Abstract
Numerous studies have attempted to identify gene expression profiles which can be utilized to predict responses to neoadjuvant chemotherapy (NAC), but their findings are not clinically applicable at present. In the present study, we sought to determine DNA copy number alterations (CNAs) in breast cancer tissues which are associated with the response to NAC. Frozen tumor tissues from 63 breast cancer patients were obtained using core needle biopsy prior to NAC (3 cycles of docetaxel plus adriamycin) and were microdissected. Array comparative genomic hybridization (array CGH) with 4,045 bacterial artificial chromosome (BAC) probes was performed to identify the CNAs. Changes in tumor size in response to NAC were measured via magnetic resonance imaging. Fluorescence in situ hybridization (FISH) was conducted to verify array CGH results and for independent validation studies. CNAs at eight chromosomal loci encompassing 24 clones were correlated with changes in tumor size after NAC (p<0.05; t-test). Two CNAs were selected, 17p12 deletion and 17q21.32-33 gain, which were significantly associated with a smaller reduction in tumor size following NAC, via prioritization of the regions containing the candidate genes. In an independent validation set of samples from 39 patients, FISH assay further showed that the 17p12 deletion was markedly associated with smaller changes in tumor size (p=0.006), while the 17q21.32-33 gain was not significant (p=0.309). In conclusion, we successfully identified a 17p12 deletion in breast cancer tissue which can be applied in predicting tumor resistance to NAC.
Keywords: breast neoplasms, comparative genomic hybridization, copy number, microarray, neoadjuvant chemotherapy, predictive marker
Introduction
Breast cancer is a leading cause of death in women worldwide. Neoadjuvant chemotherapy (NAC) is an effective way to achieve breast conservation in women with large tumors and to control micrometastases. Additionally, the in vivo response of the tumor to NAC can be observed and pathologic complete remission (pCR) is a potential surrogate marker of patient survival outcomes (1). However, due to the current lack of clinically useful predictive markers of response to NAC in breast cancer, all patients are exposed to uniform regimens of chemotherapy, leading to unnecessary toxicities in the majority of individuals (2).
Following the success of gene expression profiling in the molecular classification of breast cancer and distinguishing prognostic subgroups, a number of investigators have attempted to derive gene signatures that facilitate the prediction of response to NAC in breast cancer. Several studies have demonstrate associations between specific gene signatures and response to NAC (3,4), whereas other conflicting studies have reported no such relationship (5,6).
Since genomic DNA is more stable than mRNA and DNA copy number alterations (CNAs) in defining key genetic events driving tumorigenesis, genomic alterations serve as useful markers of subtype classification and represent potential therapeutic targets (7,8). Our group has shown that array-based comparative genomic hybridization (array CGH) is a useful tool for the prediction of tamoxifen response and prognosis, and for the detection of molecular subtype-specific genomic changes in breast cancer (9–11).
In this study, array CGH using fresh frozen microdissected gun-biopsied tissues was performed, prior to the start of chemotherapy, with a view to identify CNAs associated with NAC response in breast cancer. Significant CNAs were validated in an independent sample set using fluorescence in situ hybridization (FISH).
Materials and methods
Patients and tumor specimens
Patient enrollment and tissue sampling were conducted between February 2005 and July 2007 at the Seoul National University Hospital. The inclusion criteria were as follows: invasive ductal carcinoma, clinical stage II or III disease, eligibility for chemotherapy and informed consent. A total of 63 patients were included in the study, and the tissue samples were obtained with a 14-gauge core needle biopsy under ultrasonographic guidance, prior to NAC. Tissue samples were collected, snap-frozen in liquid nitrogen and stored at −80°C. Cancer tissue was isolated by microdissection from core biopsy specimens to reduce contamination with non-tumor tissues. The microdissection technique has been described previously (9). The proportion of tumor cells in microdissected specimens was >90%. Patients were treated with 3 cycles of docetaxel (75 mg/ m2) and adriamycin (50 mg/m2) (DA) concomitantly administered every 3 weeks. All patients underwent mastectomy or breast conserving surgery, according to the standard protocol of our institute. Immunohistochemical (IHC) tests were performed to determine tumor expression levels of estrogen receptor (ER) and progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2). The primary antibodies and IHC methods have been previously described (12).
Response evaluation
The response of the primary tumor to chemotherapy was evaluated clinically using magnetic resonance imaging (MRI) and pathology. MRI size evaluation was performed before initiation of the 1st cycle of NAC and between the last cycle of NAC and the surgery. The percentage of MRI size change was calculated as below:
The X and Y dimensions of the tumor were measured in sagittal view. The Z dimension was measured by the number of image sections that the tumor shows.
Using the t-test, MRI size changes were compared between the two groups on the basis of the CNAs identified.
Array CGH
The array used in this study consisted of 4,045 human bacterial artificial chromosomes (BACs) spaced at an ∼1 Mb interval on average across the entire genome (MacArray™ Karyo4000; Macrogen, Inc., Seoul, Korea). Details of the BAC clones, DNA labeling, hybridization and imaging have been described in a previous report (9).
Data pre-processing and analysis
Arrays were scanned and analyzed using GenePix 4200, a two-color fluorescent scanner (MDS Analytical Technologies, Sunnyvale, CA, USA). From the scanned images, signal intensity ratios (test/reference) were measured and converted to a log2 scale using MacViewer 1.6 imaging software (Macrogen, Inc.). Background-corrected signal intensity ratios were normalized using LOWESS normalization, and the normalized data points scaled via mean centering. Instead of a fixed threshold, the data were smoothed first, and subsequently a sample-specific threshold was set based on the smoothed data points. For smoothing, a moving average with a 3-Mbp sliding window was used, which progresses along the clones ordered according to base pair positions on the chromosome (13). For each sample, the means and standard deviations were estimated to define sample-specific thresholds, with the aim of determining whether specific clone signals show copy number changes. Using the estimated parameters, we applied a normal distribution to fit data points across the whole genome, which were measured over an individual. Next, the clones as gain/normal/loss were classified, based on the upper and lower thresholds of the Gaussian components (set as p<0.05), calculated from the two tails of the distribution. For each clone, we analyzed whether the changes in volume among the three groups differed statistically via simple analysis of variance (ANOVA) test.
Our data were deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), and are accessible through GEO Series accession no. GSE10129.
Fluorescence in situ hybridization (FISH) analysis
Gene copy numbers were analyzed by performing FISH on formalin-fixed paraffin-embedded (FFPE) tissues. For dual-probe hybridization FISH, BACs were labeled to generate locus-specific probes mapping to 17q21.33 and 17p12, and the chromosome enumeration probe (CEP) 17 (Macrogen, Inc.) was employed. FISH probes for each region were generated via fusion of two human BAC clones. For the 17q21.33 region, the FISH probe was manufactured by fusion of two concatenate clones in chromosome 17, with the overall start and end positions at 45,890,122 and 46,071,397, respectively (NCBI build 37). The overall start and end positions of the fused FISH probes for the 17p12 region were 12,773,307 and 12,955,283, respectively.
Briefly, 4-μm tissue sections were deparaffinized, dehydrated, immersed in 0.2 N HCl, boiled in a microwave in citrate buffer (pH 6.0) and incubated in 1 M NaSCN for 35 min at 80°C. Sections were immersed in pepsin solution, and tissues were fixed in 10% neutral-buffered formalin. The probe mixture (locus-specific probe and CEP17) was applied to the slides, which were incubated in a humidified atmosphere with Hybrite™ (Vysis) at 73°C for 5 min to simultaneously denature the probe and target DNA, and subsequently at 37°C for 19 h to achieve hybridization. Following post-hybridization washing, nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) and anti-fade compound (p-phenylenediamine). FISH signals were assessed under an Olympus BX51TRF microscope (Olympus, Japan) equipped with a triple-pass filter (DAPI/ Green/Orange; Vysis). The entire tissue area was evaluated in each case and as many non-overlapping nuclei as possible were assessed for orange (locus-specific probe, marker) and green (CEP, reference) signals by a single pathologist (Y.K.J.) blinded to clinical information of the patients. The marker-to-reference signal ratio was calculated by dividing the number of orange signals by the number of green signals. Cases were classified as a 17q21.33 copy number gain when the 17q21.33 to CEP17 ratio was ≥2.0 or ≥40% of cells displayed ≥4 copies of the 17q21.33 signal (high polysomy). A 17p12 to CEP17 ratio of <0.8 signified a 17p12 deletion.
Verification and validation sets
For technical verification of the array CGH results, the FISH assay was performed on 19 FFPE cancer tissues out of the 63 samples profiled via array CGH. For independent validation sets, FFPE tissues from 39 breast cancer patients that were not included in the array CGH experiment were selected for FISH.
Results
Clinicopathological characteristics of patients
The clinicopathological characteristics of the patients are summarized in Table I. Patients were relatively young, with a mean age of 44.8 years. The mean tumor size measured using MRI decreased from 4.6 to 2.7 cm after NAC. Following three cycles of DA chemotherapy, 4 patients (6.3%) displayed pCR. The majority of patients had partial response (PR) (60.3%). ER and HER-2 positivities were 50.8 and 33.3%, respectively.
Table I.
Clinicopathological characteristics of the patients and tumors.
| Parameters | |
|---|---|
| Mean age (range), in years | 44.8 (28–69) |
| Pre-chemotherapy MRI size, in cm median (range) | 4.6 (1.0–11.0) |
| Post-chemotherapy MRI size, in cm median (range) | 2.7 (0–6.9) |
| Histological grade, n (%) | |
| II | 25 (39.7) |
| III | 26 (41.3) |
| Unknown | 12 (19.0) |
| Endolymphatic tumor emboli, n (%) | |
| No | 17 (27.0) |
| Yes | 36 (57.1) |
| Unknown | 10 (15.9) |
| ER status, n (%) | |
| Positive | 32 (50.8) |
| Negative | 31 (49.2) |
| PgR status, n (%) | |
| Positive | 20 (31.7) |
| Negative | 43 (68.3) |
| HER-2 status, n (%) | |
| Positive | 21 (33.3) |
| Negative | 42 (66.7) |
Selection of DNA aberrations associated with chemotherapy response
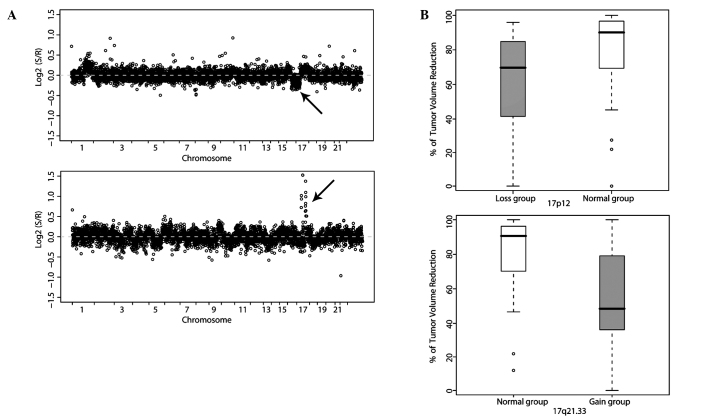
In total, 4,015 clones across the whole genome were profiled in 63 breast cancer patients and analyzed for DNA CNAs in association with NAC response. Genomic imbalances significantly associated with tumor size alterations were detected at eight chromosomal loci encompassing 24 clones. These characteristics are summarized in Table II. Clones were located in chromosomes 8, 10 and 17. Tumors with a DNA copy number gain on chromosomes 8q11.22-q24.23 and 17q21.32-q21.33, and those displaying loss of 17p12 displayed higher resistance to DA chemotherapy. However, tumors with a copy number gain on 10q26.3 were more responsive to chemotherapy. We finally selected two regions, 17p12 and 17q21.32-33 (Fig. 1A and B), as putative genomic markers for predicting breast cancer response to NAC by prioritizing regions containing candidate genes with aberrations in two or more consecutive BAC probes. 17q21.32-33 harbors candidate genes associated with the chemotherapy response, such as ABCC3 and the HOXB family, while 17p12 harbors ELAC2 (Table II).
Table II.
Regions of DNA copy number aberrations significantly associated with tumor size changes after NAC.
| Cytoband | Start position | Candidate genes | Frequencies of DNA copy number aberrations | Average percentage of tumor volume reduction according to aberration | Raw p-value | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Loss | Normal | Gain | Loss | Normal | Gain | ||||
| 8q11.22 | 52402874 | PXDNL | 0 | 51 | 12 | - | 79.7 | 59.2 | 0.0248 |
| 8q12.3 | 65039478 | 0 | 50 | 13 | - | 79.4 | 56.3 | 0.0192 | |
| 8q13.1 | 66247345 | 0 | 53 | 7 | - | 79.6 | 53.6 | 0.0286 | |
| 8q13.2 | 69455939 | C8orf34 | 0 | 54 | 9 | - | 79.6 | 48.2 | 0.0033 |
| 8q13.3 | 73392374 | 0 | 50 | 13 | - | 79.0 | 60.2 | 0.0479 | |
| 8q21.11 | 76752408 | 0 | 49 | 8 | - | 79.8 | 48.2 | 0.0077 | |
| 8q21.12 | 79843613 | IL7 | 0 | 50 | 8 | - | 81.0 | 55.1 | 0.0183 |
| 8q24.23 | 137759202 | 0 | 32 | 18 | - | 84.2 | 62.0 | 0.0131 | |
| 10q26.3 | 131211273 | MGMT | 0 | 53 | 10 | - | 72.0 | 93.2 | 0.0411 |
| 17p12 | 12773358 | ELAC2 | 11 | 52 | 0 | 58.9 | 79.8 | - | 0.0222 |
| 17p12 | 12800643 | ELAC2 | 10 | 52 | 0 | 59.3 | 79.8 | - | 0.0318 |
| 17q21.32 | 43988517 | HOXB3, HOXB4, HOXB5, HOXB6, HOXB7, HOXB8, HOXB9 | 0 | 54 | 9 | - | 78.9 | 58.4 | 0.0381 |
| 17q21.32 | 44002448 | HOXB3, HOXB4, HOXB5, HOXB6, HOXB7, HOXB8, HOXB9 | 0 | 54 | 9 | - | 78.9 | 58.4 | 0.0381 |
| 17q21.33 | 44810305 | PHB | 0 | 54 | 9 | - | 78.9 | 58.4 | 0.0381 |
| 17q21.33 | 44816025 | PHB, NGF | 0 | 54 | 9 | - | 78.9 | 58.4 | 0.0381 |
| 17q21.33 | 45062570 | SPOP, SLC35B1 | 0 | 51 | 9 | - | 81.7 | 55.0 | 0.0035 |
| 17q21.33 | 45565237 | PPP1R9B, SGCA, HILS1, COL1A1 | 0 | 53 | 10 | - | 79.9 | 56.2 | 0.0115 |
| 17q21.33 | 45661414 | TMEM92 | 0 | 53 | 10 | - | 79.9 | 56.2 | 0.0115 |
| 17q21.33 | 45890171 | CHAD, RSAD1, MYCBPAP, EPN3, SPATA20, CACNA1G | 0 | 53 | 10 | - | 79.9 | 56.2 | 0.0115 |
| 17q21.33 | 45965729 | EPN3, SPATA20, CACNA1G, ABCC3 | 0 | 53 | 10 | - | 79.9 | 56.2 | 0.0115 |
| 17q21.33 | 46222836 | WFIKKN2, TOB1 | 0 | 52 | 11 | - | 79.6 | 59.7 | 0.0294 |
| 17q21.33 | 46255459 | WFIKKN2, TOB, | 0 | 52 | 11 | - | 79.6 | 59.7 | 0.0294 |
| 17q21.33 | 46490057 | SPAG9, NME1 | 0 | 52 | 11 | - | 79.6 | 59.7 | 0.0294 |
| 17q21.33 | 46509890 | SPAG9, NME1, MBTD1 | 0 | 52 | 11 | - | 79.6 | 59.7 | 0.0294 |
Figure 1.
Genomic aberrations associated with chemotherapy response in breast cancer. (A) Scatter plots displaying array CGH results of two representative patients. The x-axis represents the genomic position and y-axis represents the log2 ratio of tumor/control DNA copy number. Arrows indicate loss of the 17p12 region (top) and gain of the 17q21.33 region (bottom). (B) Tumors with the 17p12 deletion displayed smaller volume reduction after chemotherapy than those with a normal copy number (top). Tumors with 17q21.33 gain showed a smaller volume reduction after chemotherapy than those with a normal copy number (bottom).
FISH verification of array CGH results
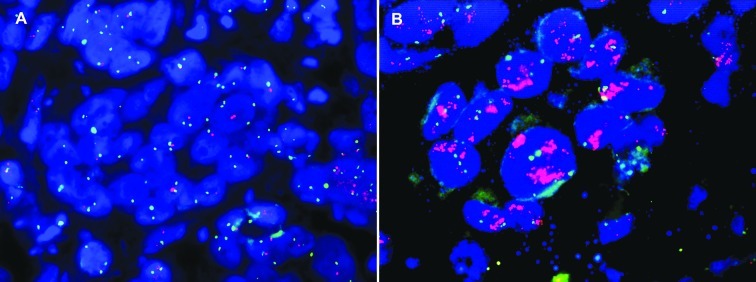
To confirm the array CGH results, multicolor FISH assay was performed on FFPE tissues from 19 of the 63 patients subjected to array CGH profiling (Fig. 2). The BAC clones used for array CGH were additionally employed as FISH probes for 17p12 and 17q21.32-33. The FISH results for 17q21.32-33 gain were identical to array CGH results in 15 out of 19 cases (79%). For the 17p12 region, we failed to obtain reliable data in 4 cases due to sample quality, and 13 of 15 cases showed deletions (86.7%), consistent with array CGH data.
Figure 2.
FISH assay showing 17p12 deletion and 17q21.33 amplification. Each test probe (orange) was manufactured by fusion of two consecutive BAC clones in the 17p12 and 17q21.33 regions. The control probe (green) was for chromosome 17 centromere. Examples of FISH assay showing (A) 17p12 deletion and (B) 17q21.33 amplification.
FISH validation of candidate genomic markers
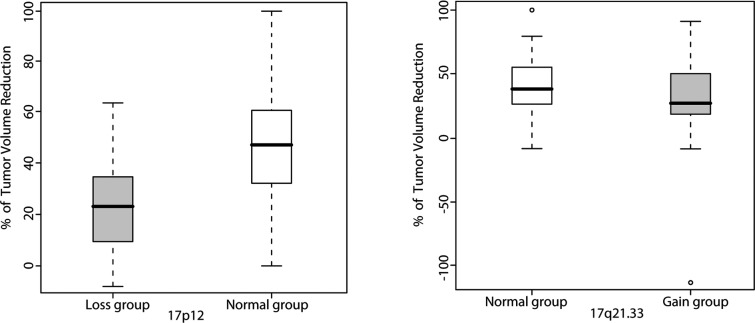
Subsequently, validation in an independent sample set from 39 breast cancer patients was performed to further evaluate the predictive performance of the two FISH probes. FISH assay protocols and probes were similar to those used for the verification study. The associations between response rate and DNA copy number aberrations detected using FISH were estimated with the t-test. The 17p12 deletion was significantly linked with lower response to NAC (p=0.0069). However, 17q21.33 gain was not significant in this validation set (p=0.3094) (Fig. 3). In an analysis based on ER status, the association between 17p12 deletion and chemotherapy resistance was significant in the ER-negative (p=0.0114), but not in the ER-positive group (p= 0.3182).
Figure 3.
Validation of the association between genomic aberrations and NAC response using the FISH assay. Tumors with a 17p12 deletion showed smaller volume reduction after chemotherapy than those with normal copy number (p=0.0069) (left). The percentage of volume reduction was not significantly different between tumors with 17q21.33 gain and the normal copy group (right).
Discussion
In this study, we showed that the 17p12 deletion is significantly associated with smaller reduction in tumor size induced by DA NAC in an array CGH study of 63 breast cancer patients and validation study using FISH for 39 independent patients. This finding is significant, since the current lack of biomarkers to predict NAC response in breast cancer is a clinically important issue (2).
Several studies have provided evidence that chromosome 17p12–13 loss or deletion is related to chemotherapy resistance in breast and other types of cancer. Kim et al showed in their array CGH study that 17p12 loss was more frequent in chemoresistant serous ovarian carcinoma than chemosensitive disease (14). Moreover, Robledo et al reported that 17p13 loss is associated with poor response to CHOP chemotherapy and short survival in diffuse large B-cell lymphoma (15).
The functional genes in this chromosome region that critically contribute to chemotherapy resistance are not known at present. In the present study, ELAC2 was the only candidate gene located within this region. Linkage analysis by Tavtigian et al (16) revealed that ELAC2 is a candidate prostate cancer susceptibility gene. ELAC2 interacts with γ-tubulin to cause a delay in G2-M progression in HeLa cells (17), and thus possibly affects response to taxane chemotherapy. However, there is no evidence of a direct relationship between ELAC2 expression and chemotherapy response at present. Functional studies for the deletion of this chromosomal region and identification of candidate genes in the BAC probe coverage and flanking regions are warranted.
ER negativity is generally associated with greater sensitivity to chemotherapy, and gene expression and CNA patterns are dominated by ER status (18). Therefore, the biomarkers identified may be an aberration simply related to ER status. However, the 17p12 deletion in our study was not significantly associated with ER status (Chi-square p= 0.8178; data not shown). The non-significant results in the ER-positive group may be attributed to small sample size.
To our knowledge, this is the first study to identify DNA copy number changes significantly associated with chemotherapy response/resistance. Notably, a similar study by Pierga et al (19) disclosed no marked correlation. The present study has several strengths. First, significant results were reproducible with an independent validation set. Second, a FISH probe that was directly applicable to clinical samples was manufactured. Third, a microdissection technique was employed to avoid contamination with DNA from non-cancer cells.
Conversely, the limitations of our study included relatively small sample size, lack of candidate genes and low resolution of BAC clone chips, compared to higher-resolution oligonucleotide chips.
In conclusion, our array CGH and FISH assay experiments collectively revealed a significant association of the 17p12 deletion with resistance to NAC in breast cancer. This experimental finding may be translated to prediction of clinical response in breast cancer patients using FISH for gun biopsy tissues before administration of NAC.
Acknowledgments
This study was supported by a grant from the Seoul National University Hospital Research Fund (0320070190), and a grant from the Korea Health 21 R&D Project, Ministry of Health and Welfare, Republic of Korea (01-PJ3-PG6-01GN07-0004 and A050558).
References
- 1.Untch M, von Minckwitz G. Recent advances in systemic therapy: advances in neoadjuvant (primary) systemic therapy with cytotoxic agents. Breast Cancer Res. 2009;11:203. doi: 10.1186/bcr2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolff AC, Berry D, Carey LA, et al. Research issues affecting preoperative systemic therapy for operable breast cancer. J Clin Oncol. 2008;26:806–813. doi: 10.1200/JCO.2007.15.2983. [DOI] [PubMed] [Google Scholar]
- 3.Gianni L, Zambetti M, Clark K, et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol. 2005;23:7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 4.Liedtke C, Hatzis C, Symmans WF, et al. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol. 2009;27:3185–3191. doi: 10.1200/JCO.2008.18.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hannemann J, Oosterkamp HM, Bosch CA, et al. Changes in gene expression associated with response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2005;23:3331–3342. doi: 10.1200/JCO.2005.09.077. [DOI] [PubMed] [Google Scholar]
- 6.Reid JF, Lusa L, de Cecco L, et al. Limits of predictive models using microarray data for breast cancer clinical treatment outcome. J Natl Cancer Inst. 2005;97:927–930. doi: 10.1093/jnci/dji153. [DOI] [PubMed] [Google Scholar]
- 7.Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 8.Chin K, deVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Han W, Jung EM, Cho J, et al. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47:490–499. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- 10.Han W, Han MR, Kang JJ, et al. Genomic alterations identified by array comparative genomic hybridization as prognostic markers in tamoxifen-treated estrogen receptor-positive breast cancer. BMC Cancer. 2006;6:92. doi: 10.1186/1471-2407-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang KT, Han W, Cho J, et al. Genomic copy number alterations as predictive markers of systemic recurrence in breast cancer. Int J Cancer. 2008;123:1807–1815. doi: 10.1002/ijc.23672. [DOI] [PubMed] [Google Scholar]
- 12.Jung SY, Han W, Lee JW, et al. Ki-67 expression gives additional prognostic information on St. Gallen 2007 and Adjuvant! Online risk categories in early breast cancer. Ann Surg Oncol. 2009;16:1112–1121. doi: 10.1245/s10434-009-0334-7. [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Erdogan F, Ropers HH, Lenzner S, Ullmann R. CGHPRO – a comprehensive data analysis tool for array CGH. BMC Bioinformatics. 2005;6:85. doi: 10.1186/1471-2105-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SW, Kim JW, Kim YT, et al. Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: potential predictive markers of chemoresistant disease. Genes Chromosomes Cancer. 2007;46:1–9. doi: 10.1002/gcc.20384. [DOI] [PubMed] [Google Scholar]
- 15.Robledo C, Garcia JL, Caballero D, et al. Array comparative genomic hybridization identifies genetic regions associated with outcome in aggressive diffuse large B-cell lymphomas. Cancer. 2009;115:3728–3737. doi: 10.1002/cncr.24430. [DOI] [PubMed] [Google Scholar]
- 16.Tavtigian SV, Simard J, Teng DH, et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet. 2001;27:172–180. doi: 10.1038/84808. [DOI] [PubMed] [Google Scholar]
- 17.Korver W, Guevara C, Chen Y, et al. The product of the candidate prostate cancer susceptibility gene ELAC2 interacts with the gamma-tubulin complex. Int J Cancer. 2003;104:283–288. doi: 10.1002/ijc.10945. [DOI] [PubMed] [Google Scholar]
- 18.Pusztai L, Ayers M, Stec J, et al. Gene expression profiles obtained from fine-needle aspirations of breast cancer reliably identify routine prognostic markers and reveal large-scale molecular differences between estrogen-negative and estrogen-positive tumors. Clin Cancer Res. 2003;9:2406–2415. [PubMed] [Google Scholar]
- 19.Pierga JY, Reis-Filho JS, Cleator SJ, et al. Microarray-based comparative genomic hybridisation of breast cancer patients receiving neoadjuvant chemotherapy. Br J Cancer. 2007;96:341–351. doi: 10.1038/sj.bjc.6603483. [DOI] [PMC free article] [PubMed] [Google Scholar]