Abstract
Cancer epithelial cells often migrate away from the primary tumor to invade into the surrounding tissues. Their migration is commonly assumed to be directed by pre-existent spatial gradients of chemokines and growth factors in the target tissues. Unexpectedly however, we found that the guided migration of epithelial cells is possible in vitro in the absence of pre-existent chemical gradients. We observed that both normal and cancer epithelial cells can migrate persistently and reach the exit along the shortest path from microscopic mazes filled with uniform concentrations of media. Using microscale engineering techniques and biophysical models, we uncovered a self-guidance strategy during which epithelial cells generate their own guiding cues under conditions of biochemical confinement. The self-guidance strategy depends on the balance between three interdependent processes: epidermal growth factor (EGF) uptake by the cells (U), the restricted transport of EGF through the structured microenvironment (T), and cell chemotaxis toward the resultant EGF gradients (C). The UTC self-guidance strategy can be perturbed by inhibition of signalling through EGF-receptors and appears to be independent from chemokine signalling. Better understanding of the UTC self-guidance strategy could eventually help devise new ways for modulating epithelial cell migration and delaying cancer cell invasion or accelerating wound healing.
Introduction
Soon after its discovery, more than 50 years ago,1 epidermal growth factor (EGF) was found to play critical roles not only in the survival,1 proliferation,1,2 and differentiation3 of epithelial cells, but also in controlling epithelial cell migration.4 Current paradigm recognizes the role of EGF gradients in guiding epithelial cells toward higher EGF concentrations,5 while it is generally assumed that a uniform concentration of EGF can only stimulate random migration (chemokinesis).6 It is also known that EGF can be internalized by epithelial cells7 and released through autocrine8 and paracrine interactions.9 While it is theoretically possible that the epithelial cell release or depletion of EGF could interfere with the concentration and distribution of EGF around the cells, the importance of these processes on cell migration remain underappreciated and their practical consequences difficult to measure.10
To better understand the implications of cell-induced changes of EGF concentration on the migration of epithelial cells, we focused our attention on the migration of cells confined inside micron-size channels. We rationalized that inside the channels, cell-induced changes could be magnified by the limited amounts of EGF available around the cells. Such experiments would not be possible using cells cultured in Petri dishes, where the supply of EGF diffusing from the culture media exceeds the rate of cellular EGF uptake and the environment around cells remains quasi-homogeneous.10 Thus, we designed micron-size channels and analyzed the directional choices that cells made when moving through channels of various configurations. We quantified the effect of various EGF concentrations and signalling inhibitors. Our results show that the combination of cellular EGF uptake and restricted EGF transport through the channels could result in significant alterations of the EGF concentration around the cells. These alterations include the formation of EGF gradients which can subsequently drive the persistent migration of the epithelial cells. This self-guidance strategy is effective in the absence of pre-existent gradients and enables epithelial cells to navigate along the shortest path through micron scale mazes.
Methods
Computational model
Diffusion and consumption rates for cells in channels were simulated using a one-dimensional finite element model, solved using the Runge–Kutta method. Fick’s second law37 was discretized as:
where c[n,t] is the oncentration at position n and time t, D is the diffusion constant, and Δt and Δx are the temporal and spatial discretization sizes, respectively. At locations where cells are present, an additional term proportional to the normalized consumption rate rc is subtracted to represent metabolism of the molecule by the cells. The path along the central well and through the micron-size channels was treated as a single dimension and it was assumed that concentration was independent of the other dimensions. At locations where the cross-sectional area undergoes a step change (e.g. from the central well to the microchannels), the concentration change in the larger volume resulting from flux across this step change was scaled by the ratio of cross-sectional areas. The location of migrating cells was treated as a continuous variable, but for the purpose of calculating consumption, the cell was assumed to be confined to a single segment of the channel.
To generate Fig. 1d in the paper, the simulation was implemented with the cell biophysical parameters in Table 1 and the following geometrical parameters, chosen to match the experimental conditions. The central well, 2 mm high, was seeded with cells covering the bottom at 50% density. Leading from this well to the external bath were 100 straight channels, 600 µm long, with a 12 × 12 µm2 cross-section. The spatial discretization was chosen to match the width of an individual microchannel. The temporal discretization was chosen so that halving the value changed the result by less than 0.1%. To facilitate the comparison of different molecular species and concentrations, all concentrations were normalized by the initial concentration. Consumption rates were normalized by the initial concentration and the cell seeding density. As a result, concentrations became unit-less, and consumption rates had units of m3 s−1; i.e., the volume of liquid containing the amount consumed per second. At time t = 0 the system was in steady-state; that is, consumption at the base of the well was balanced by flux from the well and channels, and the net flux into any portion of the system was zero. Immediately after, cells began migrating along the channel at a rate of 100 µm h−1. No attempt was made to simulate staggered entry times; however, Fig. 1d shows that for straight channels, the contribution of the migrating cells to the overall gradient at one hour was negligible. Steric hindrance by migrating cells was ignored in this analysis; experiments described in this manuscript show that this effect was not significant.
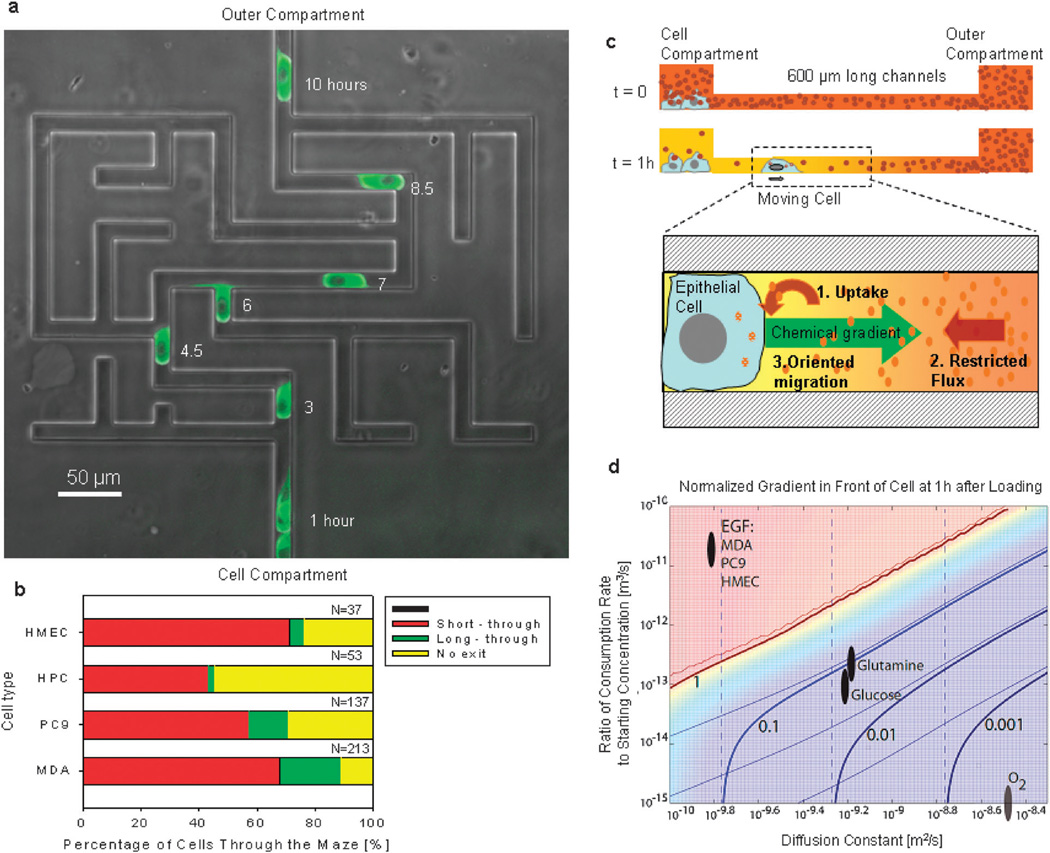
Fig. 1.
Epithelial cells can spontaneously migrate through mazes in the absence of pre-existent chemical gradients. (a) Migration of one PC9 cell through a maze. The maze combines one short path, one long path, and numerous dead end channels. Fluorescent images of GFP–tubulin PC9 cells at 1, 3, 4.5, 6, 7, 8.5 and 10 hours are overlapped on a phase contrast image of the microfluidic maze. Scale bar is 50 µm. (b) Percentages of HMEC (human mammary epithelial cells), HPC (human pancreatic cells), PC9 and MDA MB231 cell lines moving through the maze along the short (red) or the long (green) paths or getting trapped (yellow) inside the maze. N represents the number of cells observed moving through the mazes. Only the first cell entering each maze was analyzed. (c) Schematics of the self-ordination strategy of epithelial cells through mazes. Immediately after seeding the cells in the cell compartment, the concentration of EGF is uniform throughout the device (t = 0). As cells consume EGF a gradient is established between the cell and outer compartments (t = 1 h). The process depends on the consumption of EGF (1) and its diffusion through the channels (2) from the outer compartment. The gradient that is subsequently formed can guide the directional migration of cells (3) from the cell compartment to the outer compartment. (d) Simulation of normalized concentration gradient in front of a moving cell in a straight channel at t = 1 h, plotted vs. diffusion constant D and normalized consumption rate rc. The gradient was normalized so that a value of 1 indicates a linear decrease in concentration along the entire length of the channel. Thick solid lines indicate iso-gradient contours. Thin solid lines indicate similar contours when consumption by the moving cell is neglected; the similarity of these lines to the thick ones indicates that the contribution of moving cells to the overall gradient was minimal for most conditions. Dashed lines indicate contours when consumption by cells in the central well is neglected. Ovals indicate regions of D and rc corresponding to several biologically relevant molecules for the cell types used in this study. The gradient for oxygen is quite small; for glucose and glutamine, moderate; for EGF, very high.
Table 1.
Quantitative parameters for the biophysical model
| Molecular weight | Diffusivity/m2 s−1 | Concentration in media/mol L−1 | Consumption rate/mol per cell per h | References | |
|---|---|---|---|---|---|
| EGF | 6054 | 1.6×10−10 | 1.6×10−10 | 2.0×10−16 | 29–31 |
| Glucose | 180 | 7.6×10−10 | 1.0×10−03 | 2.0×10−13 | 32,33 |
| Glutamine | 146 | 6.7×10−10 | 1.0×10−03 | 1.0×10−13 | 32,34 |
| Oxygen | 32 | 3.0×10−09 | 2.5×10−03 | 7.0×10−13 | 35,36 |
The model was subsequently used to predict EGF gradients for the side-chamber experiments. EGF gradients were computed at the junction between the side chamber and the outer compartment as a function of time as a cell migrated toward the junction. The model was run for both the 1 : 1 and 3 : 1 geometries, with side chamber concentrations ranging from 1/10× to 5×. Each simulation was run twice, either including or excluding consumption of EGF by the moving cell and the cells in the cell compartment. In addition, both the initial EGF concentration within the microchannels (either the side chamber or outer compartment concentration) and the time delay before the cell entered the channel (8–24 hours) were explored, but neither variable had a significant qualitative effect on the results. The model was also used to predict EGF gradients inside devices with one bifurcation toward through and dead-end channels.
Microfluidic devices
Microfluidic devices were constructed of polydimethyl siloxane (PDMS, Dow Corning, Midland. MI) inside 24 well glassbottom plates (MatTek Co, Ashland, MA), as previously described in detail.11 Briefly, the PDMS pieces were manufactured using standard microfabrication techniques. For this, PDMS was cast as a 2 mm thick layer on a micro-patterned silicon wafer on which two layers of photoresist (SU8, Microchem, Newton, MA) were patterned using standard photolithographic technologies. The first, thinner layer (12 µm thin) defined the shape of the channels through which cells would migrate. The second, thicker layer (50 µm) was used for the reservoirs connecting all channels between the central and the outer compartments of each device. The cast PDMS was then cut using a 1.5 mm hole-punch for the central well and a 5 mm hole-punch for the device (Fig. S1, ESI†). Twenty four to forty eight PDMS pieces at a time, depending on the design of the devices, were exposed to oxygen plasma for 20 seconds, in a plasma asher (March, Concord, CA), manually placed in pairs on the glass-bottom of a 24-well plate (Mattek), and baked for 3 min at 70 °C.
Immediately following bonding, while the PDMS was still hydrophilic, 2 µL of a solution of 20 µg mL−1 collagen IV in phosphate buffer (PBS, Sigma Aldrich, St. Louis, MO) was pipetted into the center well of each device. Although no fluorescence labeling was performed, we assumed that the strong capillary force caused the capillaries to be uniformly coated. When devices with side chambers were used for experiments, to facilitate the filling of the side chambers, the entire plate was placed under vacuum for 10 minutes after loading the central well with 5 µL solution of the serum-free media with various EGF concentrations. After priming, the extra fluid in the central compartment of the device was removed, devices were washed three times with PBS, and then covered with serum-free media with nominal EGF concentration. No collagen coating was used for the experiments with side chambers. To study the migration of human-derived cells, we modified the design to replace the cell compartment with a side channel. These changes were necessary in order to maintain an overall low cell density per unit of surface, avoid inhibitory cell–cell interactions, and bring the cells closer to the entrance of the mazes at the beginning of the experiment.
Cell preparation
The human epithelial breast adenocarcinoma cell line (MDA-MB-231, American Type Culture Collection, Manassas, VA), and the non-small lung carcinoma cell line (PC9ZD, a gift from Dr Sri Sharma) were cultured in DMEM media, supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (Cellgro, Manassas, VA) in a humidified atmosphere of 5% CO2 in air. Human mammary epithelial cells (HMEC, Lonza) were cultured and handled following standard protocols, using serum-free culture media. The serum-free media were prepared using mammary epithelial growth media (MEGM, Lonza) supplemented with bovine pituitary extract, EGF, hydrocortisone, insulin and gentamicin/ amphotericin-B (Lonza). Prior to the migration assay, cells grown in cell culture flasks were rinsed with 1× PBS, detached from the flask using 3 mL of either Cell Stripper (Sigma Aldrich), or Accutase (Innovative Cell Technologies, San Diego, CA). After incubation for 3–5 min the cell release solution was neutralized by the addition of up to 6 mL culture media. The cells were then centrifuged for 5 min at 1000 rpm at 25 °C and re-suspended in the assay media at a cellular concentration of 106 cells mL−1. Suspensions of prepared cells (3–4 µL) were loaded into the center well of each collagen-coated device in a 24-well plate. Assay media (1 mL) were then added to each well to ensure complete submersion of each device in the media.
Human pancreatic cancer cells (HPC) were isolated from surgical specimens of histopathologically confirmed pancreatic ductal adenocarcinomas (PDAC), which were obtained according to Institutional Review Board-approved guidelines. A piece from the primary tumor (approximately 1 cm size) was minced with two razor blades, put into sterile, serum-free medium (RPMI 1640; Mediatech, Manassas, VA and Matrigel; BD Biosciences, San Jose, CA), and implanted subcutaneously into 6 immunodeficient nude mice nu/nu (Jackson Laboratory, Bar Harbor, Maine). When the tumor reached a size of 2 cm or more, the mice were sacrificed and the tumor harvested and re-implanted into a green fluorescent protein (GFP) nude mouse (GFP++; nu/nu, a C56BL/6 nude mouse in which homozygous GFP expression is driven by the ubiquitin C promoter). The experiments involving live animals were performed with knowledge and following the guidelines of theMassachusetts General Hospital Subcommittee on Research Animal Care (SRAC). All cells of mouse origin expressed GFP, allowing them to be reliably differentiated from the human tumor cells under fluorescent light illumination. For motility experiments, fresh pieces of PDAC tumor xenograft grown in the GFP++; nu/nu mouse were removed aseptically and transferred to 10 mL HBSS (Mediatech) with 16 mg Collagenase Type 1 (Sigma) and incubated at 37 °C for enzymatic dissociation. Every 15 minutes the cells were mechanically dissociated by pipetting with a 10 mL pipette. The collagenase was inactivated with HBSS (Mediatech) containing 10% bovine serum albumin (BSA Fraction V, Fisher Scientific, Pittsburgh, PA). At the end of the reaction cells were filtered through a 100 µm nylon mesh (Fisher) and washed three times with DPBS (Mediatech) containing 0.2% BSA (BSA Fraction V, Fisher). The cells were then re-suspended into MEGM media (Lonza) and loaded into the migration assay. The superfluous cells were transferred into cell culture dishes with DMEM/F12 50 : 50 + 10% BSA (Mediatech) and penicillin–streptomycin (Cellgro). The growth medium was replaced every 3 to 5 days.
Assay media and EGF uptake rates
For experiments targeting the role of EGF in epithelial cell migration, assay media were prepared by adding different concentrations of EGF (from 0 to 10 ng mL−1) to the epithelial growth media (MEGM, Lonza) supplemented with bovine pituitary extract, hydrocortisone, insulin and gentamicin/ amphotericin-B (Lonza). The rates of EGF uptake for HMEC, MDA and PC9 cells were measured in 96 well plates, in the presence of assay media with 10 ng mL−1 EGF, with approximately 10 000 cells per well, using a human EGF immunoassay (Quantikine, R&D Systems, Minneapolis, MN), and following the instructions from the manufacturer. For all other experiments, devices were filled with DMEM media, supplemented with 10% FBS (Atlanta Biologicals) and 1% penicillin/streptomycin (Cellgro).
Migration assay and image analysis
To record the migration of epithelial cells through the microfluidic device, time-lapse imaging was performed using a fully automated Nikon Ti-E microscope fitted with the perfect focus system and an environmental chamber maintained at 37 °C with humidified 5% CO2. For the majority of the experiments, time-lapse images were captured at 10 locations on each device, at 15 min intervals, for 24–72 h. Cells migrating through mazes were quantified for directional choices, and only the first cell entering a maze was considered for analysis. For each condition, we counted the number of cells entering the maze and the number of cells moving through the shorter and longer paths for 48 h after cell seeding. Migration time was measured from a starting point at the first bifurcation to the end at the last bifurcation.
Statistical analysis
The directional choices of cells inside bifurcating channels were discretized as binary outcomes. Cells were classified as either moving “toward side chamber” or “toward outer compartment” in the experiments using devices with side chambers. Cells were classified as either “through” (cells moving toward the outer compartment) or “side” (cells moving toward the dead-end channels) in the experiments using devices with dead-end side channels. One directional choice for one cell at a bifurcation was considered to be an independent event. Only the first cell entering a channel and reaching a bifurcation was evaluated. Cells that entered channels from the outer compartment and cells dividing inside the channels were excluded from the analysis. Results were analyzed using chi-squared statistics and differences considered statistically significant for p ≤ 0.05. The directional bias of cells moving through bifurcating channels with side chambers for all eight conditions was compared to random choices.
The migration speed of cells migrating through channels was analyzed using the manual tracking plug-in of Image J (NIH). More than 20 cells were tracked in two devices for each condition. The statistical significance of differences in average migration speed between cells under different conditions was estimated using the ANOVA test.
Results
To better understand the roles of cell-induced microenvironment changes during epithelial cell migration, we studied the motility of cells in confined environments, through micron-size channels. In implementing this strategy, one has to take into account the mechanical guidance along the axis of the channels with cross section size comparable to that of epithelial cells.11 To decouple the effects of chemical and mechanical stimuli on moving cells, we designed networks of bifurcating channels, connecting a central cell compartment and a larger outer compartment, which shared the same cell culture media. Each network of channels was configured with one inlet and one outlet, two through paths, one short and one long, and several dead end channels (Fig. 1a). We studied the migration of cells from the cell compartment to the outer compartment, through the network of channels, a distance longer than 900 µm.
We expected that in the absence of pre-existent chemical gradients, cells are guided solely by mechanical cues, and make random decisions at each of the seven bifurcations. Assuming that cells only move forward, the calculated chance of a cell to make the necessary sequence of decisions that would allow it to reach the exit would be less than three in 32. To our surprise, significantly more epithelial cells were able to navigate the maze. Within 48 hours of the start of the experiments, 76% of the human derived mammary epithelial cells (HMEC), 90% of MDA-MB-231 cells (MDA—breast adenocarcinoma cell line), 72% of PC-9ZD cells (PC9—lung carcinoma cell line), and 43% of the human-derived pancreatic carcinoma cells (HPC) entering the maze had reached the exit (Fig. 1; Movies S1–S3, ESI†). Only the first cell entering each maze was included in these calculations. A strong bias toward the short path was also obvious for all cell types tested, with the ratio of cells migrating along the short vs. long path ranging from 3 : 1 (MDA) to 21 : 1 (HPC) (Fig. 1b). This bias was present in spite of the absence of pre-existent guiding gradients. This finding and also the fact that high numbers of cells are able not only to exit the maze, but do so following the short path, suggest that moving cells employ some orientation mechanism that is not purely mechanical.
To understand the unexpected efficiency of cell orientation through the mazes, we proposed the hypothesis that a spatial chemical gradient between the center cell compartment and the outer compartment is being generated by the epithelial cells (Fig. 1c). To test this hypothesis, we first employed a biophysical model to estimate the magnitude of cell-induced gradients in our devices. Our calculations helped refine the space of conditions for biomolecule uptake or release rates, average concentration, and diffusion coefficients for which significant gradients could be generated by the cells (Table 1). We further narrowed down the list of biomolecules that could potentially form gradients by taking into consideration the surprising finding that cell orientation can also occur in serumfree, epithelial growth media. On the resulting list, EGF stood out for the magnitude of the estimated cell-induced gradients (Fig. 1d). To analyze the role of EGF in our system, we measured the EGF consumption rates in HMEC, MDA and PC9 cells at 0.4, 0.2, and 0.1 fM per cell per hour, respectively. We estimated that 1 hour after seeding 20 000 cells in the cell compartment, up to 10% EGF concentration difference can form along the first 100 µm of a 600 µm long straight-channel (Fig. 1d). The magnitude of these EGF gradients is comparable with gradients previously demonstrated to be effective for biasing the migration of epithelial cells toward the highest EGF concentrations.5 If a gradient was established by autocrine secretion, it would have the opposite orientation, i.e. from the outer compartment toward the cell compartment, preventing the cells from escaping the cell compartment. No significant gradients of oxygen, glucose, or glutamine were predicted to form along the micron-sized channels during the first hour, as a result of cell metabolism (Fig. 1d).
To test the hypothesis that a self-generated EGF gradient is guiding the moving epithelial cells, we designed experiments in which the presence or absence of EGF uptake by the cells would result in substantial differences in the steepness and orientation of EGF gradients in front of the migrating epithelial cells. Cells moving through bifurcations would follow the EGF gradients and steer toward the side with the steepest EGF gradient. The experimental system consisted of the cell and outer compartments, connected through bifurcating channels to side chambers. The size of the side chambers and the length of the channels were such that a fluorescein gradient that forms toward the compartments filled with water was stable for more than 24 hours (Fig. S2, ESI†). The length ratio between the channels toward the outer compartment and the side chamber was either 1 : 1 or 3 : 1 (Fig. 2a). The side chambers were preloaded with serum free media containing EGF in various concentrations relative to cell and outer compartments (5×, 1×, 1/2×, 1/10×).
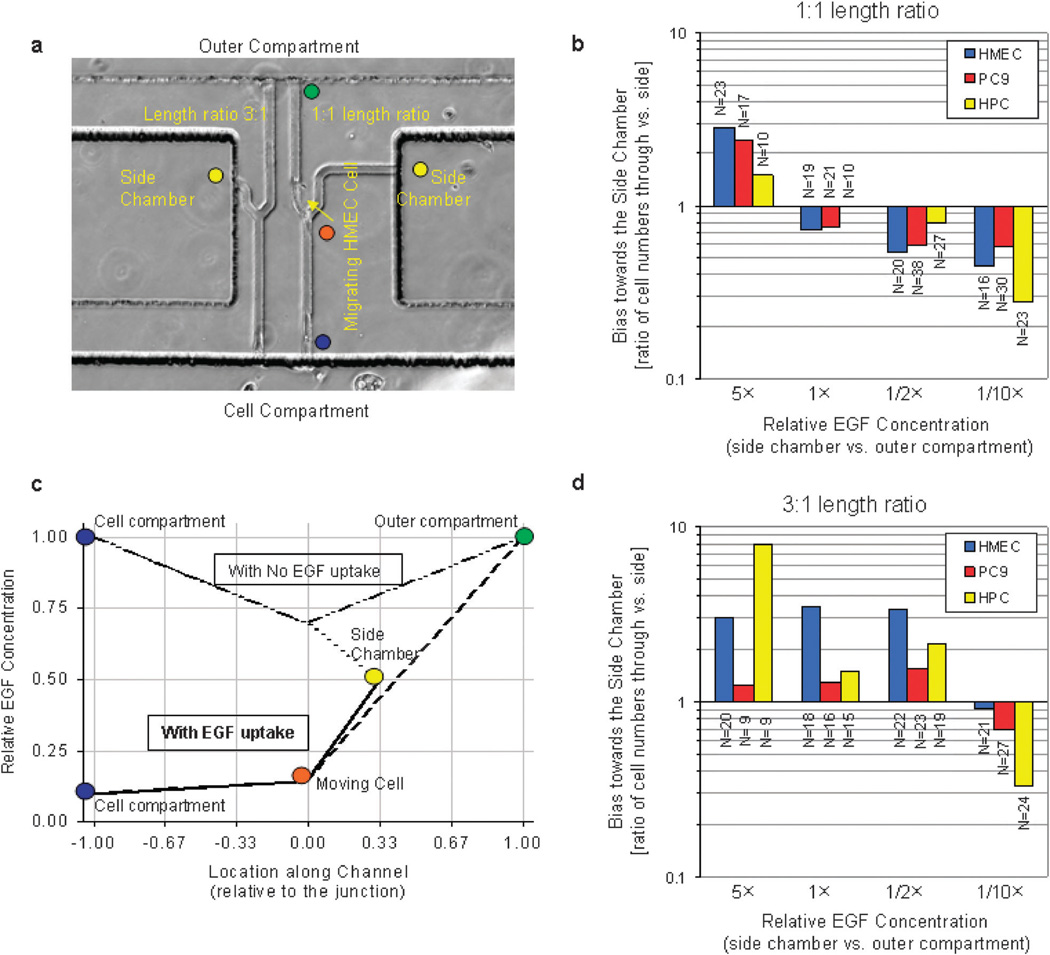
Fig. 2.
Epithelial cell EGF-uptake changes the orientation bias at bifurcations. (a) Microscopy image of one MDA cell migrating past the channel junction (orange dot), from the cell compartment (blue dot) toward the outer compartment (green dot). The ratio between the lengths of the channels toward the outer compartment and the side chamber (yellow dot) was 3 : 1 for the device to the left and 1 : 1 for the device to the right. (b) Inversion of gradient slope toward the outer compartment and the side chamber due to cellular EGF uptake, for 3 : 1 length channel ratio and 1/2× EGF concentration in the side chamber. In the absence of cellular EGF uptake, a large positive gradient toward the outer compartment (green dot) and a negative gradient toward the side chamber (yellow dot) were predicted. When cellular EGF uptake is taken into consideration, the EGF concentration in the cell compartment decreases (blue dot) and a stronger gradient toward the side chamber compared to the one toward the outer compartment develops by the time a cell arrives at the junction (orange dot). (c and d) Bias of HMEC, HPC, and PC9 cells toward side chambers with various concentrations of EGF (1× in the outer compartment vs. 5×, 1×, 1/2×, 1/10× in the side chamber) and 1 : 1 ratio between the length of the channels after bifurcation, toward the outer compartment and the side chamber (c), or 3 : 1 ratio (d). Epithelial cells consistently migrate toward the higher concentration of EGF when the ratio of the two channels is 1 : 1. Cells can migrate toward a lower concentration of EGF (1/2×) when the side chamber is positioned closer to the bifurcation (3 : 1 length ratio), suggestive of significant EGF concentration decrease at the bifurcation. N represents the number of cells observed moving through the bifurcating channels, only the first cell reaching the bifurcation was analyzed.
The directional bias of cells at the bifurcation becomes an indicator for the presence or absence of EGF uptake by the cells inside the device. In the control cases of channels with equal length (1 : 1 length ratio), the bias toward the side chamber is independent of the EGF concentration at the bifurcations and depends only on the EGF concentrations in the outer compartment and the side chamber. Steeper gradients toward the side chambers preloaded with 5× EGF, or away from the side chambers preloaded with 1/10× EGF could be predicted, independent of cellular uptake (Table 2). These controls are important to assure that the directional bias is dependent only on EGF and not the other factors present in the serum-free culture media.
Table 2.
Predicted gradients for side-chamber experiments (fractional concentration per µm × 1000). The larger gradient at the junction is outlined in bold. Note the inversion of the gradient with the presence of a cell that uptakes EGF, in the 3 : 1 geometry, and with 1/2× EGF in the side chamber
| EGR Concentrationin the Chamber | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1/10× | 1/2× | 1× | 5× | ||||||
| Channel length ration |
EGF Uptake | Outer | Side | Outer | Side | Outer | Side | Outer | Side |
| 1:1 | NO uptake | 0.4 | −2.8 | 0.3 | −1.5 | 0 | 0 | −0.8 | 13.0 |
| uptake | 3.3 | 0.1 | 3.2 | 1.4 | 3.1 | 3.1 | 2.3 | 16.0 | |
| 3:1 | NO uptake | 0.5 | −4.5 | 0.3 | −2.4 | 0 | 0 | −1.3 | 21.0 |
| uptake | 3.2 | 2.2 | 3.1 | 4.3 | 2.9 | 7.0 | 1.5 | 28.0 | |
For channels with 3 : 1 length ratio, the bias toward the side chambers becomes dependent on the EGF concentration in front of the moving cells. No directional bias would be expected in the absence of cellular EGF uptake and equal EGF concentrations (1×) preloaded in the side chamber and the outer compartment. However, it is expected that cells will be biased toward the side chamber if the EGF concentration in the channels is being reduced by the cells. A decrease in the EGF concentration in front of the cells at the bifurcation will result in steeper gradients along the shorter channel toward the side chamber (1×) than the longer channel toward the outer compartment (also 1×), biasing the direction of cell migration accordingly. Even more informative is the predicted behavior of the cells when the side chamber is loaded with 1/2× EGF when the cell and outer compartments are filled with the same 1× EGF (Fig. 2b). In the absence of EGF uptake, as well as in the presence of small decreases in EGF concentration at the bifurcation cells reaching the bifurcation should be guided toward the outer compartment. Only in the presence of substantial EGF concentration decreases at the bifurcation, steeper concentration gradients could be formed toward the side chamber, steering the cells toward the side chamber (Fig. 2b).
To verify these predictions, we observed HMEC, HPC, and PC9 cells moving through the bifurcations and quantified their directional bias toward side chambers for 1 : 1 (Fig. 2c) and 3 : 1 channel length ratio (Fig. 2d). We found that at bifurcations with 3 : 1 channel length ratio, cells preferentially steered toward the side chambers preloaded with 1× EGF, consistent with the predictions from our model for the condition of significant EGF uptake by the cells. Also consistent with cellular EGF uptake, more cells steered toward the side chambers preloaded with 1/2× EGF, suggesting a strong EGF uptake. In control experiments, we measured a significant directional bias toward the side chambers loaded with 5× EGF and significant bias toward the outer compartment when the side chambers were loaded with 1/10× EGF, regardless of the channel length. These results demonstrate the critical importance of EGF cellular-uptake during the migration of epithelial cells in confined spaces.
To further analyze the importance of the self-generated EGF gradients relative to other possible guidance strategies in a confined environment, we designed a series of experiments involving various designs of microscale channels (Fig. 3a). First, we verified that cell migration through channels is an actin-polymerization dependent process. We exposed cells to 5 µM cytochalasin B before loading them into devices and observed that they do not move through straight channels. Washing the devices with media removed most of the cytochalasin and promptly restored the directional migration of the cells (Fig. 3b).
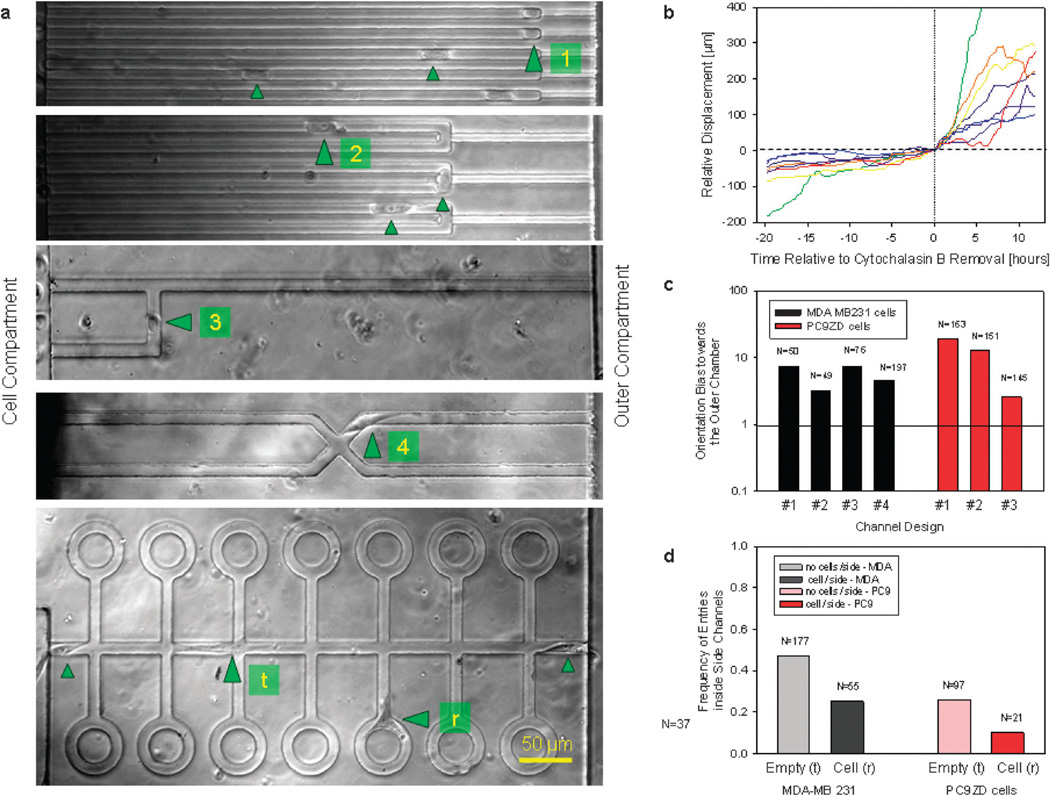
Fig. 3.
Orientation bias of MDA and PC9 cells migrating through channels of various geometries. (a) Significant orientation bias for MDA and PC9 cells toward the outer chamber. Most of the moving cells either squeeze through the smaller channel and continue moving or are “trapped” within a 50 µm region at the transition between larger and smaller channels (#l1). Similar cell migration patterns are observed even when the larger U-shaped channels are employed (#2). When migrating toward a bifurcation, cells preferentially steer toward the longer channel leading to the outer compartment rather than toward the shorter channel leading to the cell compartment (#3). At the intersection of four orthogonal channels, cells preferentially move toward the outer compartment (#4). Cells migrating through channels (#t) with “open roads” and “cul-de-sacs” bias their migration toward the open road when other cells are present inside the “cul-de-sac” (# s). (b) Pre-treatment with 5 µM cytochalasin B in media blocks the migration of MDA cells through straight channels. Removing the cytochalasin solution and replacing it with fresh media at time t=0 restores the migration of cells through channels. (c) The significant bias toward the outer compartment for the geometries in (a) suggests that the mechanical guidance inside channels is not the dominant factor during cell orientation. Only the orientation of the first cell to enter each channel and in the absence of any other cells in the channels was quantified. All cells present in the channels were counted and N represents the number of directional decisions for all cells observed. (d) The frequency of cells entering the “cul-de-sac” decreases when another cell is already present inside that channel.
To evaluate the role of mechanical guidance during the migration of the cells through channels, we analyzed the migration of MDA and PC9 cells through channels that abruptly change their cross section from 12 × 12 µm to 6 × 3 µm (Fig. 3a, cell#1). We restricted our analysis to only the channels with one cell at a time and to only the first cell entering a channel. We found that only one in 7 MDA cells and one in 19 PC9 cells reversed their direction of migration to travel more than 50 µm back to the cell compartment (Fig. 3c). Moreover, inside an U-shaped channel (Fig. 3a, cell#2), only one in 3 MDA and one in 13 PC9 cells traveled more than 50 µm back from the transition zone (Fig. 3c). These results indicate that a chemical gradient along the channels rather than mechanical cues is the dominant factor in guiding cell migration through channels.
To probe the role of the hydraulic resistance in front of the cells, we designed experiments in which moving cells arriving at a bifurcation can follow either a low hydraulic resistance path back to the cell compartment or a higher hydraulic resistance path forward toward the outer compartment (Fig. 3a, cell#3). We observed significant bias toward the longer path (7 times more MDAs and 3 times more PC9 cells turned toward the outer compartment, Fig. 3c). Moreover, we examined that migrating MDA cells are often stretched and have a cross section smaller than the cross section of the channels. These cells are capable of orienting along the shortest path even when there is enough space between the cells and the walls of the channels to relieve any pressure (Movie S2, ESI†). These observations argue against a mechanism based on differences in hydraulic resistance guiding the cells moving through channels.
Inside “crossed” channels orthogonally split toward the cell and outer compartments (Fig. 3a, cell#4), we also measured a significant bias toward the outer compartment. Five times more MDAs turned toward the outer compartment (Fig. 3c), whereas physical guidance or surface bound cues would have resulted in a more balanced distribution between the four channels. Again, we restricted our analysis to only the channels with one cell at a time and to only the first cell entering a channel.
Additional support for the role of chemical cues during cell orientation in microscale channels comes from observations of the long-distance interactions between multiple cells inside a complex network of through and dead-end channels (Fig. 3a, cell#t/r). In these experiments, we calculated the frequency with which cells passing through the main channel entered the “cul-de-sac” to the side, in the presence or absence of a second cell in the “cul-de-sac”. Different from all other experiments in this study, we followed the migration of multiple cells present in the same channels at the same time. We found that for both the MDA and PC9 cells, the frequency of a cell entering the “cul-de-sac” was significantly lower when a second cell was present in the “cul-de-sac”, suggestive for the ability of cells in channels to influence the direction of migration of distant cells (Fig. 3d). There was no significant correlation in the proportion of times a cell entered a loop and the history of cells previously occupying the loop. Cells entered with comparable frequencies in loops that were previously occupied or not by other cells (49% vs. 41%), suggesting that the differences are not due to any long lasting modifications that cells do to their local environment. Together, our results demonstrate the significant effect that biochemical gradients established around cells and diffusion from the surroundings have on cell guidance during migration and redistribution in confined environments.
To further study the role of EGF during cell migration through channels and the self-guiding abilities of cells during migration, we employed symmetrically-bifurcating channels (Fig. 4a). The smaller footprint of the bifurcating channels helped increase the number of cells observed in each experiment. This simple configuration allowed us to decouple the directionality from the speed of moving cells, and to quantify and compare the binary directional decisions under various conditions. Cells migrating from the cell compartment made one directional decision at the bifurcation, and steered either toward the “through” path leading to the outer compartment, or toward the “side” path leading to a dead-end. We found that in the presence of standard or serum-free growth media, MDA and PC9 cells steered toward the “through” path more than three times more often than toward the “side”. This result is consistent with the formation of a guiding gradient along the through channel, between the cell and outer compartments (Fig. 4b).
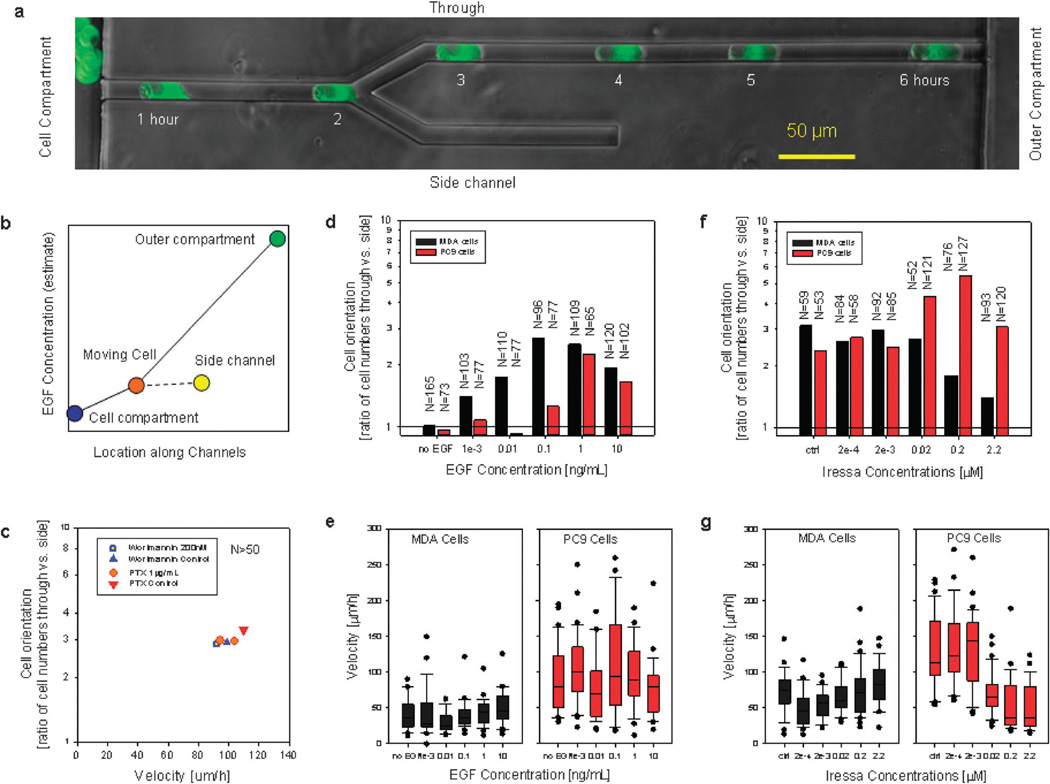
Fig. 4.
The orientation of MDA and PC9 cells migrating through bifurcating channels is influenced by EGF concentrations. (a) Migration of PC9 cells through a bifurcating channel in the presence of complete cell culture media. Fluorescent images at 1 hour intervals are overlapped with a phase contrast image of the bifurcating channel. (b) At the time when a cell reaches the bifurcation (orange dot), a gradient of EGF toward the outer compartment (green dot) could bias the direction of cell migration toward the “through” channel and away from the side channel (yellow dot). (c) The orientation of MDA cells in bifurcating channels is not altered in the presence of PI3K inhibitor wortmannin (200 nM) or G-coupled protein receptor blocker, pertussis toxin (1 µg mL−1). (d and e) Orientation and velocity of migrating MDA (black) and PC9 (red) cells in the presence of various concentrations of EGF. N represents the number of cells observed moving through the bifurcating channels, only the first cell reaching the bifurcation was analyzed. (f and g) Orientation and velocity of migrating MDA (black) and PC9 (red) cells in the presence of various concentrations of EGFR kinase inhibitor Iressa. For the box plots displaying cell velocities, the boundary of the box closest to zero indicates the 25th percentile, a line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. Whiskers above and below the box indicate the 90th and 10th percentiles, and dots represent the outliers.
Following the numerous reports suggesting a role of chemokines in cancer cell migration,12–15 we quantified the effect of various inhibitors on the migration of MDA cells through bifurcating channels. We found that the directional migration was not affected by the inhibition of G-protein coupled receptors using Pertussis Toxin (PTX) at a concentration of 1 µg mL−1 or the inhibition of PI3K activity using wortmannin (200 nM, Fig. 4c).
The guidance of MDA and PC9 cells through bifurcating channels was significantly impaired when the concentration of EGF in the serum-free media was reduced (Fig. 4d and e). MDA cells displayed significant bias toward the “through” channels at concentrations above 10 pg mL−1 with maximum orientation at 0.1 ng mL−1 EGF. PC9 cells required concentrations above 0.1 ng mL−1 for significant bias toward the “through” channels, with maximum guidance at 1 ng mL−1 EGF. At the highest EGF concentrations, cell orientation decreases, consistent with the saturation of EGF receptors. Quite remarkably, the velocity of the MDA or PC9 cells passing through the bifurcation did not change significantly with changes in EGF concentration (Fig. 4e).
In keeping with the role of EGF in self-generated guidance, inhibition of EGF receptor tyrosine kinase signaling using Gefitinib (Iressa) had a strong effect at concentrations above 200 nM on the guidance toward the “through” channels of MDA cells, an Iressa-sensitive, triple-negative breast cell line over-expressing EGF receptor16 (Fig. 4f and g). In contrast, we observed no significant effect of Iressa up to 2 µM concentration on the guidance of PC9 cells, which harbor an activating truncation mutation in the EGF receptor.17 Interestingly, an increase in guidance toward the “through” channels occurs at concentrations above 22 nM and is correlated with slower migration speed for these cells compared to those treated with lower doses of drug (Fig. 4g). All the experiments using EGFR inhibitors were performed in the presence of complete growth media.
Discussion
We discovered that epithelial cells are capable of employing a strategy for self-guidance during migration under conditions of mechanical confinement and in the absence of pre-existent gradients. This guidance strategy is dependent on the presence of EGF and three associated processes. First, epithelial cells take up significant amounts of EGF, partially depleting the cell microenvironment. Second, the consumed EGF is replenished by diffusive transport from the environment and the total EGF flux is restricted through the network of channels. Finally, epithelial cells respond to the local gradients that result from EGF uptake and flux, and move in the direction of the steepest gradients. Acting together, the three processes (uptake– transport–chemotaxis—UTC) enable the epithelial cells to overcome the absence of pre-existent gradients and to guide their migration to exit confinement along the shortest path.
The UTC self-guidance strategy does not require pre-existing chemical gradients, and hence, persistent migration is not bound by the spatial limits of any gradient. This characteristic of UTC self-guidance strategy is clearly distinct from the classical chemotaxis, which predicts that cells move directionally only in the limited area where a spatial gradient is present and switch to random movement (chemokinesis) as soon as they reach the area of highest concentration. Instead, the self-guidance strategy predicts that the gradient, once formed, will continually move with the cells. The overlap of two functions in epithelial cells (making the gradients and responding to them) creates the conditions for a positive reinforcement loop that can lock the cells in a persistent migratory phenotype.
The overlap of functions in the same cells makes the selfguidance strategy distinct from recently reported mechanisms for sharpening pre-existent gradients in tissues.18–21 Although these mechanisms rely on cells that can bind and neutralize chemokine factors, two distinct cell types are generally involved e.g. the somatic cells that enhance the gradient, and the primordial germ cells that respond to it,18,19 or the platelets in the fibrin clot and the fibroblasts.21 With two distinct cell types accomplishing separate functions, a robust control is achieved over the positioning of the moving cells. The critical difference in the case of self-guidance is that the uptake and chemotaxis functions overlap in the same cells. The robust positioning control is lost and a new behavior emerges, which drives cells toward locations with fewer cells or reduced cumulative rate of EGF uptake.
The self-guidance strategy is also distinct from autocrine or paracrine signalling,8,9 when gradients of attractant molecules are being produced by some cells and direct the migration of other cells toward the secreting cells. Paradoxically, despite relying on a classical cell-attractant cue, EGF, the UTC self-guidance strategy has a net effect on dispersing cells away from their peers rather than bringing them together. The self-guidance strategy could contribute to the uniform distribution of normal epithelial cells in monolayers and the redistribution of cells after epithelial injuries.
The functional relationship that is being established inside confined spaces between EGF uptake and the formation of chemical gradients serves to guide the migration of malignant epithelial cells. The various networks of channels in this study may be regarded as replicating conditions close to those inside tissues and around tumors, where inter-cellular spaces, conduits like the lymphatic vessels,22 perivascular spaces,23 perineural spaces24,25 are well known to facilitate the dissemination of malignant cells. Moreover, we have previously shown that the migration of cells through straight channels correlates with their ability to form lung metastases in a mouse model.26 Together, these results suggest that our experimental setup of channel networks could be a relevant in vitro model for cancer cell invasion.
Several questions related to the epithelial cell self-guidance strategy will have to be addressed in the future. Our finding, that the mechanisms of cell directionality can be impaired without affecting the migration speed, will have to be investigated further, to determine if speed and directionality, the two fundamental processes of cell migration,27,28 are under the control of independent signalling pathways. The interplay between the EGF bound to various extracellular matrix proteins and the soluble EGF gradients during self-guided migration will have to be dissected to further understand the complexities of in vivo epithelial cell migration. More precise characterization is also needed to understand the differences and similarities between the self-guidance strategies of normal and cancer cells, during wound healing and cancer invasion.
Conclusions
Normal and malignant human epithelial cells can employ a self-guidance strategy, which is dependent on the balance between EGF uptake by the cells, restricted EGF diffusion, and the ability of the cells to chemotax toward the resulting spatial gradients of EGF. The UTC self-guidance strategy does not require pre-existent external gradients, and allows epithelial cells to produce self-generated gradients guiding their persistent migration toward unoccupied spaces. Further exploration and better understanding of the directionality strategies in epithelial cells could lead to more effective therapeutic approaches to accelerate wound healing or delay cancer metastasis.
Supplementary Material
Insight, innovation, integration.
We discovered a novel strategy for the orientation of epithelial cells during migration in the absence of pre-existent gradients. To quantify the directional choices of moving epithelial cells with unprecedented precision, we designed microscale mazes and bifurcating channels with cross section smaller than the cell size. Inside these microscale channels, the competition between epidermal growth factor (EGF) uptake by the cells and the restricted diffusion of EGF results in the formation of longitudinal EGF gradients that can guide the migration of epithelial cells along the shortest path to exit confined spaces.
Acknowledgements
We thank Dr Jun Yan for help with the biochemical analysis and Miss Nil Gural for help with the image analysis. This work was supported by funds from the National Institutes of Health (CA135601, EB002503, and CA117969).
Footnotes
Electronic supplementary information (ESI) available: Four movies and two supplementary figures. See DOI: 10.1039/c2ib00106c
Notes and references
- 1.Cohen S. J Biol. Chem. 1962;237:1555–1562. [PubMed] [Google Scholar]
- 2.Cohen S, Elliott GA. J Invest. Dermatol. 1963;40:1–5. doi: 10.1038/jid.1963.1. [DOI] [PubMed] [Google Scholar]
- 3.Rheinwald JG, Green H. Nature. 1977;265:421–424. doi: 10.1038/265421a0. [DOI] [PubMed] [Google Scholar]
- 4.Westermark B, Magnusson A, Heldin CH. J Neurosci. Res. 1982;8:491–507. doi: 10.1002/jnr.490080236. [DOI] [PubMed] [Google Scholar]
- 5.Saadi W, Wang SJ, Lin F, Jeon NL. Biomed. Microdevices. 2006;8:109–118. doi: 10.1007/s10544-006-7706-6. [DOI] [PubMed] [Google Scholar]
- 6.Shibata T, Kawano T, Nagayasu H, Okumura K, Arisue M, Hamada J, Takeichi N, Hosokawa M. Tumor Biol. 1996;17:168–175. doi: 10.1159/000217979. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter G, Cohen S. J Cell Biol. 1976;71:159–171. doi: 10.1083/jcb.71.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh AB, Harris RC. Cell. Signalling. 2005;17:1183–1193. doi: 10.1016/j.cellsig.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 9.Condeelis J, Segall JE. Nat. Rev. Cancer. 2003;3:921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 10.Bailly M, Wyckoff J, Bouzahzah B, Hammerman R, Sylvestre V, Cammer M, Pestell R, Segall JE. Mol. Biol. Cell. 2000;11:3873–3883. doi: 10.1091/mbc.11.11.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irimia D, Toner M. Integr. Biol. 2009;1:506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller MM, Fusenig NE. Nat. Rev. Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 13.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi H, Wyckoff J, Condeelis J. Curr. Opin. Cell Biol. 2005;17:559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Bhowmick NA, Neilson EG, Moses HL. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okubo S, Kurebayashi J, Otsuki T, Yamamoto Y, Tanaka K, Sonoo H. Br. J. Cancer. 2004;90:236–244. doi: 10.1038/sj.bjc.6601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taguchi F, Koh Y, Koizumi F, Tamura T, Saijo N, Nishio K. Cancer Sci. 2004;95:984–989. doi: 10.1111/j.1349-7006.2004.tb03187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boldajipour B, Mahabaleshwar H, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 19.Yu SR, Burkhardt M, Nowak M, Ries J, Petrasek Z, Scholpp S, Schwille P, Brand M. Nature. 2009;461:533–536. doi: 10.1038/nature08391. [DOI] [PubMed] [Google Scholar]
- 20.Wolpert L. J Theor. Biol. 2011;269:359–365. doi: 10.1016/j.jtbi.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 21.Haugh JM. Biophys. J. 2006;90:2297–2308. doi: 10.1529/biophysj.105.077610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Histochem. Cell Biol. 2008;130:1147–1154. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 23.Lugassy C, Barnhill RL. Adv. Anat. Pathol. 2007;14:195–201. doi: 10.1097/PAP.0b013e31805048d9. [DOI] [PubMed] [Google Scholar]
- 24.Bapat AA, Hostetter G, Von Hoff DD, Han H. Nat. Rev. Cancer. 2011;11:695–707. doi: 10.1038/nrc3131. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi T, Ishikura H, Motohara T, Okushiba S, Dohke M, Katoh H. J. Surg. Oncol. 1997;65:164–170. doi: 10.1002/(sici)1096-9098(199707)65:3<164::aid-jso4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, Meyer CA, Lightcap ES, Tamayo P, Mesirov JP, Liu XS, Shioda T, Toner M, Loda M, Brown M, Brugge JS, Ramaswamy S. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janetopoulos C, Firtel RA. FEBS Lett. 2008;582:2075–2085. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambravaneswaran V, Wong IY, Aranyosi AJ, Toner M, Irimia D. Integr. Biol. 2010;2:639–647. doi: 10.1039/c0ib00011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorne RG, Hrabetova S, Nicholson C. J Neurophysiol. 2004;92:3471–3481. doi: 10.1152/jn.00352.2004. [DOI] [PubMed] [Google Scholar]
- 30.Masui H, Castro L, Mendelsohn J. J Cell Biol. 1993;120:85–93. doi: 10.1083/jcb.120.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters CM, Oberg KC, Carpenter G, Overholser KA. Biochemistry. 1990;29:3563–3569. doi: 10.1021/bi00466a020. [DOI] [PubMed] [Google Scholar]
- 32.Longsworth L. Diffusion in liquids and the Stokes–Einstein relation. New York, N.Y.: John Wiley & Sons, Inc; 1955. [Google Scholar]
- 33.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han P, Bartels D. J Phys. Chem. 1996;100:5597–5602. [Google Scholar]
- 36.Mehta G, Mehta K, Sud D, Song JW, Bersano-Begey T, Futai N, Heo YS, Mycek MA, Linderman JJ, Takayama S. Biomed. Microdevices. 2007;9:123–134. doi: 10.1007/s10544-006-9005-7. [DOI] [PubMed] [Google Scholar]
- 37.Fick A. Philos. Mag. 1855;10:30–39. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.