Abstract
The aim of this study was to investigate the relationship between the expression of activating transcription factor 5 (ATF5) and clinicopathological features in human rectal cancer. Relative quantitative real-time RT-PCR and immunohistochemical staining were used to detect ATF5 mRNA and protein expression in 92 paired samples of rectal cancer and distant normal tissues. Immunohistochemical staining of the matched rectal tissue samples revealed that the positive expression rate of the ATF5 protein in rectal cancer was significantly higher compared to that in the normal tissue. Furthermore, the expression of ATF5 in poorly differentiated cancers was higher compared to that in well to moderately differentiated cancers (P=0.013). However, there was no significant association between ATF5 protein expression and patient age, gender, histological tumor type, cell differentiation, invasive depth, lymph node metastasis or distant metastasis (P>0.05). However, to our surprise, there was no difference in the relative mRNA expression levels of ATF5 between normal tissues and rectal cancers. Our findings indicate that overexpression of ATF5 protein may be an important biomarker of the degree of malignancy, and increased expression may be related to the post-transcriptional regulation of ATF5 in rectal cancer tissues.
Keywords: activating transcription factor 5, human rectal cancer, clinicopathological feature, immunohistochemical staining, quantitative real-time polymerase chain reaction
Introduction
Colorectal cancer is the third most common cancer and the third leading cause of cancer-related death in the world, and there are approximately 1,020,000 new cases and 530,000 related deaths per year (1–3). In many Asian countries, including China, Japan, South Korea and Singapore, colorectal cancer morbidity has increased by 2–4 times during the past few decades (4). It has been proven that the carcinogenesis and development of colorectal cancer is a multi-step, multi-stage and multi-gene genetic process. Due to normal gene dysregulation, abnormality in the signal transduction network leads to the process of carcinogenesis and development. Thus, the recognition of critical genes in biological pathways may undoubtedly provide a better understanding of the carcinogenesis and development of colorectal cancer.
Activating transcription factor 5 (ATF5; also referred to as ATFX) is a member of the ATF/CREB [cyclic AMP (cAMP) responsive element binding protein] family of transcription factors and plays an essential role in regulating CRE-dependent genes (5) which interact with a variety of proteins, including E2 ubiquitin-conjugating enzyme Cdc34 (6), HTLV-1 viral protein Tax (7), GABA-β receptor (8), cyclin D3 (9), DISC1 and PRL-1 (10,11). ATF5 has been associated with cell differentiation, proliferation and survival in several tissues and cell types (12–23). It may be a potent repressor of p53 and its elevated expression may be related to enhanced malignant phenotypes (21). ATF5 has been found to be overexpressed in human glioblastomas and human and rat glioma cell lines (16,23,24). Interference of ATF5 expression in glioma cells leads to apoptotic cell death both in vitro and in vivo (16). Additionally, ATF5 overexpression in breast carcinoma cells has been detected (15,22), and interference of ATF5 expression selectively triggers their death. These observations suggest that ATF5 may be an attractive target for therapeutic intervention in such tumor types. Consistent with these findings, ATF5 is highly expressed in a wide variety of neoplasms, including renal cell cancer, lung cancer, lymphomas, seminomas, cervical cancer, prostate cancer, hematopoietic carcinoma and hepatocellular carcinoma (17,20,25,26).
Although ATF5 has emerged as an important cell survival factor and is highly expressed in various carcinomas, ATF5 expression in human rectal cancer and its role in carcinogenesis, as well as the relationships between ATF5 expression and clinicopathological factors remain unknown. In this study, mRNA and protein expression of ATF5 in matched rectal tissue samples was assessed by real-time RT-PCR and immunohistochemical staining, respectively, and the correlationship between ATF5 expression level and clinicopathological features was further investigated.
Materials and methods
Patients and tissue samples
Ninety-two paired samples of rectal cancer and distant non-tumoral rectum tissues were obtained from 92 inpatients undergoing surgery at West China Hospital of Sichuan University between March and October 2009. No patient had received pre-operative chemotherapy or radiotherapy. The rectal cancer and paired normal tissues were allocated into the cancer and control groups. Certain parts of each sample were fixed by formalin and embedded in paraffin. The histological grade was assessed according to the WHO criteria by a pathologist (27). The clinical and pathological stages were defined according to TNM classification.
qRT-PCR
Total cDNA extraction. Total RNA from rectal cancer and adjacent non-tumoral rectum tissues was extracted using TRIzol (Beyozol Co. Ltd.) method. The concentration of RNA was detected spectrophotometrically. To generate total cDNA, reverse transcription was performed according to the manufacturer's protocol (Takara Biotechnology Co.).
Primers and probes. As shown in Table I, specific primer and probes for ATF5 and β-actin genes were designed. The gene specificity of the nucleotide sequences was confirmed by BLAST searches. The primers and probe were placed at the junction between two exons and were purchased from Invitrogen Biotechnology Co. (Shanghai, China).
Table I.
Sequences of the primers and probes.
| Sequence | Length (bp) | |
|---|---|---|
| Target gene/ATF5 | 262 | |
| Forward | 5′-ACCTTCTTTCTTCAGCCGA-3′ | 19 |
| Reverse | 5′-GAGTTTCCCATAGTCTACGA-3′ | 20 |
| Probe | 5′-FAM-CTCCTGTTGCCATCAGTGCCCA-TAMRA-3′ | 22 |
| Reference gene/β-actin | 166 | |
| Forward | 5′-AAGGCCAACCGCGAGAA-3′ | 17 |
| Reverse | 5′-CCTCGTAGATGGGCACA-3′ | 17 |
| Probe | 5′-FAM-TCAACACCCCAGCCATGTACGT-TAMRA-3′ | 22 |
Real-time RT-PCR. Real-time PCR was performed using relative quantification protocol on an iCycler iQ System (Bio-Rad, USA). The optimum annealing temperature was 51°C and the following iCycler iQ run protocol was used: denaturation program (94°C, 2 min), amplification and quantification programs repeated 40 times (94°C for 20 sec, 51°C for 30 sec, 72°C for 30 sec). In addition, a no-template control (ddH2O) was analyzed for each template. Equation 1 (28), as shown below, was applied to calculate the relative expression ratio of the target gene (ATF5) in the sample vs. a control in comparison to a reference gene (β-actin).
| Equation 1: |
ΔCT is the CT (threshold cycle) deviation of the control minus the sample of the target or reference gene transcript. The PCR efficiency (E) was calculated using the iCycler iQ fluorescence PCR machine automatically from the slope of the line using Equation 2 (29): E = 10−1/slope. A ratio >1 for mRNA expression of ATF5 in the rectal cancer tissues designated up-regulation.
Immunohistochemical staining
Specimens were formalin-fixed, paraffin-embedded and consecutively sliced in 4-μm sections, which were dewaxed in xylene and then rehydrated in alcohol. The endogenous peroxidase activity was suppressed by 3% hydrogen peroxide for 15 min. After being washed twice in phosphate-buffered saline (PBS), the sections were incubated at 37°C for 30 min with normal goat serum, and then at 4°C overnight with rabbit polyclonal antibody against ATF5 (Santa Cruz Biotechnology) which was diluted to 1:400. PBS was used as a negative control. After rinsing three times in PBS, the sections were treated with biotinylated goat anti-rabbit immunoglobulin at 37°C for 40 min and then, after a washing with PBS, the sections were treated with horseradish peroxidase streptavidin complex at 37°C for 40 min, which were diluted as recommended by the manufacturer. Finally, the sections were washed three times in PBS and visualized in DBA coloration fluid. The sections were counterstained in hematoxylin.
Stained sections were reviewed and scored by two researchers, including a pathologist. Consensus between the two researchers was achieved in all cases. ATF5 expression was evaluated by an immunoreactive score (IRS) system (30). According to the staining intensity of tumor cells, the immunoreactive scores were graded as 0 (negative), 1 (faint yellow staining), 2 (brown staining) and 3 (dark brown staining). At the same time, the scores were graded as 0 (0%), 1 (0–10%), 2 (11–50%) and 3 (51–100%) according to the percentage of the positive cells. Finally, a total score was obtained by multiplying the two scores. Eight visual fields from different areas in every section were randomly selected for IRS evaluation, and the mean value of the IRS was calculated. The final extent of ATF5 immunohistochemical staining was categorized as negative and positive, corresponding to IRS values of ≤1 and >1, respectively.
Statistical analysis
Group-wise comparison and statistical analysis in mRNA relative expression was performed by the Relative Expression Software Tool (31). The categorical data were analyzed by the Chi-square test, and P<0.05 was considered statistically significant. All of the test results were analyzed using software SPSS 16.0.
Results
Clinicopathological features of the specimens
The 92 total specimens of rectal cancer tissues consisted of 85 tubular or papillary adenocarcinomas and 7 myxoadenocarcinomas. The cases included 53 men and 39 women with a mean age of 58.3 years (range 25–89). The histological differentiation of the samples consisted of 54 well to moderate and 38 poor differentiated tumors. Thirty-five samples were invasive through the muscularis and 57 through the serous membrane. More than 12 lymph nodes were obtained in every case, and 40 cases with lymph node metastasis were noted by pathologists. There were 27 cases with distant metastases.
ATF5 expression in the human rectal cancer tissues
ATF5 mRNA expression in the two tissue groups. Determining the speciality of the amplified product with 1% agrose electrophoresis and ethidium bromide (EB) staining showed the single lines of 166 and 262 bp for β-actin and ATF5 genes, respectively.
Among the 92 paired samples, the ratio of ATF5 mRNA expression was >1 for 33 (35.9%) cases and <1 in 59 (64.1%) cases. REST software analysis showed that mRNA expression of ATF5 in the cancer group was not different from that in the control group (P>0.05); thus, ATF5 mRNA expression was not up-regulated in comparison to the control group. The Ct value data are shown in Table II.
Table II.
Ct value of mRNA expression in the two groups.
| mRNA | No. | Cancer group Ct (mean ± SD) | Control group Ct (mean ± SD) | P-value |
|---|---|---|---|---|
| β-actin | 92 | 21.2±3.5 | 21.2±3.9 | |
| ATF5 | 92 | 32.6±8.7 | 31.5±9.1 | 0.363 |
SD, standard deviation.
Relationship between ATF5 mRNA expression and clinicopathological features. As shown in Table III, the comparisons indicated that the expression levels were not significantly associated with the factors of age, gender, histological type of the tumor, grade of differentiation, depth of invasion, lymph node metastasis and distant metastasis.
Table III.
Relationship between ATF5 gene expression and clinicopathological features.
| Clinicopathological features | Total no. of patients | Ratio ≤1.0 n (%) | Ratio >1.0 n (%) | P-value | IRS ≤1.0 n (%) | IRS >1.0 n (%) | P-value |
|---|---|---|---|---|---|---|---|
| Age (years) | 0.124 | 0.879 | |||||
| ≤60 | 52 | 30 (57.7) | 22 (42.3) | 19 (36.5) | 33 (63.5) | ||
| >60 | 40 | 29 (72.5) | 11 (27.5) | 14 (35.0) | 26 (65.0) | ||
| Gender | 0.382 | 0.996 | |||||
| Male | 53 | 32 (60.4) | 21 (39.6) | 19 (35.8) | 34 (64.2) | ||
| Female | 39 | 27 (69.2) | 12 (30.8) | 14 (35.9) | 25 (64.1) | ||
| Histological type | 0.675 | 0.222 | |||||
| Adenocarcinoma | 85 | 54 (63.5) | 31 (36.5) | 29 (34.1) | 56 (65.9) | ||
| Mucinous carcinoma | 7 | 5 (71.4) | 2 (28.6) | 4 (57.1) | 3 (42.9) | ||
| Differentiation grade | 0.246 | 0.013 | |||||
| Poor | 38 | 27 (71.1) | 11 (28.9) | 8 (21.1) | 30 (78.9) | ||
| Well-moderate | 54 | 32 (59.3) | 22 (40.7) | 25 (46.3) | 29 (53.7) | ||
| Invasive depth | 0.274 | 0.517 | |||||
| Ia | 35 | 20 (57.1) | 15 (42.9) | 14 (40.0) | 21 (60.0) | ||
| IIb | 57 | 39 (68.4) | 18 (31.6) | 19 (33.3) | 38 (66.7) | ||
| Lymph node metastasis | 0.879 | 0.142 | |||||
| Negative | 52 | 33 (63.5) | 19 (36.5) | 22 (42.3) | 30 (57.7) | ||
| Positive | 40 | 26 (65.0) | 14 (35.0) | 11 (27.5) | 29 (72.5) | ||
| Distant metastasis | 0.530 | 0.744 | |||||
| Negative | 65 | 43 (66.2) | 22 (33.8) | 24 (36.9) | 41 (63.1) | ||
| Positive | 27 | 16 (59.3) | 11 (40.7) | 9 (33.3) | 18 (66.7) |
Through the muscularis;
through the serous membrane. IRS, immunoreactive score.
Immunohistochemical staining for ATF5 in the human rectal cancer tissues
Immunohistochemical staining for ATF5 in the human rectal cancer and normal tissues. Representative staining of ATF5 protein expression in the normal tissues, the well and poorly differentiated cancer tissues, as well as the negative control are shown in Fig. 1. The nuclei of cells in the rectal cancer tissues were positively stained, while the cell membranes and stromal components were not. Of the 92 paired samples, 59 (64.1%) cancer tissues and 29 (31.5%) normal tissues showed positive staining, achieving significant difference (P=0.000) (Table IV).
Figure 1.
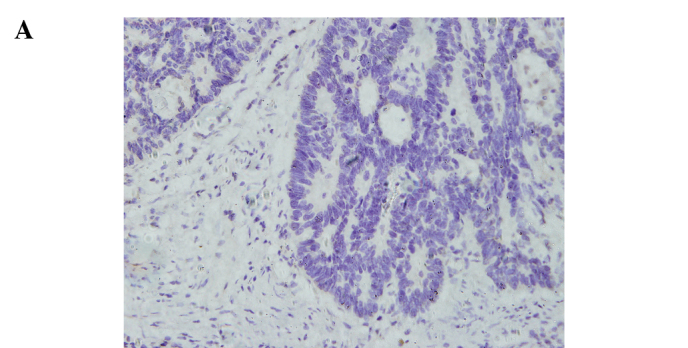
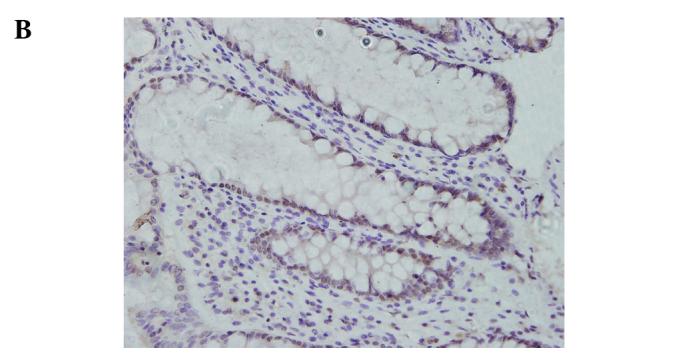
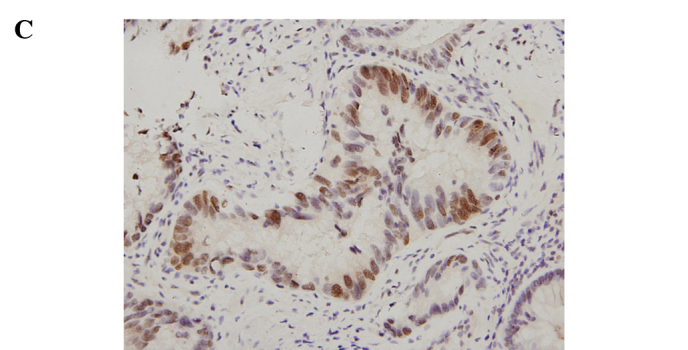
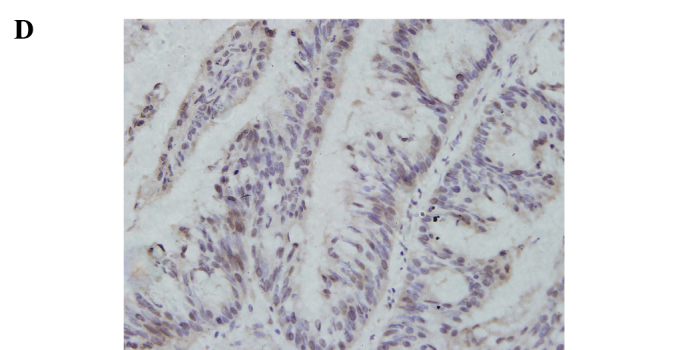
Immunohistochemical staining for ATF5 in rectal cancer and normal tissue (SP, x400). (A) Negative control slide with PBS was not positively stained. (B) Positive expression of ATF5 in normal rectal tissue. (C) Positive expression of ATF5 in rectal adenocarcinoma with low differentiation. (D) Positive expression of ATF5 in rectal adenocarcinoma with well differentiation.
Table IV.
Result of ATF5 protein expression in the normal and cancer tissues.
| Total no. of patients | IRS ≤1.0 n (%) | IRS >1.0 n (%) | χ2-value | P-value | |
|---|---|---|---|---|---|
| 19.602 | 0.000 | ||||
| Normal tissue | 92 | 63 (68.5) | 29 (31.5) | ||
| Cancer tissue | 92 | 33 (35.9) | 59 (64.1) |
Relationship between ATF5 protein expression and clinicopathological features. The intra-group comparison by clinicopathological features indicated that the ATF5 protein expression level was associated with the grade of differentiation of the rectal cancer cases (P=0.013) (Table III). Regarding the differentiation, 29 (53.7%) out of the 59 positive cancer samples were well to moderately differentiated tissues and the remaining 30 (78.9%) were poorly differentiated. IRS of the poorly differentiated tumors was significantly higher than that of the well to moderately differentiated (P<0.05) cases. The results indicate that the lower the grade of differentiation, the higher the expression rate was. Representative staining of ATF5 protein expression in the poorly and well to moderately differentiated tumor tissues is shown in Fig. 1. Additionally, there were no significant differences stratified by age (P=0.879), gender (P=0.996), histological type (P=0.222), invasive depth (P=0.517), lymph node metastases (P=0.142) and distant metastases (P=0.744).
Discussion
Transcription factors play an important role in the control of cell proliferation, cell cycle modulation, differentiation and survival and are attractive targets for cancer therapy. Transcription factor ATF5, an ATF/CREB family member and bZIP protein, functions as a cancer-specific cell survival factor and promotes survival of cancer cells (22,32). Although evidence has been presented that ATF5 is involved in the onset and progression of cancer (20–26), no study, to the best of our knowledge, has reported whether ATF5 is related to rectal cancer.
In the present study, the ATF5 protein and mRNA expression in human rectal carcinoma and normal tissue was analyzed using immunohistochemistry and RT-PCR. Immunohistochemical analysis indicated that ATF5 protein expression was significantly increased in the rectal cancer relative to the normal tissues. Moreover, immunohistochemical analysis showed that nuclear expression was increased with differentiation in the poorly differentiated tissues vs. the well to moderately differentiated tissues, which is in accordance with the fact that ATF5 is related to cell differentiation (33,34). However, there was no significant association between ATF5 expression and patient age, gender, histological type of tumor, depth of invasion, lymph node metastasis and distant metastasis. These observations suggest that ATF5 plays a role in the differentiation of rectal cancer and that ATF5 overexpression may be a marker for the degree of malignancy as poorer differentiation indicates a greater likelihood of malignant behavior.
Notably, there was no significant difference in the relative mRNA expression levels of ATF5 between the rectal cancer and normal tissues. Moreover, the ATF5 mRNA expression was not related to the clinicopathological features of the human rectal cancer cases. The different results obtained for mRNA and protein expression in this study support the hypothesis that overexpression of ATF5 protein may be related to the up-regulation of expression at the post-transcriptional level, increased protein stability and hence a longer half-life time or regulation by other related proteins in cancer tissues. These results also emphasis the importance of proteomics in the study of disease-associated gene expression at the protein level.
According to our results, the expression of ATF5 in rectal cancer was higher than that in the normal tissues. Poorly differentiated tissues had a higher level of expression than the well to moderately differentiated tumors. This indicates that ATF5 plays a role in the carcinogenesis and differentiation of rectal cancer.
In conclusion, overexpression of ATF5 protein in rectal cancer cells may be a marker for the degree of malignancy. This overexpression may be related to post-transcriptional regulation of this gene. Therefore, the role of ATF5 in rectal carcinogenesis and the application of ATF5 in the therapeutic intervention of rectal cancer require further investigation.
Acknowledgments
The authors thank Professor Zhou for guiding this study, members of the Pathological Research Institution of West China Hospital for their useful assistance and the colleagues at the Department of Gastrointestinal Surgery for providing the experimental specimens. The study was supported by the National Natural Science Foundation of China (no. 30830100).
References
- 1.Jemal A, Siegel R, Xu JQ, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. New Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 4.Sung JJ, Lau JY, Goh KL, Leung WK, Asia Pacific Working Group on Colorectal Cancer Increasing incidence of colorectal cancer in Asia: implications for screening. Lancet Oncol. 2005;6:871–876. doi: 10.1016/S1470-2045(05)70422-8. [DOI] [PubMed] [Google Scholar]
- 5.Hansen MB, Mitchelmore C, Kjaerulff KM, Rasmussen TE, Pedersen KM, Jensen NA. Mouse Atf5: molecular cloning of two novel mRNAs, genomic organization, and odorant sensory neuron localization. Genomics. 2002;80:344–350. doi: 10.1006/geno.2002.6838. [DOI] [PubMed] [Google Scholar]
- 6.Pati D, Meistrich ML, Plon SE. Human Cdc34 and Rad6B ubiquitin-conjugating enzymes target repressors of cyclic AMP-induced transcription for proteolysis. Mol Cell Biol. 1999;19:5001–5013. doi: 10.1128/mcb.19.7.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forgacs E, Gupta SK, Kerry JA, Semmes JO. The bZIP transcription factor ATFx binds human T-cell leukemia virus type 1 (HTLV-1) tax and represses HTLV-1 long terminal repeat-mediated transcription. J Virol. 2005;79:6932–6939. doi: 10.1128/JVI.79.11.6932-6939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White JH, McIllhinney RA, Wise A, Ciruela F, Chan WY, Emson PC, Billinton A, Marshall FH. The GABAB receptor interacts directly with the related transcription factors CREB2 and ATFx. Proc Natl Acad Sci USA. 2000;97:13967–13972. doi: 10.1073/pnas.240452197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu WJ, Sun MY, Jiang JH, et al. Cyclin D3 interacts with human activating transcription factor 5 and potentiates its transcription activity. Biochem Biophys Res Commun. 2004;321:954–960. doi: 10.1016/j.bbrc.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 10.Peters CS, Liang XP, Li SX, Kannan S, Peng Y, Taub R, Diamond RH. ATF-7, a novel bZIP protein, interacts with the PRL-1 protein-tyrosine phosphatase. J Biol Chem. 2001;276:13718–13726. doi: 10.1074/jbc.M011562200. [DOI] [PubMed] [Google Scholar]
- 11.Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 12.Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 13.Mora J, Cheung NKV, Chen LS, Qin J, Gerald W. Loss of heterozygosity at 19q13.3 is associated with locally aggressive neuroblastoma. Clin Cancer Res. 2001;7:1358–1361. [PubMed] [Google Scholar]
- 14.Angelastro JM, Ignatova TN, Kukekov VG, Steindler DA, Stengren GB, Mendelsohn C, Greene LA. Regulated expression of ATF5 is required for the progression of neural progenitor cells to neurons. J Neurosci. 2003;23:4590–4600. doi: 10.1523/JNEUROSCI.23-11-04590.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow LSN, Lam CW, Chan SYY, et al. Identification of RASSF1A modulated genes in nasopharyngeal carcinoma. Oncogene. 2006;25:310–316. doi: 10.1038/sj.onc.1209001. [DOI] [PubMed] [Google Scholar]
- 16.Angelastro JM, Canoll PD, Kuo J, Weicker M, Costa A, Bruce JN, Greene LA. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene. 2006;25:907–916. doi: 10.1038/sj.onc.1209116. [DOI] [PubMed] [Google Scholar]
- 17.Monaco SE, Angelastro JM, Szabolcs M, Greene LA. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int J Cancer. 2007;120:1883–1890. doi: 10.1002/ijc.22469. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Lin G, Zhang Z. ATF5 promotes cell survival through transcriptional activation of Hsp27 in H9c2 cells. Cell Biol Int. 2007;31:1309–1315. doi: 10.1016/j.cellbi.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Pascual M, Gomez-Lechon MJ, Castell JV, Jover R. ATF5 is a highly abundant liver-enriched transcription factor that cooperates with constitutive androstane receptor in the transactivation of CYP2B6: implications in hepatic stress responses. Drug Metab Dispos. 2008;36:1063–1072. doi: 10.1124/dmd.107.019380. [DOI] [PubMed] [Google Scholar]
- 20.Gho JW, Ip WK, Chan KY, Law PT, Lai PB, Wong N. Re-expression of transcription factor ATF5 in hepatocellular carcinoma induces G2-M arrest. Cancer Res. 2008;68:6743–6751. doi: 10.1158/0008-5472.CAN-07-6469. [DOI] [PubMed] [Google Scholar]
- 21.Nishioka T, Miyai Y, Haga H, et al. Novel function of transcription factor ATF5: blockade of p53-dependent apoptosis induced by ionizing irradiation. Cell Struct Funct. 2009;34:17–22. doi: 10.1247/csf.08041. [DOI] [PubMed] [Google Scholar]
- 22.Li GF, Li WH, Angelastro JM, Greene LA, Liu DX. Identification of a novel DNA binding site and a transcriptional target for activating transcription factor 5 in C6 glioma and MCF-7 breast cancer cells. Mol Cancer Res. 2009;7:933–943. doi: 10.1158/1541-7786.MCR-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greene LA, Lee HY, Angelastro JM. The transcription factor ATF5: role in neurodevelopment and neural tumors. J Neurochem. 2009;108:11–22. doi: 10.1111/j.1471-4159.2008.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong SM, Nutt CL, Betensky RA, et al. Histology-based expression profiling yields novel prognostic markers in human glioblastoma. J Neuropath Exp Neur. 2005;64:948–955. doi: 10.1097/01.jnen.0000186940.14779.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YW, Hussain M, Sarkar SH, Eliason J, Li R, Sarkar FH. Gene expression profiling revealed novel mechanism of action of Taxotere and Furtulon in prostate cancer cells. BMC Cancer. 2005;5:7. doi: 10.1186/1471-2407-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei YY, Jiang JH, Sun MY, Chen XN, Wang HZ, Gu JX. ATF5 increases cisplatin-induced apoptosis through up-regulation of Cyclin D3 transcription in HeLa cells. Biochem Biophys Res Commun. 2006;339:591–596. doi: 10.1016/j.bbrc.2005.11.054. [DOI] [PubMed] [Google Scholar]
- 27.Jass JR, Sobin LH, Watanabe H. The World Health Organization's histologic classification of gastrointestinal tumors. A commentary on the second edition. Cancer. 1990;66:2162–2167. doi: 10.1002/1097-0142(19901115)66:10<2162::aid-cncr2820661020>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 28.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard PS, Wittwer CT. Real-time PCR technology for cancer diagnostics. Clin Chem. 2002;48:1178–1185. [PubMed] [Google Scholar]
- 30.Wu CC, Shyu RY, Chou JM, Jao SW, et al. RARRES1 expression is significantly related to tumour differentiation and staging in colorectal adenocarcinoma. Eur J Cancer. 2006;42:557–565. doi: 10.1016/j.ejca.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis. 2003;8:225–228. doi: 10.1023/a:1023633704132. [DOI] [PubMed] [Google Scholar]
- 33.Mason JL, Angelastro JM, Ignatova TN, Kukekov VG, Lin G, Greene LA, Goldman JE. ATF5 regulates the proliferation and differentiation of oligodendrocytes. Mol Cell Neurosci. 2005;29:372–380. doi: 10.1016/j.mcn.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Angelastro JM, Mason JL, Ignatova TN, Kukekov VG, Stengren GB, Goldman JE, Greene LA. Downregulation of activating transcription factor 5 is required for differentiation of neural progenitor cells into astrocytes. J Neurosci. 2005;25:3889–3899. doi: 10.1523/JNEUROSCI.3447-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


