the X-ray irradiation dose. In calli irradiated with 5 Gy, DNA damage was maximal at 30 min, decreasing again at 1 h after irradiation (Figure 1b). This result shows that rice callus does not lose its DNA repair machinery following a 5 Gy dose of irradiation. To confirm that 5 Gy irradiation is sufficient to induce a DNA damage response, we analyzed the transcription level of the rice RAD51 gene. The Rad51 protein is crucial to homologous recombination repair, and transcription of RAD51 is drastically induced by DNA damage in Arabidopsis (Culligan et al., 2006). Two Rad51 orthologs, Rad51A1 (Os11g0615800) and Rad51A2 (Os12g0497300) exist in rice. Transcription of RAD51A2 (but not RAD51A1) is enhanced by DNA damage (M.E., unpublished data). OsRAD51A2 transcription was increased by 5 Gy X-ray irradiation (Figure 1c).
Figure 1.
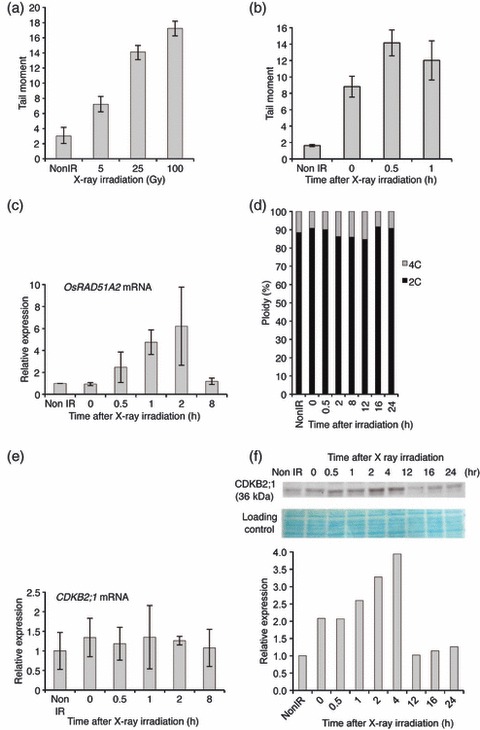
DNA damage response in rice calli following X-ray irradiation.
(a) Quantitative analysis of DSBs by the comet assay. Wild-type rice calli were irradiated with 5, 25 and 100 Gy doses of X-rays at 133 Gy h−1, and tail moment values were measured just after irradiation.
(b) Quantification of DSBs by the comet assay at various time points after irradiation of calli with a 5 Gy dose of X-rays.
(c) Transcription level of OsRAD51A2 determined by real-time quantitative PCR.
(d) Ploidy of X-ray-irradiated rice calli. The DNA content of nuclei prepared from calli was analyzed by flow cytometry at various times following irradiation.
(e) Transcription levels of CDKB2;1 determined by real-time quantitative PCR.
(f) Immunological detection of CDKB2;1. Upper panel: Western blot analysis of protein extracts from non-irradiated (Non-IR) or X-ray-irradiated rice calli using antibodies against Orysa;CDKB2;1. Middle panel: stained membrane showing equal loading of protein samples. Lower panel: quantification of CDKB2;1 protein.
Next, we analyzed ploidy levels in X-ray-irradiated rice calli to investigate the effect of DNA damage on cell-cycle progression. 2C represents G1 cells and 4C represents G2 cells. We detected no significant change in DNA ploidy distribution, and observed no 8C or 16C cells within 24 h of X-ray irradiation (Figure 1d) or later (data not shown). We also analyzed the effect of 25 and 100 Gy irradiation on ploidy but detected no polyploid cells (Figure S1a,b). From these data, we concluded that endoreduplication is not a major DNA damage response in rice. This conclusion was also supported by data obtained from bleomycin-treated calli (discussed below).
Orysa;CDKB2;1 is not down-regulated by DNA damage
A major component of the switch to endoreduplication is prevention of mitosis by reducing CDK activity to a level that does not initiate mitosis but is able to drive replication of DNA (for review, see John and Qi, 2008). Indeed, in Arabidopsis, knockdown of CDKB2 led to higher nuclear DNA content than in wild-type (Andersen et al., 2008), suggesting that DNA damage signaling could reduce the amount of CDKB2, resulting in enhanced endoreduplication in Arabidopsis. Furthermore, a recent study by Adachi et al. (2011) demonstrated this connection directly.
To analyze the relationship between DNA damage signaling and the CDKB2 expression level in rice, we analyzed transcription and protein levels of Orysa;CDKB2;1 after X-ray irradiation. Transcript levels of Orysa;CDKB2;1 were not affected significantly by X-ray irradiation (Figure 1e), but we found increased levels of CDKB2;1 protein immediately after X-ray irradiation, and levels continued to increase for at least 4 h (Figure 1f).
The expression profile of CDKB2;1 upon cell-cycle progression and DNA damage was confirmed in the rice suspension-cultured cell line OC (Baba et al., 1986) using the DNA polymerase inhibitor aphidicolin (Spadari et al., 1984) and the radiomimetic reagent bleomycin. Aphidicolin arrests cells at S phase; release from this block enables cells to progress synchronously through S, G2 and M phase. Thus, by using synchronized OC cells, we expected to observe cell cycle-dependent oscillation of CDKB2;1 expression, in addition to CDKB2;1 expression in response to DNA damage. Without bleomycin treatment, protein levels of CDKB2;1 first peaked at 2 h, decreased from 2–8 h, but started to accumulate again from 8 h after aphidicolin removal (Figure 2a, −Bleo). However, when bleomycin was added to suspension cultured cells 4 h after aphidicolin removal, protein levels of CDKB2;1 continued to increase (Figure 2a, +Bleo). We confirmed that this treatment was sufficient to induce DNA damage, which led to cell death as shown by propidium iodide (PI) staining of cells. PI is used as a marker for loss of membrane integrity and cell death, as dead cells take up PI readily, in contrast to live cells, which actively exclude it (Truernit and Haseloff, 2008). PI staining was only detected in bleomycin-treated OC cells (Figure 2b). Transcription of CDKB2;1 did not differ much between bleomycin-treated and untreated cells (Figure 2c). Taken together, these results indicate that DNA damage does not affect transcription but instead increases the amount of CDKB2;1 at the protein level.
Figure 2.
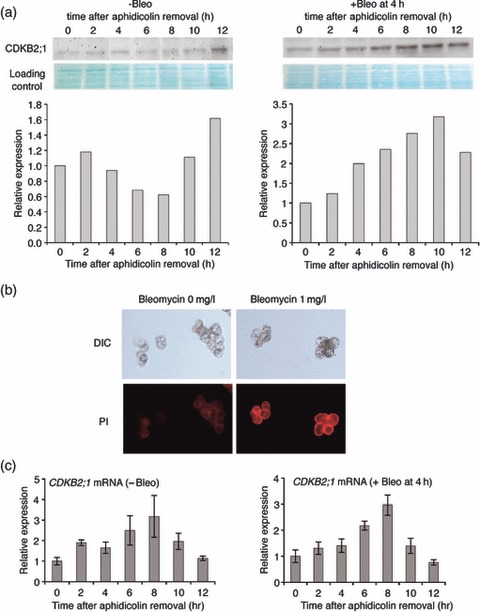
DNA damage response in rice suspension-cultured cells after bleomycin treatment.
(a) Immunoblot analysis of CDKB2;1 using rice suspension-cultured cells. Suspension-cultured cells were blocked in S phase for 24 h using aphidicolin (left panel). The DNA-damaging agent bleomycin (1 mg L−1) was added to the culture medium 4 h after aphidicolin block removal (right panel). Crude proteins were prepared at 2 h intervals after release from the aphidicolin block.
(b) PI staining of suspension-cultured cells with or without 2 h bleomycin treatment.
(c) Transcription level of CDKB2;1 determined by real-time quantitative PCR.
Knockdown of Orysa;CDKB2;1 induces polyploidy
We next speculated that the lack of CDKB2;1 down-regulation in response to DNA damage could explain why rice does not enter the endocycle. To test this hypothesis, we generated CDKB2;1 constitutive knockdown mutants (B2RNAi). We cloned an RNAi construct towards an internal 302 bp region of CDKB2;1 (Figures 3a and S2) into the pANDA vector (Miki and Shimamoto, 2004), which incorporates a maize ubiquitin promoter and intron (Christensen et al., 1992), and produces high levels of trigger dsRNA in transgenic rice (Miki and Shimamoto, 2004). We confirmed decreased expression of CDKB2;1 in B2RNAi T0 calli (Figure 3b,c). Despite a low level of sequence homology of this RNAi region with rice CDKA;1, CDKA;2 or CDKB1;1 (Figure S2), no statistically significant difference in transcription of CDKA;1, CDKA;2 and CDKB1;1 between wild-type and B2RNAi lines was seen (Figure S3). To show any effect of CDKB2;1 knockdown on DNA ploidy distribution, we analyzed ploidy in B2RNAi calli by flow cytometric analysis. In B2RNAi calli, the 4C nuclei population increased drastically at the expense of the proportion of nuclei with 2C (Figure 3d). Surprisingly, 8C and 16C fractions also appeared in B2RNAi calli. We confirmed that the CDKB2;1 protein level was severely reduced in B2RNAi calli showing polyploidy (Figure 3e). Initially, we assumed that the polyploidy detected in B2RNAi calli was due to endoreduplication, because polyploid cells are known to occur in rice endosperm (Larkins et al., 2001). Cells that enter endoreduplication cannot proliferate because the endocycle repeats DNA synthesis without mitosis. However, the cell proliferation of B2RNAi calli was comparable to that of wild-type calli, despite the high population of polyploid cells (Figure 3f). Given these results, we hypothesized that endomitosis rather than endoreduplication occurs in B2RNAi calli.
Figure 3.
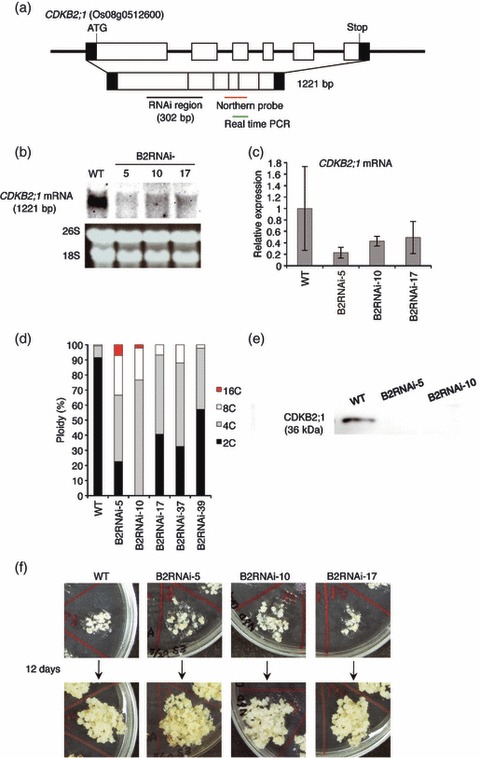
Characterization of Orysa;CDKB2;1 knockdown calli.
(a) Schematic representation of CDKB2;1 structure and RNAi silencing target region.
(b) Northern blot analysis of CDKB2;1. Upper panel: OsCDKB2;1 mRNA. Lower panel: RNAs transferred to nylon membrane to visualize equal loading of total RNA in each lane.
(c) Transcription level of CDKB2;1 determined by real-time quantitative PCR. The region amplified is indicated in (a).
(d) Ploidy of CDKB2;1 knockdown rice calli (B2RNAi) grown on selection medium for 36 days.
(e) Immunological detection of CDKB2;1.
(f) Growth of wild-type (WT) and B2RNAi calli on N6D solid medium at 0 days (upper panel) and 12 days (lower panel) after transfer.
The polyploidy in the Orysa;CDKB2;1 knockdown line is due to endomitosis
During endomitosis, chromosomes double and condense, and sister chromatids separate and return to the interphase state as in the mitotic cycle. As a result, chromosome numbers double in each cycle. On the other hand, endoreduplication involves an endonuclear chromosome duplication that occurs in the absence of any obvious condensation or decondensation steps (Lorz, 1947; Levan and Hauschka, 1953). Therefore, although DNA content doubles in each nucleus, chromosome number does not change in cells undergoing endoreduplication. To distinguish between endoreduplication and endomitosis, we analyzed the chromosome number of polyploid cells. We found nuclei with 24, 48 and 96 chromosomes in line B2RNAi-5 (Figure 4a–c), which harbors 2C, 4C, 8C and 16C nuclei (Figure 3d). In line B2RNAi-10, which did not contain 2C nuclei according to flow cytometric analysis (Figure 3d), we detected nuclei with 48 and 96 chromosomes but no nuclei with 24 chromosomes within eight analyzed nuclei (Figure S4). From these data, we conclude that the polyploid cells found in B2RNAi calli result from endomitosis. Transcription of CDKB2;1 has been reported to be limited to the dividing region of rice roots (Umeda et al., 1999a), and CDKB2;1 transcripts were abundant from G2 to M phase but disappear when cells complete mitosis at telophase (Umeda et al., 1999b). Furthermore, a CDKB2;1–GFP fusion protein was tightly associated with chromosome alignment as well as with the spindle structure during metaphase (Lee et al., 2003). During telophase, this GFP signal was localized to the spindle mid-zone and the separating sister chromosomes, and then to the phragmoplast (Lee et al., 2003). These observations strongly support the idea that CDKB2;1 plays a functional role in the separation of sister chromatids.
Figure 4.
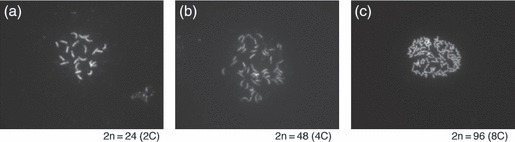
Chromosome observation of Orysa;CDKB2;1 knockdown calli. Chromosome spreads of nuclei from Orysa;CDKB2;1 knockdown line B2RNAi-5. Nuclei containing 24 (a), 48 (b) or 96 (c) chromosomes were found.
Decreased expression of Orysa;CDKB2;1 disturbs morphogenesis
Increased nuclear DNA content often occurs concomitantly with the onset of cell expansion (Sugimoto-Shirasu and Roberts, 2003). In order to investigate the effect of endomitosis on rice morphogenesis, we attempted to regenerate tetraploid plants from B2RNAi calli. No tetraploid plants were detected in 34 plants regenerated from 11 independent B2RNAi calli, despite the fact that these calli showed a high population of polyploid cells and proliferated well. Furthermore, root growth of some diploid regenerated plants ceased soon after regeneration (Figure S5a). We changed the medium from MS hormone-free solid medium to MS liquid medium because liquid medium is more easily absorbed and allows roots to grow without physical impedance. Despite this, these plants stopped growing and died (Figure S5b). Based on these results, we conclude that constitutive knockdown of CDKB2;1 induces endomitosis in calli, and that it also disturbs regeneration and growth of regenerated plants.
Although we could not regenerate tetraploid rice plants from CDKB2;1 constitutive knockdown calli (B2RNAi), we expected that tetraploid rice plants could be regenerated from calli in which endomitosis was induced temporarily. To this end, we used the XVE induction system (Zuo et al., 2000) to create CDKB2;1 knockdown plants (see Appendix S1). We observed that β-estradiol treatment increased the 4C population, and removal of β-estradiol reversed this increase in induced CDKB2;1 RNAi-transformed calli (B2RNAiID) (Figure S6). After induction of endomitosis in B2RNAiID calli by β-estradiol treatment and transfer of these calli to regeneration medium without β-estradiol, we succeeded in obtaining a tetraploid plantlet, B2RNAiID-1 (Figure 5a,b). The files of epidermal cells in B2RNAiID-1 leaves were wider than those of wild-type, and the guard cells were significantly larger (Figure 5c). Correlation between ploidy, nuclear DNA content and cell size has been reported in Arabidopsis and many other plant species (Nagl, 1978; Galbraith et al., 1991; Melaragno et al., 1993). This correlation was also seen in the tetraploid rice plant generated by CDKB2;1 inducible knockdown. Normally, epidermal cells of rice blades are arranged regularly in rows parallel to the midrib, but the files of the B2RNAiID-1 leaf epidermis were disordered. Well-organized cell division at the shoot apical meristem is necessary for orderly phyllotaxis. Rice flattened shoot meristem (fsm) mutants lacking the p150 subunit of CAF-1, which is tightly associated with cell-cycle progression, showed a defect in maintenance of the shoot apical meristem as well as irregular cell files (Abe et al., 2008). Therefore, we speculated that the morphological aberration detected in B2RNAiID plants was caused by non-cooperative cell division due to leaky expression of CDKB2;1 RNAi during regeneration. All of these results suggest that expression of CDKB2;1 must be under strict control in order for normal development of rice to occur.
Figure 5.
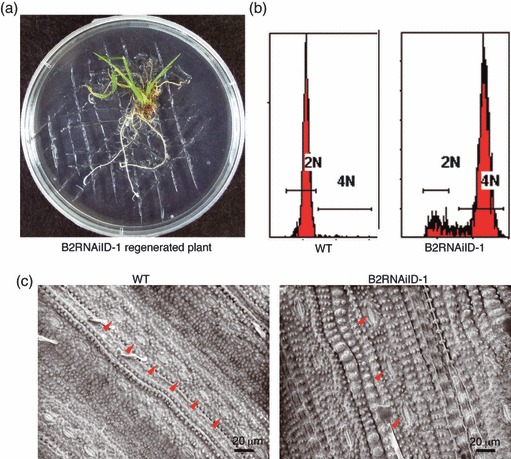
Phenotypic analysis of a regenerated plant from the Orysa;CDKB2;1 inducible knockdown line.
(a)Tetraploid rice plant regenerated from CDKB2;1 inducible RNAi transformed callus (B2RNAiID-1).
(b) Flow cytometry measurements of DNA content from leaf cells of wild-type and B2RNAiID-1.
(c) Scanning electron micrographs of the epidermal leaf surface of wild-type (diploid) and B2RNAiID-1 (tetraploid) plants. Arrowheads indicate stomata.
Decreased Orysa;CDKB2;1 expression induces DNA damage hypersensitivity
To investigate the effect of CDKB2;1 knockdown on the DNA damage response, we analyzed the X-ray sensitivity of B2RNAi plants. Although the decrease in CDKB2;1 protein levels was not great (Figure 6a), and no growth inhibition was detected under normal conditions (Figure 6b, upper panel), B2RNAi-7 plants stopped growing almost immediately after irradiation (Figure 6b, lower panel). Furthermore, cells in the root tips of the X-ray-irradiated B2RNAi-7 mutant were swollen and disorganized. The profuse production of root hairs detected in X-ray-irradiated B2RNAi-7 plants is a typical morphological phenotypic response to DNA damage in roots (Hefner et al., 2006). DNA damage hypersensitivity due to CDKB2;1 knockdown was more marked when we used calli to analyze bleomycin sensitivity. In wild-type calli, bleomycin treatment (0.5 and 2 mg L−1) disturbed cell proliferation (Figure 7a), but these treatments did not induce polyploidy or accumulation of 4C nuclei (Figure 7b). These results indicate that G2 arrest or inhibition of mitosis is not the major DNA damage response in rice. However, in CDKB2;1 knockdown calli, proliferation was severely inhibited (Figure 7a) and polyploidy was enhanced (Figure 7b) by bleomycin treatment. Endomitosis is due to inhibition of mitosis; CDKB2;1 knockdown calli may be arrested at M phase due to accumulated DNA damage at M phase, and/or CDKB2 may be involved directly in the progression of M phase in rice. Application of bleomycin may further induce DNA damage at M phase in CDKB2;1 knockdown callus, which may result in enhanced endomitosis.
Figure 6.
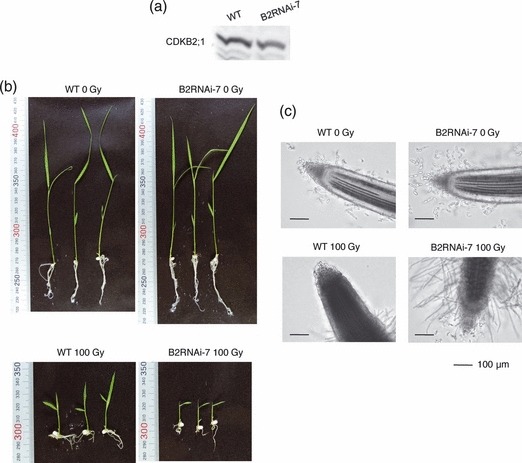
X-ray sensitivity of Orysa;CDKB2;1 knockdown plants.
(a) Western blot analysis of CDKB2;1. Crude protein was prepared from roots of non-irradiated wild-type (WT) and B2RNAi-7 plants.
(b) WT and CDKB2;1 knockdown plants (B2RNAi-7-1 and B2RNAi-16-1) grown on MS culture medium were irradiated with 100 Gy X-rays. Non-irradiated and irradiated plants were grown on MS culture medium for 5 days.
(c) Morphology of primary root tip response with or without 100 Gy X-ray irradiation.
Figure 7.
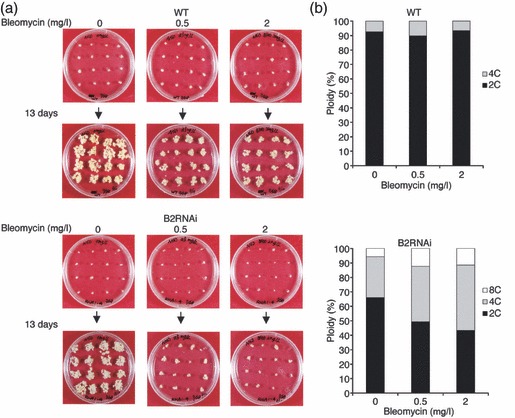
Effect of constitutive DNA damage by bleomycin on growth and ploidy of the calli.
(a) Proliferation of calli on medium supplemented with 0, 0.5 or 2mg L−1 bleomycin.
(b) Ploidy of rice calli grown on medium containing 0, 0.5 or 2 mg L−1 bleomycin for 10 days.
In mammals and yeast, there are several reports on the function of cell-cycle regulators in DNA repair. Cyclin A1 expression is reported to be induced by γ-irradiation, and cyclins A1 and A2 enhance DNA DSB repair by homologous recombination in mice (Müller-Tidow et al., 2004). Esashi et al. (2005) described CDK-dependent phosphorylation of BRCA2, which interacts directly with the essential recombination protein Rad51 in cultured human cells. Furthermore, involvement of CDK in recruitment of Rad51 to the site of DNA DSBs has been demonstrated in Saccharomyces cerevisiae (Aylon et al., 2004; Ira et al., 2004). In contrast, the function of cell cycle-related factors in the DNA damage response in plants is not clear. CDKB2;1, which is a plant-specific CDK expressed only in G2/M phase, may have a function in enhancing DNA repair at this stage in rice. Alternatively, impaired cell-cycle progression may increase cell sensitivity to DNA damage.
Concluding remarks
In contrast to Arabidopsis, which shows increased DNA ploidy in response to DNA DSBs or CDKB2 knockdown, no induction of endoreduplication or CDKB2;1 down-regulation was detected following DNA damage in rice. We speculate that these differences are related to different strategies for morphogenesis and cell survival in response to DNA damage. In Arabidopsis, DNA replication and cell division occur not only in meristematic tissues but also in other tissues (Donnelly et al., 1999). Furthermore, pavement cells of Arabidopsis leaves have a variety of sizes and shapes and form a mosaic-like structure. These morphological flexibilities may allow Arabidopsis to utilize endoreduplication in order to avoid cell death by DNA damage. In contrast to Arabidopsis, growth of rice leaves results from cell proliferation at the base of the leaf and cell elongation that takes place in the elongation zone directly above the meristem. This spatial gradient, synchronized cell division and cell expansion results in leaf growth taking place predominantly along a one-dimensional axis. Disorganized cell elongation accompanied by endoreduplication in rice would disturb normal morphogenesis and development. Thus, rice cells may need to suppress entry to the endocycle except in the endosperm.
Previously published evidence has shown that meristem cells respond differently to DNA damage compared with differentiated cells. In Arabidopsis, DNA DSBs cause cell-cycle arrest at G1/S in meristem cells but not in endoreduplicating differentiating cells (Hefner et al., 2006). Root and shoot stem cells and their early descendants are killed selectively by treatment with radiomimetic drugs, X-rays or mutations that disrupt DNA repair by non-homologous end-joining (Fulcher and Sablowski, 2009). In addition, within both the root and shoot meristem, ataxia telangiectasia mutated (ATM) and ATM/RAD3-related (ATR) protein kinase-dependent, non-apoptotic programmed cell death is used to eliminate damaged cells specifically from the population of stem cells and their early descendants (Fulcher and Sablowski, 2009). These results indicate that the response to DNA damage is cell type-dependent, and that endoreduplication is repressed in stem cells of Arabidopsis. By contrast, endoreduplication in rice plants may be actively repressed not only in stem cells but also in meristematic cells. Thus rice may not deploy mechanisms to induce endoreduplication upon DNA damage, but instead may induce early onset of cell differentiation as well as extensive repair of damaged DNA during cell-cycle arrest.
In this study, we did not detect Orysa;CDKB2;1 down-regulation in response to DNA damage. DNA damage responses are mediated by the highly conserved ATM and ATR protein kinases (Shiloh, 2006; Su, 2006; Cimprich and Cortez, 2008). Furthermore, a plant-specific transcription factor, SOG1 (suppressor of gamma response 1) has been shown to process signals associated with multiple responses to DNA damage, and to function downstream of ATM and ATR (Yoshiyama et al., 2009). Recently, Adachi et al. (2011) demonstrated that expression of Arath;CDKB2;1 was suppressed in response to DSBs at both the transcriptional and protein level, but this down-regulation was not necessarily required for endocycle induction because treatment with the DNA-damaging agent zeocin did not decrease the protein level of CDKB2;1 in root tips of an Arabidopsis atr mutant although endoreduplication occurred normally. These results indicate that reduction of the CDKB2 level is one of the factors inhibiting entry into M phase, but that other factors also contribute to down-regulating mitotic CDK activities, leading to endoreduplication in Arabidopsis. In summary, induction of DSBs up-regulates CDK inhibitors and down-regulates cyclin A/B and CDKB2 via the ATM/ATR–SOG1 signal transduction pathway, resulting in increased endoreduplication. Putative ATM and ATR homologs (Os01g0106700 and Os06g0724700, respectively) have been found in the rice genome database. However, the function of these protein kinases has not yet been analyzed. The non-degradable nature of Orysa;CDKB2;1 after DNA damage could be explained by differences in the ATM- and ATR-mediated DNA damage signal transduction pathway between rice and Arabidopsis. Alternatively, Orysa;CDKB2;1 itself may be resistant to protein degradation, in contrast to Arath;CDKB2;1.
Additionally, unlike Arath;CDKB2;1, we detected an enhanced level of Orysa;CDKB2;1 protein after DNA damage. Furthermore, Orysa;CDKB2;1 knockdown plants showed increased sensitivity to DNA damage. These data suggest the important role of Orysa;CDKB2;1 in DNA repair. In mammals, there is increasing evidence that the catalytic activities of CDKs play a critical role in the DNA damage response (for review, see Yata and Esashi, 2009). It has been observed experimentally that chemical inhibitors of CDKs sensitize cells to reagents, such as ionizing radiation and cisplatin, that create DSBs (Ongkeko et al., 1995; Maggiorella et al., 2003; Deans et al., 2006). Furthermore, analysis in yeasts and mammalian cells has revealed that CDK activity is essential for DNA resection and progression of homologous recombination repair during S and G2 phase (Caspari et al., 2002; Ferreira and Cooper, 2004; Ira et al., 2004; Deans et al., 2006; Sonoda et al., 2006). Remarkably, in most Arabidopsis mutants suffering from endogenous DNA stress, the CYCB1:1 gene encoding a G2/M phase-specific B-type cyclin is strongly induced; this gene is also induced by DSB-causing treatments (Chen et al., 2003; Culligan et al., 2006; Ricaud et al., 2007). This transcriptional induction depends on ATM. Interestingly, in ATR knockout plants, ATM-dependent CYCB1;1 induction is apparent, but the protein shows decreased stability, suggesting that ATR controls its abundance post-transcriptionally (Culligan et al., 2006). The role of CycB1;1 in the DNA stress response is unknown, but the unique behavior of CYCB1;1 hints at a specific function for this particular cyclin. Cools and De Veylder (2009) suggest that CYCB1;1 may block all cells from entering the endocycle, preventing loss of all division-competent cells, and allowing meristem reactivation after repair of DNA damage. We found that endomitosis occurs in Orysa;CDKB2;1 knockdown cells. Interestingly, the presence of non-degradable CYCB1;1 in tobacco leads to a doubled DNA content as a result of endomitosis (Weingartner et al., 2004). These results indicate that timely expression and degradation of mitotic cyclin and CDK in plants is required for reorganization of mitotic microtubules to the phragmoplast and for proper cytokinesis. In rice, CYCB1;1 has not been reported as the partner of CDKB2;1, but both CDKB2;1 and CYCB1;1 are expressed specifically during S/G2 phase. Further analyses are required to determine whether these factors work cooperatively in mitotic cell-cycle regulation, protein degradation after mitosis and the response to DNA damage.
Experimental procedures
X-ray irradiation
Rice calli on solid N6D medium were irradiated using an OM-100RS soft X-ray system (Omic) at a dose of 133 Gy/h. Exposure times of 2.5, 11 and 33 min are equivalent to irradiation of 5, 25 and 100 Gy, respectively.
Comet assay
X-ray-irradiated rice calli grown on solid N6D medium were processed as described previously (N/N protocol; Menke et al., 2001). A CCD camera was used to capture images of comets stained using SYBR Green (Lonza, http://www.lonza.com/group/en.html). Signals were quantified using Comet analyzer software (YOUWORKS, http://youworks.jp/) under conditions that excluded severely damaged nuclei. The intensity of DNA is shown in graded colors. DNA DSBs are represented as the ‘tail moment’ (Olive et al., 1990), which is defined as the tail distance multiplied by the sum of tail intensity/sum of cell intensity. Individual parameters have been defined previously (Endo et al., 2006). We analyzed 30–50 nuclei at each time point, and this experiment was repeated at least three times.
Real-time PCR
Total RNA was prepared using an RNeasy plant mini kit (Qiagen, http://www.qiagen.com/) according to the manufacturer's instructions. The RNA preparation was then treated with RNase-free DNase I (Qiagen). First-strand cDNA synthesis was performed using oligo(dT) primer and ReverTra Ace (TOYOBO, http://www.toyobo.co.jp/). Detailed conditions for real-time PCR and the primer sequences used in this experiment are given in Appendix S1.
Flow cytometric analysis
Nuclei extraction and DNA staining were performed using CvStain UV precise P (Partec, http://www.partec.com/). The filtered nuclei were subjected to flow analysis (Partec) with laser excitation at 357 nm.
Western blot analysis
Proteins extracted from calli or liquid suspension-cultured cells were used for Western blot analysis. Proteins (20 μg) were fractionated by SDS–PAGE on a 5–20% Tris/glycine SDS gradient pre-cast polyacrylamide gel (Bio-Rad, http://www.bio-rad.com/), and subjected to immunoblotting using SuperSignal West Dura maximum sensitivity substrate (Thermo Scientific, http://www.piercenet.com/). Polyclonal antibody was raised in rabbits against the C-terminal peptide PYFNDVNKELY of OsCDKB2 (Umeda et al., 1999b).
OsCDBK2 RNAi plasmid construction
The OsCDBK2 RNAi plasmid was constructed as described in Appendix S1.
Rice transformation
Agrobacterium-mediated transformation of rice (Oryza sativa L. cv. Nipponbare) was performed as described previously (Toki, 1997; Toki et al., 2006). After co-cultivation of Agrobacterium carrying the binary vector with rice scutellum-derived calli (pre-cultured for 3 weeks) for 3 days, infected calli were transferred to fresh callus induction medium (Toki, 1997) containing 50 mg L−1 hygromycin B (Wako Pure Chemicals, http://www.wako-chem.co.jp/english/) and 35 mg L−1 carbenicillin (Wako) to remove Agrobacterium. Hygromycin-tolerant cells were selected over a period of 25 days.
Scanning electron microscopy
Scanning electron micrographs of leaf epidermal cells (Figure 5c) were obtained using a VE-8800 scanning electron microscope (Keyence, http://www.keyence.com/) at 0.8 kV with a secondary electron detector.
Northern blot analysis
Northern blot analysis was performed as described in Appendix S1.
Chromosome observation
Chromosome observation was performed as described in Appendix S1.
Acknowledgments
We thank Y. Kawagoe for scanning electron micrographs, and H. Rothnie for English editing of the manuscript. This work was supported by a PROBRAIN (Program for Promotion of Basic Research Activities for Innovative Biosciences) grant to M.U. and S.T. from the Bio-Oriented Technology Research Advancement Institution (BRAIN) of Japan, and by Grants-in-Aid for Scientific Research on Innovative Areas (grant number 22119009) to M.U. from the Ministry of Education, Culture, Sports, Science and Technology of Japan. M.E. was supported by a fellowship from Grant-in-Aid for Research Activity Start-up of the Japan Society for the Promotion of Science. This work was also supported by the budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology, based on screening and counseling by the Atomic Energy Commission to S.T.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Figure S1. Effect of high dosage X-ray irradiation on the ploidy and expression of CDKB2;1 in wild-type rice calli.
Figure S2. Alignment of rice CDK mRNAs.
Figure S3. Transcription of CDK genes in Orysa;CDKB2;1 knockdown rice calli.
Figure S4. Increased chromosome number in B2RNAi-10.
Figure S5. Orysa;CDKB2;1 knockdown lines with increased ploidy levels cease growth after regeneration.
Figure S6. Orysa;CDKB2;1 inducible knockdown can induce G2 arrest.
Appendix S1. Methods for real-time PCR, plasmid construction, Northern blot analysis and chromosome observation.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Abe M, Kuroshita H, Umeda M, Itoh J, Nagato Y. The rice FLATTENED SHOOT MERISTEM, encoding CAF–1 p150 subunit, is required for meristem maintenance by regulating the cell-cycle period. Dev. Biol. 2008;319:384–393. doi: 10.1016/j.ydbio.2008.04.040. [DOI] [PubMed] [Google Scholar]
- Adachi S, Minamisawa K, Inagaki S, Okushima Y, Yoshiyama K, Kurihara D, Matsunaga S, Umeda M. Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl Acad. Sci. USA. 2011;108:10004–10009. doi: 10.1073/pnas.1103584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SU, Buechel S, Zhao Z, Ljung K, Novák O, Busch W, Schuster C, Lohmann JU. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell. 2008;20:88–100. doi: 10.1105/tpc.107.054676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba A, Hasezawa S, Syono K. Cultivation of rice protoplasts and their transformation mediated by Agrobacterium spheroplasts. Plant Cell Physiol. 1986;27:463–471. [Google Scholar]
- Caspari T, Murray JM, Carr AM. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 2002;16:1195–1208. doi: 10.1101/gad.221402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H. The transcriptional response of Arabidopsis to genotoxic stress – a high-density colony array study (HDCA) Plant J. 2003;35:771–786. doi: 10.1046/j.1365-313x.2003.01847.x. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Tyers M, Sundaresan V. Isolation and characterization of cDNA clones encoding a functional p34cdc2 homologue from Zea mays. Proc. Natl Acad. Sci. USA. 1991;88:3377–3381. doi: 10.1073/pnas.88.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T, De Veylder L. DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 2009;12:23–28. doi: 10.1016/j.pbi.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- Deans AJ, Khanna KK, McNees CJ, Mercurio C, Heierhorst J, McArthur GA. Cyclin-dependent kinase 2 functions in normal DNA repair and is a therapeutic target in BRCA1-deficient cancers. Cancer Res. 2006;66:8219–8226. doi: 10.1158/0008-5472.CAN-05-3945. [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- Endo M, Ishikawa Y, Osakabe K, et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006;25:5579–5590. doi: 10.1038/sj.emboj.7601434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 2004;18:2249–2254. doi: 10.1101/gad.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Hemerly AS, Villarroel R, Van Montagu M, Inzé D. The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell. 1991;3:531–540. doi: 10.1105/tpc.3.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert PR, Gaudin V, Lunness P, Coen ES, Doonan JH. Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell. 1996;8:1465–1476. doi: 10.1105/tpc.8.9.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R. Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl Acad. Sci. USA. 2009;106:20984–20988. doi: 10.1073/pnas.0909218106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S. Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol. 1991;96:985–989. doi: 10.1104/pp.96.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefner E, Huefner N, Britt AB. Tissue-specific regulation of cell-cycle responses to DNA damage in Arabidopsis seedlings. DNA Repair. 2006;5:102–110. doi: 10.1016/j.dnarep.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Hirt H, Páy A, Györgyey J, Bakó L, Németh K, Bögre L, Schweyen RJ, Heberle-Bors E, Dudits D. Complementation of a yeast cell cycle mutant by an alfalfa cDNA encoding a protein kinase homologous to p34cdc2. Proc. Natl Acad. Sci. USA. 1991;88:1636–1640. doi: 10.1073/pnas.88.5.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H, Páy A, Bögre L, Meskiene I, Heberle-Bors E. cdc2MsB, a cognate cdc2 gene from alfalfa, complements the G1/S but not the G2/M transition of budding yeast cdc28 mutants. Plant J. 1993;4:61–69. doi: 10.1046/j.1365-313x.1993.04010061.x. [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John PC, Qi R. Cell division and endoreduplication: doubtful engines of vegetative growth. Trends Plant Sci. 2008;13:121–127. doi: 10.1016/j.tplants.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Joubès J, Chevalier C, Dudits D, Heberle-Bors E, Inzé D, Umeda M, Renaudin JP. CDK-related protein kinases in plants. Plant Mol. Biol. 2000;43:607–620. doi: 10.1023/a:1006470301554. [DOI] [PubMed] [Google Scholar]
- Larkins BA, Dilkes BP, Dante RA, Coelho CM, Woo YM, Liu Y. Investigating the hows and whys of DNA endoreduplication. J Exp Bot. 2001;52:183–192. [PubMed] [Google Scholar]
- Lee J, Das A, Yamaguchi M, Hashimoto J, Tsutsumi N, Uchimiya H, Umeda M. Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. Plant J. 2003;34:417–425. doi: 10.1046/j.1365-313x.2003.01736.x. [DOI] [PubMed] [Google Scholar]
- Levan A, Hauschka ST. Endomitotic reduplication mechanism in ascites tumors of the mouse. J. Natl Cancer Inst. 1953;14:1–36. [PubMed] [Google Scholar]
- Lorz AP. Supernumerary chromonemal reproduction: polytene chromosomes, endomitosis, multiple chromosome complexes, polysomaty. Bot. Rev. 1947;13:597–624. [Google Scholar]
- Maggiorella L, Deutsch E, Frascogna V, Chavaudra N, Jeanson L, Milliat F, Eschwege F, Bourhis J. Enhancement of radiation response by roscovitine in human breast carcinoma in vitro and in vivo. Cancer Res. 2003;63:2513–2517. [PubMed] [Google Scholar]
- Magyar Z, Meszaros T, Miskolczi P, et al. Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell. 1997;9:223–235. doi: 10.1105/tpc.9.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell. 1993;5:1661–1668. doi: 10.1105/tpc.5.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 2003;53:423–442. doi: 10.1023/B:PLAN.0000019059.56489.ca. [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- Menke M, Chen IP, Angelis KJ, Schubert I. DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat. Res. 2001;493:87–93. doi: 10.1016/s1383-5718(01)00165-6. [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Müller-Tidow C, Ji P, Diederichs S, et al. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol. Cell. Biol. 2004;24:8917–8928. doi: 10.1128/MCB.24.20.8917-8928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl W. Endopolyploidy and Polyteny in Differentiation and Evolution. Amsterdam: Elsevier; 1978. [Google Scholar]
- Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the ‘comet’ assay. Radiat. Res. 1990;122:86–94. [PubMed] [Google Scholar]
- Ongkeko W, Ferguson DJ, Harris AL, Norbury C. Inactivation of Cdc2 increases the level of apoptosis induced by DNA damage. J. Cell Sci. 1995;108:2897–2904. doi: 10.1242/jcs.108.8.2897. [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheldt JP, Segers G, De Veylder L, Barroco RD, Casteels P, Van Montagu M, Inze D, Mironov V. A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 2001;276:36354–36360. doi: 10.1074/jbc.M011060200. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 2007;144:105–120. doi: 10.1104/pp.106.094979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montané MH. ATM-mediated transcriptional and developmental responses to γ-rays in Arabidopsis. PLoS ONE. 2007;2:e4301–e4321. doi: 10.1371/journal.pone.0000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schöbinger U, Hülskamp M. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell. 2003;15:303–315. doi: 10.1105/tpc.008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Spadari S, Pedrali-Noy G, Falaschi MC, Ciarrocchi G. Control of DNA replication and cell proliferation in eukaryotes by aphidicolin. Toxicol. Pathol. 1984;12:143–148. doi: 10.1177/019262338401200205. [DOI] [PubMed] [Google Scholar]
- Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu. Rev. Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. ‘Big it up’: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Toki S. Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Rep. 1997;15:16–21. [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47:969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Truernit E, Haseloff J. A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods. 2008;4:15. doi: 10.1186/1746-4811-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M, Iwamoto N, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H. Molecular characterization of mitotic cyclins in rice plants. Mol. Gen. Genet. 1999a;262:230–238. doi: 10.1007/s004380051079. [DOI] [PubMed] [Google Scholar]
- Umeda M, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H. Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiol. 1999b;119:31–40. doi: 10.1104/pp.119.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell. 2005;17:1723–1736. doi: 10.1105/tpc.105.032383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M, Criqui MC, Mészáros T, Binarova P, Schmit AC, Helfer A, Derevier A, Erhardt M, Bögre L, Genschik P. Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell. 2004;16:643–657. doi: 10.1105/tpc.020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yata K, Esashi F. Dual role of CDKs in DNA repair: to be, or not to be. DNA Repair (Amst) 2009;8:6–18. doi: 10.1016/j.dnarep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Yoshiyama K, Conklin PA, Huefner ND, Britt AB. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl Acad. Sci. USA. 2009;106:12843–12848. doi: 10.1073/pnas.0810304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Chua NH. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


